Abstract
Sexual development in the fission yeast Schizosaccharomyces pombe culminates in meiosis and sporulation. We used ribosome profiling to investigate the translational landscape of this process. We show that the translation efficiency of hundreds of genes is regulated in complex patterns, often correlating with changes in RNA levels. Ribosome-protected fragments show a three-nucleotide periodicity that identifies translated sequences and their reading frame. Using this property, we identified 46 novel translated genes and found that 24% of non-coding RNAs are actively translated. We also detected 19 nested antisense genes, in which both DNA strands encode translated mRNAs. Finally, we identified 1,735 translated upstream ORFs in leader sequences. In contrast with Saccharomyces cerevisiae, sexual development in S. pombe is not accompanied by large increases in upstream ORF use, suggesting that this is an organism-specific adaptation and not a general feature of developmental processes.
Introduction
Fission yeast diploid cells undergo sexual differentiation (meiosis and sporulation) upon nitrogen starvation1. This process is accompanied by a complex gene expression program, in which more than 50% of the genome is regulated. Microarray studies showed that changes in RNA levels occur in successive expression waves that coincide with major biological events: starvation and pheromone-induced genes, early genes (pre-meiotic S phase and recombination), middle genes (meiotic divisions and spore formation) and late genes (spore maturation)2. The induction of most of these groups is mediated by meiosis-specific transcription factors2-5, although regulation of mRNA decay by RNA-binding proteins is also involved6,7. More recently, RNA-seq experiments revealed that S. pombe cells express hundreds of meiosis-specific non-coding RNAs (ncRNAs)8-10. Although gene expression during sexual differentiation has been extensively studied, nothing is known about the contribution of translational control to this process.
Ribosome profiling can provide a genome-wide view of translation with single-nucleotide resolution11. This approach is based on the isolation and sequencing of ribosome-protected mRNA fragments (RPFs), and can be used to identify translated regions and to estimate mRNA translational efficiency12. This approach was recently applied to the meiotic program of S. cerevisiae, revealing extensive translational control and a meiosis-specific increase in the use of open reading frames upstream of annotated coding sequences (uORFs)13. Ribosome profiling of budding yeast and higher eukaryotes also exposed that many genes annotated as ncRNAs appear in the RPF fractions11,14. However, whether these ncRNAs are actively translated has been contentious15,16. A key advantage of ribosome profiling is that RPFs show a characteristic triplet periodicity when aligned to mRNA sequences that can be used to determine the frame in which a given region is being translated11. This property has been used to identify dually decoded regions17 and short translated ORFs18. We used it here to discover newly translated genes, to detect translated uORFs and to investigate whether genes annotated as long ncRNAs are actively translated.
We have applied ribosome-profiling to S. pombe cells growing vegetatively and undergoing sexual differentiation. We discovered dozens of new translated genes, identified translated uORFs in the 5′ leader sequences of 25% of S. pombe coding genes, and found that 24% of genes annotated as ncRNAs are actively translated. These results reveal pervasive translation of the fission yeast genome. We have also shown that sexual differentiation in S. pombe is accompanied by a complex translational program, in which the translation efficiency of hundreds of genes is regulated.
Results
Ribosome profiling in S. pombe
We carried out ribosome profiling of S. pombe diploid cells undergoing meiosis and sporulation. To achieve good synchrony we used thermo-sensitive mutants in the Pat1 meiotic inhibitor19,20. Diploid cells were blocked in G1 by nitrogen starvation, and entered meiosis synchronously upon inactivation of the Pat1 kinase (Supplementary Fig. 1A). In addition, we performed ribosome profiling in wild type haploid cells growing vegetatively. Ribosome profiling involves the purification of RPFs, which are used to generate a library containing adaptors for Illumina sequencing. In parallel, a second library is produced from fragmented mRNAs. The first library allows the identification and quantification of translated regions, while the second one is used to estimate mRNA levels. In all current protocols, the production of both libraries involves PCR-based amplification. As part of our library generation strategy, we used Unique Molecular Identifiers (UMIs)21,22. The primer employed for reverse transcription contained a random barcode sequence, which uniquely tagged each cDNA fragment. Reads that contained identical sequences with the same barcode were very likely to have originated from the same RNA fragment, and only one of them was retained. Therefore, the reads we used for analysis constituted a truly non-redundant dataset. This approach helps avoid PCR amplification and sampling artefacts, and can be used to estimate the complexity of the original library22. 83 % of reads that did not map to rRNAs were unique, indicating that the complexity of the original libraries was high (Supplementary Table 1). Overall, 707 million reads were reduced to 250 million unique ones (Supplementary Table 1). As expected, RPFs showed a strong bias in their distribution along mRNAs: After accounting for length, 92.4 % of RPFs mapped to annotated coding sequences, 6.9 % were located in 5′ leader sequences (also known as 5′ Untranslated Regions, or 5′-UTRs) and only 0.7 % in 3′-UTRs. By contrast, mRNA reads showed a more equal distribution (Supplementary Fig. 1B). The results were highly reproducible between independent biological repeats (Supplementary Fig. 2).
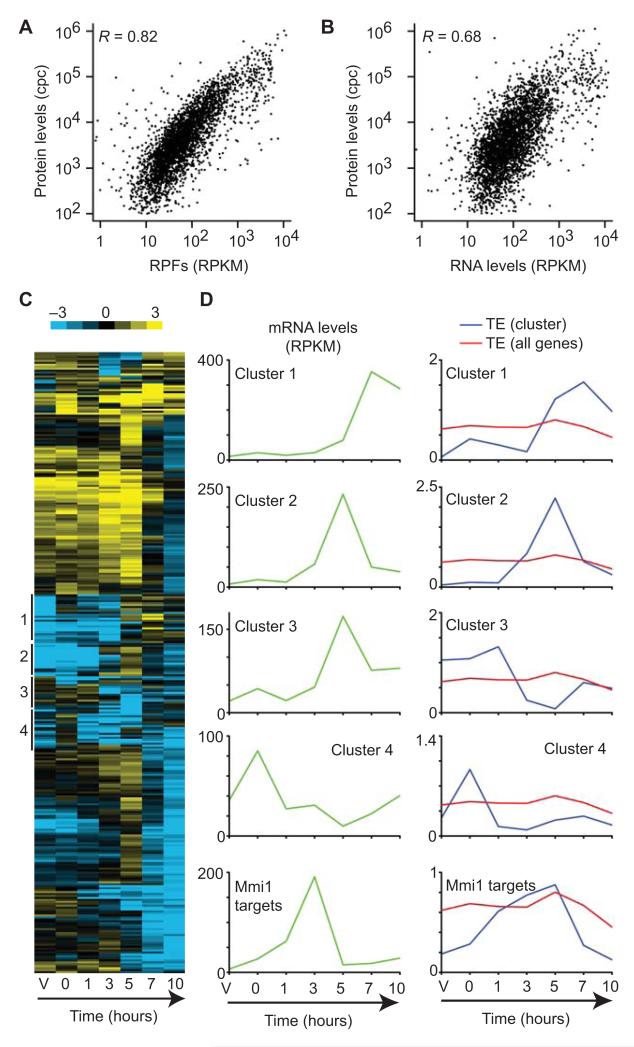
We used the densities of mRNA fragments and RPFs to estimate mRNA levels and protein synthesis rates, respectively. Both features correlated well with each other over the meiotic time course (Supplementary Table 2, average R = 0.79). Total translation rates (measured as RPF densities) are expected to be better predictors of protein abundance than mRNA levels. Consistently, protein levels estimated from a mass spectrometry study of vegetative cells23 showed higher correlation with RPF density (Fig. 1A, R = 0.82) than with RNA levels (Fig. 1B, R = 0.68). Direct comparison of RPFs and RNA levels allowed us to estimate gene-specific translation efficiencies (TEs), which were calculated by dividing the normalized number of RPFs (in reads per kilobase per million reads or RPKM) by that of mRNA fragments across coding sequences. In vegetative cells, TEs varied over a range of over 100-fold, and did not correlate with mRNA half-lives (R = -0.12) or mRNA levels (average R over the time course = -0.08, Supplementary Table 2). As expected, TEs displayed a positive correlation with the mean number of bound ribosomes determined in a microarray-based polysome profiling study (R = 0.43)24. Poor TE in vegetative cells was associated with lowly expressed genes, including all major groups of meiotic genes. Moreover, targets of the nonsense-mediated decay (NMD) pathway25 were poorly translated. This phenomenon has been previously observed in S. cerevisiae, although it is unclear whether the NMD pathway directly represses translation or whether these mRNAs contain features that make their translation inefficient26,27.
