Abstract
Low molecular weight heparins (LMWHs) are heterogeneous, polydisperse, and highly negatively charged mixtures of glycosaminoglycan chains prescribed as anticoagulants. The detailed characterization of LMWH is important for the drug quality assurance and for new drug research and development. In this study, online hydrophilic interaction chromatography (HILIC) Fourier transform mass spectrometry (FTMS) was applied to analyze the oligosaccharide fragments of LMWHs generated by heparin lyase II digestion. More than 40 oligosaccharide fragments of LMWH were quantified and used to compare LMWHs prepared by three different manufacturers. The quantified fragment structures included unsaturated disaccharides/oligosaccharides arising from the prominent repeating units of these LMWHs, 3-O-sulfo containing tetrasaccharides arising from their antithrombin III binding sites, 1,6-anhydro ring-containing oligosaccharides formed during their manufacture, saturated uronic acid oligosaccharides coming from some chain nonreducing ends, and oxidized linkage region oligosaccharides coming from some chain reducing ends. This bottom-up approach provides rich detailed structural analysis and quantitative information with high accuracy and reproducibility. When combined with the top-down approach, HILIC LC-FTMS based analysis should be suitable for the advanced quality control and quality assurance in LMWH production.
Heparin is a complex sulfated polysaccharide, known as a glycosaminoglycan, which is widely used as a clinical anticoagulant.1 Heparin, on average, is composed of ∼25 disaccharide repeating units and has an average molecular weight of ∼16 000 Da. Its major repeating disaccharide unit is trisulfated, (→4) α-L-IdoA2S (1→4) α-D-GlcNS6S (1→), where IdoA is iduronic acid, GlcN is glucosamine, and S is sulfo.1,2 In addition to these residues, heparin contains glucuronic acid (GlcA) and N-acetyl glucosamine (GlcNAc) residues having varying numbers of O-sulfo groups.2,3 Heparin also contains pentasaccharide sequences, with a central 3-O-sulfo group-containing GlcN residue, which are the primary structures responsible for heparin binding to antithrombin III (AT), resulting in inhibition of blood coagulation.4 Low molecular weight heparins (LMWHs), derived from heparin by controlled chemical or enzymatic depolymerization,5,6 have a primary structure similar to heparin but an average molecular weight of ∼4000 to ∼8000 Da, corresponding to ∼6 to ∼12 disaccharide units. Several types of LMWHs are in clinical use. The current study focuses on the most widely used LMWH, enoxaparin, which is prepared through the chemical esterification of heparin’s uronic acid carboxyl groups followed by base treatment leading to β-eliminative cleavage and ester hydrolysis.5 This chemical method used in the manufacturing of enoxaparin can modify its structure, not only changing the polydispersity and sequence heterogeneity of LMWH but also introducing unnatural saccharide residues, such as unsaturated uronic acid residues at the nonreducing ends and unnatural 1,6-anhydro ring structures at the reducing ends of some of the enoxaparin’s chains.7,8
A number of methods, including top-down and bottom-up approaches, has been applied for the structural characterization of enoxaparin sodium. In the top-down approach, the intact oligosaccharide chains are directly analyzed without further depolymerization. Nuclear magnetic resonance (NMR) spectroscopy plays a very important role in the structural identification of intact chains,9,35 and two-dimensional (2D) NMR can be used to identify LMWH type and to calculate their monosaccharide composition.10 Nonetheless, milligram amounts of pure samples are needed to acquire the data for NMR, and some relatively minor residues cannot be easily detected by NMR. Mass spectrometry