Abstract
HIP14 is the most highly conserved of 23 human palmitoyl acyltransferases (PATs) that catalyze the post-translational addition of palmitate to proteins, including huntingtin (HTT). HIP14 is dysfunctional in the presence of mutant HTT (mHTT), the causative gene for Huntington disease (HD), and we hypothesize that reduced palmitoylation of HTT and other HIP14 substrates contributes to the pathogenesis of the disease. Here we describe the yeast two-hybrid (Y2H) interactors of HIP14 in the first comprehensive study of interactors of a mammalian PAT. Unexpectedly, we discovered a highly significant overlap between HIP14 interactors and 370 published interactors of HTT, 4-fold greater than for control proteins (P = 8 × 10−5). Nearly half of the 36 shared interactors are already implicated in HD, supporting a direct link between HIP14 and the disease. The HIP14 Y2H interaction set is significantly enriched for palmitoylated proteins that are candidate substrates. We confirmed that three of them, GPM6A, and the Sprouty domain-containing proteins SPRED1 and SPRED3, are indeed palmitoylated by HIP14; the first enzyme known to palmitoylate these proteins. These novel substrates functions might be affected by reduced palmitoylation in HD. We also show that the vesicular cargo adapter optineurin, an established HTT-binding protein, co-immunoprecipitates with HIP14 but is not palmitoylated. mHTT leads to mislocalization of optineurin and aberrant cargo trafficking. Therefore, it is possible that optineurin regulates trafficking of HIP14 to its substrates. Taken together, our data raise the possibility that defective palmitoylation by HIP14 might be an important mechanism that contributes to the pathogenesis of HD.
INTRODUCTION
Huntington disease (HD) is an adult-onset neurodegenerative disorder caused by a CAG trinucleotide repeat expansion in the huntingtin (HTT) gene, resulting in loss of motor control and severe cognitive and psychiatric disturbances (1). HTT is a scaffold protein that is involved in hundreds of interactions (2–6). One of these is with Huntingtin Interacting Protein 14 (HIP14, or ZDHHC17), a highly conserved palmitoyl acyltransferase (PAT) (7,8) that was first discovered in a yeast two-hybrid (Y2H) screen for proteins that interact with HTT (8,9). HIP14 has been implicated in the pathogenesis of HD through multiple lines of evidence and aberrant palmitoylation contributes to the pathogenesis of several neuropsychiatric diseases in addition to HD (reviewed in (7)).
HIP14 and HTT co-localize in the medium spiny neurons of the striatum in mice, the most severely affected and earliest target of neurodegeneration in HD (8,10,11). HIP14 modifies HTT post-translationally by adding the fatty acid palmitate to internal cysteine residues, primarily at Cys214, in a process known as palmitoylation, influencing trafficking and aggregation of HTT (12–14). Any process underlying the pathogenesis of HD should be altered in the presence of the causative mutation (mHTT). Indeed HTT's interaction with and palmitoylation by HIP14 are reduced in the presence of mHTT in vitro and in vivo (8,12,15). When mHTT is rendered palmitoylation-resistant (C214S-HTT-128Q), inclusion formation and toxicity are enhanced in neurons compared with those expressing fully palmitoylatable mHTT (HTT-128Q) (13).
In fact, the functions of HIP14 and HTT are interdependent. The activity of a PAT is dependent on palmitoylation of the PAT itself (16) and the palmitoylation of HIP14 is highly correlated with levels of wild-type HTT (wtHTT) (12). wtHTT promotes palmitoylation of the HIP14 substrate SNAP25 in vitro and loss of wtHTT leads to reduced palmitoylation of SNAP25 and GluR1 in vivo (12). HIP14's PAT activity is also altered in the presence of mHTT, with HIP14 isolated from brains of the YAC128 mouse model of HD (17) having a reduced ability to palmitoylate SNAP25 (15). Thus, HIP14 palmitoylation of HTT influences HTT function, and HTT modulates HIP14's activity toward its substrates.
HIP14 is the most highly conserved of 23 mammalian PATs (7) suggesting it has significant functional importance. There are 13 known substrates and three additional known interactors of HIP14 in mammals (Supplementary Material, Table S1). HIP14 palmitoylates and/or interacts with proteins involved in neurotransmission and neuronal development (14,15,18–24) as well as with proteins involved in signal transduction (24–27) and transcriptional regulation (28). Proper palmitoylation is critical for trafficking and membrane localization of many neuronal proteins and has roles in neuronal development and in synaptic transmission and plasticity (29). Hip14-deficient mice recapitulate some cardinal features of HD, such as reduced striatal volume and neuronal number, and motor deficits (15). We hypothesize that HIP14 dysfunction contributes to the phenotype in HD through a direct effect on HTT functions as well as an indirect effect of the reduced ability of HIP14 to palmitoylate key substrates, resulting in their mislocalization and altered function.
Here we report the first comprehensive study of interactors of a mammalian PAT. Our most surprising finding was that a significantly high proportion of known HTT interactors also interact with HIP14 and nearly half of the 36 shared interactors have previously been implicated in HD. The evidence presented here underlines the extensive functional connection between HIP14 and HD and highlights potential mechanisms for how an altered HIP14–HTT interaction and altered palmitoylation of HTT and other HIP14 substrates might contribute to the pathogenesis of HD. All protein interactions have been submitted to the IMEx Consortium (http://www.imexconsortium.org) through IntAct (30) and assigned the identifier IM- 21827.
RESULTS
Identification of HIP14 interactors by automated yeast two-hybrid interaction screening
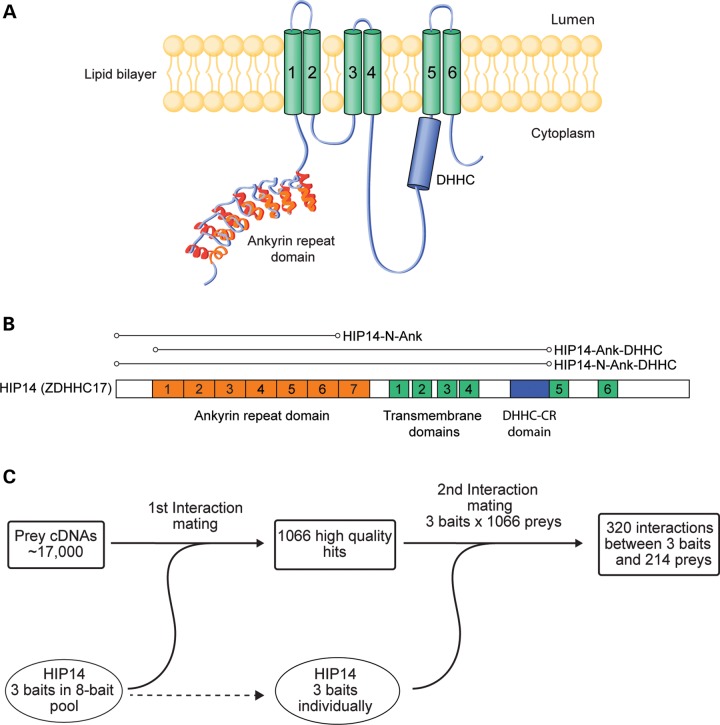
To identify HIP14 interactors, we conducted a Y2H screen using HIP14 as bait against a set of ∼17 000 prey proteins primarily from the human ORFeome collection (31,32). The preys were supplemented with HTT subclones that together spanned its entire length (33). Three HIP14 baits were constructed with a focus on the ANK domain, a key domain for interaction with HTT, that is unique to HIP14 and HIP14L (ZDHHC13) (Fig. 1A and B) (12,34). HIP14-N-Ank included the N-terminus and the ANK domain. HIP14-N-Ank-DHHC included the N-terminus, ANK domain, four transmembrane domains and the full 51 amino acid DHHC cysteine-rich (DHHC-CR) domain. HIP14-Ank-DHHC included the ANK domain, four transmembrane domains and the DHHC-CR domain but lacked the N-terminal segment (Fig. 1B). These baits were also included as preys in the screen.
Figure 1.
Yeast two-hybrid screen for HIP14 interactors. (A) HIP14 functional domains include a seven-repeat ankyrin domain (orange), six transmembrane domains (green, 1–6) and a 51 amino acid catalytic DHHC cysteine-rich domain (blue). (B) Three HIP14 baits were used in the Y2H screen. HIP14-N-Ank includes amino acids 1–257 spanning the N-terminus to the end of ankyrin repeat 6. HIP14-N-Ank-DHHC includes amino acids 1–487 spanning the N-terminus to the end of the 51 amino acid catalytic DHHC-CR domain. HIP14- Ank-DHHC includes amino acids 64–487 spanning the middle of ankyrin repeat 1 to the end of the catalytic DHHC-CR domain. Amino acid numbering is according to accession NP_056151. (C) Three HIP14 baits in an 8-bait pool were screened for interactions with 17 000 human prey cDNAs in the first Y2H interaction mating. This first interaction mating yielded 1066 high quality hits. To confirm bait–prey interactions, a second interaction mating was conducted between pairs of individually arrayed HIP14 baits and the 1066 high quality preys. The second interaction mating yielded 320 interactions between 3 HIP14 baits and 214 preys.
The Y2H screen was conducted as described previously (35) (Fig. 1C). For the initial interaction screening, the three HIP14 baits were pooled with five additional baits that have other neuronal functions, as part of a larger screen. Pooled baits were mated with prey clones in four replicates. One thousand and sixty-six preys interacted with the bait-pool in three or more replicates. In a second test to confirm HIP14-specific interactions, each of the three HIP14 baits was individually mated with each of the 1066 preys identified in the first interaction screen. This approach yielded a total of 320 interactions between baits and preys involving 214 non-redundant HIP14-interacting proteins. These are referred to as the HIP14 Y2H protein–protein interaction (PPI) set (Supplementary Material, Table S3).
We found that HIP14 baits interacted with HTT N-terminal fragments and also interacted with itself in the Y2H assay, but did not interact with any other PATs including its close relative, HIP14L, despite the presence of 19 of the 23 mammalian PATs as preys in the screen (ZDHHC1, ZDHHC8, ZDHHC14 and ZDHHC18 were not represented). This suggests that HIP14 might form homodimers in mammalian cells (36).
The HIP14 Y2H PPI set included proteins involved in trafficking including Golgi-, motor- and microtubule-associated proteins, membrane-bound proteins such as channels and transporters, proteins involved in signal transduction, protein translation, regulated protein phosphorylation, ubiquitin-related and nuclear processes (transcription factors, RNA processing factors, histones, DNA repair proteins, chromatin remodeling factors), as well as proteins with unknown functions (Supplementary Material, Table S3). The PPI set included 17 synaptic proteins according to SynSysNet (37) (Supplementary Material, Table S3). We probed the functional features of the PPI set with DAVID (http://david.abcc.ncifcrf.gov/), a bioinformatics resource for analyzing the biological information in large lists of genes (38). The DAVID Functional Annotation Chart did not identify significant enrichment, after Bonferroni correction, for any biological process or molecular function when the analysis was done using default parameters (Supplementary Material, Table S2). The PPI set was, however, enriched for the cysteine-rich Sprouty domain (InterPro IPR007875; P = 0.016, Bonferroni corrected) that is present in only seven human proteins: the Sprouties, SPRY1–4 and the Sprouty-related, EVH1 domain-containing proteins, SPRED1–3. HIP14 interacted with four of these in the Y2H screen: SPRED1, SPRED2, SPRY2 and SPRY4. All of the known Sprouty domain-containing proteins except for SPRED3 were included as preys in the experiment. SPRY1 and SPRY3 were included as preys but were not identified as interactors in the Y2H screen.