Figure 1. Quantification of translation using ribosome profiling.
(A) Comparison of RPF levels (this work) with protein levels (Marguerat el al. 2012). RPF levels are presented in reads per kilobase per million reads (RPKM), and protein levels are measured in copies per cell (cpc). The Pearson correlation is indicated. (B) Comparison of RNA levels to protein amounts. Labelling as in A. (C) Cluster analysis of the 318 genes that showed changes in TE of at least 10-fold across meiotic time points. Columns represent experimental points (V corresponds to vegetative cells and the numbers denote hours after meiotic induction), and rows represent genes. The colors show the TE (log2 scale, see bar at the top). The position of selected clusters discussed in the text is shown on the left. (D) Changes in TE over meiosis for selected clusters. The plots on the left (green) represent median mRNA levels for the indicated clusters. The graphs on the right show the median TE as a function of time for genes in the indicated cluster (blue) or for all genes (red).
The translational program of meiosis
We used hierarchical clustering to investigate the changes in RPF levels across the meiotic time course. Translation patterns were dynamic and generally correlated with changes in RNA expression (Supplementary Fig. 3A). Changes in RNA levels were similar to those reported in a microarray study, providing independent validation of our data (Supplementary Fig. 3A)2. As in vegetative cells, TEs varied over a wide range during meiosis, and were dynamically regulated. 25.8% of all coding genes showed changes of 5-fold of more in TE, and 6.7% of more than 10-fold. We looked for patterns in TE regulation by clustering the 318 genes that showed the strongest variation (Fig. 1C). Several groups of clustered genes were enriched in co-expressed genes. For example, a subset of late genes (cluster 1) was poorly translated until 7 hours into meiosis, when its TE increased (Fig. 1D). This group was enriched in late meiotic genes regulated by the Atf21 and Atf31 transcription factors. Similarly, a group of middle genes was only translated efficiently at 5 hours (cluster 2), and a cluster enriched in genes expressed in response to nitrogen starvation (cluster 4) showed enhanced TE after nitrogen removal (Fig. 1D). In all three cases the peak of TE coincided with that of mRNA levels. Increases in TE coupled to those in RNA levels (potentiation) have been observed before during responses to stress.28-30. A cluster that showed a strong drop in TE at 5 hours despite high mRNA levels (cluster 3) was enriched in targets of the Meu5 RNA-binding protein (Fig. 1D). Meu5 binds to a subset of the middle genes (so called ‘late-decay’) and stabilizes their transcripts6. Although the RNA levels of many of these genes peaked at 5 hours, the accumulation of RPFs was delayed compared to that of mRNA fragments. This suggested that translation of some Meu5 targets genes was repressed to delay the production of the corresponding protein. We checked this hypothesis by looking at published protein levels during a meiotic time course for Meu5 targets that did or did not show the decrease in TE (see Supplementary Fig. 11 in reference 6). In all three cases, the peak of protein expression corresponded to that of RPF accumulation. Finally, we noticed that targets of the Mmi1 protein, an RNA-binding protein that promotes the degradation of a group of meiotic RNAs in vegetative cells7, showed strong changes in TE, with high efficiency correlated with the expression of their mRNAs and a strong repression in vegetative cells, when expression of these genes is toxic for the cell (Fig. 1D). These results suggest that the regulation of TE and the control of mRNA levels are coordinated during the meiotic process. However, this relationship appears to be complex: in some cases it caused increased translation at peaks of RNA levels (potentiation), whereas in others (some Meu5 targets) it led to delays in protein production with respect to RNA accumulation.