is a particularly useful technique for the structural characterization of LMWHs, providing both high sensitivity and high resolution. Powerful separation techniques,35 particularly when combined with MS, can enhance the structural identification of heterogeneous and polydisperse LMWHs. Reversed-phase ion-pairing liquid chromatography (RPIP)-ESI-MS has been widely applied to the analysis of glycosaminoglycan-derived oligosaccharides,11−13,36 and Chi et al.14 has recently utilized this method to profile the intact LMWHs and identify more than 200 intact components in enoxaparin sodium. Ultraperformance size exclusion chromatography/electrospray quadruple time-of-flight-mass spectrometry (UPSEC/Q-TOF-MS) has also been applied to identify more than 70 intact components in enoxaparin sodium, including oligosaccharides with unnatural structures, such as 1,6-anhydro rings, and saturated uronic acid residues and odd-numbered oligosaccharides at the nonreducing end.15 Our group has elucidated and semiquantified nearly 300 intact chains in LMWHs (made up of a smaller number of glycosidically linked disaccharide and oligosaccharide components) using hydrophilic interaction chromatography (HILIC)-Fourier transform-(FT) electrospray ionization-mass spectrometry (ESI-MS).16
In bottom-up approaches, oligosaccharides are typically prepared through the digestion of LMWH with heparin lyases (I, II, and/or III) and then analyzed by strong anion exchange chromatography (SAX), RPIP chromatography, and capillary electrophoresis (CE) and then detected by their UV absorbance or through MS.17−22,36,42 Disaccharide compositional analysis is one of the primary ways to characterize LMWH composition.19,20,42 In addition to disaccharides, oligosaccharides are formed that include 3-O-sulfo group-containing tetrasaccharides,23 1,6-anhydro ring-containing oligosaccharides, oligosaccharides with a saturated uronic acid residue, and oligosaccharides with an odd number of saccharide units, arising from a nonreducing end of the chain.15 Many of the reported structures can be related to the activity and pharmacology of LMWHs.8,24,25 Therefore, there is an urgent need to develop improved bottom-up methods to profile all these oligosaccharides.
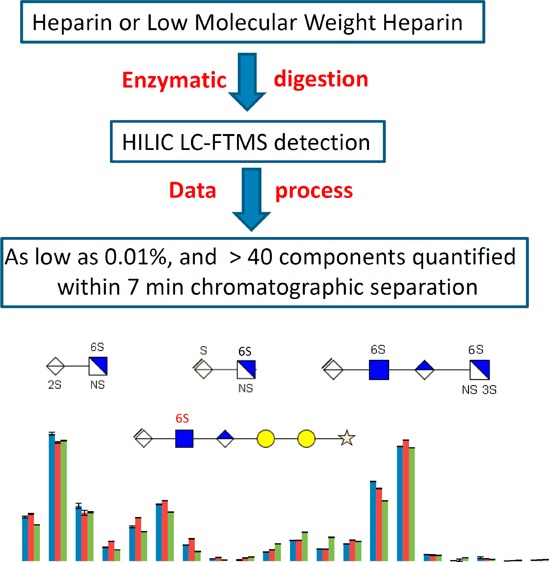
In this paper, we report a rapid analytical method for the characterization of enoxaparin-type LMWHs using a bottom-up approach. This method relies on HILIC LC-Fourier transform mass spectrometry (FTMS) analysis combined with heparin lyase II digestion. Data analysis utilizes a newly developed bioinformatics software package, GlycReSoft.29 The oligosaccharides (dp2 to dp6) generated from LMWHs prepared by three different manufacturers are compared.
Experimental Section
Materials
LMWHs, of enoxaparin sodium from Sanofi-Aventis (Lovenox) and enoxaparin sodium from Sandoz (a generic version of Lovenox), were obtained from hospital pharmacies (3 lots of each) and were freeze-dried prior to analysis. A bulk-lyophilized LMWH prepared using a process similar to that used to prepare enoxaparin sodium was a gift from Celsus Laboratories Inc. (Cincinnati, Ohio). E. coli expression and purification of the recombinant F. heparinum heparin lyase II (EC# 4.2.2.X) was performed in our laboratory as described.26 Acetonitrile, ammonium acetate, and water were of HPLC grade (Sigma-Aldrich, St. Louis, MO).
Enzymatic Digestion of LMWHs
Three lots of desalted LMWH samples (100 μg) from each manufacturer were dissolved in 100 μL of distilled water and completely digested by heparin lyase II (20 mU) at 35 °C for 2 h (longer digestion times of up to 12 h gave similar results). Aliquots were immediately heated in a 100 °C water bath to stop the reaction and were then spun down at 12 000 rpm for 5 min; supernatants were used directly for LC-MS analysis.