HIP14 Y2H interactors are enriched for palmitoylated proteins
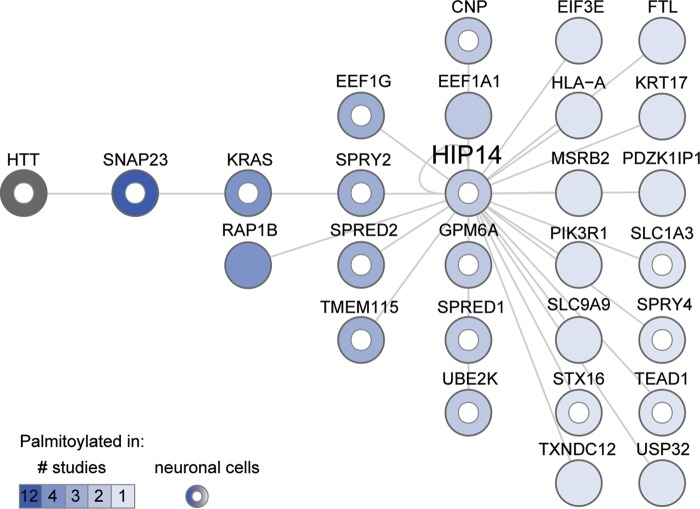
To determine whether any of the interactors represent potential novel substrates of HIP14, we compared the PPI set to 9 datasets of palmitoylated proteins comprising 13 independent mammalian proteomic studies published between 2008 and 2013. These palmitoyl-proteomes were derived from a variety of tissues and species: rat cultured neuronal cells and brain synaptosomal fraction (39); four studies from mouse: neuronal stem cells (40), T-cell hybridoma cells (41), macrophages (42) and dendritic cells (43); and eight studies from human: Jurkat T cells (44,45), B lymphocytes (46), endothelial cells (47), resting platelets (48), HeLa cells (49), HEK293 cells (50) and a prostate cancer cell line (51). Twenty-seven of the HIP14 interactors, as well as HIP14 itself, were reported to be palmitoylated in these mammalian palmitoyl-proteome studies (Fig. 2, Supplementary Material, Table S4). HTT palmitoylation was not observed in these high-throughput studies, but is well established (12,13,15,52). This represents a significant enrichment when compared with the total 1513 palmitoylated proteins in the nine datasets, against a background population of 20 774 protein-coding genes in the human genome (Ensembl release 72) (53) (P = 0.002, hypergeometric test). Palmitoylation of 16 HIP14 interactors, including HTT, has been observed in neuronal cells (39,40) and half of the palmitoylated interactors (14/27) have been observed in more than one palmitoyl-proteome study.
Figure 2.
Twenty-eight HIP14 Y2H interactors are palmitoylated. HIP14-interacting proteins are sorted in columns and shaded according to the number of published proteomic studies (1, 2, 3, 4 or 12) in which their palmitoylation has been detected. HTT palmitoylation was not detected in those proteomic studies but is well established. A white inner circle marks proteins whose palmitoylation has been detected in neuronal cells. For each protein, Supplementary Material, Table S4 provides Entrez Gene ID, description and citations for studies in which its palmitoylation was detected.
The palmitoylated HIP14 interactors in Figure 2 include known substrates (HIP14 and HTT), and putative novel substrates of HIP14. The synaptosomal protein SNAP23 is likely to be a bona fide substrate, as its membrane association is enhanced by HIP14 (20). Its close functional relative SNAP25 is a well-established neuronal substrate of HIP14 whose palmitoylation is reduced in the presence of mHTT (14,15,20,22,24,54,55). Ion channels and transporters, such as the HIP14 potassium channel substrate STREX-BK, are commonly palmitoylated to control channel maturation, trafficking and regulation (56), and represent a specific class of potential novel substrates. These include the glial high affinity glutamate transporter SLC1A3 (also known as GLAST or EAAT1) discussed below and the sodium/hydrogen exchanger SLC9A9 (NHE9) (Fig. 2).
All four cysteine-rich Sprouty domain-containing proteins, SPRED1, SPRED2, SPRY2 and SPRY4, have been observed as palmitoylated proteins in neuronal cells (Fig. 2; Supplementary Material, Table S4) (39,40). The amino acid sequence model for the Sprouty domain family (PfamID PF05210) (57) shows that the locations of at least eight cysteines are conserved and these represent potential palmitoylation sites. Thus, the SPREDs and SPRYs represent a special class of potential novel substrates of HIP14 for which no PAT has previously been identified.
HIP14 and HTT share an unusually large number of interactors
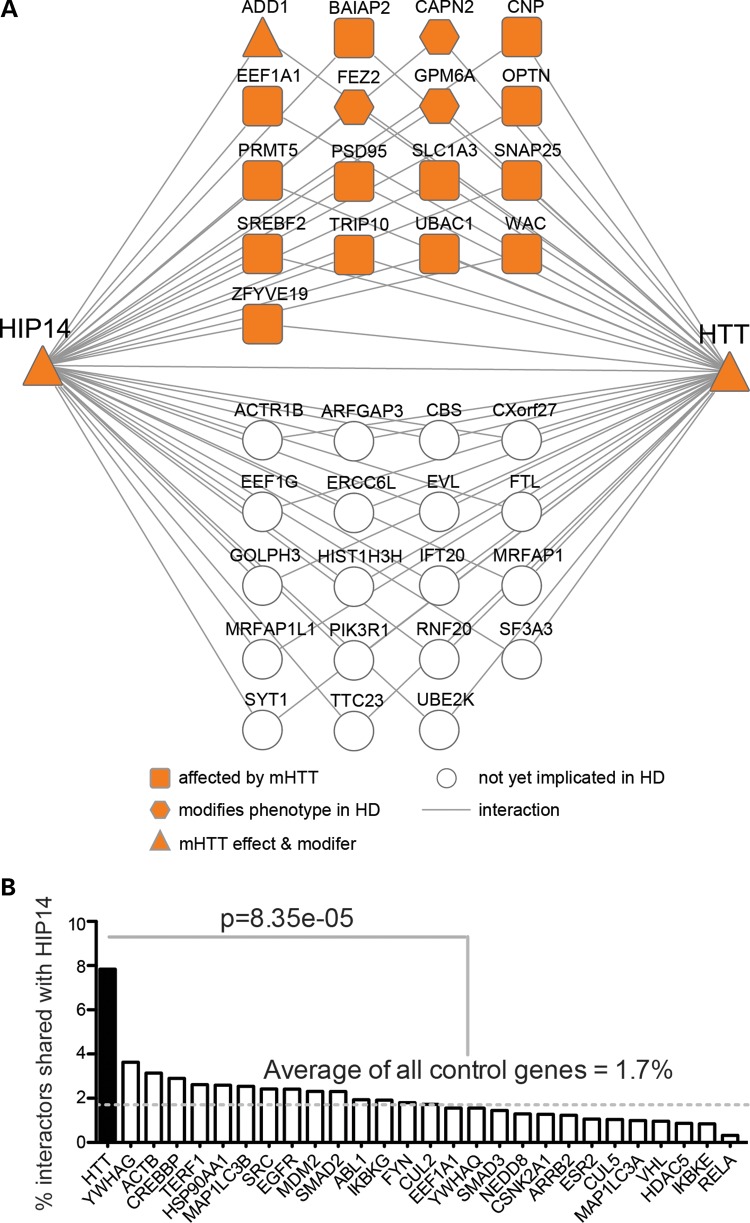
The ability of HTT to modulate HIP14-mediated palmitoylation of SNAP25 might be explained in part if HTT is acting as a scaffold to bring HIP14 together with its substrates (12). Reduced interaction between mHTT and HIP14 might be responsible for reduced palmitoylation and subsequent mislocalization of some HIP14 substrates. HTT interacts with the HIP14 substrates SNAP25 (3,4), PSD95 (DLG4) (14,15,22,58) and SYT1 (4,14). To determine whether our Y2H screen identified additional interactors that are shared with HTT, we compared the HIP14 PPI set plus the 16 published HIP14 substrates and interactors (Supplementary Material, Table S1) to all HTT interactors published to date. This included the exhaustive list curated in the HIPPIE database (59) and two recent quantitative affinity purification mass spectrometry (AP-MS) based studies of proteins that are physically associated with full-length HTT in vivo in mouse brain (2,3). In total, HIP14 and HTT shared 36 interactors, 32 of which are novel HIP14 interactors identified in this study (Fig. 3A; Supplementary Material, Table S6). These included HTT-associated proteins from both Y2H and AP-MS based studies (2–5,8,9,33,60–62).
Figure 3.
HIP14 and HTT share an unusually large number of interactors and many are implicated in HD. (A) Thirty-six HIP14 interactors also interact with HTT. Seventeen shared HIP14-HTT interactors have already been implicated in HD. Table 1 provides Entrez Gene ID, description and citations for these and Supplementary Material, Table S6 provides this information for all 36 shared interactors. (B) Plot of the percentage of a protein's interactors that also interact with HIP14. HTT had 370 interactors in HIPPIEdb. The 27 proteins that had the closest number of total interactors to that of HTT—between 300 and 440—were selected from HIPPIEdb as controls for HTT. Protein names are labeled on the x-axis. Numbers of HIPPIEdb interactors and UniProt IDs for each protein are provided in Supplementary Material, Table S5. HTT has four times more interactors in common with HIP14 than the control proteins, on average (P = 8.35 × 10−5, Fisher's exact test).
Given the fact that HTT interacts with hundreds of proteins, it is statistically likely to share interactors with other proteins. Therefore, we sought to determine whether this large overlap with HIP14 interactors is unusual. HTT has 370 interactions in HIPPIEdb (release date December 12, 2012). There are 27 other proteins with a similar number of interactions (between 300 and 440) in HIPPIEdb, and we considered these to be negative controls (Supplementary Material, Table S5). We extracted the interactors for each of the 27 control proteins and compared them to the HIP14 Y2H PPI set plus the 16 published HIP14 substrates and interactors (Supplementary Material, Table S1). For HTT, 29 of its 370 interactors (7.8%) were also HIP14 interactors (Fig. 3B). This is more than two times higher than the next closest protein, YWHAG, for which 14 of its 386 interactors (3.6%; P = 0.02, Fisher's exact test) were also HIP14 interactors. On average, HTT has four times more interactors in common with HIP14 than the control proteins do (1.7%; P = 8.35 × 10−5, Fisher's exact test).
Is this large number of shared interactors unique to HIP14's relationship with HTT, or could this be a feature of other polyglutamine-containing proteins? A recent study of HTT and Ataxin-1 (ATXN1) Y2H interactors (60) that were deposited into HIPPIEdb allowed us to make a direct comparison. A polyglutamine-encoding CAG trinucleotide repeat expansion in ATXN1 causes the neurodegenerative disorder Spinocerebellar Ataxia 1 (63). ATXN1 has 272 interactors in HIPPIEdb (release date December 12, 2012), and just 10 of these are shared with HIP14. This overlap (3.7%) is two times less than that between HTT and HIP14 (P = 0.03, Fisher's exact test), leading to the conclusion that sharing interactors with HIP14 is not a generic feature of polyglutamine-containing proteins like ATXN1. The significant number of shared HIP14-HTT interactors highlights the specific functional connection between these proteins.
Proteins that interact with both HIP14 and HTT are implicated in HD
What is the biological relevance of the overlap between proteins that interact with both HIP14 and HTT? Although HTT may act as a scaffold that links HIP14 to its substrates, not all of the shared interactors are likely to be palmitoylated. While a DAVID Functional Annotation analysis (38) did not reveal enrichment of any one function or process among the set of 36 shared HIP14-HTT interactors (data not shown), we noted that a subset of these proteins are involved in trafficking, echoing HTT's role in connecting cargoes to molecular motors to mediate intracellular protein transport (64,65). HTT and HIP14 might form a complex with these cargo adaptors to achieve proper intracellular transport of HIP14 to its substrates, which is consistent with the ability of HTT to modulate the activity of HIP14. Indeed, FEZ2 and optineurin, which are both cargo adaptors important in post-Golgi trafficking, are functionally linked to HTT. Optineurin shows decreased association with mHTT in striatal neurons (66) and its mRNA shows a significant neuron-specific decrease in the caudate nucleus of patients with early grade HD relative to controls (67,68).
