Abstract
The d-mannonate dehydratase (ManD) subgroup of the enolase superfamily contains members with varying catalytic activities (high-efficiency, low-efficiency, or no activity) that dehydrate d-mannonate and/or d-gluconate to 2-keto-3-deoxy-d-gluconate [Wichelecki, D. J., et al. (2014) Biochemistry53, 2722–2731]. Despite extensive in vitro characterization, the in vivo physiological role of a ManD has yet to be established. In this study, we report the in vivo functional characterization of a high-efficiency ManD from Caulobacter crescentus NA1000 (UniProt entry B8GZZ7) by in vivo discovery of its essential role in d-glucuronate metabolism. This in vivo functional annotation may be extended to ∼50 additional proteins.
The enolase superfamily (ENS) has been the subject of function discovery-based research since it was demonstrated that its homologous members catalyze different chemical reactions.1 Although all members of the ENS catalyze a common partial reaction (general base-catalyzed abstraction of a proton α to a carboxylate group to form a Mg2+-stabilized enediolate intermediate), their overall reactions and chemistries can be quite diverse (e.g., β-elimination and 1,1-proton transfer reactions).2,3 Nevertheless, the members of the ENS share significant structural homology: they share a (β/α)7(β)-barrel domain for acid/base catalysis and an (α+β) capping domain for substrate specificity.4−9
When the d-mannonate dehydratase (ManD) subgroup of the ENS was discovered in 2007, all of its members were assumed to catalyze the dehydration of d-mannonate to 2-keto-3-deoxy-d-gluconate.10 The Asp, Glu, and Glu ligands for the essential Mg2+ are located at the ends of the third, fourth, and fifth β-strands, respectively, of the barrel domain. The general base catalyst is a Tyr-Arg dyad located at the start of the “150–180s” loop (between the second and third β-strands); the general acid catalyst is a His located at the end of the third β-strand.10
A large-scale in vitro survey of sequence–function space in the ManD subgroup was supported by the Enzyme Function Initiative (EFI; U54GM093342), a large-scale, collaborative, project focused on devising tools and strategies for addressing and correcting the widespread non- and misannotation present in today’s automatically curated protein databases.3,11 For confident functional and physiological assignment, the EFI seeks not only in vitro enzymatic activity (values of kcat and kcat/KM) but also in vivo metabolic function.
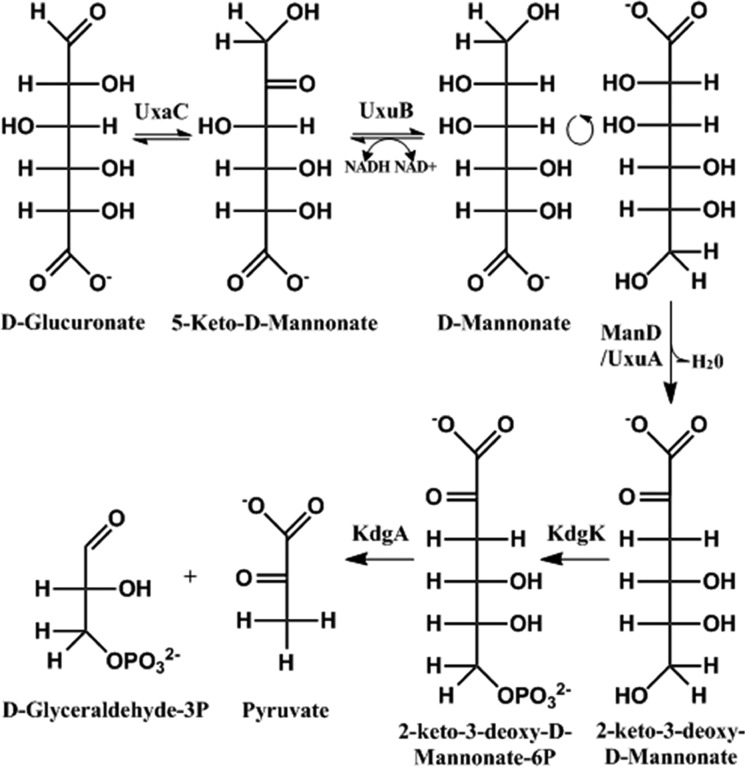
The ManD subgroup of the ENS (in Pfam families PF02746 and PF13378) initially was thought to be isofunctional, but the survey of the sequence space revealed dehydration activity with d-mannonate and/or d-gluconate.12 The members of the ManD subgroup have a range of catalytic activities and efficiencies: high-efficiency (specific to d-mannonate with a kcat/KM of 103–104 M–1 s–1), low-efficiency (d-mannonate and/or d-gluconate with a kcat/KM of 10–102 M–1 s–1), and no activity.12 However, the in vivo physiological function had not been assigned to any member of the ManD subgroup. The only metabolic pathway in which dehydration of d-mannonate is known to occur is the catabolism of d-glucuronate (Figure 1), in which the ManD reaction is performed by the well-characterized UxuA (utilization of hexuronate A)13,14 that is included in Pfam family PF03786.