HILIC LC ESI-LTQ-Orbitrap-FTMS Analysis of Digested LMWHs
A Luna HILIC column (2.0 × 50 mm, 200 Å, Phenomenex, Torrance, CA) was used to separate the LMWHs. Mobile phase A was 5 mM ammonium acetate prepared with HPLC grade water. Mobile B was 5 mM ammonium acetate prepared in 98% HPLC grade acetonitrile with 2% of HPLC grade water. The gradient was used from 5% A to 70% A in 7 min then reset to 5% A at a flow rate of 250 μL/min. The LC column was directly connected online to the standard ESI source of LTQ-Orbitrap XL FT MS (Thermo Fisher Scientific, San-Jose, CA). The source parameters for FTMS detection were optimized using Arixtra (a synthetic ultra LMWH from Sanofi-Aventis, Paris, France) to minimize the insource fragmentation and sulfate loss and maximize the signal/noise in the negative-ion mode. The optimized parameters, used to prevent in-source fragmentation, included a spray voltage of 4.2 kV, a capillary voltage of −40 V, a tube lens voltage of −50 V, a capillary temperature of 275 °C, a sheath flow rate of 30 L/min, and an auxiliary gas flow rate of 6 L/min. External calibration of mass spectra routinely produced a mass accuracy of better than 3 ppm. All FT mass spectra were acquired at a resolution 60 000 with 200–2000 Da mass range.
Bioinformatics
Charge deconvolution was autoprocessed by DeconTools software (web source from PNNL at http://omics.pnl.gov/). LMWH structural assignment was done by either manual or automatic processing using GlycReSoft 1.0 software developed at Boston University (http://code.google.com/p/glycresoft/downloads/list).29 For automatic processing, GlycReSoft 1.0 parameters were set as Minimum Abundance, 1.0; Minimum Number of Scans, 1; Molecular Weight Lower Boundary, 200 Da; Molecular Weight Upper Boundary, 6000 Da; Mass Shift, ammonium; Match Error (E_M), 5.0 ppm; Grouping Error (E_G), 80 ppm; Adduct Tolerance (E_A), 5.0 ppm. For LMWH digested components identification, the theoretical database was generated by GlycReSoft 1.0 for different structures. All of the quantitative data were normalized to the total identified oligosaccharides peak area (in the format of percentage, %).
Results and Discussion
Depolymerization of LMWHs Using Heparin Lyase II
LMWHs, such as enoxaparin sodium, are more complicated in structure than the precursor heparin since their chemical fractionation introduces new structural artifacts.5,6 These chemical processes differ from manufacturer-to-manufacturer and can uniquely modify the structure of each LMWH. The resulting products are polydisperse mixtures of chains, that while retaining most of heparin’s natural structure also contain unnatural, unsaturated uronate residues at the nonreducing ends of some of their chains and unnatural 1,6-anhydro ring structures at the reducing ends of some of their chains. The characterization of the detailed structural information requires a bottom-up approach in LMWH analysis. Enzymes are the crucial tool for achieving such bottom-up analyses of LMWHs. Heparin lyases are eliminases that act on the glycosidic linkage between IdoA or GlcA and GlcN residues affording disaccharide and oligosaccharide products having an unsaturated uronic acid residue (ΔUA) at their nonreducing ends. Heparin lyase II is a heparinase with broad specificity that acts at linkages containing either (1 → 4)-α-L-IdoA or (1 → 4)-β-D-GlcA residues and accommodates many different sulfation patterns in these polysaccharides.27,28 Oligosaccharides of heparins generated by digestion of heparin lyase II have been mapped.23 Tetrasaccharides containing 3-O-sulfo groups are observed in the products of heparin digested by heparin lyase II.38 Therefore, we selected heparin lyase II and developed a quick and exhaustive digestion method to prepare oligosaccharides from LMWH samples for bottom-up analysis.