Systematic identification of novel translated regions
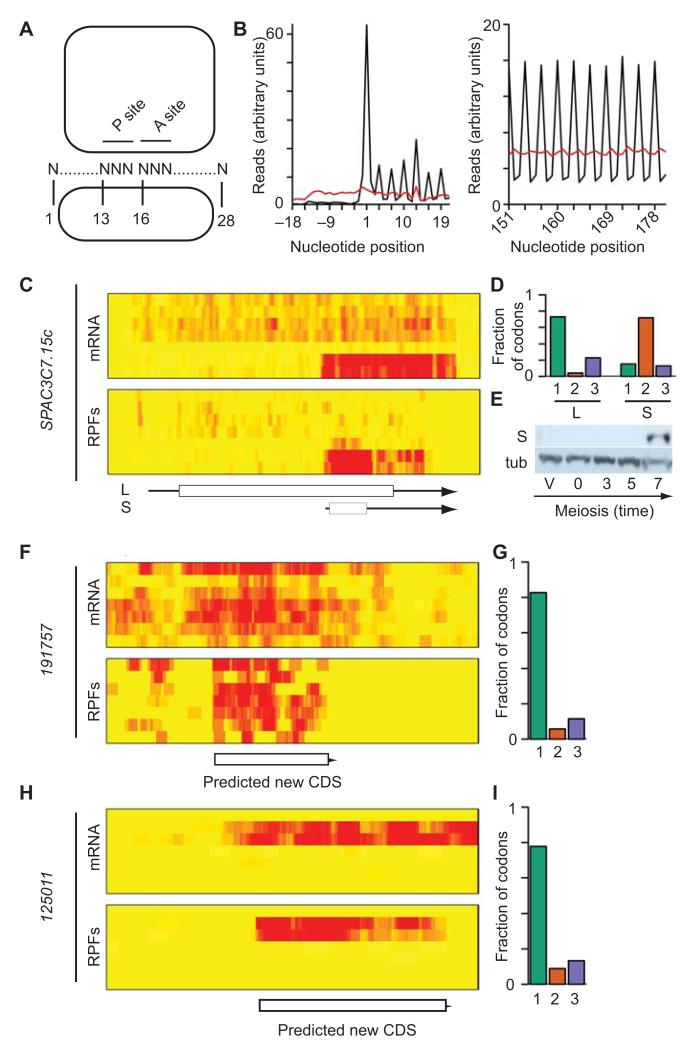
In a 28-nucleotide long RPF, nucleotide 13 will typically correspond to the first nucleotide of the codon located at the P site (Fig. 2A)11. Consistently, we found that nucleotide 13 mapped to the first nucleotide within a codon in 75 % of the 28-nucleotide RPFs, while mRNA fragments were equally distributed across all three nucleotides (Supplementary Table 3). When the data were aggregated for all annotated coding sequences, this effect led to a 3-nucleotide periodic pattern, which was observed at all positions (Fig. 2B). As previously noted, there was an accumulation of RPFs at the initiation codon (Fig. 2B) and at the last codon11,14. By contrast, mRNA fragments did not show a periodic behavior (Fig. 2B). As long as the RPF coverage is high enough, this feature allows the detection of any translated genomic region, as well as the identification of its reading frame. Although translated sequences can be identified simply by the accumulation of RPFs, the use of periodicity allows the distinction between translated regions and contaminants in the RPF sample. We therefore used read periodicity to define translated regions in parts of the genome with no annotated features (‘intergenic regions’), in genes annotated as non-coding RNAs, and in 5′ leader sequences. We note that the experimental approach and subsequent analyses are strand-specific, so regions of the genome containing a feature on one strand were still analyzed for the other one. For all genomic regions we followed the same experimental strategy: First, we defined all possible ORFs starting with AUG as well as rarer initiation codons. Second, each ORF was screened for the presence of periodic signals from RPFs. Third, a false discovery rate (FDR) was estimated based on a randomization test, and used to fine-tune the threshold values used to designate an ORF as translated.
Figure 2. Periodic triplet behavior of mRNA reads reveals novel genes.
(A) Expected position of RPFs relative to the P and A sites of the ribosome. (B) Number of reads against nucleotide position for all ORFs (black: RPFs; red: mRNAs). (C) Overlapping transcripts in SPAC3C7.15c. The heat maps show mRNA (top) and RPF levels (bottom) across the meiotic time course, starting with vegetative cells and progressing downwards. The brightness of the red color reflects the number of reads. The scheme shows the position of UTRs (arrows) and ORFs (boxes). L corresponds to the annotated gene and S to the novel gene. (D) Triplet periodicity for the L and S forms of SPAC3C7.15c. The graphs show the fraction of codons in which most reads map to nucleotides 1, 2 or 3 for the S and L ORFs. Reading frames are calculated relative to that of L. (E) Expression of the S form of SPAC3C7.15c. The S ORF was tagged with a TAP epitope and detected by Western blot (S) during a meiotic time course. A second blot was performed with the same samples and probed with antibodies against tubulin (tub). Full blots are presented in Supplementary Fig. 8. (F) Translation of the 191757 ORF. Heat map as in C. (G) Triplet periodicity for the 191757 ORF, labelling as in D. (H) Translation of the 125011 ORF. Heat map as in C. (D) Triplet periodicity of the 125011 ORF, labelling as in D. Note there are no annotated features overlapping 191757 and 125011.
To validate this approach we calculated periodicity scores, which measure the fraction of codons translated in each reading frame, for all annotated S. pombe coding sequences (using both RPF and mRNA data). The periodicity score discriminated clearly between both datasets (Supplementary Fig. 4A), and confirmed the translation in the predicted frame of 4,923 out of 5,102 annotated high-confidence coding sequences (96.5%)31. We manually examined genes that displayed periodicity in unexpected frames. This led to the discovery of several mis-annotated genes (Supplementary Table 4A), in which translation took place in a frame different from the annotated one or in which exon annotation was incorrect (Supplementary Fig. 4B-D). We also found a clear case of alternative intron retention, which is very rare in S. pombe (Supplementary Fig. 4D). A total of 71 coding genes in S. pombe are annotated as dubious31, indicating that the evidence for their existence is poor. Inspection of their translation profiles revealed that only 11 appeared to be clearly translated as predicted (15%), and 3 were translated in frames different from those annotated. In addition, in 25 cases (35%) the mRNA was well expressed but no translation was observed, suggesting that these genes may not be translated (Supplementary Table 4B). In the remaining genes, expression levels were not sufficient to allow the evaluation of translation. These results validate, refine and improve the annotation of the fission yeast genome.
This analysis also revealed instances of overlapping coding sequences in the same strand (dually decoded regions). For example, SPAC3C7.15c was translated from a long mRNA in the predicted frame during early meiosis. In late meiosis, a short transcript appeared that was completely enclosed within the long form of the mRNA but was translated in a different frame (Fig. 2C-2D). We epitope-tagged the predicted short form of the protein and observed a meiosis-specific polypeptide of the expected size, confirming that translation of the short ORF leads to the production of a stable polypeptide (Fig. 2E). The sensitivity of the approach was also exemplified by the fact that it allowed the discovery of changes in translation caused by single-nucleotide polymorphisms in our strains compared to the reference strain. We detected translation of the C terminal part of the Nup184 protein, which was not predicted to be translated in S. pombe due to an in-frame stop codon created by a single nucleotide deletion32. Translation of this region in the strains we used was explained by the presence of single nucleotide insertion that reverted the effect of the deletion in the reference sequence (Supplementary Fig. 5A and 5B).
