Abstract
Background
Only a minority of the genetic component of prostate cancer (PrCa) risk has been explained. Some observed associations of single nucleotide polymorphisms (SNPs) with PrCa might arise from associations of these SNPs with circulating prostate specific antigen (PSA) because PSA values are used to select controls.
Methods
We undertook a genome-wide association study (GWAS) of screen detected PrCa (ProtecT 1146 cases and 1804 controls); meta-analysed the results with those from the previously published UK Genetic Prostate Cancer Study (1854 cases and 1437 controls); investigated associations of SNPs with PrCa using either ‘low’ (PSA <0.5ng/ml) or ‘high’ (PSA ≥3ng/ml, biopsy negative) PSA controls; and investigated associations of SNPs with PSA.
Results
The ProtecT GWAS confirmed previously reported associations of PrCa at 3 loci: 10q11.23, 17q24.3 and 19q13.33. The meta-analysis confirmed associations of PrCa with SNPs near 4 previously identified loci (8q24.21,10q11.23, 17q24.3 and 19q13.33). When comparing PrCa cases with low PSA controls, alleles at genetic markers rs1512268, rs445114, rs10788160, rs11199874, rs17632542, rs266849 and rs2735839 were associated with an increased risk of PrCa, but the effect-estimates were attenuated to the null when using high PSA controls (p for heterogeneity in effect-estimates<0.04). We found a novel inverse association of rs9311171-T with circulating PSA.
Conclusions
Differences in effect estimates for PrCa observed when comparing low vs. high PSA controls, may be explained by associations of these SNPs with PSA.
Impact
These findings highlight the need for inferences from genetic studies of PrCa risk to carefully consider the influence of control selection criteria.
Keywords: Prostate Specific Antigen, research design, prostate cancer, case-control studies, genome wide association studies
Introduction
Prostate cancer (PrCa) is the leading cause of cancer in men in developed countries, accounting for 25% of new cancer cases. Prostate-specific antigen (PSA) testing is currently the most widely used screening test for PrCa, but it is limited because current thresholds defining raised levels (typically circulating PSA levels > 3 or 4 ng/ml) do not distinguish clinically important from indolent cancer, and lower levels do not exclude PrCa(1).
Epidemiological studies suggest a significant genetic component in the aetiology of the disease, but only 30% of the estimated heritability has so far been explained (2), highlighting the need for more studies to identify the full genetic profile of PrCa. The majority of genome-wide association studies (GWAS) of PrCa have been based on clinically-detected cases (3-11), with some studies specifically focusing on young age at onset, and/or familial or aggressive PrCa (i.e. pathological stage T3/T4,N+,M+, Gleason score ≥ 7, grade III, metastases, androgen ablation therapy, or PSA at least 10-50 ng/ml) (4-6, 8-10, 12). Most studies have compared cases to controls that have been selected on the basis of PSA levels under a certain threshold (5, 6, 10, 12-17); including some studies which have used controls with extremely low PSA levels (<0.5ng/ml; “supernormal” controls) (3, 5, 6, 12, 15). Such control selection strategies can make it difficult to interpret associations between a genetic variant and disease, since such associations may result from a relationship between the variant and PSA levels rather than PrCa. A limited number of GWAS have attempted to clarify this relationship by analysing associations of PSA levels with genetic markers identified using PSA screened controls (2, 5, 6, 15, 18, 19). It has been shown that SNPs at 13 known prostate cancer susceptibility loci (rs6869841, rs1270884, rs17632542, rs2242652, rs6983561, rs620861, rs10090154, rs7837688, rs12500426, rs7127900, rs10993994, rs2659056, rs2735839, rs5945619) are also associated with PSA concentration in blood in controls (2, 5, 6, 15, 19). As controls were defined by a PSA cut-off, however, associations between genotypes and the full distribution of PSA in men without PrCa could not be assessed. Only one study specifically investigated the relationship between genetic variants and circulating PSA level in men without detected PrCa (19). The Prostate Testing for cancer and Treatment study (ProtecT) contributed replication data to this study for the significant SNPs identified in the discovery GWAS. Genome wide significant associations between PSA levels and markers at 6 different loci were reported (19).
Our investigation had three aims. Firstly, to conduct a GWAS in a population-based screen detected PrCa population (ProtecT)(20). Secondly to conduct a meta-analysis to combine the results of the ProtecT GWAS with the previously published UK Genetic Prostate Cancer Study (UKGPCS)(6). Thirdly, to investigate whether previously identified associations of genetic variants with PrCa could be explained by associations with PSA levels, rather than cancer per se. For the third aim we: i) identified previously published PrCa risk single nucleotide polymorphisms (SNPs) and compared their association with PrCa in two case-control studies nested within ProtecT; the first compared PrCa cases (diagnosed following a PSA ≥3 ng/ml and a positive biopsy) with ‘low’ PSA controls (PSA <0.5 ng/ml) and the second compared PrCa cases with ‘high’ PSA controls (PSA ≥3 ng/ml and a negative biopsy); our hypothesis was that if the genetic marker is associated with PSA level and not PrCa, then the effect estimates would be greatest when using the low PSA controls and close to the null when using the high PSA controls; and ii) examined the relationship of each of the identified genetic markers with PSA level as a continuous variable in controls selected independently of PSA level (“unrestricted” controls), including those with PSA levels ≥3 ng/ml who were biopsy negative, as well as those with levels below 3 ng/ml.