HILIC LC ESI-LTQ-Orbitrap-FTMS Analysis of Digested Products of LMWHs
Luna-HILIC separation is based on analyte polarity and affords good separation of glycans.16 The high-resolution and high mass accuracy of ESI-FTMS make this method suitable for glycan analysis. HILIC-ESI-FTMS proved to be a powerful platform in the top-down approach with the high resolution separation and analysis of intact LMWH chains.16 We elucidated and semiquantified nearly 300 of oligosaccharide chains of LMWH samples using this method.16 Here, a similar method was applied in the bottom-up approach to characterize disaccharides and oligosaccharides prepared from LMWHs using heparin lyase II, except HILIC separation relied on a shorter column (5 cm), greatly reducing the analytical run time, to 7 min. The total ion chromatograms (TIC) of digested LMWH (in triplicate) using HILIC-ESI-FTMS are shown in Figure 1A, and the typical unsaturated oligosaccharide quantification comparison after digestion of LMWHs is shown in Figure 1B. Figure 1A demonstrates excellent run-to-run reproducibility of this method and the separation of different oligosaccharides. The major TIC peaks observed in Figure 1A were disaccharides and tetrasaccharides (degree of polymerization (dp)2 and (dp)4). Nevertheless, the mass spectrum for each peak was still too complex to be completely interpreted, as each peak contained a mixture of multiple components having various sequences and degrees of sulfation. Therefore, the raw data from the HILIC LC-FTMS was deconvoluted using Decon Tools, and then, the output of Decon Tools was processed by GlycResoft to generate matching structures and to provide quantitative information.29 Quantitative results on the major unsaturated oligosaccharide of three different LMWHs samples are shown in Figure 1B. Approximately 23 unsatutated oligosaccharide structures were matched by GlycResoft including 6 disaccharides, 4 trisaccharides, 11 tetrasaccharides, and 2 pentasaccharides in Figure 1A. The majority of these oligosaccharides were even-numbered, although odd-numbered oligosaccharides were also identified. As expected, the trisulfated disaccharide, corresponding to ΔUA2S-GlcNS6S, was the major component in each sample.
Figure 1.
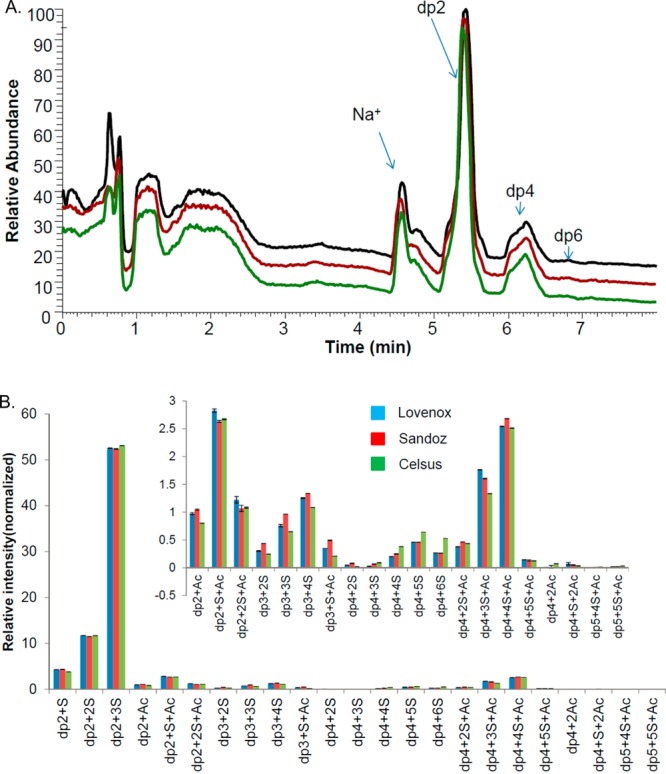
HILIC LC-FTMS total ion chromatogram (TIC) and major heparin lyase II-derived oligosaccharides quantified from the LMWHs of three different manufacturers. (A) TIC of digested LMWH, reproducibility of the HILIC LC-FTMS method followed by enzyme digestion. Triplicated sample preparations were analyzed on the same LC-MS batch. (B) Typical unsaturated oligosaccharide products quantification comparison after digestion of LMWHs from three different manufacturers. Figure inset is the magnified profile of low abundance oligosaccharides.
