Abstract
The use of DNA to deliver vaccine antigens offers many advantages, including ease of manufacture and cost. However, most DNA vaccines are plasmids and must be grown in bacterial culture, necessitating elements which are either unnecessary for effective gene delivery (e.g. bacterial origins of replication) or undesirable (e.g. antibiotic resistance genes). Removing these elements may improve the safety profile of DNA for the delivery of vaccines. Here we describe a novel, double-stranded, linear DNA construct produced by an enzymatic process that solely encodes an antigen expression cassette, comprising antigen, promoter, polyA tail and telomeric ends. We compared these constructs (called ‘Doggybones’ because of their shape) with conventional plasmid DNA. Using luciferase-expressing constructs, we demonstrated that expression levels were equivalent between Doggybones and plasmids both in vitro and in vivo. When mice were immunized with DNA constructs expressing the HIV envelope protein gp140, equivalent humoral and cellular responses were induced. Immunizations with either construct type expressing haemagluttinin were protective against H1N1 influenza challenge. This is the first example of an effective DNA vaccine which can be produced on a large scale by enzymatic processes.
Keywords: DNA Vaccine, Gene Delivery, Novel Construct
Introduction
In the 1990s it was demonstrated that when antigen-encoding nucleic acid is introduced into the body it can invoke an immune response against the encoded antigen(1). DNA vaccines have many advantages over traditional strategies: they are relatively safe compared to attenuated vaccines, they are stable and can be stored frozen or lyophilized, they are easy to manufacture and can stimulate cell mediated immune responses(2). Whilst there are DNA vaccines licensed for veterinary use(3) and DNA vaccines have been shown to be effective in numerous models, currently there are no DNA vaccines licensed for human use(4, 5). The main hurdle has been poor immunogenicity in humans and but there are also concerns over genomic integration and induced allergy to DNA. However the FDA has recently relaxed its regulations regarding the latter two points(6).
Conventionally DNA vaccines are produced as plasmids grown in genetically modified bacteria, normally Escherichia coli. Such plasmids must contain, alongside the gene of interest, a bacterial origin of replication and a selective gene, normally encoding antibiotic resistance, to maintain the persistence of the plasmid in the bacterium(7). Whilst DNA vaccines have proven to be safe in a number of animal models and early phase clinical trials, there is a concern that the antibiotic resistance genes in the DNA vaccine may be conveyed to pathogenic bacteria or microflora. The use of antibiotic resistance genes has previously been negated through the use of RNAi ‘RNA-OUT’ as a counter selectable marker or auxotrophic selection(8, 9). Such methods can increase production efficiency by decreasing the metabolic burden on the bacteria(10). However the reliance on bacteria as a method to produce the vaccine means there is a small chance of recombination events within the plasmid, leading to the loss of the antigenic determinant(11). To avoid problems with recombination events, extensive quality controls at a molecular level need to be installed on a batch by batch basis. Additionally, the presence of endotoxin within the system means that, after extraction, DNA must be cleaned and further quality control (QC) measures applied(12).
The use of minimal DNA constructs for example minicircles, small circular fragments of DNA derived from a larger plasmid(13) or minimalistic immunologically defined expression (MIDGE) vectors(14), that only encode an antigen expression cassette (promoter, antigen and polyA region), goes some the way to solve the problem of extraneous elements. These vectors have been shown to be immunogenic, inducing both a cellular and humoral response, but both of these vectors require a bacterial fermentation step(15). Synthesizing the vaccine entirely in vitro, with the absence of a bacterial step, would ensure uniformity between batches and increase their readiness for good manufacturing practice (GMP) production. Recently the molecular tools by which to do this have become available in the form of enzymes derived from bacteria and bacteriophages. Using a proprietary method from Touchlight Genetics, covalently closed linear DNA constructs can be made synthetically which permits rapid vaccine design and manufacture. The DNA vectors produced by the process are referred to as ‘Doggybones’ (DB) due to their proposed shape and are synthesized in controlled batch reactions.
The aim of this study was to characterize Doggybone constructs for use as DNA vaccines and to compare them to plasmids. We demonstrated that Doggybones are comparable to conventional plasmids in terms of expression and immunogenicity. This study therefore concludes that Doggybones could be a suitable replacement for plasmid DNA for gene delivery in the context of DNA vaccines.
