Abstract
Self-organization of cytoskeletal proteins such as actin and tubulin into filaments and microtubules is frequently assisted by the proteins binding to them. Formins are regulatory proteins that nucleate the formation of new filaments and are essential for a wide range of cellular functions. The vertebrate inverted formin 2 (INF2) has both actin filament nucleating and severing/depolymerizing activities connected to its ability to encircle actin filaments. Using atomic force microscopy, we report that a formin homology 2 (FH2) domain-containing construct of INF2 (INF2-FH1-FH2-C or INF2-FFC) self-assembles into nanoscale ringlike oligomeric structures in the absence of actin filaments, demonstrating an inherent ability to reorganize from a dimeric to an oligomeric state. A construct lacking the C-terminal region (INF2-FH1-FH2 or INF2-FF) also oligomerizes, confirming the dominant role of FH2-mediated interactions. Moreover, INF2-FFC domains were observed to organize into ringlike structures around single actin filaments. This is the first demonstration that formin FH2 domains can self-assemble into oligomers in the absence of filaments and has important implications for observing unaveraged decoration and/or remodeling of filaments by actin binding proteins.
Introduction
Self-assembly and regulated assembly of proteins into nanostructures are ubiquitous in biology and disease and among the most intriguing features of biological systems.1 Proteins may assemble to form various nanostructures like nanotubes, vesicles, helical ribbons, filaments, and fibrous scaffolds. Some well-known examples of such assembly include cytoskeletal filaments such as actin and tubulin,2 amyloid fibril formation,3,4 chromatin assembly,5 and phospholipid6 membrane self-assembly. Intriguingly, some of the protein regulators of cytoskeletal filaments can also form higher-order assembly structures. The morphology and function of these structures are yet to be elucidated. Some examples of such self-assembly are actin binding proteins tropomyosin and drebrin that can form oligomeric structures in vivo. Oligomers of tropomyosin have been shown to be isoform specific, and their level is drastically altered in malignant tumor cells.7 Drebrin was also shown to form higher-order oligomers named drebrosomes.8,9 It was hypothesized that such structures allowed for maintaining a high local concentration of these regulatory proteins in the needed regions in cells.
Focusing on the actin-based elements of cytoskeleton, we found the in vitro studies of self-assembly of purified actin alone, and in complex with actin binding proteins, offer a simple route to understanding how these proteins interact and function under controlled conditions. To this end, complementary experimental methods such as electron microscopy, cryo-electron microscopy (cryo-EM), and fluorescence microscopy have been extensively used to study the assembly and disassembly processes with actin binding proteins and the structures of the assembled complexes. Total internal reflection fluorescence (TIRF) microscopy has emerged as an important method for visualizing single molecules and single filaments.10 TIRF has been used to study the actin polymerization time course,10 kinetics of filament barbed end capping,11 and processive growth mediated by formins,12,13 yet TIRF can provide only limited structural information, within the resolution of the TRIF field (<200 nm). In the cryo-EM area, there have been recent advances in near atomic resolution analysis of protein structures by single-particle electron microscopy, using direct electron sensing cameras.14 However, large data sets of molecules captured in random orientations are needed for their frequently complex structural analysis. In the case of actin filaments, the averaging of data does not shed light on the heterogeneity of individual filaments or their segments or on the local changes in their twist.15
Though still a relatively new application to the study of actin binding proteins and F-actin, atomic force microscopy (AFM)16 is a powerful technique for nanoscale characterization of biological structures.17−23 It offers unique capability for direct three-dimensional (3D) imaging of single actin filaments without electron dense staining, fixation, or extreme temperatures, and with an imaging resolution comparable to that of negatively stained electron microscopy samples. Several proteins and protein conformational changes have been investigated at submolecular resolution using AFM. AFM imaging does not suffer from diffraction limits and allows exceptionally high signal-to-noise subnanometer lateral resolution and up to 1 Å vertical resolution.20,24−26 Among biomolecules, membrane proteins in their native state have been imaged most extensively.20,26 Besides isolated viruses and phages,27,28 specific substructures like viral capsomeres, bacteriophage connectors, and tails29,30 have been imaged at a resolution comparable to that of EM. AFM has been used successfully also to image high-resolution structures of F-actin31 and the structure of complexes of F-actin with its binding proteins, such as drebrin and cofilin.32 This method can reveal the structural heterogeneity of filament populations as well as analyze local changes in segments of individual filaments.