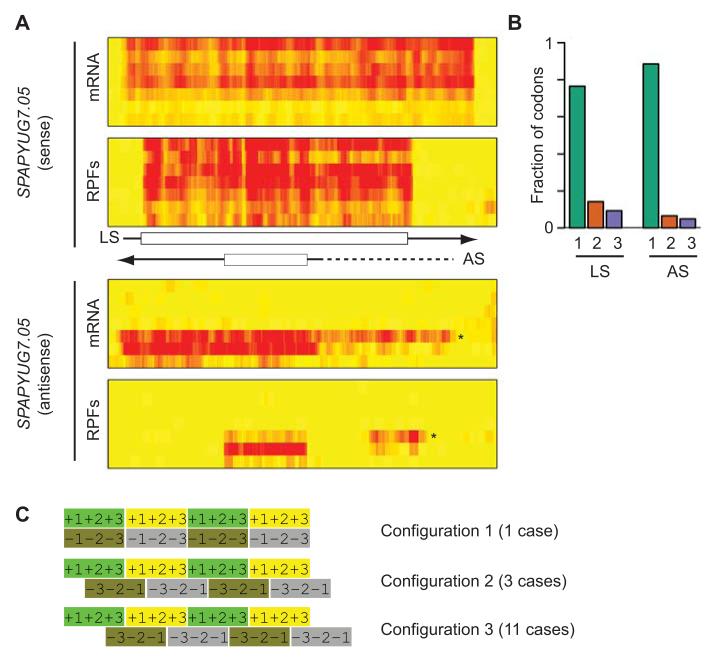
We then scanned intergenic regions systematically, and identified 715 translated ORFs with an estimated FDR of 10.3 % (Supplementary Table 5). Most of these regions were short (Supplementary Fig. 5C) and had a tendency to use AUG as initiation codon (90.6%, Supplementary Table 6). We manually inspected the 46 translated ORFs of 45 codons or more. Examination of the corresponding mRNA data suggested that 39 of them (85%) were transcribed as independent units (Fig. 2F-H). Of the remaining 6, 2 appeared to be extensions to annotated 5′ leaders, 3 were present in the 3′-UTRs of highly translated genes (possibly reflecting leaky termination of translation) and one was located downstream of the nup184 gene (discussed above). 15 translated ORFs did not overlap with annotated coding sequences (in either strand). Their sequences were generally not conserved, but one of them displayed homology to the N-terminal part of a protein present in multiple copies in S. cryophilus and S. octosporus. Surprisingly, we found 14 cases in which the newly-discovered translated ORF were antisense to an annotated coding sequence (completely overlapping in 10 cases, and more than 80% in the other 4) (Fig. 3A-3B, Supplementary Fig. 6A-B). In addition, we found a similar situation (with complete overlap) in 5 annotated antisense ncRNAs. These exonic Nested Antisense Genes (eNAGs) are extremely rare in eukaryotic cells, with a single case described in S. cerevisiae (the NAG1 gene)33. Simultaneous coding on both strands imposes strict constraints on the evolution of both proteins. The nature of these limitations depends on the relative frame of the sense and antisense coding sequences, which can adopt three different arrangements (Fig. 3C). We examined this configuration in all 15 S. pombe eNAGs that were completely nested within the major coding sequence, and found that in 11 cases arrangement 3 was preferred (Fig. 3C). In the case of S. cerevisiae NAG1, configuration 3 was also used. This organization causes very specific dependencies between the sequences encoded in both strands, as nucleotides 1 and 2 of every codon on both strands are encoded by the same DNA sequence. Because codons 1 and 2 possess most information content, the encoding of a particular amino acid on one strand can determine the nature of the amino acid on the antisense strand. For example, in arrangement 3, the presence of a CCN codon (proline) necessarily implies a GGN codon (glycine) in the antisense gene (and vice versa). Our results suggest that a specific arrangement of reading frames is evolutionary favored when DNA both strands are translated.
Figure 3. Identification of translated nested antisense genes.
(A) Nested antisense gene in SPAPYUG7.05. The heat maps show mRNA (top) and RPF levels (bottom) across the meiotic time course, starting with vegetative cells and progressing downwards. The brightness of the red color is proportional to the number of reads. The scheme shows the position of UTRs (arrows) and ORFs (boxes). Data for the sense and antisense strands are displayed separately. LS corresponds to the annotated ORF and AS to the newly discovered antisense transcript. The arrows indicate the direction of transcription. Note the presence of a 5′ extension (star on the heat map and dotted line on the scheme) that appears at the 5 hour time point and whose translation correlates inversely with that of the downstream ORF. (B) Triplet periodicity for the sense and antisense ORFs of SPAPYUG7.05. The graphs show the fraction of codons in which the majority of reads map to nucleotides 1, 2 or 3 for the S and L ORFs. (C) Possible relative configurations of reading frames in exonic nested antisense genes. For each configuration the top strand represents translation in the 5′ to 3′ direction, and the bottom one 3′ to 5′. The numbers represent the position of nucleotide 1, 2 and 3 for each codon.
Translation of annotated non-coding RNAs
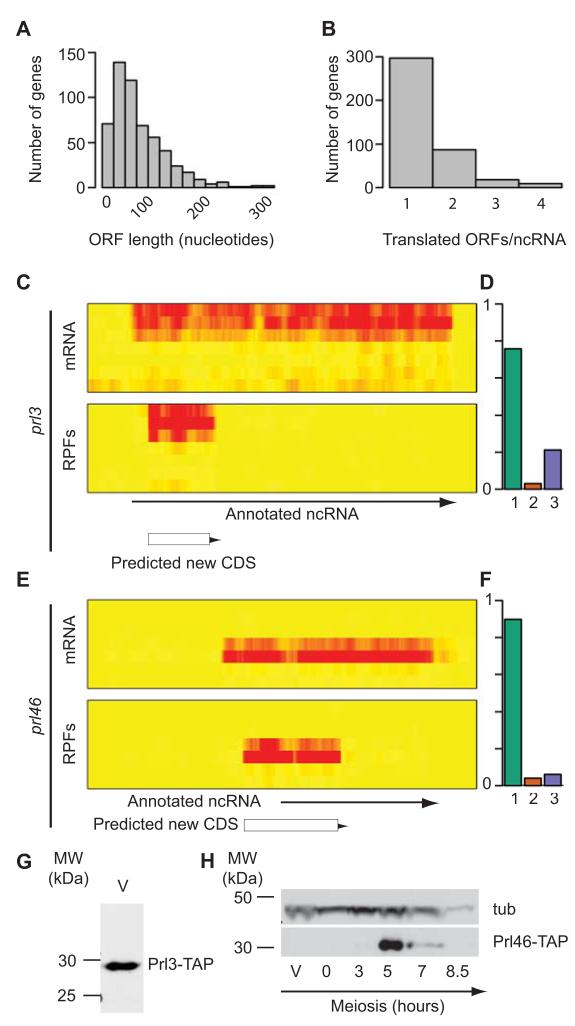
The S. pombe genome contains 1,571 annotated long non-coding RNAs (ncRNAs)9,10,31. Visual inspection of our data revealed the presence of numerous meiosis-specific novel genes. In vegetative cells and during most of meiosis, reads mapped to ncRNAs accounted for less than 3.5% of all reads. However, in mid-meiosis (3 and 5 hours) this number increased to 11%, suggesting that ncRNA function may be especially important during cellular differentiation. Recent work in several eukaryotes has revealed that ncRNAs are often present in ribosomal fractions, although it is unclear to which extent this association represents active translation11,14-16. We used triplet periodicity to address this question. We identified 499 translated regions in 375 genes (FDR 7.5%, Supplementary Table 7). These regions had a tendency to start with the canonical AUG (96.8%, Supplementary Table 6). Their median length was 21 codons, and 37 were longer than 45 (Fig. 4A). 28% of translated ncRNAs contained more than one translated ORF (Fig. 4B). Some of these translated regions were reminiscent of upstream ORFs, with a longer coding sequence preceded by several short uORFs. ORFs in ncRNAs have the potential to produce short polypeptides. To investigate if this was the case, we epitope-tagged two predicted peptides from the prl3 and prl46 ncRNA genes. The prl3 gene was well expressed in vegetative cells, whereas prl46 gene was meiotic-specific. Both contained ORFs that appeared to be highly translated (Fig. 4C-4F). Tagging of the corresponding peptides allowed the detection of proteins of the predicted molecular weight, confirming that both genes encoded expressed polypeptides (Fig. 4G-H). The majority of the translated sequences did not have homologs in other organisms, although there were some exceptions: The Prl46 protein and those encoded by SPNCRNA.557 and SPNCRNA.1597 were conserved in other Schizosaccharomyces species.