Materials and Methods
Samples
Two study populations were used in this analysis. Firstly participants from the Prostate Testing for cancer and Treatment (ProtecT) study(20), and secondly the UK Genetic Prostate Cancer Study stage I (UKGPCS), both of European ancestry(6). The ProtecT study recruited all men aged 50-69 years between 2001-2009 from 337 general practices in nine UK centres (Birmingham, Bristol, Cambridge, Cardiff, Edinburgh, Leeds, Leicester, Newcastle and Sheffield). All men were invited to a prostate check clinic and were offered a PSA test: approximately 110,000 attended. For those with a PSA level of ≥3ng/ml, a transrectal ultrasound guided prostate biopsy was conducted. All detected tumours were histologically confirmed and assigned a Gleason score by a specialist uropathologist following a standard proforma. Tumours were categorised as low (score <7), mid (score=7) or high (score>7) grade. Clinical staging was assigned using the TNM system(21) as either localised (T1-T2) or advanced (T3-T4) (although most in the latter category were locally advanced and few tumours had metastasized distally). Approximately 3000 men were diagnosed with PrCa between 2001 and 2009. All men diagnosed with PrCa after attending a prostate check clinic before end November 2006 (and their matched controls) were eligible for inclusion in the current ProtecT GWAS (n=1215 cases).
Men with no evidence of PrCa were eligible for selection as controls (approximate n=107,000); these were men with a PSA<3ng/ml or a PSA≥3ng/ml with the most recent biopsy being negative. Controls were stratum matched to the genotyped PrCa cases by age, GP practice and calendar time period, in two distinct rounds of matching. In the first round a random sample of 6 controls per case were selected from all eligible controls in each age and GP practice strata (“unrestricted” controls); as the prostate check clinics were completed in one GP practice before moving to the next, matching on GP practice also matched on calendar time period. The second round randomly selected 1 control with a PSA<0.5ng/ml (and with whole blood collected) per case from the same age and GP practice strata as the index case (‘supernormal controls’). A total of 1925 matched controls were eligible for inclusion in the current GWAS. Trent Multicenter Research Ethics Committee (MREC) approved the ProtecT study (MREC/01/4/025) and the associated ProMPT study which collected biological material (MREC/01/4/061), and written informed consent was obtained.
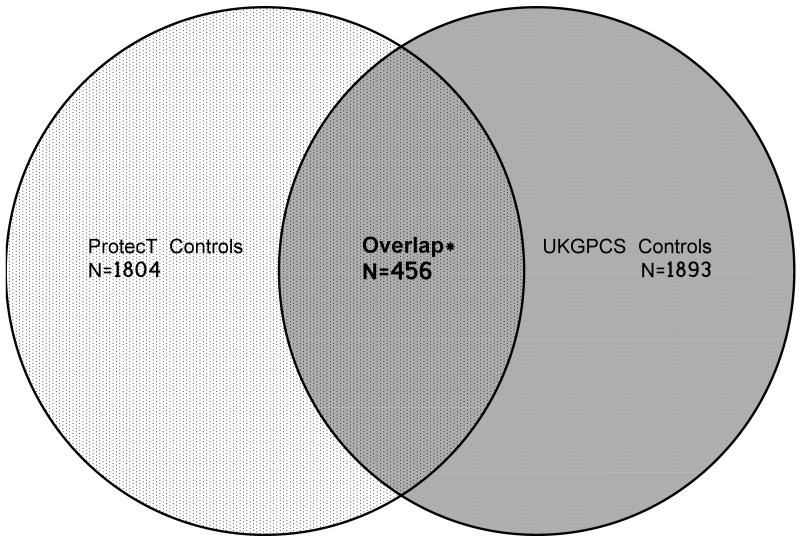
The UKGPCS (stage I) dataset has been previously described (6). Briefly the cases (n=2017) were clinically detected and selected if the man had a diagnosis at ≤60 years of age or a first/second degree family history of PrCa. Self-reported “non-white” men were excluded, as were those who were diagnosed through asymptomatic screening. UKGPCS controls (total n = 1893) were solely selected from men in the ProtecT study who had a PSA <0.5ng/ml. In the current meta-analysis, we excluded from the UKGPCS results those men who were in the ProtecT GWAS control series described above (n=456) so that the populations in the pooled analysis were independent samples (Figure 1).
Figure 1.
Venn Diagram showing the inclusion of controls in the ProtecT and UKGPCS GWAS
* Controls overlapping between control populations were excluded from the UKGPCS GWAS
Genotyping in ProtecT
The genotyping was performed at the Center National de Genotypage, Evry, France using the Illumina Human660W-Quad_v1_A array (Illumina, Inc., San Diego, CA). Quality control measures included exclusions for sex-mismatches, minimal (<0.325) or excessive heterozygosity (>0.345), cryptic relatedness as estimated by proportion of loci identical by descent (IBD>0.1), and disproportionate levels of individual missingness (>3%). Individuals were then checked for evidence of population stratification by multidimensional scaling analysis and compared with HapMap II (release 22) European descent (CEU), Han Chinese (CHB), Japanese (JPT) and Yoruba (YRI) reference populations. Individuals showing evidence of non-European ancestry were removed. SNPs with a minor allele frequency of below 1%, a call rate of <95% or evidence for violation of Hardy-Weinberg equilibrium (p<5 × 10−7) were discarded.