The real potential for a contribution of HIP14 interactors to the pathogenesis of HD lies in the evidence that 17 of the 36 shared HIP14-HTT interactors have already been implicated in HD (Fig. 3A; Table 1) (3,4,15,54,58,66,67,69,70). Some, like optineurin, have altered interactions with mHTT compared with wtHTT. Others, like SNAP25 and SLC1A3, show reduced palmitoylation in brains of the YAC128 mouse model of HD, or, like the palmitoylated protein GPM6A, are genetic modifiers of HD-related phenotypes. Given the demonstrated relevance of these shared HIP14-HTT interactors for HD as well as the significant correlation of HTT protein abundance with that of HIP14 interactors GPM6A, SLC1A3 and PRMT5 in mouse brain (2), we suggest that the other interactors warrant investigation for a contribution to the pathogenesis of this disease.
Table 1.
Seventeen HIP14 interactors that interact with HTT and are implicated in HD
| Gene | Description | Effect of mHTT | Modifies phenotype in models of HD |
|---|---|---|---|
| HIP14 interactors—published | |||
| PSD95 | Post-synaptic density protein 95, alias DLG4 | 16Q < 56Q in vitro (58); reduced palmitoylation in brains of YAC128 mouse model of HD (54) | Not tested |
| SNAP25 | Synaptosomal-associated protein, 25 kDa | Reduced palmitoylation in vivo (15); reduced palmitoylation in brains of YAC128 mouse model of HD (54) | Not tested |
| SREBF2 | Sterol regulatory element binding transcription factor 2 | 23Q > 55Q in vitro (4) | Not tested |
| HIP14 interactors—this study | |||
| ADD1 | Adducin 1 (alpha) | 23Q > 55Q in vitro (ADD3; (4)) | Three loss alleles suppress fly eye degeneration (4) |
| BAIAP2 | BAI1-associated protein 2 | 23Q < 55Q in vitro (4) | Not tested |
| CAPN2 | Calpain 2, (m/II) large subunit | Not tested | Knockdown reduces aggregation and cell death (69) |
| CNP | 2′,3′-Cyclic nucleotide 3′ phosphodiesterase | 7Q < 140Q, in vivo (3); reduced palmitoylation in brains of YAC128 mouse model of HD (54) | Not tested |
| EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | Aggregate-interacting protein (70) | Not tested |
| FEZ2a | Fasciculation and elongation protein zeta 2 (zygin II) | Not tested | Three loss alleles enhance fly eye degeneration (4) |
| GPM6Ab | Glycoprotein M6A | Not tested | One loss allele suppresses fly eye degeneration; 1 gain allele enhances fly eye degeneration (4) |
| OPTN | Optineurin, alias HYPL | mHTT delocalizes OPTN/Rab8 complex from Golgi (66); neuron-specific mRNA decrease in HD caudate (67,68); 23Q = 55Q in vitro (4) | Not tested |
| PRMT5b | Protein arginine methyltransferase 5 | 7Q < 140Q, in vivo (3) | Not tested |
| SLC1A3b | Solute carrier family 1 (glial high affinity glutamate transporter), member 3, alias EAAT1, GLAST | Reduced palmitoylation in brains of YAC128 mouse model of HD (54) | Not tested |
| TRIP10 | Thyroid hormone receptor interactor 10 | 18Q < 128Q in vitro; no difference in HD brain coIP (61) | Not tested |
| UBAC1 | UBA domain containing 1 | 23Q > 55Q in vitro (4) | Not tested |
| WAC | WW domain-containing adaptor with coiled-coil | 23Q > 55Q in vitro (4) | Not tested |
| ZFYVE19 | Zinc finger, FYVE domain containing 19 | 23Q > 55Q in vitro (4) | Not tested |
aOrtholog interaction.
bProtein levels correlated with full-length HTT in (2).
Investigating selected Y2H interactions in mammalian cells
We next selected a broad representation of different types of interactors to examine their interaction with HIP14 in the high-throughput Luminescence-based Mammalian IntERactome co-immunoprecipitation (LUMIER) assay in HEK293 cells (33,71,72) (Table 2). These included putative substrates, known HTT interactors, proteins that have been implicated in HD, proteins with roles in trafficking, and potential regulators of HIP14. Full-length HIP14 and the ANK-focused HIP14 baits from the Y2H screen (Fig. 1B) were used as baits in the LUMIER assay. Eighteen of the 43 Y2H interactors tested interacted with full-length HIP14 in the LUMIER assay (42%) and 37 out of 43 interacted with at least one HIP14 bait (87%). Six of the 43 did not interact with any HIP14 bait (13%) (Table 2). Of the 15 palmitoylated proteins tested, 12 interacted with at least one HIP14 bait and out of 14 shared HIP14-HTT interactors, 11 interacted with at least one HIP14 bait. We considered preys that interacted with at least one HIP14 bait as valid interactors. Thus, 37 proteins have been shown to interact with HIP14 in independent assays in yeast and in mammalian cells.
Table 2.
Selected HIP14 Y2H interactors tested in LUMIER assay of interaction in HEK293 cells
| HIP14 full-length | HIP14-N-Ank | HIP14-N-Ank-DHHC | HIP14-Ank-DHHC | Gene | Entrez ID | Description | Palmitoylated | HTT interactor |
|---|---|---|---|---|---|---|---|---|
| + | + | + | + | BAIAP2 | 10458 | BAI1-associated protein 2 | Y | |
| + | + | + | + | GOLPH3L | 55204 | Golgi phosphoprotein 3-like | Y | |
| + | + | + | + | GPM6A | 2823 | Glycoprotein M6A | Y | Y |
| + | + | + | + | KLK8 | 11202 | Kallikrein-related peptidase 8 | ||
| + | + | + | + | PDPK1 | 5170 | 3-Phosphoinositide-dependent protein kinase-1 | ||
| + | + | + | LASP1 | 3927 | LIM and SH3 protein 1 | |||
| + | + | + | PLEKHB1 | 58473 | Pleckstrin homology domain containing, family B (evectins) member 1 | |||
| + | + | + | RAB39B | 116442 | RAB39B, member RAS oncogene family | |||
| + | + | + | SPRED1 | 161742 | Sprouty-related, EVH1 domain containing 1 | Y | ||
| + | + | + | SPRY2 | 10253 | Sprouty homolog 2 (Drosophila) | Y | ||
| + | + | + | USP32 | 84669 | Ubiquitin-specific peptidase 32 | Y | ||
| + | + | SETDB1 | 9869 | SET domain, bifurcated 1 | ||||
| + | + | SNAP23 | 8773 | Synaptosomal-associated protein, 23 kDa | Y | |||
| + | + | SPRY4 | 81848 | Sprouty homolog 4 (Drosophila) | Y | |||
| + | + | WASF2 | 10163 | WAS protein family, member 2 | ||||
| + | + | + | PPP2R5E | 5529 | Protein phosphatase 2, subunit B′, epsilon isoform | |||
| + | + | PIK3R1 | 5295 | Phosphoinositide-3-kinase, subunit 1 (alpha) | Y | Y | ||
| + | + | SLC9A9 | 285195 | Solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 | Y | |||
| + | + | + | GSK3A | 2931 | Glycogen synthase kinase 3 alpha | |||
| + | + | ATF2 | 1386 | Activating transcription factor 2 | ||||
| + | + | EVL | 51466 | Enah/Vasp-like | Y | |||
| + | + | FEZ2 | 9637 | Fasciculation and elongation protein zeta 2 (zygin II) | Y | |||
| + | + | IFT20 | 90410 | Intraflagellar transport 20 homolog (Chlamydomonas) | Y | |||
| + | + | IFT57 | 55081 | Intraflagellar transport 57 homolog (Chlamydomonas), alias HIPPI | ||||
| + | + | OPTN | 10133 | Optineurin | Y | |||
| + | + | GOLGA2 | 2801 | Golgin A2, alias GM130 | ||||
| + | EIF3E | 3646 | Eukaryotic translation initiation factor 3, subunit E | Y | ||||
| + | + | CAPN2 | 824 | Calpain 2, (m/II) large subunit | Y | |||
| + | APBB1IP | 54518 | Amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | |||||
| + | ARFGAP3 | 26286 | ADP-ribosylation factor GTPase activating protein 3 | Y | ||||
| + | CSNK1D | 1453 | Casein kinase 1, delta | |||||
| + | EEF1A1 | 1915 | Eukaryotic translation elongation factor 1 alpha 1 | Y | Y | |||
| + | FKBP6 | 8468 | FK506 binding protein 6, 36 kDa | |||||
| + | PDE4D | 5144 | Phosphodiesterase 4D, cAMP-specific | |||||
| + | DNAH11 | 8701 | Dynein, axonemal, heavy chain 11 | |||||
| + | SPRED2 | 200734 | Sprouty-related, EVH1 domain containing 2 | Y | ||||
| + | STX16 | 8675 | Syntaxin 16 | Y | ||||
| CNP | 1267 | 2′,3′-Cyclic nucleotide 3′ phosphodiesterase | Y | Y | ||||
| DAXX | 1616 | Death-domain associated protein | ||||||
| SLC25A31 | 83447 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 | ||||||
| SNW1 | 22938 | SNW domain containing 1 | ||||||
| TRIP10 | 9322 | Thyroid hormone receptor interactor 10 | Y | Y | ||||
| WAC | 51322 | WW domain-containing adaptor with coiled-coil | Y | Y |
‘+’ Indicates interaction detected with specific HIP14 bait; empty cell indicates no interaction detected.
‘Y’ indicates interactors that are palmitoylated or are HTT interactors.
SPRED1, SPRED3 and GPM6A are novel substrates of HIP14
We next selected several palmitoylated interactors to determine whether they were substrates of HIP14. The HIP14 Y2H PPI set was enriched for the cysteine-rich Sprouty domain and four of the seven known Sprouty domain-containing proteins interacted with HIP14 in both yeast and mammalian cell assays: SPRED1, SPRED2, SPRY2 and SPRY4 (73–75) (Table 2). None of these have an established connection with HTT but the SPREDs contain an EVH1 polyproline-binding domain that could theoretically interact with the proline-rich region of HTT. Palmitoylation of the SPREDs and SPRYs in neuronal cells has been observed in multiple studies, but their PATs are unknown (39,40) SPRED1, which interacted with full-length HIP14 in the LUMIER assay, was selected for further analysis. SPRED3, while not present in the Y2H prey set, was included in this analysis because it contains the Sprouty domain common to this group of interactors and is specifically expressed in the brain (76).
The neuronal membrane glycoprotein M6A (GPM6A), which interacted with full-length HIP14 and all three HIP14 fragment baits in the LUMIER assay, was also selected for further study. This synaptic protein interacts with HTT in human and mouse brain, its protein levels are correlated with those of full-length HTT across striatum, cortex and cerebellum in mouse brain, and gain and loss of function alleles in Drosophila modify the HTT-fragment induced neurodegeneration phenotype in the eye (2–4). Palmitoylation of GPM6A has been observed in rat neuronal cultures and synaptosomal fractions and in mouse neuronal stem cells (39,40), but its PAT is unknown. Thus it is a potential novel substrate of HIP14.