Figure 1.
Degradation pathway of d-glucuronate in Escherichia coli. The dehydration of d-mannonate to 2-keto-3-deoxy-d-gluconate is performed by UxuA.
One high-efficiency ManD (Uniprot entry B8GZZ7; Protein Data Bank entries 3VCN and 4GME) was shown to be specific for d-mannonate with a catalytic efficiency of 1.2 × 104 M–1 s–1.12 The UxuAs in d-glucuronate catabolism have catalytic efficiencies of ∼103 M–1 s–1. Interestingly, low-efficiency and no activity ManDs are encoded by genomes that encode a member of the UxuA family; however, genomes that encode a high-efficiency ManD do not encode a UxuA.12 This suggested that high-efficiency ManDs are analogues for UxuAs, with low-efficiency and no activity ManDs likely having different physiological functions.
Of the high-efficiency ManDs that were characterized in vitro, we selected B8GZZ7 for in vivo study because its encoding organism, Caulobacter crescentus NA1000, is amenable to genetic manipulation.15C. crescentus is an important organism in the study of asymmetric cell division and prokaryotic development because it can differentiate into mobile swarmer cells and immobile stalk cells.15 Species of Caulobacter utilize aquatic vegetation as a carbon source in freshwater environments;15d-glucuronate makes up a large part of plant cell-wall biomass.16 Therefore, we hypothesized that C. crescentus NA1000 could utilize d-glucuronate as a carbon source via B8GZZ7, a high-efficiency ManD, despite the absence of UxuA.
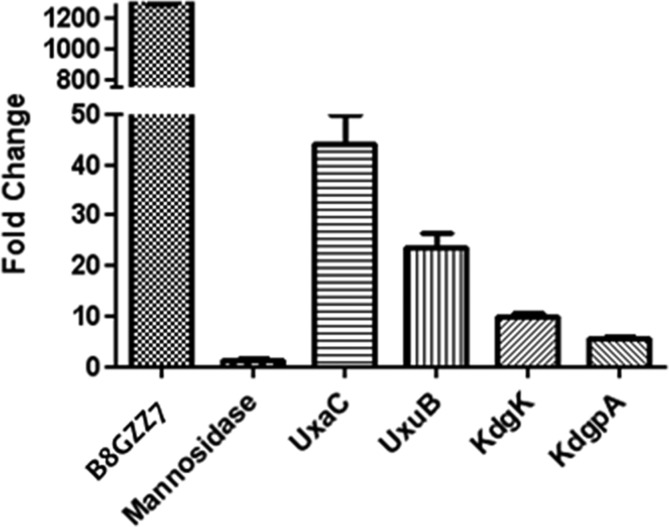
The gene encoding B8GZZ7 is located in a genome neighborhood that is not indicative of d-glucuronate metabolism. Nearby genes encode catabolic enzymes for N-acetylglucosamine 6-phosphate, a mannosidase, and a GntR transcriptional regulator (Figure 2). Nevertheless, the ability of d-glucuronate to upregulate the gene encoding B8GZZ7 was investigated via quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Expression of the gene encoding B8GZZ7 was induced >1000-fold when C. crescentus NA1000 was grown on d-glucuronate as a carbon source (relative to growth on d-xylose) (Figure 3). Importantly, the gene encoding the mannosidase immediately upstream of the gene encoding B8GZZ7 was not upregulated (Figure 3); therefore, the gene encoding B8GZZ7 is transcribed separately from its genome neighbors and is likely not involved in N-acetylglucosamine catabolism. The absence of cotranscription of the genome neighborhood was further verified by the lack of a PCR product when cDNA was amplified using primers inside of the genes encoding both B8GZZ7 and the mannosidase (such that the potential product would span the gap between the two genes). Thus, we conclude that genomic context can be misleading for functional assignment in the absence of definitive information about regulation and the identities of transcriptional units.