Results
Production of linear closed DNA construct
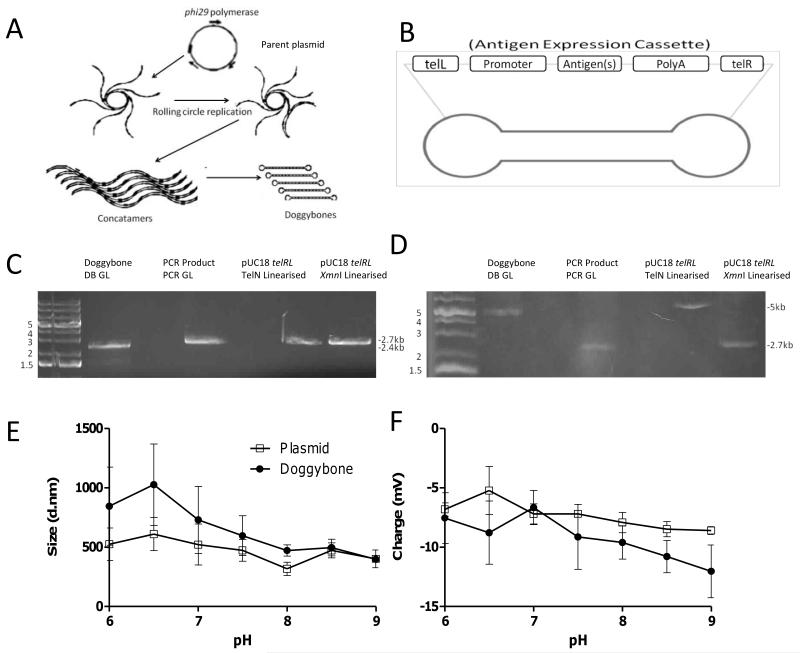
Linear DNA constructs were produced using the outlined mechanism (Fig. 1A)(16). The process is composed of two steps; firstly plasmid DNA which has the sequence for the antigen flanked by telRL sites is amplified by rolling circle replication using phi29 DNA polymerase from Bacillus subtilis phage phi29, resulting in the production of long concatamers. The protelomerase TelN (from E. coli phage N15) then cleaves the concatamers into strands containing a single cassette and seals the ends with a short hairpin loop(17). The constructs are composed of a linear double stranded region with an antigen expression cassette, encoding the sequences for the CMV Immediate Early promoter plus enhancer, the gene of interest and the SV40 late poly A tail, flanked by single stranded telomere ends (Fig. 1B). In the initial round of amplification, plasmid DNA is used as a template, but this is then selectively digested with restriction enzymes and then exonuclease III. In subsequent rounds of amplification the Doggybone itself can be used as the template.
Figure 1. Characterisation of Doggybone constructs.
Doggybones were produced using the method outlined by(16), a summary of the process is shown (A). Rolling circle replication takes place from a starting plasmid resulting in concatamers which are then resolved through the actions of TelN on the telRL sites included in the sequence. Addition of restriction enzymes and exonuclease removes any contamination from the plasmid backbone sequences to leave the cassette Doggybone only. The end product is a linear dsDNA construct flanked by ssDNA hairpins (Doggybones) (B). To confirm the Doggybone structure constructs were run on a native (C) and a denaturing agarose gel (D). The following were run: Doggybone DB GL (lane 1 created from pGL DOG (Luciferase cassette)), the PCR GL insert only obtained through PCR (lane 2), pUC18 telRL plasmid linearised with TelN (lane 3) or XmnI (lane 4). The size (E) and charge (F) of Doggybone (●) or plasmid (□) were assessed by dynamic light scattering, points represent mean of n=3 +/− SEM.
To confirm that hairpin loops had formed, DNA migration was compared on denaturing or native gels. A representative Doggybone construct (DB GL derived from pGL DOG containing a luciferase cassette) was compared with linear DNA (PCR GL) encoding the same sequence but derived through PCR. The only structural difference is that the Doggybones contain covalently closed telomere ends whereas the PCR products contained open ends. On the native gel the constructs migrated at a similar speed reflecting the similarity in size (2.4 kb for DB GL and 2.7kb for PCR GL) (Fig. 1C). However, on a denaturing gel the Doggybone became a large open single stranded circular structure and thus appeared larger than the equivalent linear PCR construct which became single DNA strands (Fig. 1D). A similar observation was made when a plasmid containing the telRL sites was linearised by protelomerase TelN - resulting in hairpin ended DNA or restriction endonuclease - resulting in open ended DNA (Fig. 1C-D).
The plasmid and Doggybone constructs that were used in subsequent imaging studies, were characterized using dynamic light scattering over a pH gradient. Both plasmid and Doggybone show similar trends and were at their largest (Fig. 1E) and most cationic (Fig. 1F) at low pH. They became more condensed and more negatively charged as the solution became more basic. The two constructs behaved remarkably similarly given the differences in structure and molecular weight.