In this work, we apply AFM imaging to study in vitro the self-assembly of formin and its complexes with F-actin. Formins are actin assembly proteins that play essential roles in fundamental cellular processes such as polarity, adhesion, and cytokinesis.33−35 The actin regulatory activity of formins is mediated through the conserved formin homology 2 (FH2) domain.36−38 Formins exert multiple effects on actin polymerization in vitro (nucleation, elongation, and anticapping) mediated through the activity of the FH2 domain.37 The vertebrate specific inverted formin 2 (INF2) domain has the additional ability to sever filaments,39,40 which involves both the FH2 domain and an actin monomer binding motif at its C-terminus.
Structurally, the ∼400-amino acid FH2 domain is a donut-shaped head-to-tail dimeric structure consisting of two rigid hemispherical halves connected by a flexible linker.36,41−43 The FH2 dimer is very stable in some formins but has the ability to dissociate in others.44,45 Formin FH2 domains remain bound to actin filament barbed ends during filament elongation, moving processively as new actin monomers add through a “stair-stepping” mechanism, although the exact sequence of steps remains uncertain.36,38,42,46 It is predicted that the donut-shaped FH2 dimer (∼11 nm diameter) partially encircles the 7 nm diameter barbed end, without the individual domains needing to dissociate and reassociate. In addition to barbed end binding, INF2 has the ability to bind filament sides. Side binding is accomplished by FH2 domain encirclement of the filament, wherein the FH2 domain adopts binding interactions similar to those used at the barbed end. To accomplish this feat, the FH2 domain presumably must dissociate from its dimeric state, because the alternative mechanism of sliding along several micrometers of filament is unlikely. However, the ability of any FH2 dimer to dissociate and recombine has not been observed directly.
As an approach to improving our understanding of the FH2 homodimer actin regulatory function and to overcome the existing limitations of structure determination, we obtained the structure of INF2-FH1-FH2-C (INF2-FFC) (Figure 1) alone and in complex with single actin filaments. We present the first nanoscale structure and self-organization of INF2 alone and the side binding structure of INF2 in complex with actin filaments at the single-filament level using tapping mode AFM imaging.
Figure 1.
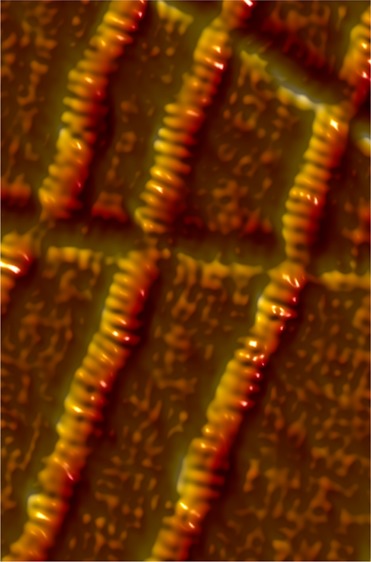
Schematic representation of the domain organization of inverted formin 2 (INF2) showing regions important for actin binding and protein–protein interactions. Formin constructs used in this study include INF2-FH1-FH2-C, also termed INF2-FFC, and INF2-FH1-FH2 (INF2-FF). N and C denote the N- and C-termini, respectively, of protein sequences. Other abbreviations are DID (diaphanous inhibitory domain), FH1 (formin homology 1), FH2 (formin homology 2), and DAD (diaphanous autoregulatory domain).
Experimental Section
Protein Preparation
Skeletal actin was purified using the method of Spudich and Watt.47 Human INF2-FFC (amino acids 469–1249) and INF2-FF (amino acids 469–941) constructs were expressed and purified as previously described.39 Rabbit skeletal Ca-ATP-G-actin was converted to Mg-ATP-G-actin by being incubated with 0.4 mM EGTA and 50 μM MgCl2 for 3 min at room temperature, and then polymerized by addition of 10× KMEH buffer [final concentrations of 50 mM KCl, 1 mM MgCl2, 0.4 mM EGTA, and 10 mM Hepes (pH 7)]. The final F-actin preparation contained also 0.2 mM ATP and 1 mM DTT and was supplemented with phalloidin (1:1 molar ratio) and 10–25 mM phosphate. INF2 constructs were dialyzed overnight into 1× KMEH buffer and then prespun at 4 °C in a TLA100 rotor, for 20 min at 80000 rpm, to remove any aggregates. The protein concentration was measured by the Bradford assay.