Figure 2.
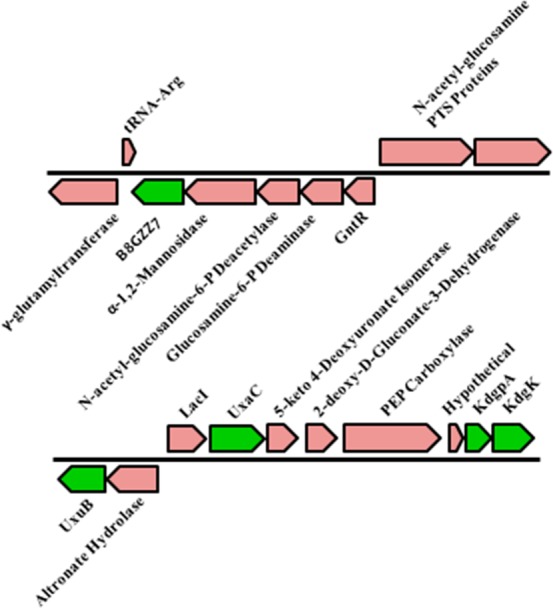
Genome neighborhoods of B8GZZ7, a high-efficiency ManD, (top) and canonical d-glucuronate catabolism genes (bottom). The genes directly involved in d-glucuronate metabolism are colored green.
Figure 3.
Upregulation of the genes encoding B8GZZ7 and the d-glucuronate catabolism genes shown in Figure 2.
ThermoFluor17 ligand binding studies were performed on the GntR transcriptional regulator in the genome neighborhood of B8GZZ7 using a library of acid sugars, including d-glucuronate and d-mannonate, as well as other metabolites (Table S2 of the Supporting Information). This assay yielded no stabilizing ligands, providing further evidence that the gene encoding B8GZZ7 is regulated separately.
The genes encoding the remaining enzymes in the canonical d-glucuronate pathway are located elsewhere in the genome, closely clustered with other sugar-metabolizing genes (Figure 2). The genes encoding d-glucuronate isomerase (UxaC), fructuronate reductase (UxaB), 2-keto-3-deoxy-d-gluconate kinase (KdgK), and 2-keto-3-deoxy-d-gluconate-6-phosphate aldolase (KdgA) were upregulated when they were grown on d-glucuronate (relative to growth on d-xylose) (Figure 3). These qRT-PCR data provide compelling evidence that B8GZZ7 is performing the ManD reaction in the canonical d-glucuronate catabolic pathway.