Figure 4. Identification of translated regions in annotated ncRNAs.
(A) Histogram showing the length distribution of all translated uORFs (measured in nucleotides). (B) Number of translated ORFs per ncRNA. (C) Expression and translation of the prl3 gene. The heat maps show mRNA (top) and RPF levels (bottom) across the meiotic time course, starting with vegetative cells at the top and progressing downwards. The brightness of the red color is proportional to the number of reads. The arrow shows the position of the annotated ncRNA, and the box that of the translated ORF. (D) Triplet periodicity for the translated ORF in prl3. The graph shows the fraction of codons in which a majority of the reads map to nucleotides 1, 2 or 3 for the ORF. (E) Expression and translation of the prl46 gene. Labelling as in C. (F) Triplet periodicity for the translated ORF in prl46. Labelling as in D. (G) Expression of the Prl3 protein. The translated ORF in prl3 was tagged with a TAP epitope and detected by Western blot in vegetative cells (V). (H) Expression of the Prl46 protein. The translated ORF in prl46 was tagged with TAP and detected by Western blot. A single band of the expected molecular weight was observed at 5 hours into meiosis, coinciding with the peak of translation. The blot was also probed with antibodies against tubulin (tub). Full blots are presented in Supplementary Fig. 8.
These results show that 24% of annotated ncRNAs are translated to produce short peptides. However, the majority of ncRNAs associate with ribosomes to translate ORFs of very few codons, indicating that the distinction between coding and non-coding RNAs is not clear-cut. The functional importance of these observations is unclear, although several not mutually exclusive explanations are possible. First, short polypeptides encoded by ncRNAs may have biological activity. For example, peptides of 11 to 32 amino acids from the Drosophila tarsal-less (tal) gene regulate embryonic development34,35, peptides shorter than 30 amino acids modulate cardiac function in Drosophila36, and the S. pombe Mat-Mi protein, consisting of only 42 amino acids, is a key regulator of meiosis that functions as a transcription factor37. Second, translation could be used to target ncRNAs to polysomes, where they might be degraded (through NMD) or perform functions in translational control. Finally, translation of short ORFs in ncRNAs may reflect pervasive translation in which capped and polyadenylated sequences that reach the cytoplasm would be translated to some degree, even if their translation was not functionally important.
Translation of 5′ leader sequences
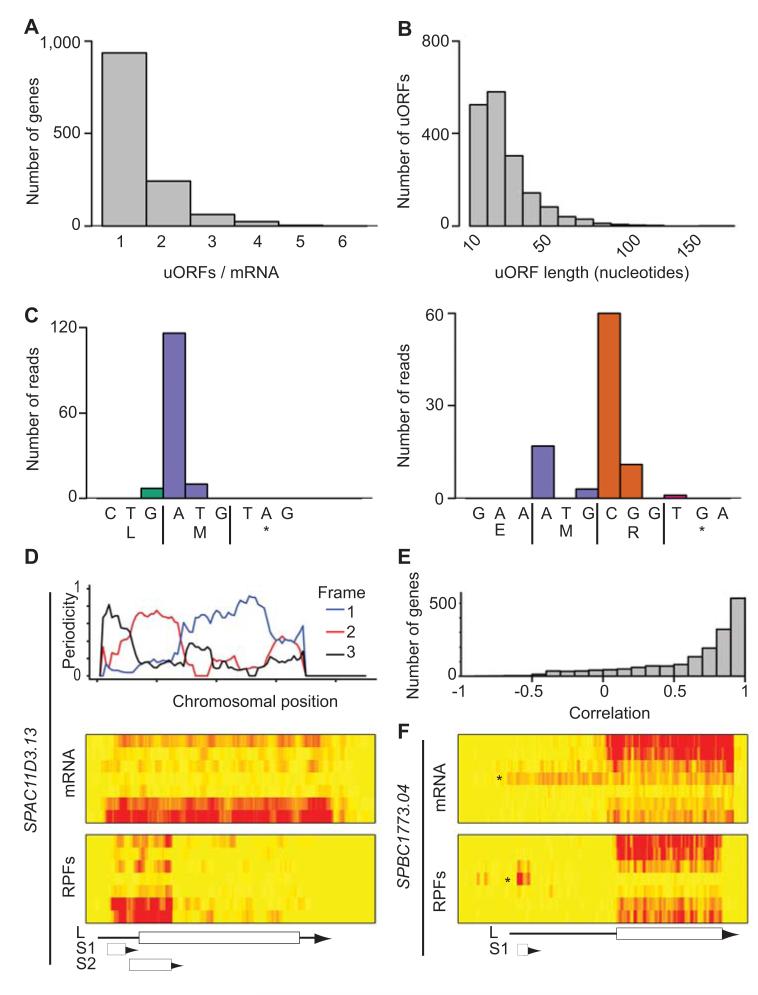
6.9 % of all RPF reads mapped to 5′ leader sequences. Translated ORFs in 5′ leaders (upstream ORFs, or uORFs) have the potential to regulate translation, although their effect can be neutral, positive or negative38. Although translation of a uORF by a ribosome may down-regulate translation by preventing it from reaching a downstream ORF, ribosome small subunits have the potential to perform scanning after termination and may recognize downstream initiation codons (reinitiation)39. In addition, uORFs can encode short peptides that are stably expressed40. We examined all predicted ORFs in annotated 5′ leaders for periodic footprint patterns, and identified 1,735 translated uORFs in 1,272 genes, with an estimated FDR of 10.0% (Supplementary Table 8). 26% of the genes contained more than a single uORF, and 7% had 3 or more (Fig. 5A). The latter group was enriched in genes encoding transcription factors, periodically expressed genes, middle meiotic genes and genes induced in response to nitrogen starvation. Interestingly, mRNAs encoding four key regulators of meiosis contained four or more uORFs: pat1 and mei2, which encode a kinase and an RNA-binding protein, respectively, that control entry into meiosis19,20,41, ste11, encoding a transcription factor that mediates pheromone communication5, and atf21, which codes for a transcription factor responsible for the induction of late meiotic genes2. uORFs had a mean length of 13.1 codons, and a median of 10 (Fig. 5B). Some were extremely short, with 6.8% containing a single AUG codon (Fig. 5C). 5′ leader sequences contained less ORFs starting with AUG compared to other genomic regions (Supplementary Table 6), and translated uORFs used AUG as a start codon less frequently (Supplementary Table 6).