AFM Imaging of Formin and Actin–Formin Complexes
Different concentrations of formin solutions (5.0 μL) in buffer were added to freshly cleaved mica, incubated for 1 min, gently rinsed with dilution buffer three times (to remove unbound actins and proteins), and allowed to air-dry. In the case of formin–actin complexes, formin was allowed to bind F-actin in solution; 5.0 μL of the solution was added to freshly cleaved mica substrates, incubated for 1 min, gently rinsed with dilution buffer three times (to remove unbound actin and protein), and allowed to air-dry. Dimension 5000 AFM (Bruker Scientific) under tapping mode was used to image formin and actin–formin complexes with OTESP probes (Bruker Scientific). Topographic height images were recorded at 1024 pixels × 1024 pixels at 1 Hz. SPIP was used for image processing, which includes zero-order flattening and band-pass filtering. While sample air-drying and AFM tip may result in flattening of the structures, the dimeric formin rings are consistent with overall structures of the INF2 molecules measured by other independent techniques and reported previously.36,41−43 Similar ringlike structures were observed around polymerized actin filaments, as well (Figure 4), which provides further evidence of the lack of significant structural perturbation. Additionally, we confirmed previously32 (also see Figure S1 of the Supporting Information) that the helical pitch of bare F-actin as well as cofilin-decorated filaments measured by AFM (under our conditions) was in excellent agreement with the EM-based measurements, validating our assumption that F-actin filaments under ambient AFM imaging conditions are not perturbed significantly.
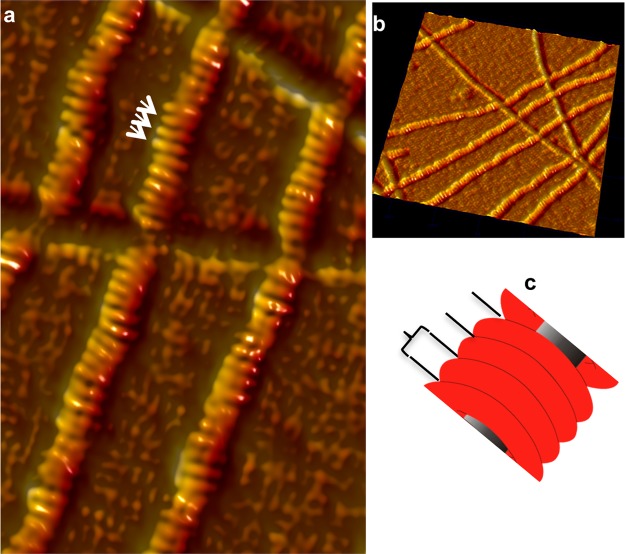
Figure 4.
Structure of INF2-FFC and F-actin complexes. AFM image of INF2-FFC and F-actin complexes (at a 1:1 molar ratio) revealing ringlike binding around actin filaments. (a) Close-up of the INF2-FFC–F-actin complex displaying detailed orientations of adjacent INF2-FFC homodimer rings assembled around F-actin. Notably, there is 5–10° occasional bending of adjacent units, indicating flexibility or strain across the bound complex. (b) 3D side view (500×, 500 nm) showing the stacklike arrangement of INF2-FFC over F-actin. (c) Tubular structure spacing shown schematically.
Results and Discussion
Structure of Free INF2-FFC Homodimers and Oligomers
FH2 domain dimerization has been proposed to be essential for the actin polymerization activity of formins,41,44,48,49 yet there are several gaps in our current understanding of the assembly and actin regulation via FH2 dimerization. Crystal structures of the yeast Bni1 FH2 domain36 and mammalian formin FH2 domains42,43,50 indicate that the two monomers are connected by flexible tethers to form a dimeric “ring”. Some of these FH2 dimers, such as in FMNL1 and mDia2, appear to be able to dissociate and reassociate.45 In addition, the apparently stable Bni1p FH2 can also assemble into oligomers36 around nonhelical actin structures. Other formins appear to be strictly dimeric. For example, despite containing two different dimerization domains (the N-terminal DD and the C-terminal FH2), mDia1-FL does not form higher-order oligomers but remains as dimers in the extended and activated state and closed and autoinhibited state.51
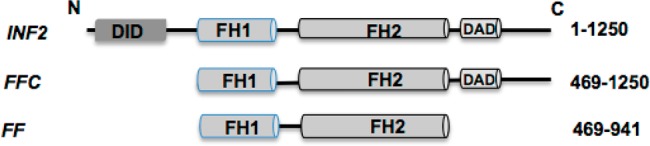
INF2’s FH2 domain has been shown to bind actin filament sides through a mechanism by which its dimer dissociates.40 There are two possible modes by which the INF2 FH2 domain might bind actin filaments in this manner. The individual FH2 dimers may re-form dimeric rings around F-actin but not associate with each other (Figures 2 and 6). Alternatively, FH2 domains may rebind the filament in a circular pattern, with oligomerization (Figure 6) occurring through the FH2 dimerization sites, similar to the crystal structure of Bni1p FH2 bound to actin.36 To differentiate between these two topologically distinct binding modes, i.e., dimeric rings or circular spirals over F-actin (Figure 6), we explored the structural characteristics of INF2-FFC alone to determine if it could assemble into high-order oligomers. We obtained AFM 3D images of free INF2-FFC constructs over freshly cleaved mica substrates at varying protein concentrations. At low concentrations (0.02 μM), isolated donut-shaped dimers were observed, in either hollow “open” or “closed” conformations (Figure 2). The two conformations, either hollow open (Figure 2a,c,d) or closed (Figure 2b), are consistent with cryo-EM structures of full-length formin mDia1 showing two distinct open and closed structures.51 Compared to a value of ∼10 nm for the crystal structure of the Bni1p FH2 domain,41 our AFM images showed diameters of 22 ± 0.1 nm (closed ring) and 27 ± 0.4 nm (open ring). The apparent increase in the width of the formin molecules in AFM images is likely the result of an AFM tip broadening effect (see Supporting Information) as well as the interactions of the protein with mica surfaces.