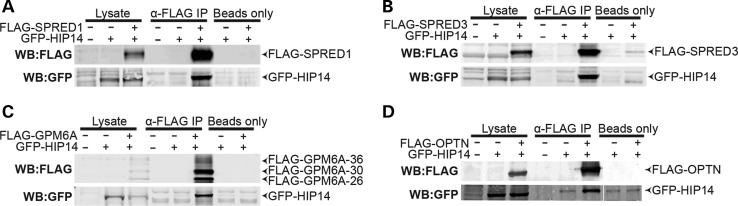
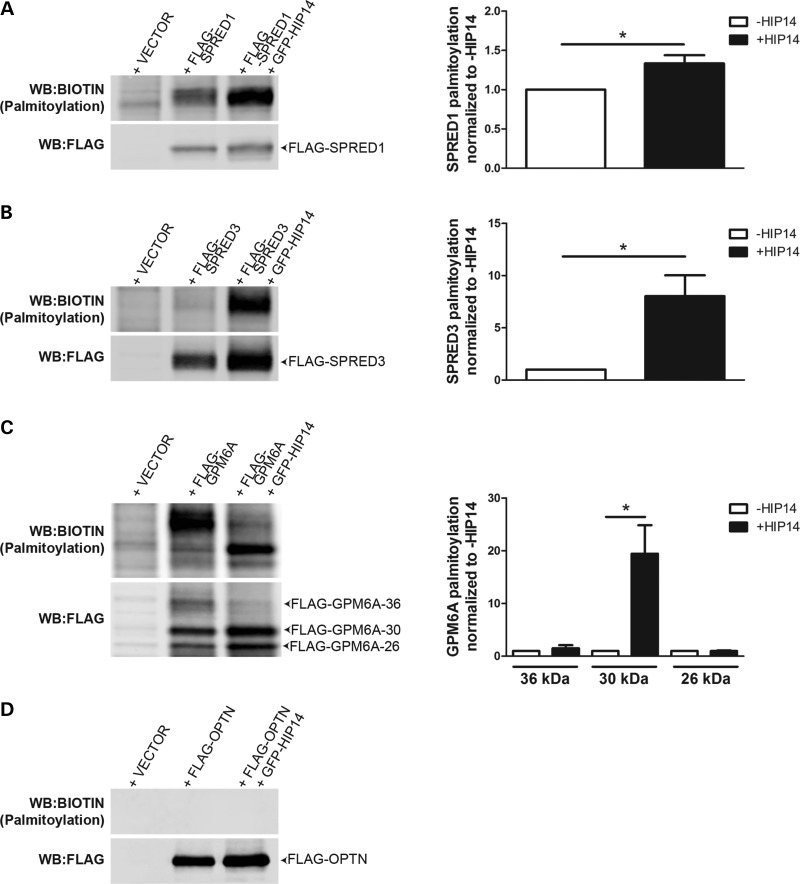
To define their relationships with HIP14, each putative substrate, SPRED1, SPRED3, and GPM6A, was tested for interaction with full-length HIP14 by co-immunoprecipitation. HIP14-GFP was co-expressed in HEK293 cells with each of the FLAG-tagged putative interactors. Following FLAG immunoprecipitation from cell lysates, western blots confirmed that HIP14-GFP was co-immunoprecipitated in each case, indicating that full-length HIP14 can interact with SPRED1, SPRED3 and GPM6A (Fig. 4). To determine whether HIP14 is a PAT for any of these novel interactors, HEK293 cells transiently expressing each FLAG-tagged interactor with or without HIP14-GFP were metabolically labeled with the palmitate analog 17-octadecyonic acid (alkyne-C18; 17-ODYA). Immunoprecipitated FLAG proteins were subjected to Click chemistry to label each 17-ODYA molecule with biotin, and the palmitoylation signal was normalized to protein levels detected by western blot. HIP14 increased mSPRED1 palmitoylation by 1.35-fold (P = 0.03; Wilcoxon signed-rank test) and mSPRED3 palmitoylation by 8.0-fold (P = 0.03; Wilcoxon signed-rank test), suggesting that these interactors are substrates of HIP14 (Fig. 5). FLAG-GPM6A presented as a triplet by western blot (26, 30 and 36 kDa), and HIP14 increased palmitoylation of the 30 kDa band by 19.5-fold (P = 0.03; Wilcoxon signed-rank test) suggesting that GPM6A is also a HIP14 substrate (Fig. 5C). There was no significant change in palmitoylation of the other two bands (36 and 26 kDa), indicating that HIP14 may be responsible for palmitoylating a specific form of GPM6A.
Figure 4.
HIP14 coimmunoprecipitates with SPRED1, SPRED3, GPM6A and OPTN. Co-expression of HIP14-GFP (NM_015336) with (A) FLAG-SPRED1 (mouse NM_033524), (B) FLAG-SPRED3 (mouse NM_182927), (C) FLAG-GPM6A (human NM_005277), or (D) FLAG-OPTN (human NM_001008211) in HEK-293 cells followed by FLAG immunoprecipitation results in detection of HIP14 by immunoblot. Fig. 4D is a composite of multiple lanes from a single western blot. n = 3 for SPRED1, SPRED3, OPTN; n = 4 for GPM6A.
Figure 5.
HIP14 palmitoylates SPRED1, SPRED3 and GPM6A but not OPTN. FLAG-tagged HIP14 interactors expressed alone or co-expressed with HIP14-GFP in HEK-293 cells and labeled with 17-ODYA were immunoprecipitated and subjected to Click chemistry to detect palmitoylation levels (calculated as the ratio of signal from the Biotin western blot over the signal from the FLAG western blot). Palmitoylation of (A) FLAG-SPRED1 and (B) FLAG-SPRED3 was increased 1.3-fold and 8.0-fold, respectively, in the presence of HIP14 (P = 0.03; Wilcoxon signed-rank test; n = 6 independent experiments). (C) FLAG-GPM6A-36 (36 kDa) is palmitoylated in the absence of HIP14, while palmitoylation of FLAG-GPM6A-30 (30 kDa) is increased 19.5-fold by HIP14 (P = 0.03; Wilcoxon signed-rank test; n = 6 independent experiments). (D) FLAG-OPTN is not palmitoylated. (P = 0.03; Wilcoxon signed-rank test; n = 6 independent experiments).
GPM6A is glycosylated at position N164 (77) and therefore we hypothesized that the triplet observed by western blot represents differentially glycosylated forms of the protein. GPM6A immunoprecipitated from transiently transfected HEK293 cells was subjected to glycosidase treatment to determine the modification state of each band (Supplementary Material, Fig. S1A). Treatment with peptide N-glycosidase (PNGase)-F, which recognizes and cleaves all N-linked carbohydrates, reduced the GPM6A 36, 30 and 26 kDa bands to a single 26 kDa band (Supplementary Material, Fig. S1B). Treatment with endoglycosidase H (EndoH), which cleaves core (high-mannose) N-linked oligosaccharides present in the ER or cis-Golgi but not hybrid or complex N-glycans, reduced only the 30 kDa band to the 26 kDa position (Supplementary Material, Fig. S1B). These results indicated that the 30 kDa band is a high-mannose form of GPM6A that might be palmitoylated by HIP14 in the cis-Golgi (8,14).
HIP14 alters the subcellular localization of SPRED1 and SPRED3
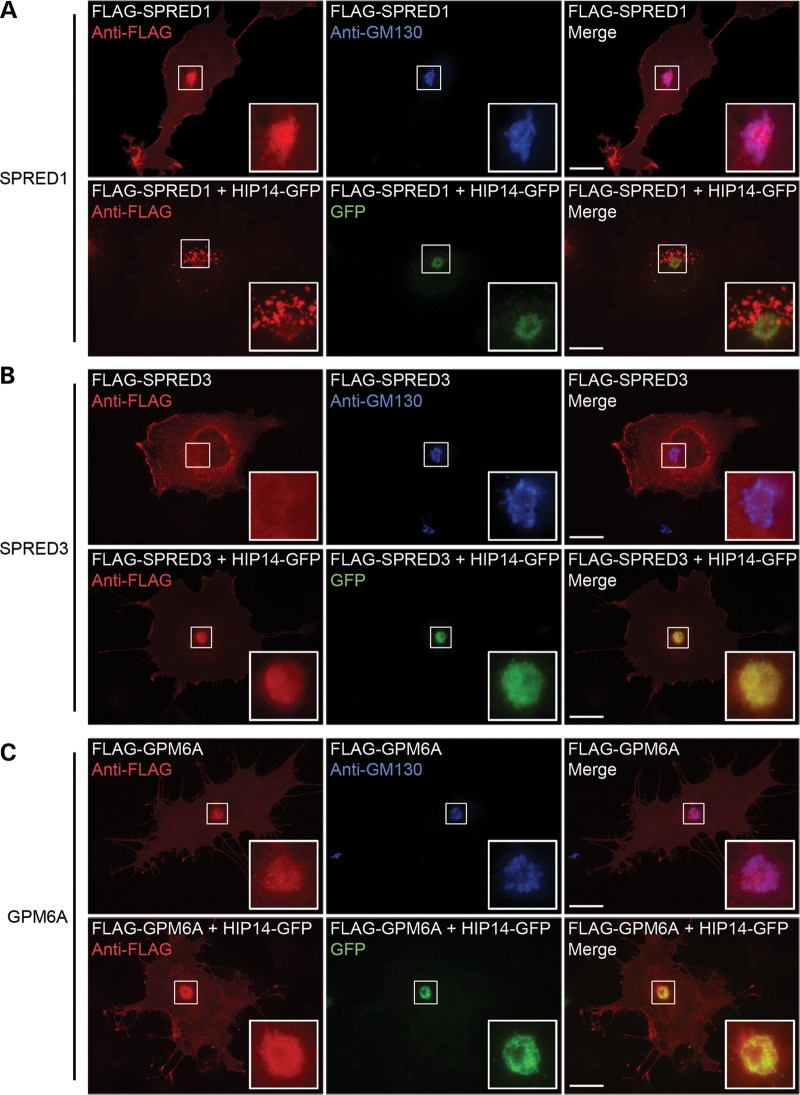
Consistent with the role of palmitoylation in protein trafficking and membrane localization, overexpression of HIP14 has been shown to dramatically alter the subcellular localization of its confirmed substrates HTT, PSD95 and SNAP25 (13,14). Therefore, we investigated the effect of exogenous HIP14 on the localization of SPRED1, SPRED3 and GPM6A. As previously observed (14,22), HIP14-GFP transiently expressed in COS-7 cells co-localized with the cis-Golgi marker GM130 by immunofluorescence microscopy (data not shown). FLAG-mSPRED1 expressed in COS-7 cells showed localization to the Golgi and to the plasma membrane. When co-expressed with HIP14-GFP, FLAG-mSPRED1 redistributed to numerous vesicular structures proximal to the Golgi (Fig. 6A). FLAG-mSPRED3 localized mainly to the cytosol and plasma membrane in COS-7 cells, however, it strongly localized to the Golgi when co-expressed with HIP14-GFP (Fig. 6B).
Figure 6.
HIP14 influences subcellular localization of SPRED1 and SPRED3 but not GPM6A in COS-7 cells. HIP14 substrates expressed alone or with HIP14-GFP in COS-7 cells were visualized by immunofluorescence microscopy. (A) FLAG-SPRED1 localized to the plasma membrane and colocalized with the Golgi marker GM130. Co-expression with HIP14-GFP promoted relocalization of FLAG-SPRED1 to vesicles. (B) Overexpression of FLAG-SPRED3 showed cytosolic and plasma membrane staining with no Golgi localization. Co-expression with HIP14-GFP promoted its colocalization in the Golgi. (C) FLAG-GPM6A localizes to the Golgi and to filopodia. No change was seen with HIP14-GFP co-expression; both proteins colocalized to the Golgi. Scale bar = 20 μm.
Unlike peripheral membrane proteins SPRED1 and SPRED3, GPM6A is an integral membrane protein. However, because it was palmitoylated by HIP14 its localization pattern was also investigated using immunofluorescence microscopy. Expression of FLAG-GPM6A in COS-7 cells resulted in its localization to both the Golgi and to filopodia, consistent with its role in the formation of filopodial projections in this cell type (78). Co-expression of FLAG-GPM6A with HIP14-GFP did not alter this localization pattern to any observable degree, indicating that palmitoylation by HIP14 may play an alternate role in the function of GPM6A compared with that of the SPREDs or that interactions with other proteins control the ultimate fate of palmitoylated GPM6A (Fig. 6C).