Imputation of autosomal genotypic data used Markov Chain Haplotyping software (MACH v.1.0.16(22)) and phased haplotype data from CEU individuals (HapMap release 22, Phase II NCBI B36, dbSNP 126) on 514,432 autosomal SNPs. All SNPs with poor imputation quality (r2 hat <0.3) were removed. The final working dataset included 2950 individuals (cases n=1146, controls n=1804). A total of 1272 controls were included from the first round of “unrestricted” control selection (PSA <0.5ng/ml n=238; PSA ≥0.5 and PSA<3ng/ml n=941; PSA≥3ng/ml n=93) and a further 532 from the second round (“supernormal” control selection).
UKGPCS genotyping
This has been reported elsewhere (6). Briefly genotypes were generated using the Illumina Infinium HumanHap550 array, and we only used genotypes with a call rate of >97%. Related samples and those with Asian/African ancestry were excluded. After exclusions, 1,854 cases and 1,437 controls (independent of ProtecT – see Figure 1) were used in the current meta-analysis.
Statistical Analysis
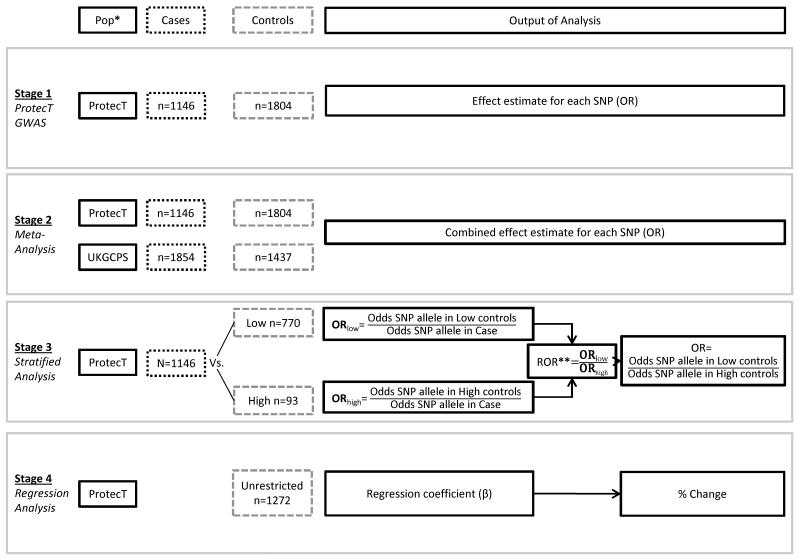
The statistical analysis was undertaken in four stages (see Figure 2). In the first stage, a genome-wide association analysis was performed on the ProtecT samples using the software package MACH2DAT(22, 23), employing a logistic regression model based on an expected allelic dosage model for SNPs. Associations between each SNP and disease were assessed by a Wald test and a p-value of < 5 × 10−8 was considered genome-wide significant. All estimates are reported in the direction of the risk allele.
Figure 2.
Analysis strategy
* Population
** Ratio of Odds Ratios
In the second stage, we meta-analysed the top SNPs identified in ProtecT with the UKGPCS dataset. A total of 381 SNPs reaching a threshold for suggestive significance of 5 × 10−5 in the ProtecT GWAS were included in the meta-analysis. The threshold level of 5 × 10−5 was a pragmatic choice, based on similar thresholds for suggestive significance cited in the literature, because of the relatively small sample size in ProtecT. Using a fixed effect inverse variance model, the β-coefficients and standard errors from ProtecT and UKGPCS were combined using the metan command in STATA. A p-value of 5 × 10−8 was used to identify genome-wide significant SNPs in the meta-analysis. Heterogeneity between the ProtecT and UKGPCS studies was evaluated using Cochran‘s Q statistic and the I2 value(24). The two studies had the same strand orientation and used the same effect alleles. Associations showing moderate-high levels of heterogeneity (I2>50) were investigated further to determine whether the heterogeneity could be explained by the difference in control or case populations used in ProtecT and UKGPCS. We did this in two stages: first, we used only the low PSA (<0.5 ng/ml) controls in ProtecT to generate new effect estimates and re-ran the meta-analysis for those SNPs showing moderate-high levels of heterogeneity in the original analysis; second, for those SNPs which still showed moderate-high levels of heterogeneity, we restricted the ProtecT cases to include only young cases (≤60 years of age) and/or those with a family history and re-ran the meta-analysis.