Figure 2.
Structural evidence of self-assembly of INF2-FFC at various concentrations. At low concentrations (0.02 μM), INF2-FFC forms ring-shaped homodimers. Representative AFM images of free INF2-FFC showing self-assembled round structures likely formed by dimerization of FH2 domains of INF2-FFC. Schematic representations are shown in the insets. (a) Single INF2-FFC dimer. (b) Compact and closed round dimers were observed also without a hollow core. (c) The central core region attached to the dimeric ring periphery via two small extensions (diamond arrows), which corresponds potentially to the unstructured FH1 domain. (d) An example of the INF2-FFC dimer in the open configuration. The FH2 domain within the INF2-FFC dimer can potentially dissociate (open) and reassociate (close) to form trimers as well as oligomers [at a higher surface concentration (0.2 μM)]. (e) AFM image of INF2-FFC in oligomeric configurations (0.2 μM INF2-FFC). Representative images of the INF2-FFC dimer, trimer, and tetramer (f–h, respectively) and their corresponding schematic representations (i–iii, respectively). At higher concentrations (2 μM), INF2-FFC assembles into tubular ringlike structures, as marked with a white arrow in part i. Other close-up views are shown in parts i–iv of panel j.
Figure 6.
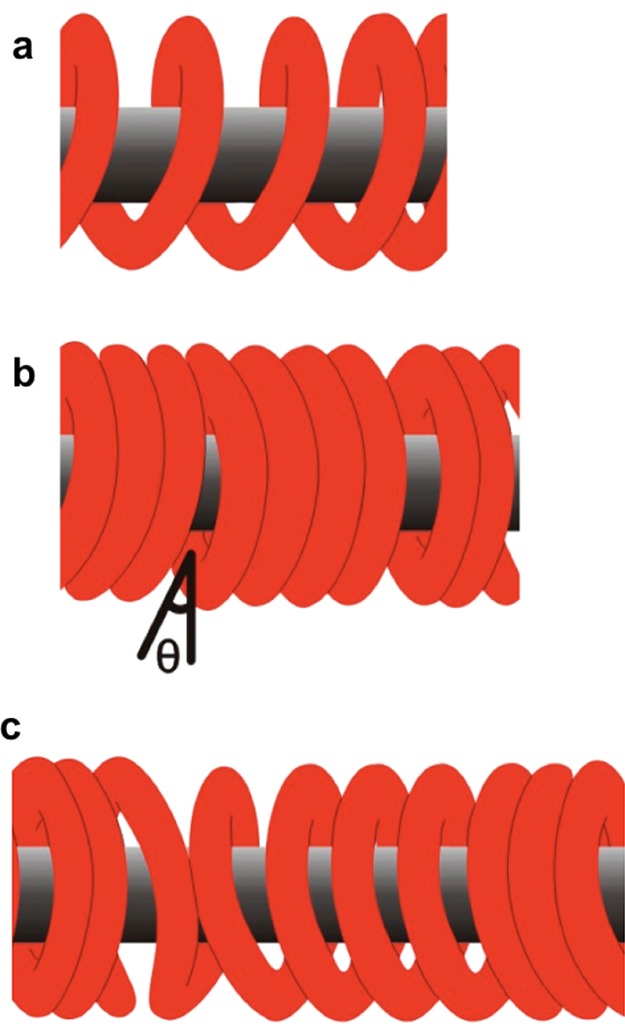
Schematic representation of plausible orientations of INF2-FFC homodimers over F-actin. (a) Adjacent INF2-FFC dimeric rings interact in an end-to-end manner to form a spiral coil structure. (b) Neighboring homodimers make longitudinal contact with adjacent F-actin-bound dimers to form continuous structure without any gaps or with small angle gaps and/or openings. (c) Combinations of regions as in panels a and b, along the length of F-actin. Alternatively, dimers may reassociate to form individual dimer rings stacked along the length of the filament with or without gap openings.