HIP14 interacts with but does not palmitoylate optineurin
Optineurin is known to form a complex with both HTT and RAB8 that links post-Golgi vesicles to Myosin VI motors to promote proper trafficking of cargoes from the Golgi to the plasma membrane and lysosomes. In the brains of HD patients, optineurin mRNA is significantly reduced and in striatal neurons and the absence of Golgi-localized HTT destabilizes the optineurin/Rab8 complex, leading to aberrant cargo trafficking (66–68). Optineurin has not previously been observed to be palmitoylated. In the co-immunoprecipitation experiments described above, optineurin interacted with full-length HIP14 in HEK293 cells (Fig. 4D). In the Click palmitoylation assay, however, FLAG-optineurin presented no detectable palmitoylation signal alone or in the presence of HIP14 (Fig. 5D), indicating that optineurin is not palmitoylated and suggesting this interaction is independent of an enzyme–substrate relationship and may serve an alternate function.
DISCUSSION
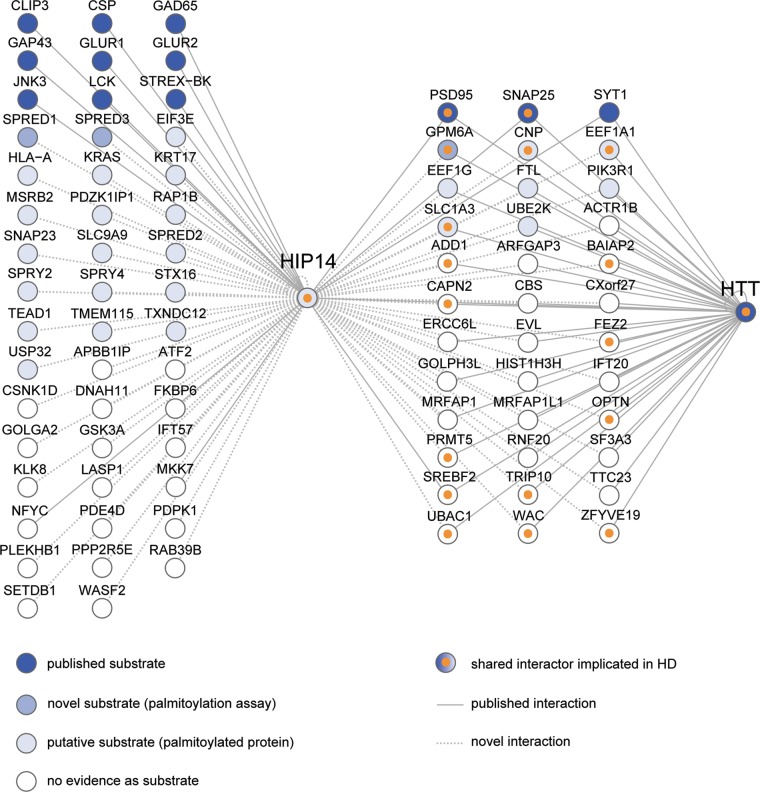
Understanding the scope of HIP14's contribution to the pathogenesis of HD will require the identification of the full range of proteins that interact with HIP14. Prior to this study, HIP14 was known to have 16 substrates and interactors and our results increase this total number to 229. Figure 7 summarizes our expanded knowledge of HIP14-interacting proteins.
Figure 7.
Summary of HIP14 interactors characterized to date. HIP14-interacting proteins are depicted as nodes (circles) that are colored from dark blue to light blue according to their status as potential HIP14 substrates: published substrates, novel substrates confirmed by palmitoylation assay in this study and palmitoylated proteins that are putative substrates. Interactors with no evidence of palmitoylation are colored white. Shared HIP14-HTT interactors that are already implicated in HD are marked with orange inner circles. Interactions between two nodes are depicted as edges (lines) between nodes: published interactions with HIP14 or HTT (solid lines); novel interactions identified in this study by one or more assays including Y2H, LUMIER and co-immunoprecipitation (dotted lines). The following are official gene names for interactors identified by common names in the figure: CSP (DNAJC5), GAD65 (GAD2), GLUR1 (GRIA1), GLUR2 (GRIA2), JNK3 (MAPK10), STREX-BK (KCNMA1), PSD95 (DLG4) and MKK7 (MAP2K7).
HIP14 and HTT share an unusually large number of interactors and many are implicated in HD
Our most provocative finding is the high degree of overlap between HIP14 and HTT interactors. HTT shares 7.8% of its interactors with HIP14 and several lines of evidence suggest this overlap is specific to HIP14's relationship to HTT. First, the number of shared interactions is 4-fold greater than that seen for 27 matched control proteins (P = 8 × 10−5). Furthermore, ATXN1, whose CAG-expansion causes a neurodegenerative disorder similar to HD, shares only 3.7% of its interactors with HIP14. The striking overlap between the interactors of HIP14 and HTT suggests that HIP14 is critically linked to HTT through its interactions. In total, 36 HIP14 interactors are also known to interact with HTT (Fig. 7). The fact that 17 of these, including the novel synaptic substrate GPM6A and the potential HIP14 regulator optineurin, have previously been implicated in HD, provides strong evidence that HIP14 is a crucial link to the disease. These findings warrant closer investigation of HIP14 interactors that have not yet been linked to HD. We discuss several mechanisms below through which disturbed function of HIP14 interactors in the presence of mHTT may contribute to the pathogenesis of HD.
The vesicular cargo adaptor optineurin is a potential regulator of HIP14
Some of the 36 shared HIP14-HTT interactors have roles that relate to HTT's function as a scaffold that facilitates protein trafficking. HIP14's interaction with optineurin, FEZ2, RAB39B, the intraflagellar transport proteins IFT20 and IFT57 and the dynein motor component DNAH11 could reflect a role for these proteins in directing HIP14 trafficking (64,79–81). Another PAT, ZDHHC2, dynamically cycles between the plasma membrane and endocytic compartments to mediate PSD95 palmitoylation at the synapse (82–84). While HIP14 localizes mainly in the Golgi in mammalian cells, it has been observed at additional locations including cytoplasmic/synaptic vesicles and nerve terminals (8,14,85). It is possible that, like ZDHHC2, HIP14 dynamically cycles between different cellular compartments through binding these trafficking proteins to mediate palmitoylation of localized substrates.
One of these trafficking proteins, optineurin, interacts directly with HTT to link post-Golgi vesicles to myosin motors at synapses and mHTT leads to aberrant cargo trafficking by optineurin (62,86). We found that optineurin interacts with HIP14 but is not palmitoylated and therefore is not a HIP14 substrate. Instead, HIP14 might be a cargo of post-Golgi vesicles destined for the plasma membrane or synapses. In HD, mislocalization of optineurin by mHTT might impair proper HIP14 trafficking in a dominant-negative manner, thereby reducing the synaptic localization of HIP14 and affecting the palmitoylation of synaptic substrates.
Neuronal glycoprotein GPM6A is a novel HIP14 substrate
GPM6A is genetic modifier of neurodegeneration in a Drosophila model of HD and its protein levels are significantly correlated with those of HTT across striatum, cortex and cerebellum in mouse, suggesting that GPM6A and HTT participate in a functional complex (2–4). We confirmed that GPM6A interacts with full-length HIP14 and that HIP14 increased the palmitoylation of GPM6A by 20-fold, providing evidence that it is a novel HIP14 substrate and identifying for the first time a PAT for GPM6A. It remains possible that co-expression of HIP14 with potential substrates in HEK-293 cells affects another factor that in turn directly palmitoylates these proteins. In order to further confirm GPM6A as a substrate of HIP14, its palmitoylation could be assessed in cell-free assays using purified proteins.
Palmitoylation of the known HIP14 substrate PSD95 promotes its clustering at synapses (87), and it is possible that palmitoylation of GPM6A plays a similar role. Alternatively, palmitoylation may induce a conformational change, promoting altered protein interactions or other functional consequences. GPM6A overexpression promotes filopodia and neurite formation in rat hippocampal neurons and its knockdown by siRNA results in reduced spine and synapse numbers (78). Both the YAC128 mouse model of HD and Hip14−/− mice have significantly reduced numbers of excitatory synapses (15). Palmitoylation of GPM6A may be important for its normal role in filopodia outgrowth and synapse formation, and aberrant palmitoylation in the presence of mHTT could potentially contribute to the synapse abnormalities observed in HD. The biological significance of palmitoylation of GPM6A by HIP14 should be assessed in vivo in HIP14-deficient mice, for example.
Other shared HIP14-HTT interactors might contribute to the pathogenesis of HD
Many interactors that are common to HIP14 and HTT have already been implicated in HD, so careful consideration of the functions of the other shared interactors may shed light on the processes through which HIP14 dysfunction contributes to the pathogenesis of the disease. Excitotoxicity, the overstimulation of neurons by glutamate, is believed to play a key role in neuronal cell death in HD and other neurodegenerative disorders (88,89). Excitotoxicity can occur through increased pre-synaptic glutamate release, increased numbers of post-synaptic glutamate receptors, or reduced glutamate uptake from the synapse by glutamate transporters (90,91). We identified the glial high affinity glutamate transporter SLC1A3 as a candidate substrate of HIP14. Protein levels of SLC1A3 are significantly correlated with those of full-length HTT across the striatum, cortex and cerebellum in mice, suggesting that SLC1A3 and HTT participate in a functional complex (2). SLC1A3 is localized primarily in the plasma membrane of astrocytes and clears extracellular glutamate from excitatory neuronal synapses. In patients with Episodic ataxia, mutations in SLC1A3 are associated with reduced glutamate uptake (92,93).
Palmitoylation of SLC1A3 is significantly reduced (−14%, P = 0.044) in brains of 12-month old mice in the YAC128 mouse model of HD and shows a trend toward a reduction (−7%, P = 0.058) in brains of Hip14−/− mice (54). We have hypothesized that in the presence of mHTT, HIP14 is less able to palmitoylate key synaptic substrates, thus contributing to synaptic dysfunction in HD. The observation that palmitoylation of SLC1A3 is reduced in an HD mouse model provides a direct link between SLC1A3 palmitoylation and HD. Reduced palmitoylation may lead to reduced glutamate uptake by SLC1A3 by prevention of forward trafficking of SLC1A3 from the Golgi to the plasma membrane, improper targeting to plasma membrane microdomains, or by altering other post-translational modifications of SLC1A3 leading to decreased function.
The cysteine-rich sprouty domain-containing proteins SPRED1 and SPRED3 are novel HIP14 substrates
Recent technical advances in identification of S-acylated proteins have resulted in rich datasets of candidate substrates for PATs. Comparison of the HIP14 PPI set to the supplemental data from 13 published mammalian palmitoyl-proteomes (39–51) revealed a significant enrichment for palmitoylated proteins and identified 27 novel candidate substrates of HIP14 (Fig. 7). One compelling new class of HIP14 substrates identified here is the set of proteins that contain a cysteine-rich Sprouty domain. SPRY2, SPRY4, SPRED1 and SPRED2 were identified in the Y2H screen, and confirmed in the LUMIER assay. An interaction between the Drosophila orthologs of HIP14 and SPRY2 was previously reported (94). The Sprouty domain is thought to be the site of their palmitoylation and is important for both membrane localization and function (73,76,95–97). Palmitoylated forms of all four Sprouty domain-containing interactors have been detected in neuronal cells (39,40).
We provide evidence that SPRED1 and the brain-specific isoform SPRED3 interacted with and were palmitoylated by HIP14. It is possible that overexpression of HIP14 with SPRED1 or SPRED3 in HEK-293 cells may affect another factor that in turn directly palmitoylates these proteins. In order to further confirm SPRED1 and SPRED3 as substrates of HIP14, their palmitoylation could be assessed in cell-free assays using purified proteins and in vivo in HIP14-deficient mice. HIP14 promoted the relocalization of SPRED1 from the Golgi to observable vesicular structures, and the relocalization of SPRED3 from the cytosol to the Golgi, as is often observed with overexpression of HIP14 with its substrates, such as HTT (13). It is possible that palmitoylation by HIP14 promotes the exit and forward trafficking of SPRED1 from the Golgi in this system. These lines of evidence support the hypothesis that palmitoylation by HIP14 may be necessary for proper endomembrane targeting of its novel substrates SPRED1 and SPRED3.