Tetrasaccharides with 3-O-sulfo groups, generated by heparin lyase II digestion, derived from heparin’s AT-binding sites, can be applied to evaluate the anticoagulant activity of each LMWH.23,24 Four major 3-O-sulfated tetrasaccharides were reported previously and are also detected in these LMWH samples. Their extracted ion chromatograms (EICs) and high-resolution FTMS spectrum examples are shown in Figure 2. Ions of m/z 477.034 (ΔUA-GlcNAc6S-GlcA-GlcNS3S), m/z 517.013 (ΔUA-GlcNAc6S-GlcA-GlcNS3S6S), m/z 535.985 (ΔUA-GlcNS6S-GlcA-GlcNS3S6S), and m/z 437.055 (ΔUA-GlcNAc-GlcA-GlcNS3S) correspond to the major 3-O-sulfo group-containing tetrasaccharides after heparin lyase II digestion. Eight additional low abundant tetrasaccharides were also profiled and quantified (Figure 1B). On the basis of the known specificity heparin lyase II, these digestion-resistant tetrasaccharides may represent putative 3-O-sulfo group-containing structures. However, because of their low abundance and difficulties in separating these tetrasaccharides from one another, their structures could not be confirmed by NMR. Thus, we cannot definitively establish that all these tetrasaccharides contain 3-O-sufate groups.
Figure 2.
Example of extracted ion chromatograms (EICs) and high resolution mass spectrogram of four major 3-O-sulfo group-containing tetrasaccharides derived from LMWHs on heparin lyase II treatment.
Terminal 1,6-anhydro-amino sugars, formed during the β-eliminative cleavage of heparin in the manufacturing process, are typical structural moieties found within LMWHs. These 1,6-anhydro-amino residues can reportedly decrease the anticoagulant activity of LMWHs.8 Since anhydo-components correspond to the loss of additional water, oligosaccharides of compositions [ΔHexA = 0, HexA, GlcN, Ac, SO3] and [ΔHexA = 1, HexA, GlcN, Ac, SO3] were added to our database to allow their identification. By matching the results generated to the GlycoResoft database, the 1,6-anhydro oligosaccharides were identified and quantified in all three of the LMWHs being studied (Figure 3A). On the basis of these results, all of the three different LMWH products examined afforded similar profiles of the 1,6-anhydro-components on heparin lyase II treatment. Some minor differences were observed in the distribution of the 1,6-anhydro oligosaccharides between the dp2 and dp4 fractions formed from the three LMWH products being studied.
Figure 3.
Quantitative comparison of identified rare heparin oligosaccharides from three commercialized LMWH products digested by heparin lyase II. (A) Quantitative comparison of anhydro oligosaccharides identified in three LMWHs; (B) quantitative comparison of nonreducing end oligosaccharides identified in three LMWHs; (C) quantitative comparison of linkage region oligosaccharides identified in three LMWHs.
Most of the LMWH chains contain 4,5-unsaturated uronic acid residues at the nonreducing end. Minor oligosaccharide structures, with saturated uronic acid residues at their nonreducing end are present in chains of LMWH that had been derived from the original nonreducing end of the parent heparin. These saturated oligosaccharide products were also identified and quantified by GlycResoft based on the composition, [ΔHexA = 0, HexA, GlcN, Ac, SO3]. Most of the heparin lyase II-derived oligosaccharide products were trisulfated disaccharides, and the normalized percentage of nonreducing end-derived oligosaccharides obtained from the LMWHs prepared by Sandoz and Celsus were higher than those obtained from Lovenox (Figure 3B), suggesting that the parent heparins for these LMWH products were slightly different in their molecular weight and composition. These differences between commercial LMWH nonreducing ends were not as obvious using top-down analysis,16 suggesting an advantage of this bottom-up approach. The current specifications of heparin sodium, the parent for each LMWH product, are significantly different from those used earlier during the market approval process for Lovenox.