In stage 3 we investigated how the choice of controls in case-control GWAS studies may have impacted on associations of SNPs with PrCa, using two control populations from the ProtecT study: 770 low PSA controls (PSA <0.5 ng/ml, from both the “unrestricted” control selection, n=238, and “supernormal” control selection, n=532); and 93 ‘high’ PSA controls (PSA ≥3ng/ml). We also did a sensitivity analysis relaxing the ‘high’ PSA control threshold to a PSA level ≥ 2ng/ml, resulting in 250 ‘high’ PSA controls. We did this in order to increase power and move our high control population towards a more general population. We generated a list of SNPs to be tested by downloading all published SNPs associated with both PrCa risk and PSA level from the catalogue of Published Genome-wide Association Studies (http://www.genome.gov/gwastudies/: downloaded 21/11/2012). We supplemented this list with additional SNPs associated with PrCa in a recent meta-analysis (2), resulting in a total of 83 SNPs. Using this list we extracted the individual genotypic data for all participants in our ProtecT dataset, although two SNPs were neither genotyped nor imputed (rs7210100 and rs16902094). Using the genotypic data we generated effect estimates for each SNP using multinomial logistic regression, where each control group (low and high) was compared to cases separately (Figure 2). To test the null hypothesis, that there was no difference between the two control groups when used to estimate the odds ratios for the associations of SNPs with PrCa, we carried out a separate logistic regression model that tested the association of each SNP with control type (low vs. high); this is same as testing whether the ratio of odds ratios using the two control groups were equivalent (i.e. equal to 1), as the base case group would be cancelled out (Figure 2).
Finally we conducted a regression analysis of SNPs on PSA level within the “unrestricted” control population (controls selected independent of PSA level). PSA values were log-transformed to approximate a normal distribution. To account for the multiple comparisons between PSA levels and SNP markers, we only highlight findings which showed evidence of an association in both the stratified analysis and a complementary association in the regression analysis.
The anti-log of the β was taken to estimate the percentage change in PSA level by increasing allele dose. In order to incorporate the uncertainty of the imputation process, expected allelic dosage data were used for the analysis. Genotypic dosages are the expected number of copies of an allele which ranges from 0-2. For genotyped SNPs the exact number of copies of an allele is known, but for imputed SNPs the estimate maybe less precise.
Results
Genome-wide association study in ProtecT (stage 1)
The study characteristics of those individuals (cases n=1146, controls n=1804) included in the ProtecT GWAS are presented in Supplementary Table 1. Supplementary Figure 1 shows the quantile-quantile (Q-Q) plot of the distribution of test statistics for comparison of genotype frequencies in cases versus controls. The lambda inflation factor was 1.025. In the GWAS, 381 SNPs were associated with PrCa at P<5 × 10−5 (Supplementary Figure 2 and Supplementary Table 2). We detected a genome-wide significant association (P<5 × 10−8) between PrCa and SNPs at 3 previously identified loci: 10q11.23 (rs10993994), 17q24.3(rs7222314) and 19q13.33(rs1058205) (Table 1 and Locus Zoom plots in Supplementary Figures 3-5).
Table 1.
Results of independent SNPs reaching genome wide significance (P<5 × 10−8) in the ProtecT GWAS
| Loci | SNP | Chromosome | Position | Alleles | β a | SEb | ORc | P-value | EAFd | RSQRe |
|---|---|---|---|---|---|---|---|---|---|---|
| 10q11.23 | rs10993994 | 10 | 51219502 | T/C | 0.33 | 0.06 | 1.39 | 1.58 × 10−09 | 0.5983 | 0.9793 |
| 17q24.3 | rs7222314 | 17 | 66616533 | A/G | 0.32 | 0.06 | 1.38 | 8.59 × 10−09 | 0.4772 | 0.9295 |
| 19q13.33 | rs1058205 | 19 | 56055210 | T/C | 0.40 | 0.07 | 1.49 | 4.63 × 10−08 | 0.1828 | 1 |
Correlation coefficient
Standard Error
Effect estimate (odds ratio)
Effect Allele Frequency
Imputation accuracy
Meta-analysis of ProtecT and UKGPCS studies (stage 2)
All SNPs at P<5 × 10−5 in ProtecT were meta-analysed with results from the UKGPCS. The total number of individuals included was 6241 (cases: ProtecT n=1146, UKGPCS n= 1854; controls: ProtecT n=1804, UKGPCS n=1437). A total of 175 SNPs reached genome wide significance (p < 5×10−8) for an association with PrCa in the meta-analysis and were all located in or near genes across 4 previously identified loci (Table 2), with 6 independent signals. There was evidence of significant heterogeneity between the studies for the rs1447295 (8q24.21) marker (I2=86.1%, p-value for heterogeneity = 0.01). There was also evidence of moderate-high levels of heterogeneity for rs6983267, rs10993994 and rs17632542. The heterogeneity of rs6983267, rs10993994 and rs17632542 was greatly attenuated when the analysis was repeated using the ProtecT effect estimates from a restricted control population (low PSA controls <0.5 ng/ml) (Supplementary table 3). The heterogeneity observed for rs1447295, however, remained high (I2=83.1%, p-value for heterogeneity = 0.01). When the analysis was repeated using ProtecT effect estimates based on young and/or familial cases and low PSA controls, the heterogeneity was only slightly reduced and remained statistically significant (I2= 78%, p-value for heterogeneity = 0.03).
Table 2.