SPRED1 inhibits the RhoA-induced activation of ROCK (97), a rho-associated kinase that phosphorylates profilin, myosin and cofilin to influence cytoskeletal remodeling and neurite retraction (98,99). Chemical inhibition of ROCK promotes the clearance of mHTT exon 1 aggregates in various cell lines (100–102). Given the established roles for palmitoylation and membrane localization in SPRED function, it is possible that palmitoylation by HIP14 may be imperative for the SPREDs′ inhibitory function of the ROCK pathway. This suggests a potential role for the SPREDs and their palmitoylation in the clearance of misfolded polyglutamine proteins.
CONCLUSIONS
The HIP14 Y2H study described here, the first for a mammalian PAT, revealed a striking and specific overlap between interactors of HIP14 and HTT. Nearly half of the HIP14-HTT shared interactors, including GPM6A and optineurin, are implicated in HD, providing a further molecular rationale for the similarities between phenotypes of Hip14−/− mice and the YAC128 mouse model of HD. The confirmed interaction between HIP14 and optineurin reveals a potential mechanism of HIP14 regulation that in the presence of mHTT might lead to reduced substrate palmitoylation. Taken together, our data suggest involvement of multiple candidate HIP14 substrates and regulators in HD, highlighting the significance of palmitoylation by HIP14 as a key contributor to pathogenesis of the disease.
MATERIALS AND METHODS
Yeast two-hybrid screen
An automated Y2H screen was performed as described previously (35). In brief, cDNAs encoding three HIP14 bait protein fragments N-Ank, Ank-DHHC, N-Ank-DHHC (Fig. 1B) were subcloned into the yeast expression vector pBTM116-D9 for synthesis of LexA DNA binding domain fusions and the resulting plasmids were transformed into the MATa yeast strain L40ccua. Bait constructs were tested for auto-activation of reporter genes HIS3, URA3, and lacZ after mating with a MATα strain expressing a Gal4 activation domain protein. None of the baits were auto-activating and were therefore suitable for interaction mating assays with prey proteins. To create a prey-matrix for interaction mating, the MATα yeast strain L40ccα was individually transformed with pACT4-DM-based plasmids for Gal4 activation domain fusions with prey proteins.
For interaction mating, liquid cultures of MATα yeast strains (preys) were replicated in 384-well microtiter plates using a pipetting robot (Tecan, Freedom EVOware®), mixed with MATa strains (baits) and transferred onto YPD agar plates. Diploid yeast carrying both bait and prey vectors were selected on SDII (-Leu-Trp) media and protein–protein interactions were identified by growth on SDIV (-Leu-Trp-Ura-His) selective plates and by positive lacZ reporter gene activation assays. The SDIV and lacZ assays were done in four replicates. Positive clones were identified by activation of the HIS3 and URA3 reporters on SDIV selective plates and/or lacZ reporter gene activation. An interaction was scored when ≥3 positives were detected (sum of SDIV and LacZ signals in the four replicate experiments).
The initial mating was conducted with the three HIP14 baits in an eight-bait pool together with other constructs. To determine specific bait–prey interactions, these baits were individually arrayed in 96-well microtiter plates and mated with the positive preys identified in the first mating screen. The second interaction mating was done in three replicates and an interaction was scored when at least one positive signal was detected; this lower threshold was used in the second interaction screen so that transient but potentially functional interactions were less likely to be excluded. After exclusion of auto-activating preys, this yielded a total of 460 bait–prey interactions (described in (35)). To maximize the specificity of these interactions for HIP14, we selected only those preys that did not interact with all baits from the original pool.
All protein interactions have been submitted to the IMEx Consortium (http://www.imexconsortium.org) through IntAct (30) and assigned the identifier IM- 21827.
Gene list annotation
To obtain functional information about each gene, the HIP14 Y2H interactors were annotated using the NCBI Entrez Gene and BioMart web services in Cytoscape (version 2.8. 3, accessed December 11, 2012) using the Entrez GeneID as the primary identifier (103–105). The following annotations were imported: Official Gene Symbol, Description, RefSeq Summary, Aliases, Gene Ontology terms for Cellular Component, Biological Process and Molecular Function (106), Pathways from KEGG (107) and Reactome (108) and PfamIDs for protein domains (57) (Supplementary Material, Table S3). Five interacting preys annotated as pseudogenes or non-coding RNAs were excluded from further analysis (Entrez GeneIDs: 100128252, 653188, 554206, 90133 and 65996).
Palmitoylated protein datasets
A total of 1513 non-redundant mammalian palmitoylated proteins were collected from supplemental data in nine papers comprising 13 individual published datasets. Wilson et al. (44) included palmitoylated proteins from their own study as well as from four other studies and these were obtained from Supplementary Material, Table S3, Sheet ‘All studies’ high and medium confidence lists. All matches between genes from the HIP14 PPI set and those from publications whose data were compiled in Wilson et al. were manually confirmed in the source datasets. Additional palmitoylated proteins were obtained from Ivaldi et al. (46) Supplementary Material, Table S3, Marin et al. (47) Supplementary Material, Table S1, Dowal et al. (48) Supplementary Material, Table S3 only for proteins with enrichment in hydroxylamine sensitive treatment at P < 0.05, Forrester et al. (50) Supplementary Material, Table S1, Martin et al. (41) Supplementary Material, Table S2, Li et al. (40) Supplementary Material Dataset S1, Supplementary Material, Table 5 only for 17-ODYA-modified and hydroxylamine sensitive proteins labeled ‘BOTH’, Merrick et al. (42) Supplementary Material, Table S10 and Yount et al. (43) Supplementary Material, Table S1 and S2. Where publishers provided data in PDF format, Excel formatted data were requested from and provided by the authors. ID conversions and comparisons were done as described below.
Note that proteins reported as palmitoylated in studies using methyl methanethiosulfonate (MMTS) in palmitoylation assays to block free thiols on cysteine residues, should be confirmed by an independent assay, because unlike N-ethylmaleimide (NEM), the disulfide bonds formed by MMTS are readily cleaved by the hydroxylamine treatment, resulting in potential false positives.
HTT interactor datasets
HTT interactors were obtained from the Human Integrated Protein–Protein Interaction rEference (HIPPIE) database (http://cbdm.mdc-berlin.de/tools/hippie; Release date December 12, 2012; downloaded in ‘hippie’ format) (59) and from two quantitative in vivo AP-MS based studies of full-length HTT-associated proteins (2,3). HIPPIEdb contains human Entrez GeneID-indexed HTT interaction data from seven key publications (4,5,9,60–62,109). From the AP-MS study by Culver et al. (3) we obtained IPI protein identifiers from Supplementary Material, Table S3 (S2 cytoplasmic 7Q and 140Q high confidence candidate interacting proteins) and Supplementary Material, Table S4 (P2 membranous 7Q and 140Q high confidence candidate interacting proteins). From the AP-MS study by Shirasaki et al. (2) we obtained UniProt identifiers from Supplementary Material, Table S7, Sheet ‘All Proteins’ for those proteins detected under two or more immunoprecipitation conditions. ID conversions and comparisons were done as described below.
Gene and protein ID conversions and comparisons
Comparison of the HIP14 PPI set to lists of palmitoylated proteins and HTT interactors from other sources required conversions of gene and protein identifiers. Sources provided one or more types of identifiers: Entrez Gene IDs (e.g. 3064), IPI protein IDs (e.g. IPI00107625), protein GI numbers (e.g. 18765733), UniProt protein Accession numbers, (e.g. P60880), RefSeq protein IDs (e.g. NP_003072), Ensembl Gene IDs (e.g. ENSG00000132639) and/or gene name and description. Use of gene names as the primary identifier was avoided where possible due to the potential for ambiguity. The Synergizer web tool (http://llama.mshri.on.ca/synergizer/translate/; accessed June 6, 2013) was used to translate identifiers from one type to another using NCBI or Ensembl Genes release 65 as the authority (53,104). Ambiguous conversions were manually assigned. For comparison of human genes from this study to mouse genes from other studies, Entrez GeneIDs for mouse orthologs of the HIP14 Y2H interactors were obtained using Ingenuity mappings (http://www.ingenuity.com/) (Supplementary Material, Table S3). List comparisons were done using Microsoft Excel with care to avoid errors in its automatic conversion of some gene names to dates (e.g. March 5 and September 11) (110). Both Entrez GeneIDs and gene symbols were compared with confirm matches whenever possible.
Generation of gateway entry plasmids
cDNAs encoding human HIP14 fragments HIP14A (HIP14-N-Ank; amino acids 1–257), HIP14B (HIP14-Ank-DHHC; amino acids 64–487), and HIP14C (HIP14-N-Ank-DHHC; amino acids 1–487) as well as full-length HIP14 (amino acids 1–632) were PCR amplified using High Fidelity DNA polymerase (Roche) from the plasmid pGAD HIP14 (NM_015336, NP_056151) and cloned into the Gateway plasmid pDONR221 (Invitrogen). Oligonucleotides (HIP14_A_C_FL_for, HIP14_B_for, HIP14_A_rev, HIP14_B_C_rev and HIP14_FL_rev, Supplementary Material, Table S7) were purchased from Invitrogen Canada. PCR reactions (50 µl volume) contained 50 ng plasmid DNA, 125 pmol primer oligonucleotides, 200 μm dNTPs and 2.5 U DNA polymerase. Fragments were amplified in 35 cycles with the following profile: 1 min denaturation at 95°C, 1 min annealing at 55°C and 2 min extension at 68°C. Amplified DNA fragments were purified using 30% polyethylene glycol 8000/30 mm MgCl2 and combined with the pDONR221 plasmid (Invitrogen) at a 1:1 ratio in the presence of BP clonase (Invitrogen). After overnight incubation at 25°C the DNA of entry plasmids was transformed into competent OmniMAX 2-T1 cells (Invitrogen). The identity of the DNA fragments was verified by DNA sequencing.
LUMIER assay of interactions in mammalian cells
LUMIER assays were performed as described previously (33,71). Briefly, protein A (PA)-Renilla luciferase (RL)-tagged HIP14 fusion proteins were co-produced with firefly luciferase (FL)-tagged putative interactor proteins in HEK293 cells. Both orientations were tested for each interaction. After 48 h, protein complexes were co-immunoprecipitated from cell extracts with IgG coated magnetic beads (Dynabeads®; Invitrogen); interactions between bait (PA-RL fusions) and prey proteins (FL fusions) were monitored by quantification of firefly luciferase activities. Quantification of Renilla luciferase activity was used to confirm that PA-RL-tagged bait proteins was successfully immunoprecipitated from cell extracts. To detect Renilla and firefly luciferase based luminescence in samples with fusion proteins the Dual-Glo Luciferase Kit (Promega) was used. Bioluminescence was quantified in a luminescence plate reader (TECAN Infinite M1000). To assess the specificity of an interaction between the proteins X and Y the protein pairs (A) PA-RL-X/FL-Y, (B) PA-RL/FL-Y and (C) PA-RL-X/FL were individually co-produced in HEK293 cells. R_op and R_ob binding ratios were obtained by dividing the firefly luminescence activity measured in sample A by activities found in samples B and C. Low R_op values are an indication for non-specific prey protein interactions, while low R_ob values indicate non-specific bait protein interactions. Based on empirical studies with a set of well-characterized positive and negative interaction pairs (not shown), we defined that R_op and R_ob binding ratios of >1.5 indicate reliable, specific protein–protein interactions.
cDNAs
FLAG-optineurin was generated by PCR amplification from optineurin cDNA (human, NM_001008211) (Openbiosystems) and subcloning into EcoRI and XhoI sites of pCMV-FlagNT (gift from Dr Stefan Taubert, the University of British Columbia). FLAG-GPM6A (human, NM_05277) was purchased from OriGene. FLAG-Spred1 (NM_033524) and FLAG-Spred3 (NM_182927) mouse cDNA clones were a gift from A. Yoshimura, Keio University School of Medicine.