Figure 5. Identification and characterization of uORFs.
(A) Number of translated uORFs per mRNA. (B) Length distribution of all translated uORFs (in nucleotides). (C) Examples of very short highly translated uORFs. The x axes show the DNA and protein sequences of a uORF consisting of a single codon (left) or two codons (right). The y axis represents the number of reads at each nucleotide. (D) Translation of SPAC11D3.13. The top graph shows the triplet periodicity across the gene calculated for a running window of 35 codons. The heat maps show mRNA (top) and RPF levels (bottom) across the meiotic time course, starting with vegetative cells at the top and progressing downwards. The brightness of the red color is proportional to the number of reads. The scheme shows the position of the UTRs (arrows) and the ORFs (boxes). L corresponds to the annotated gene, S1 and S2 to predicted uORFs. Note the change in reading frame as the window moves from one ORF to the next one. S2 overlaps with L and is more heavily translated, and is thus the dominant frame. (E) Correlation coefficients between uORFs and their corresponding ORFs. (F) Translation of the SPBC1773.04 locus. Labelling as in D. L corresponds to the annotated gene. Note the presence of a longer form of the mRNA (star on the heat map that appears at the 5 hour time point and whose translation (box S1) correlates inversely with that of the downstream ORF.
We also identified 175 uORFs that overlapped partially with the main ORF (in a different reading frame) (Supplementary Table 9). These may be particularly important as repressor elements, as they would not allow reinitiation of translation in the frame of the main ORF39. Among them, there were three cases in which a strongly translated uORF overlapped substantially with a downstream ORF that was actively but poorly translated (Fig. 5D and Supplementary Fig. 6C-D). In the most extreme case (SPCC1235.01) the uORF was 347-codon long, starting 68 nucleotides upstream of the ORF (Supplementary Fig. 6C). Interestingly, the amino acid sequence encoded at the 5′ end of the uORF (that does not overlap with the ORF) was conserved in other Schizosaccharomyces species, suggesting that both reading frames may produce functional proteins. SPAC11D3.13 (Fig. 5D) and SPAC6G9.05 (Supplementary Fig. 6D) presented a similar structure, although the length of the uORFs was shorter. SPAC11D3.13 had an additional upstream uORF, in an arrangement reminiscent of the regulatory uORFs of the mammalian ATF4 and ATF5 mRNAs, which are only translated in stress situations (Fig. 5D)39.
In S. cerevisiae, there is an increase in the use of uORFs during meiosis13. We examined if a similar phenomenon takes place in S. pombe by comparing the ratio between the TEs of every uORF and its corresponding ORF for every time point (Supplementary Fig. 3B). Although we observed increases towards the end of meiosis, the changes were generally small, indicating that in S. pombe neither nitrogen starvation nor the meiotic program caused a large rise in the use of uORFs.
To investigate global effects of uORFs on translation, we compared the TE of every uORF to that of its downstream ORF. In general, the TEs of uORFs and main coding sequences did not correlate (average R = 0.20), consistent with the complex functions of uORFs38. We then examined whether changes in TE of uORFs during meiosis correlated with variations in TE of downstream coding regions. We calculated the correlation between the TE of every uORF and their downstream ORFs across all time points (see examples in Supplementary Fig. 3). The majority of genes showed strong positive correlations (Fig. 5E), although a very small subset displayed a negative relationship. In SPBC1773.04, a meiosis-specific 5′ extension to the mRNA contained two uORFs whose translation was negatively correlated with that of the main ORF, suggesting that the uORFs competed for translation with and down-regulated translation of the downstream ORF (Fig. 5F). Another example is displayed in Fig. 3A, in this case concerning an antisense transcript. Although temporal regulation of TE through changes in the use of uORFs is common in S. cerevisiae meiosis13, it appears to be rare in S. pombe.
Discussion
Our results reveal pervasive translation of the S. pombe, including dually decoded regions, exonic nested antisense genes and frequent translation of annotated ncRNAs. Overall, we found 917 translated ORFs of 20 codons or longer (in 5′ leader sequences, annotated ncRNAs, and novel translated regions), suggesting the existence of a large repertoire of small peptides with potential biological functions. We have experimentally validated the expression of some of these peptides, demonstrating that their translation results in real changes to the proteome. In addition, we have observed substantial use of TE regulation during meiosis, including homodirectional changes in RNA levels and TE (potentiation) and the use of TE changes to delay protein accumulation. The existence of a previous dataset from S. cerevisiae allowed us to address if properties of the translational programs of cellular differentiation processes are general or organism-specific. Both S. pombe and S. cerevisiae use extensive translational control, as demonstrated by the widespread and dynamic changes in TEs. In contrast to S. cerevisiae, we did not detect a switch to the use of uORFs and unconventional initiation mechanisms, suggesting that this is not a general feature of meiosis or cellular differentiation processes.
Online Methods
General methods
Standard methods and media were used for fission yeast growth. Wild type and pat1-driven meiosis were induced as described2. Vegetative cells were grown in rich medium at 32°C. Proteins were tagged with TAP. Tagged proteins were detected by Western blot using peroxidase–anti-peroxidase-soluble complexes (Sigma P1291) diluted 1:10,000, and alpha-tubulin with mouse monoclonal antibodies (Sigma, clone B-5-1-2) diluted 1:20,000. For protein detection of Prl3 and Prl46, 50 ml of cells (3-8 × 106 cells/ml) were treated with 1 mM PMSF for 5 min before centrifugation at 4 °C and freezing. Cells were later thawed, washed with cold lysis buffer (20 mM Tris-HCl pH 8.0, 140 mM KCl, 1.8 mM MgCl2 and 0.1% NP-40), resuspended in 200 μl of lysis buffer containing 1 mM PMSF and 1:100 protease inhibitor cocktail (Sigma P8340), and lysed using a bead beater (Fastprep, MP Biomedicals) at level 6 for 13 seconds. Extracts were cleared by centrifugation at 7,600 g for 5 minutes at 4°C and used for Western blotting. The short peptide overlapping SPAC3C.15c was not visible under these conditions. In order to detect it, cells were washed with cold RIPA buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Triton X100, 0.1% SDS), resuspended in 200 μl of RIPA buffer, boiled for 5 minutes and frozen. Samples were then processed as described above except that RIPA buffer was used and the extracts were not precleared by centrifugation. Original images of blots used in this study can be found in Supplementary Fig. 8.