Summary results of top independent SNPS in known loci associated with PrCa identified by meta-analysis of ProtecT with UKGPCS
| ProtecT | UKGPCS | Combined (Meta-Results) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chroma | Position | Gene | EAF | β | SE | EAFb | P-value | β | SE | EAF* | P-value | β | (95% CI) | P-value | I^2c | P-Hetd |
| rs12682344 | 8 | 128175966 | SRRM1P1 -POU5F1B | G | 0.57 | 0.13 | 0.04 | 1.28 × 10−05 | 0.79 | 0.14 | 0.04 | 9.66 × 10−09 | 0.67 | (0.48,0.86) | 4.69 × 10−12 | 21.7% | 0.26 |
| rs6983267 | 8 | 128482487 | SRRM1P1 -POU5F1B | G | 0.22 | 0.05 | 0.52 | 4.25 × 10−05 | 0.36 | 0.05 | 0.53 | 2.30 × 10−12 | 0.29 | (0.22,0.36) | 4.04 × 10−15 | 72.0% | 0.06 |
| rs1447295 | 8 | 128554220 | POU5F1B -MYC | A | 0.35 | 0.08 | 0.11 | 2.36 × 10−05 | 0.67 | 0.08 | 0.12 | 1.20 × 10−16 | 0.51 | (0.40,0.63) | 5.61 × 10−18 | 86.1% | 0.01 |
| rs10993994 | 10 | 51219502 | MSMB | T | 0.33 | 0.06 | 0.40 | 1.58 × 10−09 | 0.46 | 0.05 | 0.40 | 2.15 × 10−19 | 0.40 | (0.33,0.47) | 3.48 × 10−26 | 65.8% | 0.09 |
| rs4793529 | 17 | 66630231 | CALM2P1 -SOX9 | T | 0.33 | 0.06 | 0.47 | 1.88 × 10−09 | 0.23 | 0.05 | 0.49 | 8.11 × 10−06 | 0.28 | (0.20,0.35) | 1.78 × 10−13 | 49.8% | 0.16 |
| rs17632542 | 19 | 56053569 | KLK3 | T | 0.60 | 0.11 | 0.91 | 1.28 × 10−08 | 0.82 | 0.08 | 0.89 | 4.74 × 10−23 | 0.73 | (0.6,0.86) | 2.34 × 10−28 | 62.7% | 0.10 |
Chrom - Chromosome
Effect Allele Frequency
% of variation between study-specific effect estimates which is due to heterogeneity
Tests the hypothesis that there is no difference in the study-specific effect estimates
Stratified analysis using different control populations in ProtecT and PSA regressed in the unrestricted controls (stages 3 and 4)
Of the 81 SNPs examined, SNPs showing evidence of heterogeneity (p < 0.05) in effect estimates quantifying the difference in risk between cases and low or high PSA controls, and/or SNPs that showed at least nominal association with PSA levels in unrestricted controls (p < 0.05) are shown in Table 3. When comparing cases with low PSA controls, alleles at the genetic markers rs1512268 (8p21.2), rs445114 (8q24.21), rs10788160 (10q26.12), rs11199874 (10q26.12), rs17632542 (19q13.33), rs266849 (19q13.33) and rs2735839 (19q13.33) were strongly associated with an increased risk of PrCa (all p≤0.01), with odds ratios ranging from 1.24 to 2.73. The effect-estimates, however, were attenuated to the null when using high PSA controls (p-values for heterogeneity between estimates all <0.04). There was also some evidence of associations of these SNPs with PSA levels in the “unrestricted” control population (in particular, rs17632542 (19q19.33) associated with a 32% increase in PSA per allele), which suggests that the difference observed in the stratified analysis maybe explained by an association of these SNPs with PSA level. Only associations of the following SNPs with PSA levels were statistically significant (P≤0.05) in the “unrestricted” control population: rs445114, rs17632542, rs2735839 and rs266849. A further SNP (rs3850699) showed a similar association with PSA, with the effect estimate attenuating towards the null in the high PSA controls, but in the “unrestricted” control population the allele only increased PSA levels by 1% (p=0.73).
Table 3.
Comparison of cases with supernormal or high PSA controls and PSA regressed on SNPs in unrestricted controls *
| Stratified analysis comparing cases with two different control groups | PSA regressed on SNPs in unrestricted controls (n=1272) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Low PSA controls (n=770) | High PSA controls(n=93) | p het b | |||||||||
| Chroma | SNP | Putative Gene | Risk Allele | OR (SE) | p | OR (SE) | p | β | % PSA Change | p | |
| 3 | 189311171 | CTDSPL | T | 1.07 (0.10) | 0.49 | 1.79 (0.34) | 0.002 | 0.01 | −0.09 | −9% | 0.04 |
| 3 | rs7611694 | SIDT1 | A | 1.09 (0.63) | 0.19 | 0.87 (0.19) | 0.40 | 0.17 | 0.07 | 8% | 0.02 |
| 5 | rs6869841 | FAM44B (BOD1) | T | 1.35 (0.11) | 3.44×10−4 | 1.22 (0.24) | 0.29 | 0.63 | 0.08 | 9% | 0.03 |