Co-immunoprecipitation
HEK293 cells were harvested 24 h post-transfection and lysates incubated 16 h at 4°C with PureProteomeTM Protein G magnetic beads (Millipore) bound to IP antibody [Mouse Anti-FLAG, 4 μl per sample (OriGene)]. Beads were washed and eluted samples separated on a 4–12% Bis–Tris gel in 4-Morpholinepropanesulfonic acid (MOPS)-sodium dodecyl sulphate (SDS) running buffer (Invitrogen). Proteins were transferred to nitrocellulose and analyzed by immunoblotting with primary antibodies [Mouse Anti-FLAG, 1:1000 (OriGene) and Rabbit Anti-GFP, 1:5000 (Eusera)] and secondary antibodies [Alexa Fluor® 680 Goat Anti-Rabbit, 1:5000 and Alexa Fluor® 800 Goat Anti-Mouse, 1:5000]. Blots were scanned using the Li-Cor Odyssey® Infrared Imaging System.
Click palmitoylation assays
Metabolic labeling of HEK293 cells with 17-ODYA and subsequent Click chemistry were performed as described previously (45,111). Briefly, 24 h post-transfection, HEK293 cells were labeled with 100 μm 17-ODYA for 5 h. Cells were harvested and lysed in modified radioimmunoprecipitation assay (RIPA) buffer containing 0.1% SDS and complete protease inhibitor (Roche). Lysates were incubated for 2 h at room temperature with PureProteomeTM Protein G magnetic beads (Millipore) bound to mouse anti-FLAG [4 μl per sample (OriGene)]. Beads were washed and incubated for 30 min at 37°C in 30 μl of Click chemistry buffer (100 μm tris(benzyltriazolylmethyl)amine, 1 mm CuSO4·5H2O, 1 mm tris(2-carboxyethyl)phosphine, 100 μm biotin-azide in RIPA buffer with no protease inhibitor). Samples were eluted from beads and separated on a 4–12% Bis–Tris gel in MOPS-SDS running buffer. Proteins were transferred to nitrocellulose and blotted with primary antibody [Rabbit anti-FLAG, 1:1000 (Sigma-Aldrich)] and secondary antibody [Alexa Fluor® 800 Goat anti-Rabbit, 1:5000 (Invitrogen) and Alexa Fluor® 680 Streptavidin, 1:5000 (Invitrogen)] to detect biotin levels and total protein amounts. Blots were scanned and bands quantified using the Li-Cor Odyssey® Infrared Imaging System.
Immunofluorescence
COS-7 cells grown on glass coverslips were washed 24 h post-transfection and fixed in 4% paraformaldehyde (PFA)/PBS for 15 min. Cells were permeabilized with 0.1% Triton/1% PFA/phosphate buffered saline for 1.5 min and blocked in 0.2% Gelatin/PBS for 15 min. Coverslips were incubated for 1 h with primary antibody [Rabbit anti-FLAG, 1:400 (Sigma-Aldrich) or Mouse Anti-GM130, 1:100 (BD Transduction Laboratories)] diluted in 0.2% Gelatin/PBS. Cells were washed and incubated in the same way with secondary antibody [Alexa Fluor® 594 Goat anti-Rabbit, 1:400 (Invitrogen) or Alexa Fluor® 350 Goat anti-Mouse, 1:100 (Invitrogen)]. Washed coverslips were mounted on microscope slides with 1 drop of ProLong® Gold Antifade Reagent (Invitrogen), and viewed using a conventional wide-field fluorescence microscope. Images were captured and processed using MetaMorph® Image Analysis Software. All steps were performed at room temperature.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Canadian Institutes of Health Research (CIHR, http://www.cihr-irsc.gc.ca/, grant number GPG-102165 to M.R.H. and E.C.); the Cure Huntington Disease Initiative (CHDI, http://chdifoundation.org/ to M.R.H.); the Deutsche Forschungsgemeinschaft (DFG, http://www.dfg.de, SFB740 grant number 740/2-11, SFB618 grant number 618/3-09); the Bundesministerium für Bildung und forschung (BMBF, http://www.bmbf.de, the NGFN-Plus, NeuroNet grant number 01GS08169-73, MooDS grant number 01GS08150); the Huntington′s Disease Society of America (HDSA, http://www.hdsa.org) and the Helmholtz Association (http://www.helmholtz.de, MSBN, HelMA grant number HA-215) to E.E.W. This work was supported by Michael Smith Foundation for Health Research (MSFHR) and CIHR fellowships to S.S.S.
Supplementary Material
ACKNOWLEDGEMENTS
We thank CMMT Medical Illustrator Interns Erin Kenzie and Melissa Cory for help with figures, Chris Kay (MRH laboratory) for help with statistics, Dr Martin Schaefer (Centre for Genomic Regulation, Barcelona) for help with HIPPIEdb, Dr Akihiko Yoshimura (Keio University) for cDNA clones of FLAG-Spred1 and FLAG-Spred3, Drs Nicholas Davis (Wayne State University), Agnès Journet (CEA), Howard Hang (The Rockefeller University), and Ethan Marin (Yale University) for providing spreadsheets of published palmitoyl-proteome supplemental data files, and Dr James Wang (CHDI) for providing mouse orthologs of the human HIP14 Y2H interactors. MRH is a University Killam Professor and holds a Canada Research Chair in Human Genetics.
Conflict of Interest statement. None declared.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Shirasaki D.I., Greiner E.R., Al-Ramahi I., Gray M., Boontheung P., Geschwind D.H., Botas J., Coppola G., Horvath S., Loo J.A., et al. Network organization of the huntingtin proteomic interactome in Mammalian brain. Neuron. 2012;75:41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Culver B.P., Savas J.N., Park S.K., Choi J.H., Zheng S., Zeitlin S.O., Yates J.R., Tanese N. Proteomic analysis of wild-type and mutant huntingtin associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.359307. 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K.S., Knoblich M., Haenig C., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Harjes P., Wanker E.E. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 7.Young F.B., Butland S.L., Sanders S.S., Sutton L.M., Hayden M.R. Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog. Neurobiol. 2012;97:220–238. doi: 10.1016/j.pneurobio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Singaraja R.R., Hadano S., Metzler M., Givan S., Wellington C.L., Warby S., Yanai A., Gutekunst C.-A., Leavitt B.R., Yi H., et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 9.Kalchman M.A., Graham R.K., Xia G., Koide H.B., Hodgson J.G., Graham K.C., Goldberg Y.P., Gietz R.D., Pickart C.M., Hayden M.R. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J. Biol. Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- 10.Aylward E.H., Sparks B.F., Field K.M., Yallapragada V., Shpritz B.D., Rosenblatt A., Brandt J., Gourley L.M., Liang K., Zhou H., et al. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- 11.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Neuropathological classification of Huntington’s disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Huang K., Sanders S.S., Kang R., Carroll J.B., Sutton L., Wan J., Singaraja R., Young F.B., Liu L., El-Husseini A., et al. Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanai A., Huang K., Kang R., Singaraja R.R., Arstikaitis P., Gan L., Orban P.C., Mullard A., Cowan C.M., Raymond L.A., et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R.R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C.-A., et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Singaraja R.R., Huang K., Sanders S.S., Milnerwood A.J., Hines R., Lerch J.P., Franciosi S., Drisdel R.C., Vaid K., Young F.B., et al. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell D.A., Mitchell G., Ling Y., Budde C., Deschenes R.J. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.-Z., et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 18.Ren W., Sun Y., Du K. DHHC17 palmitoylates ClipR-59 and modulates ClipR-59 association with the plasma membrane. Mol. Cell Biol. 2013;33:4255–4265. doi: 10.1128/MCB.00527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rush D.B., Leon R.T., McCollum M.H., Treu R.W., Wei J. Palmitoylation and trafficking of GAD65 are impaired in a cellular model of Huntington’s disease. Biochem. J. 2012;442:39–48. doi: 10.1042/BJ20110679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaves J., Gorleku O.A., Salaun C., Chamberlain L.H. Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases. J. Biol. Chem. 2010;285:24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian L., McClafferty H., Jeffries O., Shipston M.J. Multiple palmitoyltransferases are required for palmitoylation-dependent regulation of large conductance calcium- and voltage-activated potassium channels. J. Biol. Chem. 2010;285:23954–23962. doi: 10.1074/jbc.M110.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M.R., El-Husseini A. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greaves J., Salaun C., Fukata Y., Fukata M., Chamberlain L.H. Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J. Biol. Chem. 2008;283:25014–25026. doi: 10.1074/jbc.M802140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukata M., Fukata Y., Adesnik H., Nicoll R.A., Bredt D.S. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Yang G., Zhou X., Zhu J., Liu R., Zhang S., Coquinco A., Chen Y., Wen Y., Kojic L., Jia W., et al. JNK3 couples the neuronal stress response to inhibition of secretory trafficking. Sci. Signal. 2013;6:ra57. doi: 10.1126/scisignal.2003727. [DOI] [PubMed] [Google Scholar]
- 26.Yang G., Cynader M.S. Palmitoyl acyltransferase zD17 mediates neuronal responses in acute ischemic brain injury by regulating JNK activation in a signaling module. J. Neurosci. 2011;31:11980–11991. doi: 10.1523/JNEUROSCI.2510-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada T., Matsuzaki O., Hayashi H., Sugano S., Matsuda A., Nishida E. AKRL1 and AKRL2 activate the JNK pathway. Genes Cells. 2003;8:493–500. doi: 10.1046/j.1365-2443.2003.00650.x. [DOI] [PubMed] [Google Scholar]
- 28.Bandyopadhyay S., Chiang C.-Y., Srivastava J., Gersten M., White S., Bell R., Kurschner C., Martin C.H., Smoot M., Sahasrabudhe S., et al. A human MAP kinase interactome. Nat. Methods. 2010;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukata Y., Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 30.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamesch P., Li N., Milstein S., Fan C., Hao T., Szabó G., Hu Z., Venkatesan K., Bethel G., Martin P., et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rual J.-F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 33.Palidwor G.A., Shcherbinin S., Huska M.R., Rasko T., Stelzl U., Arumughan A., Foulle R., Porras P., Sanchez-Pulido L., Wanker E.E., et al. Detection of alpha-rod protein repeats using a neural network and application to huntingtin. PLoS Comput. Biol. 2009;5:e1000304. doi: 10.1371/journal.pcbi.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao T., Collins R.E., Horton J.R., Zhang X., Zhang R., Dhayalan A., Tamas R., Jeltsch A., Cheng X. The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding. Proteins. 2009;76:772–777. doi: 10.1002/prot.22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 36.Lai J., Linder M.E. Oligomerization of DHHC Protein S-acyltransferases. J. Biol. Chem. 2013 doi: 10.1074/jbc.M113.458794. 10.1074/jbc.M113.458794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Eichborn J., Dunkel M., Gohlke B.O., Preissner S.C., Hoffmann M.F., Bauer J.M.J., Armstrong J.D., Schaefer M.H., Andrade-Navarro M.A., Le Novere N., et al. SynSysNet: integration of experimental data on synaptic protein–protein interactions with drug–target relations. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 39.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A.O., Thompson J.X., Roth A.F., Drisdel R.C., Mastro R., et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Martin B.R., Cravatt B.F., Hofmann S.L. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J. Biol. Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin B.R., Wang C., Adibekian A., Tully S.E., Cravatt B.F. Global profiling of dynamic protein palmitoylation. Nat. Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merrick B.A., Dhungana S., Williams J.G., Aloor J.J., Peddada S., Tomer K.B., Fessler M.B. Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase 3. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006007. M110.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yount J.S., Moltedo B., Yang Y.-Y., Charron G., Moran T.M., López C.B., Hang H.C. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson J.P., Raghavan A.S., Yang Y.-Y., Charron G., Hang H.C. Proteomic analysis of fatty-acylated proteins in mammalian cells with chemical reporters reveals S-acylation of histone H3 variants. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.001198. M110.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin B.R., Cravatt B.F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivaldi C., Martin B.R., Kieffer-Jaquinod S., Chapel A., Levade T., Garin J., Journet A. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS ONE. 2012;7:e37187. doi: 10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marin E.P., Derakhshan B., Lam T.T., Davalos A., Sessa W.C. Endothelial cell palmitoylproteomic identifies novel lipid-modified targets and potential substrates for protein acyl transferases. Circ. Res. 2012;110:1336–1344. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowal L., Yang W., Freeman M.R., Steen H., Flaumenhaft R. Proteomic analysis of palmitoylated platelet proteins. Blood. 2011;118:e62–e73. doi: 10.1182/blood-2011-05-353078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Planey S.L., Ceballos C., Stevens S.M., Keay S.K., Zacharias D.A. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol. Cell. Proteomics. 2008;7:1378–1388. doi: 10.1074/mcp.M800069-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., Casey P.J. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M.R. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutton L.M., Sanders S.S., Butland S.L., Singaraja R.R., Franciosi S., Southwell A.L., Doty C.N., Schmidt M.E., Mui K.K.N., Kovalik V., et al. Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease. Hum. Mol. Genet. 2013;22:452–465. doi: 10.1093/hmg/dds441. [DOI] [PubMed] [Google Scholar]