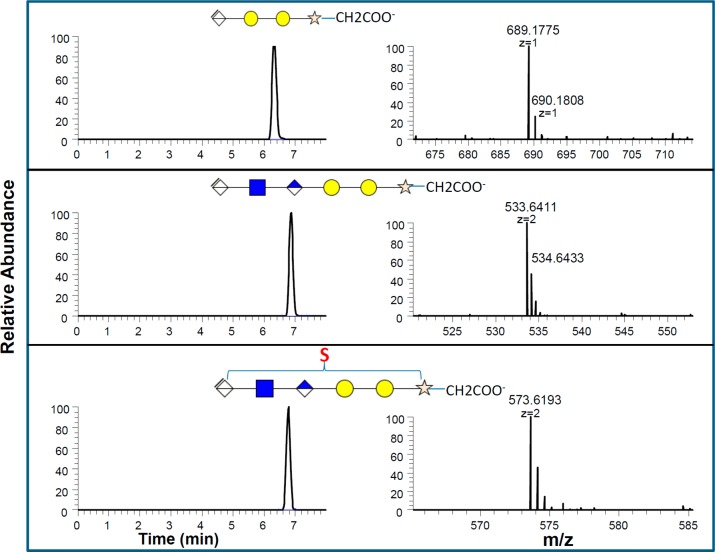
Heparin chains are biosynthesized starting from the common carbohydrate–protein linkage region.30 Nonsulfated oligosaccharide sequence ΔUA-Gal-Gal-Xyl-O-Ser (where Gal is galactose, Xyl is xylose, and Ser is serine) derived from the core-protein linkage region is usually present in the commercial heparin.31 Highly sulfated sequences have been identified that elicit the biological activities of heparin. Other activities, including antiangiogenic and antiproliferative properties, may also involve less sulfated sequences encompassing the linkage region.32−34 These studies suggest that linkage region may play a role in modulating some of the activities of heparin. Therefore, the quantification of the linkage region is necessary to completely profile a LMWH in the bottom-up approach. In the LC-MS chromatogram of the digested LMWH, we observed predominantly oxidized Ser (SerOX) at the reducing end linkage, instead of Ser (Figures 4 and 5), which is different from the reported heparin reducing end linkage. Serine is known to be sensitive to radical based oxidation.39 Reagents, such as hydrogen peroxide or peroxyacids,37,38 and permanganate oxidation40 are commonly used conditions in the processing of the commercial heparins and are also used to prepare LMWHs.41 The linkage region-O-SerOX has recently been reported in studies on glycol-split periodate oxidized LMWH products.22 MS/MS provides more detailed structures, including the location of sulfo groups provided in the linkage, corresponding to the structures, ΔUA-GlcNAc-GlcA-Gal-Gal-Xyl-O-SerOX and ΔUA-GlcNAc6S-GlcA-Gal-Gal-Xyl-O-SerOX (Figure 5A,B). The linkage region oligosaccharides of three LMWHs samples were quantified and shown in Figure 3C. The unsulfated hexasaccharide (ΔUA-GlcNAc-GlcA-Gal-Gal-Xyl-O-SerOX) was the major linkage region component, consistent with the linkage region reported in a previous study.36 However, the ratio and content of the three linkage region oligosaccharides obtained from the LMWH samples in the current study showed obvious differences. The amount of three linkage region oligosaccharides derived from the Lovenox sample was much lower than that from the other two samples. These differences might give some clue to the oxidative conditions used to process the parent heparin and to prepare these LMWH products.
Figure 4.
Example of extracted ion chromatograms (EICs) and high resolution mass spectrograms of the linkage region oligosaccharides analysis of LMWHs treated with heparin lyase II.
Figure 5.
Accurate MS and MS/MS sequence analysis of the linkage region dp6 oligosaccharides. (A) The MS/MS analysis of LR dp6 with one sulfate (m/z = 573.619, doubly charged ions in negative mode) indicated the sulfation was located at N-acetylglucosamine. (B) The MS/MS sequence analysis of LR dp6 without sulfate modifications (m/z = 533.641, doubly charged ions in negative ion mode).
Conclusions
In this study, online HILIC LC-FTMS combined heparin lyase II digestion offers a new method for the bottom-up analysis of LMWHs. This method is so sensitive that it can detect and identify as little as 0.01% components and affords a dynamic range of >5000 within about a 7 min chromatographic separation window. The current method has relatively high throughput but compromised some LC resolution. The solvent system and sample preparation were carefully selected to reduce the matrix effect on ionization and promote sensitivity. On the basis of this method, more than 40 digested components were observed. The primary unsaturated oligosaccharides, 3-O-sulfo containing tetrasaccharides, 1,6-anhydro rings, saturated uronic acid at the nonreducing end, and odd-numbered saccharide units were used to compare three different sources of LMWHs. All of the LMWHs examined in this study were very similar in structure and composition. This study provides a useful method with relatively high throughput and excellent reproducibility for the bottom-up analysis of LMWHs and is suitable for quality control and/or validation of a LMWH production, in conjunction with other methods such as LC-MS analysis42 and capillary electrophoresis43 with absorbance detection.