| 6 | rs1983891 | FOXP4 | T | 1.10 (0.08) | 0.22 | 0.96 (0.16) | 0.80 | 0.43 | 0.08 | 8% | 0.03 |
| 8 | rs1512268 | SLC25A37 -NKX3-1 | T | 1.30 (0.09) | 9.51×10−5 | 0.83 (0.13) | 0.24 | 0.004 | 0.06 | 6% | 0.07 |
| 8 | rs445114 | SRRM1P1 -POU5F1B | T | 1.46 (0.10) | 8.15×10−8 | 0.92 (0.16) | 0.60 | 0.01 | 0.08 | 8% | 0.03 |
| 8 | rs6983267 | SRRM1P1 -POU5F1B | G | 1.29 (0.05) | 8.51×10−5 | 1.15 (0.13) | 0.36 | 0.44 | 0.08 | 9% | 0.01 |
| 10 | rs11199874 | RPL19P16 - FGFR2 | A | 1.24 (0.06) | 0.01 | 0.78 (0.21) | 0.14 | 0.01 | 0.07 | 7% | 0.07 |
| 10 | rs10788160 | RPL19P16 - FGFR2 | A | 1.24 (0.06) | 0.01 | 0.80 (0.21) | 0.18 | 0.01 | 0.06 | 6% | 0.08 |
| 10 | rs3850699 | TRIM8 | A | 1.17 (0.06) | 0.03 | 0.82 (0.22) | 0.28 | 0.048 | 0.01 | 1% | 0.73 |
| 10 | rs10993994 | MSMB | T | 1.61 (0.11) | 4.54×10−12 | 1.45 (0.23) | 0.02 | 0.51 | 0.08 | 8% | 0.02 |
| 13 | rs9600079 | FABP5L1 - KLF12 | G | 1.07 (0.06) | 0.33 | 0.99 (0.16) | 0.97 | 0.65 | 0.06 | 6% | 0.048 |
| 19 | rs17632542 | KLK3 | T | 2.73 (032) | 1.40×10−17 | 0.76 (0.27) | 0.44 | 1.94 × 10−4 | 0.28 | 32% | 5.49×10−7 |
| 19 | rs2735839 | KLK3 - KLK2 | G | 2.13 (0.20) | 1.90×10−16 | 1.29 (0.28) | 0.26 | 0.02 | 0.21 | 23% | 0.004 |
| 19 | rs266849 | KLK15 -KLK3 | A | 1.76 (0.05) | 8.62×10−12 | 1.15 (0.17) | 0.49 | 0.03 | 0.12 | 13% | 2.40×10−6 |
SNPs showing evidence of heterogeneity (p < 0.05) in effect estimates quantifying the difference in risk between cases and low or high PSA controls, and/or SNPs that showed at least nominal association with PSA levels in unrestricted controls (p < 0.05).
Chrom - Chromosome
Tests the hypothesis that there is no difference between the effect estimate of the cases vs. low PSA controls and cases vs. high PSA controls
There was also evidence that a further six SNP alleles (rs7611694, rs6869841, rs1983891, rs6983267, rs10993994 and rs9600079) were associated with increasing circulating PSA level (p<0.05), and that the use of high PSA controls attenuated effect estimates of these SNPs with PrCa towards the null. The difference in estimates, however, was not significant (all p-values>0.4). Relaxing the high PSA threshold to ≥2 ng/ml, we observed a statistically significant difference when comparing the estimates generated using low vs. high PSA controls for rs6869841-T (p=0.01) and rs10993994-T (p=0.04) (Supplementary table 4).
The T-allele at 3p22.2 (rs9311171) was associated with a more pronounced increase in risk of PrCa when cases were compared with high PSA controls (79% increase) as opposed to low PSA controls (7% increase) (p-value for heterogeneity = 0.01). There was also evidence of an association between the rs9311171-T allele and decreased PSA levels (p=0.035). In the sensitivity analysis (high PSA controls ≥2ng/ml), rs4775302 (15q21.1) was associated with a larger increase in risk of PrCa when cases were compared to the high PSA controls (29% increase), as opposed to the low PSA controls (2% increase) (p-value for heterogeneity=0.03) (Supplementary table 4). This was supported by the regression analysis of PSA in the “unrestricted” control population, which showed an association in the opposite direction (5% decrease p=0.09).
Discussion
Our GWAS and meta-analysis replicate previous findings for the following loci: 10q11.23 (rs10993994 - MSMB), 17q24.3 (rs7222314 - CALM2P1 - SOX9) and 19q13.33(rs1058205 - KLK3) in the ProtecT GWAS; and 8q24.21 (rs12682344,rs6983267 and rs1447295 - intergenic), 10q11.23 (rs10993994 - MSMB), 17q24.3 (rs4793529 - CALM2P1 - SOX9)and 19q13.33 (rs17632542 –KLK3) in the meta-analysis. We also demonstrated a novel association of one SNP with circulating PSA level: rs9311171-T (inversely associated with PSA). The allele at the genetic marker rs9311171 is located near a microRNA (MIR26A1) and a protein linked to oncogenesis (CTDSPL). This SNP (rs9311171) had a larger positive association with PrCa when cases were compared with high PSA, as opposed to low PSA controls. This suggests that this marker is directly associated with both PSA level and PrCa risk.