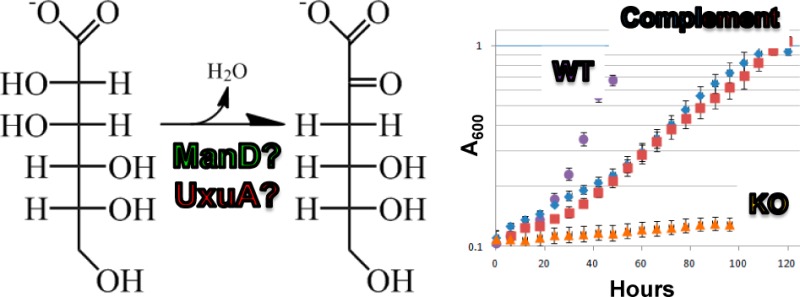
The knockout of the gene encoding B8GZZ7 showed no growth on d-glucuronate (Figure S1 of the Supporting Information). Complementation of the knockout with an isopropyl β-d-1-thiogalactopyranoside-inducible gene encoding B8GZZ7 (pSRK-Kan)18 restored growth on d-glucuronate. Although the growth rate of the complemented knockout strain was slower than that of wild-type C. crescentus NA1000 on d-glucuronate, the wild-type strain transformed with the same complementation vector showed a similar growth defect (Figure S1 of the Supporting Information), likely because of the toxicity of plasmid-based expression of B8GZZ7. The requirement of B8GZZ7 for growth on d-glucuronate and complementation of the growth defect by supplying B8GZZ7 in trans verifies that B8GZZ7 is the ManD for d-glucuronate metabolism in C. crescentus NA1000.
We visualize functional diversity in the ManD subgroup using sequence similarity networks.12 Using an E-value threshold of 10–190, corresponding to ∼75% identity, the clusters are isofunctional as judged by a sequence alignment.12 B8GZZ7 is a member of an isofunctional cluster that includes 55 additional proteins (Figure S2 and Table S3 of the Supporting Information). None of the organisms that encode these proteins encode the UxuA in d-glucuronate catabolism, although the genes encoding all of the other enzymes needed for catabolism of d-glucuronate via the pathway shown in Figure 1 are present in the genomes.
The genomes of a few of these organisms, including C. crescentus NA1000, encode two high-efficiency ManDs. In C. crescentus NA1000, the second ManD (UniProt entry B8H1R9) is not involved in d-glucuronate catabolism: the knockout of the gene encoding the B8H1R9 retains the ability to utilize d-glucuronate as a carbon source (Figure S1 of the Supporting Information); also, the gene encoding B8H1R9 is not upregulated by d-glucuronate. Apparently, the ManD activity of B8H1R9 is essential for another metabolic function that is currently unknown.
We are confident that we can transfer the now verified physiological function of B8GZZ7, the ManD in the catabolic pathway for d-glucuronate, to the high-efficiency ManDs that are encoded by the organisms with a single orthologue (Table S3 of the Supporting Information); all of these organisms encode the other enzymes in d-glucuronate catabolism. For those organisms that encode two high-efficiency ManDs, genome context is sufficient to assign function in some cases, e.g., Sphingomonas wittichii and Sphingomonas sp. MM-1; in other cases, such as C. crescentus NA1000 described here, transcriptomic and genetic experiments are needed to assign the physiological function.
Previously, no physiological function for a ManD in the ENS had been determined. We now have used in vivo approaches to establish that B8GZZ7, a high-efficiency ManD in C. crescentus NA1000, participates in d-glucuronate catabolism. Thus, we conclude that the high-efficiency ManDs in the ENS are examples of convergent evolution of function with the ManDs in the UxuA family.
This study demonstrates that (1) in vitro enzymatic activity alone cannot be used to infer in vivo physiological function for an “unknown” enzyme discovered in a genome project and (2) genome context also is not sufficient to infer the in vivo physiological function. In due course, we will report the in vivo physiological functions for members of the ManD subgroup that have low catalytic efficiencies for dehydration of d-mannonate and d-gluconate.
Supporting Information Available
Growth curves, ThermoFluor library, Uniprot entries of orthologous ManDs, and all experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
This manuscript describes characterization of in vivo function of Uniprot entry B8GZZ7.