At higher protein concentrations (0.2 μM), oligomers that were similar in height to dimers (0.3 nm) but larger in diameter were observed. Representative images in Figure 2e–h display free INF2-FFC forming trimers, tetramers, and larger oligomers. The mean diameters of these oligomers were measured as 45 ± 2 nm (trimers) and 54.6 ± 0.8 nm (tetramers) (Table 1). Similar cross sectional height profiles (0.2 nm) of dimers as well as tetramers preclude possible aggregation. The self-organizing ability of the free INF2-FFC construct is further exemplified in the case of higher (10-fold) protein concentrations adsorbed over mica substrates. At 2 μM, large aggregates of INF2-FFC were observed. Most notably, stacked tubelike structures likely formed by stacking of dissociated, and then reassembled INF2-FFC rings were observed even in the absence of F-actin (Figure 2i,j). Previous studies proposed that INF2’s FH2 domain was capable of dissociation.46 Our AFM images provide structural evidence and confirm the ability of INF2-FFC to reorganize and assemble into oligomeric forms at higher protein densities.
Table 1. AFM Measurements of Free INF2-FFC and INF2-FF Self-Assembly.
| INF2-FFC | INF2-FF | ||||
|---|---|---|---|---|---|
| concentration (μM) | 0.02 | 0.2 | 2 | 0.02 | 0.2 |
| organization | dimers | oligomers | oligomers or stacked dimers | dimers | oligomers |
| width (nm) | 22 ± 0.1 (closed) | 45 ± 2 | – | 22 ± 0.1 (closed) | 44 ± 1 to 56 ± 2 |
| 27 ± 0.4 (open) | 54.6 ± 0.8 | 26 ± 1.6 (open) | |||
| cross-section height (nm) | 0.2 | 0.3 | ≥0.3 | 0.2 | 0.3 |
Structure of Free INF2-FF Homodimers and Oligomers
The effects of the C-termini of many formins on actin have been investigated previously.52−54 A region immediately C-terminal to the FH2 domain of INF2 is known to strongly affect its activity on actin.39,40,54 To test the role of the C-terminal region in INF2 self-assembly and oligomerization, we deleted it and probed the structure of free INF2-FF at concentrations of 0.02 and 0.2 μM. Deletion of the C-terminal region did not inhibit the self-assembly of FH2 into dimers (at 0.02 μM) or oligomers (at 0.2 μM). Figure 3 shows representative images of free INF2-FF self-organization configurations. The central core region observed as a dense mass (dashed white arrows) within the oligomeric ring periphery in Figure 3a–c appears to be similar to that observed for INF2-FFC dimers and oligomers, suggesting that it comes from the unstructured FH1 domain. The corresponding sizes for dimers and oligomers measured from AFM images were 22 ± 0.1 and 45 ± 2 nm for INF2-FFC and 22 ± 0.1 and 44 ± 1 nm for INF2-FF, respectively (Table 1). Furthermore, the central dense region within the donut-shaped structure seen in hollow dimer configurations (Figure 2c) presumably represents the flexible FH1 sequences lacking tertiary structure.46 The fact that this central core is present in INF2-FF images (Figure 3) suggests that it is not the C-terminus.
Figure 3.
AFM images showing free INF2-FF self-organization configurations. The central core region observed as a dense mass (dashed white arrows) within the oligomeric ring periphery in panel a–c. The central dense core appears to be similar to that observed for INF2-FFC dimers and oligomers, suggesting that it comes from the unstructured FH1 domain.
Structure of F-Actin–INF2-FH1-FH2-C (INF2-FFC) Complexes
Recently, helical reconstruction of filaments from EM images40 has revealed details of the INF2-FFC-bound F-actin architecture at 20 Å resolution, providing the “average” structure for the entire F-actin filament population or subpopulations.55 The previous successes of tapping mode AFM imaging in unraveling the structure and mechanics of F-actin–ABP (actin binding protein) complexes32,56 prompted us to apply this technique to seek the single-filament level structure of INF2-FFC and F-actin assembly at the nanoscale level. Figure 4 shows a typical AFM tapping mode image of a 1:1 ratio of INF2-FFC and F-actin complexes revealing a ringlike arrangement of formin FH2 homodimers encircled around actin filaments (Figure 4a,b). INF2-FFC-bound actin filaments show a mean width of ∼30 nm and a height of ∼2.8 nm. Considering that bare F-actin filaments in AFM images are 1.8 nm tall and ∼18.4 nm wide, there is an estimated 1 nm increase in the height and an ∼10 nm increase in the width of the filaments upon binding with INF2-FFC.