We also found evidence that the association of PrCa risk of seven other SNPs at loci 8p21.2, 10q26.12, and 19q13.33 could be confounded by the variant-PSA association(rs1512268-T, rs10788160-A, rs445114-T, rs11199874-A, rs17632542-T, rs2735839-G and rs266849-A). The association between PrCa and the respective SNP allele was present in the comparison of cases with low-PSA controls, but this association disappeared when compared to high-PSA controls. There was also evidence of associations between these SNPs and PSA levels in the “unrestricted” control population. Associations of rs1512268-T, rs10788160-A and rs11199874-A with PSA levels were only statistically significant (p<0.05) in the stratified analysis. Whilst associations of SNPs with PSA levels were not statistically significant in the regression analysis, the direction of the effect provides some support for the conclusion that these SNP alleles are associated with increased circulating PSA level. Eeles et al (2009)(5) and Gudmundsson et al(2010) (19) previously observed a positive association of PSA with rs1512268-T (near NKX3.1) and rs10788160-A (FGFR2), respectively. The allele at the genetic marker rs445114 (intergenic) was previously shown to have a positive association with PSA level, albeit non-genome wide significant (p=1.27 × 10−2)(19). This is consistent with the findings of Al Olama et al (2009)(18), who found that a SNP (rs620861) in strong linkage disequilibrium with rs445114 was positively associated with PSA level (p=4.8 × 10−8).Whilst rs11199874-A has not previously been tested for a relationship with PSA, it is in strong linkage disequilibrium with rs10788160-A (r2=0.94) (FGFR2), which has been shown to be associated with increasing PSA levels. Our analysis suggests that rs10788160-A is associated with higher PSA levels, whilst rs10788160-G is associated with lower levels. A previous study reported a positive association between rs11199874-G and PrCa (25). This association could be driven by the large proportion (>50%) of high PSA (4-10 ng/ml) controls used rather than reflecting a true association because this high PSA enriched control sample is likely to have a higher proportion of individuals with the allele rs11199874-A; this could in turn account for the positive association of PrCa with rs11199874-G (the opposite allele to rs11199874-A). Two markers, in strong linkage disequilibrium (rs17632542-T and rs2735839-G), located near the kallikrein-related peptidase protein coding genes, have previously been observed to be positively associated with PSA level (19). SNP rs266849 is also located near, but is independent of this set of PSA- associated proteins; a previous study reported no association of rs266849 with PSA in controls with a PSA <10ng/ml (6). When we reduced our threshold for defining high PSA controls (PSA≥2 ng/ml) in the stratified analysis, we observed similar findings in the same direction to those that were found to be significant using the higher PSA threshold (PSA≥3ng/ml). In addition, in this sensitivity analysis three other SNPs were shown to be associated with PSA.
Impact of control selection
Our analysis suggests that the selection of controls for GWAS below a certain PSA threshold results in associations with PrCa, when the actual association is with PSA level (e.g. rs6869841). Additionally, the selection of these controls can lead to the masking of a PrCa association (e.g. rs9311171). To avoid these difficulties, an optimal design for a GWAS would be to select controls who have PSA levels at or above the threshold for biopsy, and who are biopsy negative. These criteria ensure that the cases and controls are comparable in terms of PSA level and therefore associations of SNP markers with PrCa are not confounded by associations with PSA.
Strengths and limitations
This study was based on a population sample of men who underwent PSA testing, providing findings relevant to screen-detected cancer, and controls that were not selected according to PSA level. We also conducted a meta-analysis using 6241 individuals (3000 cases and 3241 controls), but we were only able to confirm previous GWAS findings rather than show novel associations. There was significant heterogeneity between the ProtecT and UKGPCS studies for associations of markers at 8q24.21, 10q11.23 and 19q13.33 with PrCa. These differences were partly explained by the difference in control ascertainment between studies (Supplementary table 3). We were, however, unable to explain the substantial heterogeneity (I2=78%) for rs1447295. We had a relatively small number of high PSA controls (n=93), so our stratified analysis will have been underpowered, explaining why we were unable to detect some previously published associations of SNPs with PSA. To increase power we carried out a sensitivity analysis by relaxing the high PSA control threshold to ≥2ng/ml (n=250), which resulted in 3 additional SNPs being associated with PSA level (rs6869841, rs10993994 and rs4775302).
To avoid potential false positive results arising from multiple comparisons, we only highlight findings which showed evidence of an association in both the stratified and regression analyses. A Bonferroni corrected threshold significance level is a p-value of ≤ 0.0006(0.05/81 tests). However, this correction assumes that the tests were independent of each other, which in this case may not be true because some SNPs were correlated. Hence a rigid application of the Bonferroni corrected p-value could lead to an erroneous rejection of genuine loci (false-negatives). Nevertheless, we have presented exact p-values, allowing comparison with the Bonferroni corrected p-value. Whilst all controls with an elevated PSA level were biopsy negative, there is the possibility that some controls with a PSA level <3ng/ml had undetected PrCa, as evidenced by a study (2004)(1) showing that 14% of men with a PSA level ≤3ng/ml had PrCa. The majority of controls in our study had a PSA level ≤1ng/ml, which was associated with less than a 9% chance of undetected PrCa (1). It is also possible that up to a quarter of men with an elevated PSA level (≥3ng/ml) and a negative biopsy had undetected PrCa (26), which could explain why we did not observe associations of SNPs with PrCa when using the high PSA control group.
Conclusion
We have confirmed associations of PrCa risk with four previously identified loci (8q24.21,10q11.23, 17q24.3 and 19q13.33) and associations of seven markers with PSA level. We also found new evidence of an association of genetic variation at 3p22.2 with circulating PSA levels. We have highlighted that the method of selecting controls in case-control studies of associations of genetic variation with prostate cancer can influence the results suggesting that inferences made in these studies should carefully consider control selection.