Ribosome profiling, library preparation and sequencing
3×108 exponentially growing wild type haploid cells, or between 3×108 and 12×108 meiotic cells, were incubated for 5 minutes with 100 μg/ml cycloheximide, pelleted at 4°C and frozen in liquid N2. Each culture was split for footprint isolation and mRNA fragmentation. We generally followed a published protocol11 with the modifications stated below. Cells were resuspended in 100 μl of lysis buffer (20 mM Tris-HCl pH 8.0, 140 mM KCl, 5 mM MgCl2, 1 % Triton X-100) with 1 g of chilled glass beads (Biospec) and lysed using a Fastprep 24 bead-beater at level 6 for 13 seconds. The extract was diluted with 400 μl of lysis buffer and cleared by centrifugation in two steps at 4°C at 16,000 g (5 minutes followed by 15 minutes). For footprint isolation, 600 A260 units of wild type vegetative cell extract were digested with 750 Units of RNase I (Life Technologies) for 30 minutes, or 800 units of pat1 diploid extract were treated for 10 min with 1,500 units of RNase I. Reactions were quenched with 600 units of SUPERaseIn (Life Technologies). Digested extracts in 500 μl were loaded onto an 14 ml linear 10-50% (w/v) sucrose gradient prepared with a Gradient Master (Biocomp), and separated by centrifugation for 160 min at 35,000 rpm in a SW 40Ti rotor (Beckman). The gradients were then fractionated by upward displacement with 55% (w/v) sucrose, and fractions containing monosomes selected for further processing. RNAs were then purified by phenol extraction, passed through a YM-100 column (Millipore), and run on 15% TBE-urea gels (Life Technologies). Fragments of around 28 nucleotides were extracted from the gel. For the preparation of mRNA fragments, total RNA was prepared by phenol extraction, and polyadenylated RNA was purified from 150 μg of total RNA (vegetative cells), or 300-450 μg (pat1 cells) using oligo-dT25 magnetic beads (Life Technologies) following the manufacturer’s instructions. Purified mRNA was fragmented by mixing 20 μl of mRNA with 20 μl of 2X alkaline fragmentation buffer (2 mM EDTA, 100 mM Na2CO3) followed by incubation for 15 minutes at 95°C. Samples were run on 15% TBE-urea gels (Life Technologies), and fragments of around 28 nucleotides were extracted from the gel. From this point, both mRNA and ribosomal footprint samples were processed identically. RNA samples were purified using Purelink RNA microcolumns (Life Technologies) as described by the manufacturer, except that the samples were initially passed through the column in the presence of 70% ethanol (to favour binding of small RNAs). Samples were then treated with polynucleotide kinase (PNK, Fermentas) as described11 and polyadenylated using 12 units of poly-(A) polymerase (NEB) at 37°C for 45 minutes. Reverse transcription reactions were performed using custom primers containing an anchored oligo(dT), 4 nucleotides of known sequence used for multiplexing, and 5 random nucleotides that serve as unique molecular identifiers (see below)22. All primers were synthesized by Integrated DNA Technologies (IDT) and are listed in Supplementary Table 10. Reverse transcription products were gel-purified and circularized using CircLigase II (Epicentre), and amplified by PCR with custom library primers (P3 and P5)22 for 12 or 15 cycles. Libraries were sequenced on an Illumina Genome Analyzer II (a subset of the first biological repeat of vegetative haploids), on a Next Seq 500 sequencer (total RNA), or on a HiSeq 2000 platform (all other samples) using standard Illumina primers. We performed two biological replicates for vegetatively growing cells and for key time points of the pat1 time course (3, 5 and 7 hours). Note that these are completely independent repeats, with cultures grown on different days, and library preparation and sequencing performed separately. The results were highly reproducible (Supplementary Fig. 2), with average correlation coefficients between replicates of 0.97 (mRNA), 0.98 (RPFs) and 0.90 (TEs). To evaluate the effect of mRNA purification on the protocol we sequenced total RNA from vegetative cells and compared the results with those using oligo(dT) purified RNA. The correlation between both samples (read densities across ORFs) was 0.88, indicating that oligo(dT)-purified RNA provides a reasonable estimate of mRNA amounts.
Bioinformatic analyses
All data processing was performed with custom scripts written in Perl (www.perl.org) and all downstream statistical analysis used R (www.r-project.org/). For all analyses, S. pombe annotations and sequences available from GeneDB (http://old.genedb.org/), now PomBase (http://www.pombase.org/), on May 9, 2011 were used31.
The RT primers (Supplementary Table 10) include a 4 nucleotide barcode that allows multiplexing, which was used to allocate reads from different samples to separate files. The RT primers also contain a 5 nucleotide random sequence that serves as a Unique Molecular Identifier (UMI). Reads that contain the same UMI followed by an identical sequence are highly likely to have arisen from the same RNA molecule, and only one of them is retained. This step creates a non-redundant dataset, thus avoiding sampling biases and PCR amplification artefacts21,22. Non-redundant reads are then processed to remove adenosine residues at their 3′ ends. Reads are mapped to the S. pombe rDNA genome32 with Tophat 242 and the following parameters: --min-intron-length 29 --max-intron-length 819 --zpacker 0 --splice-mismatches 0 --max-multihits 1. Unmapped reads were recovered and aligned to the full S. pombe genome32 with Tophat 2 and the following settings: --min-intron-length 29 --max-intron-length 819 --zpacker 0 --splice-mismatches 0. A GFF file containing annotation of the S. pombe genome31 was provided as a source of exon-exon junction data for Tophat. Aligned data were visualized using the Integrated Genome Viewer43. RPF libraries are expected to contain a higher fraction of contaminating rRNA than mRNA libraries, as the latter are oligo(dT)-selected. Consistently, 81.7 % of reads from RPF-derived and 26.5 % from mRNA-derived libraries mapped to the rDNA genome (median from all experiments, Supplementary Table 1). UMIs were used to calculate the fraction of unique reads for those reads mapping to rRNA and for the remaining set. rRNA reads in the RPF samples typically showed low complexity (median unique 10.1 %, Supplementary Table 1). This may be caused by the fact that the majority of these reads originate from a small pool of sequences (presumably due to the sequence preferences of RNase I). As the UMI length is 5 nucleotides, there are 1,024 different UMIs. If the number of fragments derived from the same sequence largely exceeds this figure, the likelihood that independent fragments with the same sequence share the same UMI increases and artefactually decreases the observed number of unique sequences. This is unlikely to be a problem for rRNA reads in the poly(A)-purified sample (median unique 73%, Supplementary Table 1) or for non-rRNA reads (median unique 83.4%, Supplementary Table 1), which are evenly distributed along the RNAs.
For clustering of RPFs (Supplementary Fig. 3A), the 1,719 genes showing strongest changes in gene expression were selected. All the data were normalized to expression levels in vegetative cells of the corresponding time course, and RPF values were used for clustering. For clustering of TEs (Fig. 1C), the 418 genes that showed the largest variations in TE across the time course were chosen. Clustering was performed with Cluster 3.044,45, filtering out data with more than 20% values missing, log-transforming the data, using Pearson correlation and creating and average-linked tree. Clusters were visualized with Treeview46.