This research was supported by a program project grant (P01GM071790) and three cooperative agreements with the National Institutes of Health (U54GM093342, U54GM074945, and U54GM094662).
The authors declare no competing financial interests.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Neidhart D. J.; Kenyon G. L.; Gerlt J. A.; Petsko G. A. (1990) Nature 347, 692–694. [DOI] [PubMed] [Google Scholar]
- Babbitt P. C.; Gerlt J. A. (1997) J. Biol. Chem. 272, 30591–30594. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A.; Allen K. N.; Almo S. C.; Armstrong R. N.; Babbitt P. C.; Cronan J. E.; Dunaway-Mariano D.; Imker H. J.; Jacobson M. P.; Minor W.; Poulter C. D.; Raushel F. M.; Sali A.; Shoichet B. K.; Sweedler J. V. (2011) Biochemistry 50, 9950–9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Babbit P. C.; Rayment I. (2005) Arch. Biochem. Biophys. 433, 59–70. [DOI] [PubMed] [Google Scholar]
- Gulick A. M.; Hubbard B. K.; Gerlt J. A.; Rayment I. (2000) Biochemistry 39, 4590–4602. [DOI] [PubMed] [Google Scholar]
- Schmidt D. M. Z.; Hubbard B. K.; Gerlt J. A. (2001) Biochemistry 40, 15707–15715. [DOI] [PubMed] [Google Scholar]
- Yew W. S.; Fedorov A. A.; Fedorov E. V.; Rakus J. F.; Pierce R. W.; Almo S. C.; Gerlt J. A. (2006) Biochemistry 45, 14582–14597. [DOI] [PubMed] [Google Scholar]
- Yew W. S.; Fedorov A. A.; Fedorov E. V.; Wood B. M.; Almo S. C.; Gerlt J. A. (2006) Biochemistry 45, 14598–14608. [DOI] [PubMed] [Google Scholar]
- Yew W. S.; Fedorov A. A.; Fedorov E. V.; Almo S. C.; Gerlt J. A. (2007) Biochemistry 46, 9564–9577. [DOI] [PubMed] [Google Scholar]
- Rakus J. F.; Fedorov A. A.; Fedorov E. V.; Glasner M. E.; Vick J. E.; Babbitt P. C.; Almo S. C.; Gerlt J. A. (2007) Biochemistry 46, 12896–12908. [DOI] [PubMed] [Google Scholar]
- Schnoes A. M.; Brown S. D.; Dodevski I.; Babbitt P. C. (2009) PLoS Comput. Biol. 5, e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichelecki D. J.; Balthazor B. M.; Chau A. A.; Vetting M. W.; Fedorov A. A.; Fedorov E. V.; Lukk T.; Patskovsky Y. V.; Stead M. B.; Hillerich B. S.; Seidel R. D.; Almo S. C.; Gerlt J. A. (2014) Biochemistry 53, 2722–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell G. (1962) Methods Enzymol. 5, 190. [Google Scholar]
- Zhang Q.; Gao F.; Peng H.; Cheng H.; Liu Y.; Tang J.; Thompson J.; Wei G.; Zhang J.; Du Y.; Yan J.; Gao G. F. (2009) J. Bacteriol. 191, 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes A. K.; Meewan M.; Yang D.; Arana N.; Romero P.; McAdams H. H.; Stephens C. (2004) J. Bacteriol. 186, 1448–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G. J. (2004) Curr. Opin. Plant Biol. 7, 277–284. [DOI] [PubMed] [Google Scholar]
- Pantoliano M. W.; Petrella E. C.; Kwasnoski J. D.; Lobanov V. S.; Myslik J.; Graf E.; Carver T.; Asel E.; Springer B. A.; Lane P.; Salemme F. R. (2001) J. Biomol. Screening 6, 429–440. [DOI] [PubMed] [Google Scholar]
- Khan S. R.; Gaines J.; Roop R. M. II; Farrand S. K. (2008) Appl. Environ. Microbiol. 74, 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.