Supplementary Material
Acknowledgments
The authors thank the tremendous contribution of all members of the ProtecT study research group, and especially the following who were involved in this research (Athene Lane, Prasad Bollina, Sue Bonnington, Lynn Bradshaw, James Catto, Debbie Cooper, Michael Davis, Liz Down, Andrew Doble, Alan Doherty, Garrett Durkan, Emma Elliott, David Gillatt, Pippa Herbert, Peter Holding, Joanne Howson, Mandy Jones, Roger Kockelbergh, Howard Kynaston, Teresa Lennon, Norma Lyons, Hing Leung, Malcolm Mason, Hilary Moody, Philip Powell, Alan Paul, Stephen Prescott, Derek Rosario, Patricia O’Sullivan, Pauline Thompson, Sarah Tidball). We thank Gemma Marsden, who processed the blood samples at the biorepository and Dr Chris Metcalfe for providing statistical advice. We also would like to thank the Center National de Genotypage, Evry, France for genotyping the ProtecT samples.
Financial Support: The ProtecT study is funded by the U.K. Health Technology Assessment (HTA) Programme of the NIH Research (HTA 96/20/99; ISRCTN20141297). D.W. Knipe is funded by the Wellcome Trust 4-year studentship (WT099874MA). D.M. Evans, J.P. Kemp, G. Davey-Smith, and R.M. Martin work in an MRC Unit that is supported by the UK Medical Research Council (MC_UU_12013/1-9) and the University of Bristol. D. F. Easton is supported by a Cancer Research UK Grant (C1287/A10118). R.A. Eeles and Z.Kote-Jarai are supported by Cancer Research UK Grant (C5047/A7357) and support from the NIHR to the Biomedical Research Centre at The Institute of Cancer Research and Royal Marsden NHS Foundation Trust. J. F.C. Hamdy, D.E. Neal, and J.L. Donovan are NIHR Senior Investigators. The authors thank the provision of the additional epidemiological data by the NHS R&D Directorate supported Prodigal study and the ProMPT (Prostate Mechanisms of Progression and Treatment) collaboration which is supported by the National Cancer Research Institute (NCRI) formed by the Department of Health, the Medical Research Council and Cancer Research UK (G0500966/75466).
Footnotes
Conflict of Interest
The authors declare no conflicts of interest
References
- 1.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004 May 27;350:2239–46. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 2.Eeles RA, Al Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013 Apr;45:385–91. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng I, Chen GK, Nakagawa H, He J, Wan P, Laurie CC, et al. Evaluating Genetic Risk for Prostate Cancer among Japanese and Latinos. Cancer Epidemiol Biomarkers Prev. 2012 Nov;21:2048–58. doi: 10.1158/1055-9965.EPI-12-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007 Dec 19;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 5.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, et al. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009 Oct;41:1116–21. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–21. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009 Oct;41:1122–6. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007 May;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007 Aug;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 10.Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, et al. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009 Jan 1;69:10–5. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010 Sep;42:751–4. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 12.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011 Oct 1;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FitzGerald LM, Kwon EM, Conomos MP, Kolb S, Holt SK, Levine D, et al. Genome-wide association study identifies a genetic variant associated with risk for more aggressive prostate cancer. Cancer Epidemiol Biomarkers Prev. 2011 Jun;20:1196–203. doi: 10.1158/1055-9965.EPI-10-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011 Jun;43:570–3. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011 Aug;43:785–91. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam RK, Zhang WW, Trachtenberg J, Seth A, Klotz LH, Stanimirovic A, et al. Utility of Incorporating Genetic Variants for the Early Detection of Prostate Cancer. Clin Cancer Res. 2009 Mar 1;15:1787–93. doi: 10.1158/1078-0432.CCR-08-1593. 2009. [DOI] [PubMed] [Google Scholar]
- 17.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008 Mar;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 18.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009 Oct;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 19.Gudmundsson J, Besenbacher S, Sulem P, Gudbjartsson DF, Olafsson I, Arinbjarnarson S, et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci Transl Med. 2010 Dec 15;2:62ra92. doi: 10.1126/scitranslmed.3001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane JA, Hamdy FC, Martin RM, Turner EL, Neal DE, Donovan JL. Latest results from the UK trials evaluating prostate cancer screening and treatment: The CAP and ProtecT studies. Eur J Cancer. 2010;46:3095–101. doi: 10.1016/j.ejca.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Ohori M, Wheeler TM, Scardino PT. The New American Joint Committee on Cancer and International Union Against Cancer TNM classification of prostate cancer. Clinicopathologic correlations. Cancer. 1994;74:104–14. doi: 10.1002/1097-0142(19940701)74:1<104::aid-cncr2820740119>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using Sequence and Genotype Data to Estimate Haplotypes and Unobserved Genotypes. Genet Epidemiol. 2010 Dec;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Willer C, Sanna S, Abecasis G. Genotype Imputation. Annual Review of Genomics and Human Genetics. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003 Sep 6;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam RK, Zhang W, Siminovitch K, Shlien A, Kattan MW, Klotz LH, et al. New variants at 10q26 and 15q21 are associated with aggressive prostate cancer in a genome-wide association study from a prostate biopsy screening cohort. Cancer Biol Ther. 2011 Dec 1;12:997–1004. doi: 10.4161/cbt.12.11.18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zackrisson B, Aus G, Lilja H, Lodding P, Pihl CG, Hugosson J. Follow-up of men with elevated prostate-specific antigen and one set of benign biopsies at prostate cancer screening. Eur Urol. 2003 Apr;43:327–32. doi: 10.1016/s0302-2838(03)00044-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.