Further structural details illustrated in the close-up of INF2-FFC–F-actin complexes (Figure 4c) show INF2-FFC dimers to be 5.8–7 nm tall rings (see Figure 4a, white arrows), with approximately four or five dimeric rings bound per helical F-actin twist. The INF2-FFC rings bind along the actin filament length (as marked in Figure 4c). Frequently, 5–10° deviations in binding orientations of adjacent homodimers were also observed, suggesting a level of flexibility of the dimers, or strain developed along F-actin upon INF2-FFC binding. Because EM images provide averaged structures of the F-actin–INF2 complexes, such details on the individual filament level and deviations in binding orientations could not be observed previously.40 A similar assembly of INF-FF was also observed in complex with F-actin. Figure 5 represents a typical AFM image obtained for INF2-FF and F-actin complexes, revealing ringlike binding around actin filaments. The unique INF2-FFC and F-actin assembly reported here represents, to the best of our knowledge, the first three-dimensional AFM nanoscale structure obtained for any formin–F-actin complexes.
Figure 5.
Structure of INF2-FF and F-actin complexes. AFM image of a 1:1 ratio of INF2-FF and F-actin complexes revealing ringlike binding around actin filaments similar to that of INF2-FFC and F-actin complexes.
In summary, we have presented structural evidence confirming the ability of INF2-FFC to reorganize and assemble into oligomeric forms at higher protein concentrations, unrelated to its C-terminal region. Within the cellular context, higher-order INF2–F-actin structures may allow diverse functional roles under specific cellular conditions. Transitions between different oligomeric states may also be important in cooperative binding properties. On the basis of the AFM structures obtained for F-actin–INF2-FFC complexes, it is likely that formin undergoes dissociation and reassociation of the FH2 domains to allow its encircling of actin filaments (Figure 6). In addition, oligomerization may allow formins to generate higher-order structures that may provide greater stability, while the reduced surface area of the dimer in a complex can offer protection against severing by other actin binding proteins.
Conclusions
The self-assembly of a formin FH2 domain into oligomeric structures has been clearly resolved using AFM imaging of INF2-FFC and F-actin complexes. In the presence of actin filaments, INF2 organizes along the filament helix and creates discontinuities that might favor severing. Our data reveal significant structural insights into the possible dissociation and reassociation of INF2-FFC to yield circular binding over F-actin. From the nanotechnology perspective, engineering the self-assembly of nanostructures such as formin complexes (hollow formin nanotubes or formin nanotubes over actin core) is likely to provide new opportunities for novel molecular designs and programmed assembly for a variety of bionanotechnology applications.
Acknowledgments
Support for this work from MEXT WPI Program International Center for Materials Nanoarchitectonics (MANA), Japan, is acknowledged. P.S.G. and H.N.H. were supported by National Institutes of Health (NIH) Grant R01 GM069818, and E.E.G. and E.R. were supported by NIH Grant R01 GM077190. We acknowledge the use of the NPC core facility supported by CNSI at UCLA.