Acknowledgments
The work was supported by Grants from the National Institutes of Health in the form of Grants GM38060, GM090127, HL096972, and HL10172. G.L. was supported by the China Scholarship Council and Program for Changjiang Scholars and Innovative Research Team in University (IRT1188).
Supporting Information Available
Detailed structural information for the quantified oligosaccharides, Table S1. This material is available free of charge via the Internet at http://pubs.acs.org/.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Linhardt R. J. J. Med. Chem. 2003, 46, 2551–2554. [DOI] [PubMed] [Google Scholar]
- Nader H. B.; Dietrich C. P. In Heparin chemical and biological properties, clinical applications; Lane D. A., Lindahl U., Eds.; CRC: Boca Raton, 1989; pp 115–133. [Google Scholar]
- Rabenstein D. L. Nat. Prod. Rep. 2002, 19, 312–331. [DOI] [PubMed] [Google Scholar]
- Petitou M.; Casu B.; Lindahl U. Biochimie 2003, 85, 83–89. [DOI] [PubMed] [Google Scholar]
- Linhardt R. J.; Gunay N. S. Semin. Thromb. Hemostasis 1999, 25Suppl 35–16. [PubMed] [Google Scholar]
- Bhaskar U.; Sterner E.; Hickey A. M.; Onishi A.; Zhang F.; Dordick J. S.; Linhardt R. J. Appl. Microbiol. Biotechnol. 2012, 93, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascellani G.; Guerrini M.; Torri G.; Liverani L.; Spelta F.; Bianchini P. Carbohydr. Res. 2007, 342, 835–842. [DOI] [PubMed] [Google Scholar]
- Guerrini M.; Elli S.; Gaudesi D.; Torri G.; Casu B.; Mourier P.; Herman F.; Boudier C.; Lorenz M.; Viskov C. J. Med. Chem. 2010, 53, 8030–8040. [DOI] [PubMed] [Google Scholar]
- Jones C. J.; Beni S.; Limtiaco J. F. K.; Langeslay D. J.; Larive C. K. Annu. Rev. Anal. Chem. 2011, 4, 439–465. [DOI] [PubMed] [Google Scholar]
- Keire D. A.; Buhse L. F.; Al-Hakim A. Anal. Methods 2013, 5122984–2994. [Google Scholar]
- Patel R. P.; Narkowicz C.; Jacobson G. A. Anal. Biochem. 2009, 387, 113–121. [DOI] [PubMed] [Google Scholar]
- Thanawiroon C.; Rice K. G.; Toida T.; Linhardt R. J. J. Biol. Chem. 2004, 279, 2608–2615. [DOI] [PubMed] [Google Scholar]
- Wang B.; Buhse L. F.; Al-Hakim A.; Li M. T. B.; Keire D. A. J. Pharm. Biomed. Anal. 2012, 67–68, 42–50. [DOI] [PubMed] [Google Scholar]
- Li D.; Chi L.; Jin L.; Xu X.; Du X.; Ji S.; Chi L. Carbohydr. Polym. 2014, 99, 339–344. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Chen X.; Zhu Z.; Zhan X.; Wu Y.; Song L.; Kang J. Anal. Chem. 2013, 8531819–1827. [DOI] [PubMed] [Google Scholar]
- Li L.; Zhang F.; Zaia J.; Linhardt R. J. Anal. Chem. 2012, 84208822–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeotti F.; Volpi N. J. Chromatogr., A 2013, 1284, 141–147. [DOI] [PubMed] [Google Scholar]
- Galeotti F.; Volpi N. Anal. Chem. 2011, 83, 6770–6777. [DOI] [PubMed] [Google Scholar]
- Chang Y.; Yang B.; Zhao X.; Linhardt R. J. Anal. Biochem. 2012, 427191–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B.; Weyers A.; Baik J. Y.; Sterner E.; Sharfstein S.; Mousa S. A.; Zhang F.; Dordick J. S.; Linhardt R. J. Anal. Biochem. 2011, 415, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustkern A. M.; Buhse L. F.; Nasr M.; Al-Hakim A.; Keire D. A. Anal. Chem. 2010, 82, 9865–9870. [DOI] [PubMed] [Google Scholar]