To calculate overall periodicity and to calibrate reads, we counted the number of reads in which position 13 of a read maps to the first, second, or third position of every codon for all annotated coding sequences of the S. pombe genome (Supplementary Fig. 7A). Note that the choice of nucleotide 13 is arbitrary, and that similar results would be obtained with a different nucleotide. The advantage of nucleotide 13 is that for a ribosome starting translation at the initiation codon, it corresponds to the position of the AUG on the mRNA. This was done for mRNA and RPFs, and analysed separately for reads of lengths between 25 and 32 nucleotides (Supplementary Fig. 7B). No bias was observed in any position within a codon for the mRNA fragments. By contrast, enrichments in position 1 were observed for RPFs of 28, 29 and 30 nucleotides (Supplementary Fig. 7B). The fact that fragments longer than 28 nucleotides show periodicity is unexpected, and suggests that some feature of the ribosome allows more precise cutting of RNase I at the 5′ than at the 3′ of the protected fragments. Reads between 28 and 30 nucleotides were selected for the discovery of translated features (such as uORFs and ncRNAs), while all reads were used to calculate translational efficiencies.
For the identification of novel translated regions, annotated coding sequences, non-coding RNAs, uORFs and intergenic regions were analyzed separately. Open reading frames were defined as follows. First, selected regions were scanned in each frame until an AUG was encountered. This was defined as the start of the ORF, which was elongated until a stop codon was found. Second, the process was repeated as above for UUG, except than when an ORF overlapped with a previously defined one in the same frame it was discarded. The process was then performed as above for CUG and GUG. A total of 384,032 ORFs were defined in intergenic regions, 50,574 ORFs in ncRNAs, and 31,996 in 5′ leader sequences. Of these, 49.2%, 49.2% and 27.2% started with an AUG codon in intergenic regions, ncRNAs, and 5′ leaders, respectively (Supplementary Table 6). This indicates that AUG ORFs are depleted in 5′ leader sequences compared to other parts of the genome. The resulting ORFs were analysed for read periodicity by quantifying the fraction of codons showing enrichment of reads in the first nucleotide. For every codon, the enriched nucleotide (if existing) was defined as that having at least 60% of all the reads that mapped to the codon. In this way, all codons have equal contributions regardless of the total number of reads that map to each of them, thus avoiding biases created by small number of codons with very high number of reads. The fraction of codons enriched in nucleotide 1 within an ORF was defined as the ORF periodicity score. An ORF was defined as translated when its periodicity score was >= 0.6. To avoid noise from lowly expressed genes, a total number of 10 reads was required for a region to be considered as translated. False Discovery Rates (FDRs) were calculated by randomizing the position of the reads within each codon as follows: if a codon contains a1, a2, a3 reads in positions 1, 2 and 3, respectively, the numbers a1, a2, and a3 were randomly assigned to a position within a codon (this creates 3!=6 possibilities per codon). This was followed by the analysis of periodicity as described above. A p-value was also calculated for each feature by assuming a binomial distribution for the fraction of codons enriched at position 1. Note that under this very conservative assumption the smallest possible p-value for an ORF of a single codon is 0.33. However, given that our threshold for calling a feature as translated requires at least 10 reads and that more than 60% of the reads map to the first nucleotide, the probability of passing the threshold for such an ORF is lower than 0.0035 (based on the less conservative assumption that reads are randomly distributed within the codon).
Supplementary Material
Acknowledgements
We thank Jürg Bähler and Richard Jackson for comments on the manuscript. This work was funded by Biotechnology and Biological Sciences Research Council (UK) research grant BB/J007153/1 (JM).
Footnotes
Accession codes All sequencing raw data have been deposited in ArrayExpress with the following accession numbers: E-MTAB-2176 (vegetative haploid cells), E-MTAB-2179 (pat1 meiosis, replicate 1), E-MTAB-2265 (pat1 meiosis, replicate 2) and E-MTAB-2470 (total RNA, vegetative cells).
References
- 1.Yamamoto M, Imai I, Watanabe Y. S. pombe mating and sporulation. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Life Cycle and Cell Biology. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1997. pp. 1035–1106. [Google Scholar]
- 2.Mata J, Lyne R, Burns G, Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 3.Horie S, et al. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol. 1998;18:2118–29. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mata J, Wilbrey A, Bähler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8:R217. doi: 10.1186/gb-2007-8-10-r217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991;5:1990–9. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- 6.Amorim MJ, Cotobal C, Duncan C, Mata J. Global coordination of transcriptional control and mRNA decay during cellular differentiation. Mol Syst Biol. 2010;6:380. doi: 10.1038/msb.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harigaya Y, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 8.Bitton DA, et al. Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Mol Syst Biol. 2011;7:559. doi: 10.1038/msb.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhind N, et al. Comparative Functional Genomics of the Fission Yeasts. Science. 2011 doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 11.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel AM, Baranov PV. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdiscip Rev RNA. 2013;4:473–90. doi: 10.1002/wrna.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–7. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew GL, et al. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140:2828–34. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel AM, et al. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012;22:2219–29. doi: 10.1101/gr.133249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bazzini AA, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. Embo J. 2014;33:981–93. doi: 10.1002/embj.201488411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 20.Nurse P. Mutants of the fission yeast Schizosacharomyces pombe which alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497. [Google Scholar]
- 21.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mata J. Genome-wide mapping of polyadenylation sites in fission yeast reveals widespread alternative polyadenylation. RNA Biol. 2013;10:1407–14. doi: 10.4161/rna.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–83. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lackner DH, et al. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–55. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matia-Gonzalez AM, Hasan A, Moe GH, Mata J, Rodriguez-Gabriel MA. Functional characterization of Upf1 targets in Schizosaccharomyces pombe. RNA Biol. 2013;10:1057–65. doi: 10.4161/rna.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. Nonsense-mediated decay targets have multiple sequence-related features that can inhibit translation. Mol Syst Biol. 2010;6:442. doi: 10.1038/msb.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lackner DH, Schmidt MW, Wu S, Wolf DA, Bahler J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 2012;13:R25. doi: 10.1186/gb-2012-13-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacKay VL, et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3:478–89. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol. 2003;10:1039–47. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- 31.Wood V, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–9. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Dobry CJ, Krysan DJ, Kumar A. Unconventional genomic architecture in the budding yeast Saccharomyces cerevisiae masks the nested antisense gene NAG1. Eukaryot Cell. 2008;7:1289–98. doi: 10.1128/EC.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo T, et al. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–5. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 36.Magny EG, et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;341:1116–20. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 37.Kelly M, Burke J, Smith M, Klar A, Beach D. Four mating-type genes control sexual differentiation in the fission yeast. Embo J. 1988;7:1537–47. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol. 2009;63:385–409. doi: 10.1146/annurev.micro.62.081307.162835. [DOI] [PubMed] [Google Scholar]
- 39.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slavoff SA, et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoda C, et al. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J Bacteriol. 1987;169:93–6. doi: 10.1128/jb.169.1.93-96.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 42.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 45.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.