- 53.Flicek P., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan J., Savas J.N., Roth A.F., Sanders S.S., Singaraja R.R., Hayden M.R., Yates J.R., Davis N.G. Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington’s disease. Chem. Biol. 2013 doi: 10.1016/j.chembiol.2013.09.018. 10.1016/j.chembiol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greaves J., Prescott G.R., Fukata Y., Fukata M., Salaun C., Chamberlain L.H. The hydrophobic cysteine-rich domain of SNAP25 couples with downstream residues to mediate membrane interactions and recognition by DHHC palmitoyl transferases. Mol. Biol. Cell. 2009;20:1845–1854. doi: 10.1091/mbc.E08-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shipston M.J. Ion channel regulation by protein palmitoylation. J. Biol. Chem. 2011;286:8709–8716. doi: 10.1074/jbc.R110.210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Y., Savanenin A., Reddy P.H., Liu Y.F. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-d-aspartate receptors via post-synaptic density 95. J. Biol. Chem. 2001;276:24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- 59.Schaefer M.H., Fontaine J.-F., Vinayagam A., Porras P., Wanker E.E., Andrade-Navarro M.A. HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS ONE. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suter B., Fontaine J.-F., Yildirimman R., Rasko T., Schaefer M.H., Rasche A., Porras P., Vázquez-Álvarez B.M., Russ J., Rau K., et al. Development and application of a DNA microarray-based yeast two-hybrid system. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holbert S., Dedeoglu A., Humbert S., Saudou F., Ferrante R.J., Néri C. Cdc42-interacting protein 4 binds to huntingtin: neuropathologic and biological evidence for a role in Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2003;100:2712–2717. doi: 10.1073/pnas.0437967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Faber P.W., Barnes G.T., Srinidhi J., Chen J., Gusella J.F., MacDonald M.E. Huntingtin interacts with a family of WW domain proteins. Hum. Mol. Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 63.Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J., Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P., Zoghbi H.Y. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 1993;4:221–226. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 64.Caviston J.P., Zajac A.L., Tokito M., Holzbaur E.L.F. Huntingtin coordinates the dynein-mediated dynamic positioning of endosomes and lysosomes. Mol. Biol. Cell. 2011;22:478–492. doi: 10.1091/mbc.E10-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caviston J.P., Holzbaur E.L.F. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.del Toro D., Alberch J., Lázaro-Diéguez F., Martín-Ibáñez R., Xifró X., Egea G., Canals J.M. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuhn A., Thu D., Waldvogel H.J., Faull R.L.M., Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat. Methods. 2011;8:945–947. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- 68.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 69.Williams A., Sarkar S., Cuddon P., Ttofi E.K., Saiki S., Siddiqi F.H., Jahreiss L., Fleming A., Pask D., Goldsmith P., et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat. Chem. Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitsui K., Nakayama H., Akagi T., Nekooki M., Ohtawa K., Takio K., Hashikawa T., Nukina N. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J. Neurosci. 2002;22:9267–9277. doi: 10.1523/JNEUROSCI.22-21-09267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petrakis S., Rasko T., Russ J., Friedrich R.P., Stroedicke M., Riechers S.-P., Muehlenberg K., Möller A., Reinhardt A., Vinayagam A., et al. Identification of human proteins that modify misfolding and proteotoxicity of pathogenic ataxin-1. PLoS Genet. 2012;8:e1002897. doi: 10.1371/journal.pgen.1002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrios-Rodiles M., Brown K.R., Ozdamar B., Bose R., Liu Z., Donovan R.S., Shinjo F., Liu Y., Dembowy J., Taylor I.W., et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 73.Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 74.de Maximy A.A., Nakatake Y., Moncada S., Itoh N., Thiery J.P., Bellusci S. Cloning and expression pattern of a mouse homologue of Drosophila sprouty in the mouse embryo. Mech. Dev. 1999;81:213–216. doi: 10.1016/s0925-4773(98)00241-x. [DOI] [PubMed] [Google Scholar]
- 75.Hacohen N., Kramer S., Sutherland D., Hiromi Y., Krasnow M.A. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 76.Kato R., Nonami A., Taketomi T., Wakioka T., Kuroiwa A., Matsuda Y., Yoshimura A. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem. Biophys. Res. Commun. 2003;302:767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- 77.Huang K.-Y., Chen G.-D., Cheng C.-H., Liao K.-Y., Hung C.-C., Chang G.-D., Hwang P.-P., Lin S.-Y., Tsai M.-C., Khoo K.-H., et al. Phosphorylation of the zebrafish M6Ab at serine 263 contributes to filopodium formation in PC12 cells and neurite outgrowth in zebrafish embryos. PLoS ONE. 2011;6:e26461. doi: 10.1371/journal.pone.0026461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alfonso J., Fernández M.E., Cooper B., Flugge G., Frasch A.C. The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc. Natl. Acad. Sci. USA. 2005;102:17196–17201. doi: 10.1073/pnas.0504262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finetti F., Paccani S.R., Rosenbaum J., Baldari C.T. Intraflagellar transport: a new player at the immune synapse. Trends Immunol. 2011;32:139–145. doi: 10.1016/j.it.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Keryer G., Pineda J.R., Liot G., Kim J., Dietrich P., Benstaali C., Smith K., Cordelières F.P., Spassky N., Ferrante R.J., et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J. Clin. Invest. 2011;121:4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giannandrea M., Bianchi V., Mignogna M.L., Sirri A., Carrabino S., D'Elia E., Vecellio M., Russo S., Cogliati F., Larizza L., et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am. J. Hum. Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukata Y., Dimitrov A., Boncompain G., Vielemeyer O., Perez F., Fukata M. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 2013;202:145–161. doi: 10.1083/jcb.201302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greaves J., Chamberlain L.H. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 84.Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J. Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stowers R.S., Isacoff E.Y. Drosophila huntingtin-interacting protein 14 is a presynaptic protein required for photoreceptor synaptic transmission and expression of the palmitoylated proteins synaptosome-associated protein 25 and cysteine string protein. J. Neurosci. 2007;27:12874–12883. doi: 10.1523/JNEUROSCI.2464-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sahlender D.A., Roberts R.C., Arden S.D., Spudich G., Taylor M.J., Luzio J.P., Kendrick-Jones J., Buss F. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Craven S.E., El-Husseini A.E., Bredt D.S. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 88.Raymond L.A., André V.M., Cepeda C., Gladding C.M., Milnerwood A.J., Levine M.S. Pathophysiology of Huntington’s disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 2011;198:252–273. doi: 10.1016/j.neuroscience.2011.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau A., Tymianski M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflugers Arch. 2010;460:525–542. doi: 10.1007/s00424-010-0809-1. [DOI] [PubMed] [Google Scholar]
- 90.Schwarcz R., Whetsell W.O., Mangano R.M. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science. 1983;219:316–318. doi: 10.1126/science.6849138. [DOI] [PubMed] [Google Scholar]
- 91.McGeer E.G., McGeer P.L. Duplication of biochemical changes of Huntington’s chorea by intrastriatal injections of glutamic and kainic acids. Nature. 1976;263:517–519. doi: 10.1038/263517a0. [DOI] [PubMed] [Google Scholar]
- 92.de Vries B., Mamsa H., Stam A.H., Wan J., Bakker S.L.M., Vanmolkot K.R.J., Haan J., Terwindt G.M., Boon E.M.J., Howard B.D., et al. Episodic ataxia associated with EAAT1 mutation C186S affecting glutamate reuptake. Arch. Neurol. 2009;66:97–101. doi: 10.1001/archneurol.2008.535. [DOI] [PubMed] [Google Scholar]
- 93.Jen J.C., Wan J., Palos T.P., Howard B.D., Baloh R.W. Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology. 2005;65:529–534. doi: 10.1212/01.wnl.0000172638.58172.5a. [DOI] [PubMed] [Google Scholar]
- 94.Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mardakheh F.K., Yekezare M., Machesky L.M., Heath J.K. Spred2 interaction with the late endosomal protein NBR1 down-regulates fibroblast growth factor receptor signaling. J. Cell Biol. 2009;187:265–277. doi: 10.1083/jcb.200905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.King J.A.J., Straffon A.F.L., D'Abaco G.M., Poon C.L.C., Stacey T.T. I., Smith C.M., Buchert M., Corcoran N.M., Hall N.E., Callus B.A., et al. Distinct requirements for the Sprouty domain for functional activity of Spred proteins. Biochem. J. 2005;388:445–454. doi: 10.1042/BJ20041284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyoshi K., Wakioka T., Nishinakamura H., Kamio M., Yang L., Inoue M., Hasegawa M., Yonemitsu Y., Komiya S., Yoshimura A. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene. 2004;23:5567–5576. doi: 10.1038/sj.onc.1207759. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y., Yang F., Fu Y., Huang X., Wang W., Jiang X., Gritsenko M.A., Zhao R., Monore M.E., Pertz O.C., et al. Spatial phosphoprotein profiling reveals a compartmentalized extracellular signal-regulated kinase switch governing neurite growth and retraction. J. Biol. Chem. 2011;286:18190–18201. doi: 10.1074/jbc.M111.236133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leung T., Chen X.Q., Manser E., Lim L. The p160 RhoA-binding kinase ROK alpha is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol. Cell Biol. 1996;16:5313–5327. doi: 10.1128/mcb.16.10.5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bauer P.O., Wong H.K., Oyama F., Goswami A., Okuno M., Kino Y., Miyazaki H., Nukina N. Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J. Biol. Chem. 2009;284:13153–13164. doi: 10.1074/jbc.M809229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shao J., Welch W.J., Diamond M.I. ROCK and PRK-2 mediate the inhibitory effect of Y-27632 on polyglutamine aggregation. FEBS Lett. 2008;582:1637–1642. doi: 10.1016/j.febslet.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shao J., Welch W.J., Diprospero N.A., Diamond M.I. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell Biol. 2008;28:5196–5208. doi: 10.1128/MCB.00079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kinsella R.J., Kähäri A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Croft D., O'Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boutell J.M., Wood J.D., Harper P.S., Jones A.L. Huntingtin interacts with cystathionine beta-synthase. Hum. Mol. Genet. 1998;7:371–378. doi: 10.1093/hmg/7.3.371. [DOI] [PubMed] [Google Scholar]
- 110.Zeeberg B.R., Riss J., Kane D.W., Bussey K.J., Uchio E., Linehan W.M., Barrett J.C., Weinstein J.N. Mistaken identifiers: gene name errors can be introduced inadvertently when using Excel in bioinformatics. BMC Bioinformatics. 2004;5:80. doi: 10.1186/1471-2105-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yap M.C., Kostiuk M.A., Martin D.D.O., Perinpanayagam M.A., Hak P.G., Siddam A., Majjigapu J.R., Rajaiah G., Keller B.O., Prescher J.A., et al. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J. Lipid Res. 2010;51:1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.