Supporting Information Available
AFM imaging data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Whitesides G. M.; Grzybowski B. Self-assembly at all scales. Science 2002, 29555642418–2421. [DOI] [PubMed] [Google Scholar]
- Kueh H. Y.; Mitchison T. J. Structural Plasticity in Actin and Tubulin Polymer Dynamics. Science 2009, 3255943960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamcik J.; Lara C.; Usov I.; Jeong J. S.; Ruggeri F. S.; Dietler G.; Lashuel H. A.; Hamley I. W.; Mezzenga R. Measurement of intrinsic properties of amyloid fibrils by the peak force QNM method. Nanoscale 2012, 4154426–4429. [DOI] [PubMed] [Google Scholar]
- Jang H.; Teran Arce F.; Ramachandran S.; Kagan B. L.; Lal R.; Nussinov R. Disordered amyloidogenic peptides may insert into the membrane and assemble into common cyclic structural motifs. Chem. Soc. Rev. 2014, DOI: 10.1039/C3CS60459D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin Structure: Repeating Unit of Histones and DNA. Science 1974, 1844139868–871. [DOI] [PubMed] [Google Scholar]
- Spector M. S.; Singh A.; Messersmith P. B.; Schnur J. M. Chiral self-assembly of nanotubules and ribbons from phospholipid mixtures. Nano Lett. 2001, 17375–378. [Google Scholar]
- Grenklo S.; Hillberg L.; Zhao Rathje L. S.; Pinaev G.; Schutt C. E.; Lindberg U. Tropomyosin assembly intermediates in the control of microfilament system turnover. Eur. J. Cell Biol. 2008, 8711905–920. [DOI] [PubMed] [Google Scholar]
- Peitsch W. K.; Hofmann I.; Pratzel S.; Grund C.; Kuhn C.; Moll I.; Langbein L.; Franke W. W. Drebrin particles: Components in the ensemble of proteins regulating actin dynamics of lamellipodia and filopodia. Eur. J. Cell Biol. 2001, 809567–579. [DOI] [PubMed] [Google Scholar]
- Peitsch W. K.; Hofmann I.; Endlich N.; Pratzel S.; Kuhn C.; Spring H.; Grone H. J.; Kriz W.; Franke W. W. Cell biological and biochemical characterization of drebrin complexes in mesangial cells and podocytes of renal glomeruli. J. Am. Soc. Nephrol. 2003, 1461452–1463. [DOI] [PubMed] [Google Scholar]
- Kuhn J. R.; Pollard T. D. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 2005, 8821387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. R.; Pollard T. D. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 2007, 2823828014–28024. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D.; Jaiswal R.; Bombardier J. P.; Gould C. J.; Gelles J.; Goode B. L. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 2012, 33660851164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R.; Pollard T. D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. U.S.A. 2004, 1014114725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Mooney P.; Zheng S.; Booth C. R.; Braunfeld M. B.; Gubbens S.; Agard D. A.; Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 2013, 106584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C. M.; Penczek P. A. Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM. Curr. Opin. Struct. Biol. 2009, 195623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G.; Quate C. F.; Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986, 569930–933. [DOI] [PubMed] [Google Scholar]
- Radmacher M.; Tillamnn R. W.; Fritz M.; Gaub H. E. From molecules to cells: Imaging soft samples with the atomic force microscope. Science 1992, 25750781900–1905. [DOI] [PubMed] [Google Scholar]
- Bustamante C.; Vesenka J.; Tang C. L.; Rees W.; Guthold M.; Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry 1992, 31122–26. [DOI] [PubMed] [Google Scholar]
- Hansma H. G.; Hoh J. H. Biomolecular imaging with the atomic force microscope. Annu. Rev. Biophys. Biomol. Struct. 1994, 23, 115–139. [DOI] [PubMed] [Google Scholar]
- Muller D. J.; Schabert F. A.; Buldt G.; Engel A. Imaging purple membranes in aqueous solutions at sub-nanometer resolution by atomic force microscopy. Biophys. J. 1995, 6851681–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D. J.; Dufrene Y. F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 2008, 35261–269. [DOI] [PubMed] [Google Scholar]
- Sharma S.; Gimzewski J. K.. Atomic Force Microscopy for Medicine. In Life at the nanoscale: Atomic force microscopy of live cells; Dufrene Y., Ed.; Pan Stanford Publishing: Singapore, 2011; pp 421–436. [Google Scholar]
- Sharma S.; Gillespie B. M.; Palanisamy V.; Gimzewski J. K. Quantitative nanostructural and single-molecule force spectroscopy biomolecular analysis of human-saliva-derived exosomes. Langmuir 2011, 272314394–14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D.; Jeno P.; Mini T.; Wirtz S.; Muller S. A.; Fraysse L.; Kjellbom P.; Engel A. Structural characterization of two aquaporins isolated from native spinach leaf plasma membranes. J. Biol. Chem. 2001, 27631707–1714. [DOI] [PubMed] [Google Scholar]
- Schabert F. A.; Henn C.; Engel A. Native Escherichia coli OMPF Porin Surfaces Probed by Atomic-Force Microscopy. Science 1995, 268520792–94. [DOI] [PubMed] [Google Scholar]