- Alekseeva A.; Casu B.; Torri G.; Pierro S.; Naggi A. Anal. Biochem. 2013, 434, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z.; Tappen B. R.; Ly M.; Zhao W.; Canova L. P.; Guan H.; Linhardt R. J. J. Med. Chem. 2010, 54, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt R. J.; Rice K. G.; Kim Y. S.; Lohse D. L.; Wang H. M.; Loganathan D. Biochem. J. 1988, 254, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiguzel C.; Jeske W. P.; Hoppensteadt D.; Walenga J. M.; Bansal V.; Fareed J. Clin. Appl. Thromb./Hemostasis 2009, 15, 137–144. [DOI] [PubMed] [Google Scholar]
- Su H.; Blain F.; Musil R. A.; Zimmermann J. J.; Gu K.; Bennett D. C. Appl. Environ. Microbiol. 1996, 62, 2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai U. R.; Wang H.; Linhardt R. J. Arch. Biochem. Biophys. 1993, 306, 461–468. [DOI] [PubMed] [Google Scholar]
- Desai U. R.; Wang H.; Linhardt R. J. Biochemistry 1993, 32, 8140–8145. [DOI] [PubMed] [Google Scholar]
- Maxwell E.; Tan Y.; Tan Y.; Hu H.; Benson G.; Aizikov K.; Conley S.; Staples G. O.; Slysz G. W.; Smith R. D.; Zaia J. PloS One 2012, 79e45474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U.; Rodén L. J. Biol. Chem. 1965, 240, 2821–2826. [PubMed] [Google Scholar]
- Lane D. A.; Björk I.; Lindahl U.. Heparin and Related Polysaccharides; Plenum Press: New York, 1992. [Google Scholar]
- Lindahl U.; Lidholt K.; Spillmann D.; Kjellén L. Thromb. Res. 1994, 75, 1–32. [DOI] [PubMed] [Google Scholar]
- Hanhenberger R.; Jakobson Å. M.; Ansari A.; Wehler T.; Svahn C. M.; Lindahl U. Glycobiology 1993, 3, 567–573. [DOI] [PubMed] [Google Scholar]
- Sugahara K.; Yamada S.; Yoshida K.; de Waard P.; Vliegenthart J. F. G. J. Biol. Chem. 1992, 267, 1528–1533. [PubMed] [Google Scholar]
- Ye H.; Toby T. K.; Sommers C. D.; Ghasriani H.; Trehy M. L.; Ye W.; Kolinski R. E.; Buhse L. F.; Al-Hakim A.; Keire D. A. J. Pharm. Biomed. Anal. 2013, 85, 99–107. [DOI] [PubMed] [Google Scholar]
- Brustkern A. M.; Buhse L. F.; Nasr M.; Al-Hakim A.; Keire D. A. Anal. Chem. 2010, 82, 9865–9870. [DOI] [PubMed] [Google Scholar]
- Li G.; Yang B.; Li L.; Zhang F.; Xue C.; Linhardt R. J. Anal. Biochem. 2014, http://dx.doi.org/10.1016/j.ab.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toennies G.; Homiller R. P. J. Am. Chem. Soc. 1942, 64, 3054–3056. [Google Scholar]
- Stadtman E. R.; Levine R. L. Amino Acids 2003, 25, 207–218. [DOI] [PubMed] [Google Scholar]
- Halligudi N. N.; Desai S. M.; Mavalangi S. K.; Nandibewoor S. T. Monatsh. Chem. 2000, 131, 321–332. [Google Scholar]
- Linhardt R. J.; Gunay N. S. Semin. Thromb. Hemostasis 1999, 25, 5–16. [PubMed] [Google Scholar]
- Wang B.; Buhse L. F.; Al-Hakim A.; Boyne M. T. II; Keire D. A. J. Pharmaceut. Biomed. Anal. 2012, 67–68, 42–50. [DOI] [PubMed] [Google Scholar]
- Desai U. R.; Wang H. M.; Ampofo S. A.; Linhardt R. J. Anal. Biochem. 1993, 213, 120–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.