- Scheuring S.; Ringler P.; Borgnia M.; Stahlberg H.; Muller D. J.; Agre P.; Engel A. High resolution AFM topographs of the Escherichia coli water channel aquaporin Z. EMBO J. 1999, 18184981–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov Y.; Gershon P. D.; McPherson A. Atomic force microscopy investigation of vaccinia virus structure. J. Virol. 2008, 82157551–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov Y. G.; Victoria J. G.; Robinson W. E.; McPherson A. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-Infected lymphocytes. J. Virol. 2003, 772211896–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikai A.; Yoshimura K.; Arisaka F.; Ritani A.; Imai K. Atomic-Force Microscopy of Bacteriophage-T4 and Its Tube-Baseplate Complex. FEBS Lett. 1993, 3261-339–41. [DOI] [PubMed] [Google Scholar]
- McPherson A.; Kuznetsov Y. G. Atomic Force Microscopy Investigation of Viruses. Methods Mol. Biol. 2011, 736, 171–195. [DOI] [PubMed] [Google Scholar]
- Shao Z. F.; Shi D.; Somlyo A. V. Cryoatomic force microscopy of filamentous actin. Biophys. J. 2000, 782950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Grintsevich E. E.; Phillips M. L.; Reisler E.; Gimzewski J. K. Atomic force microscopy reveals drebrin induced remodeling of F-actin with subnanometer resolution. Nano Lett. 2011, 112825–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G.; Welch M. D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 114237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M. A.; DuPage A. G.; Goode B. L. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 2010, 11162–74. [DOI] [PubMed] [Google Scholar]
- DeWard A. D.; Eisenmann K. M.; Matheson S. F.; Alberts A. S. The role of formins in human disease. Biochim. Biophys. Acta 2010, 18032226–233. [DOI] [PubMed] [Google Scholar]
- Otomo T.; Tomchick D. R.; Otomo C.; Panchal S. C.; Machius M.; Rosen M. K. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 2005, 4337025488–494. [DOI] [PubMed] [Google Scholar]
- Higgs H. N. Formin proteins: A domain-based approach. Trends Biochem. Sci. 2005, 306342–353. [DOI] [PubMed] [Google Scholar]
- Goode B. L.; Eck M. J. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007, 76, 593–627. [DOI] [PubMed] [Google Scholar]
- Chhabra E. S.; Higgs H. N. INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 2006, 2813626754–26767. [DOI] [PubMed] [Google Scholar]
- Gurel P. S.; Ge P.; Grintsevich E. E.; Shu R.; Blanchoin L.; Zhou Z. H.; Reisler E.; Higgs H. N. INF2-Mediated Severing through Actin Filament Encirclement and Disruption. Curr. Biol. 2014, 24, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Moseley J. B.; Sagot I.; Poy F.; Pellman D.; Goode B. L.; Eck M. J. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell 2004, 1165711–723. [DOI] [PubMed] [Google Scholar]
- Thompson M. E.; Heimsath E. G.; Gauvin T. J.; Higgs H. N.; Kull F. J. FMNL3 FH2-actin structure gives insight into formin-mediated actin nucleation and elongation. Nat. Struct. Mol. Biol. 2013, 201111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M.; Higashi T.; Suetsugu S.; Sato Y.; Ikeda T.; Shirakawa R.; Kita T.; Takenawa T.; Horiuchi H.; Fukai S.; Nureki O. Crystal structure of human DAAM1 formin homology 2 domain. Genes Cells 2007, 12111255–1265. [DOI] [PubMed] [Google Scholar]
- Moseley J. B.; Sagot I.; Manning A. L.; Xu Y.; Eck M. J.; Pellman D.; Goode B. L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 2004, 152896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. S.; Higgs H. N. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006, 406, 190–214. [DOI] [PubMed] [Google Scholar]
- Paul A. S.; Pollard T. D. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 2009, 668606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. A.; Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971, 246154866–4871. [PubMed] [Google Scholar]
- Harris E. S.; Li F.; Higgs H. N. The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 2004, 2791920076–20087. [DOI] [PubMed] [Google Scholar]
- Li F.; Higgs H. N. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 2005, 28086986–6992. [DOI] [PubMed] [Google Scholar]
- Lu J.; Meng W.; Poy F.; Maiti S.; Goode B. L.; Eck M. J. Structure of the FH2 domain of Daam1: Implications for formin regulation of actin assembly. J. Mol. Biol. 2007, 36951258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S.; Michelot A.; Gould C.; Blanchoin L.; Sokolova O.; Goode B. L. Structure and activity of full-length formin mDia1. Cytoskeleton 2012, 696393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould C. J.; Maiti S.; Michelot A.; Graziano B. R.; Blanchoin L.; Goode B. L. The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 2011, 215384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsath E. G. Jr.; Higgs H. N. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 2012, 28753087–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V.; Gurel P. S.; Higgs H. N. Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J. Biol. Chem. 2012, 2874134234–34245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Zhu H.; Grintsevich E. E.; Reisler E.; Gimzewski J. K. Correlative nanoscale imaging of actin filaments and their complexes. Nanoscale 2013, 5, 5692–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.; Grintsevich E. E.; Hsueh C.; Reisler E.; Gimzewski J. K. Molecular cooperativity of drebrin(1–300) binding and structural remodeling of F-actin. Biophys. J. 2012, 1032275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.