Comparative expression from Plasmid or Doggybone constructs
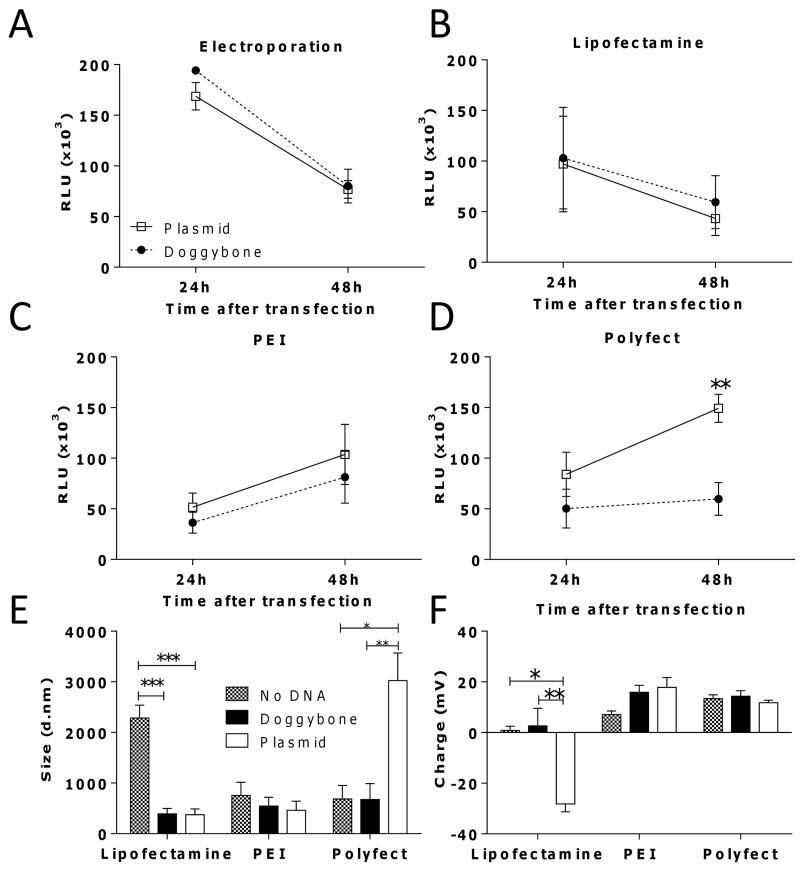
To compare expression levels we made plasmid and Doggybone constructs that encoded the firefly luciferase gene and transfected CHO-K1 cells. In tests to compare delivery of both DNA constructs by electroporation, both the Doggybone and the plasmid were transfected with equal efficiency (Fig. 2A). Due to the novelty of the Doggybone structure we wished to determine whether there were differences in the mechanisms by which transfection reagents package the DNA. We compared a liposome formulation (Lipofectamine), a polycationic complex formulation (PEI) and a branched dendrimer formulation (Polyfect). We reasoned that, having been optimized for circular supercoiled plasmids, the reagents may compact linear DNA constructs differently, leading to complexes with differing physical parameters and capacity to transfect. While Lipofectamine and Polyfect have manufacturer optimized protocols, PEI mediated transfection was optimized to balance nitrogen to phosphate ratios, an 8:1 PEI:DNA ratio gave optimum expression with both constructs (data not depicted). We observed comparable levels of expression of luciferase with Doggybone and plasmid with either Lipofectamine (Fig. 2B) or PEI (Fig. 2C) but when Polyfect was used as the transfection reagent, plasmid DNA had significantly higher transfection efficiency then Doggybone at 48 hours (p<0.01, Fig. 2D). To define why different reagents behaved differently, we measured the size (Fig. 2E) and charge (Fig. 2F) of the DNA-transfection reagent complexes using dynamic light scattering. Lipofectamine-plasmid constructs had a significantly more negative charge than Lipofectamine-Doggybone constructs (p<0.01), but this appears not to alter the capacity to transfect cells, however Polyfect-plasmid constructs were significantly larger (p<0.01) than Polyfect-Doggybone constructs suggesting, for this reagent, the secondary structure of DNA had an important role. We wished to confirm the expression data using a different gene readout system and observed similar transfection efficiency between Doggybone and plasmids expressing the red-fluorescent protein tdTomato (Supp Fig. 1).
Figure 2. Doggybone and plasmid have similar expression levels in vitro.
The efficiency of transfection of 1 μg Doggybone (●) or plasmid (□) encoding luciferase in CHO-K1 using electroporation (A), Lipofectamine (B), PEI (C) or Polyfect (D) was tested. Light emission was recorded as relative light units (RLU).The size (E) and the charge (F) of complexes formed by each transfection reagent at pH 7 was characterised by dynamic light scattering. Bars/points represent mean of n=3 replicates +/− SEM * p<0.05, **p<0.01, ***p<0.001, calculated using ANOVA and Bonferroni’s post test.
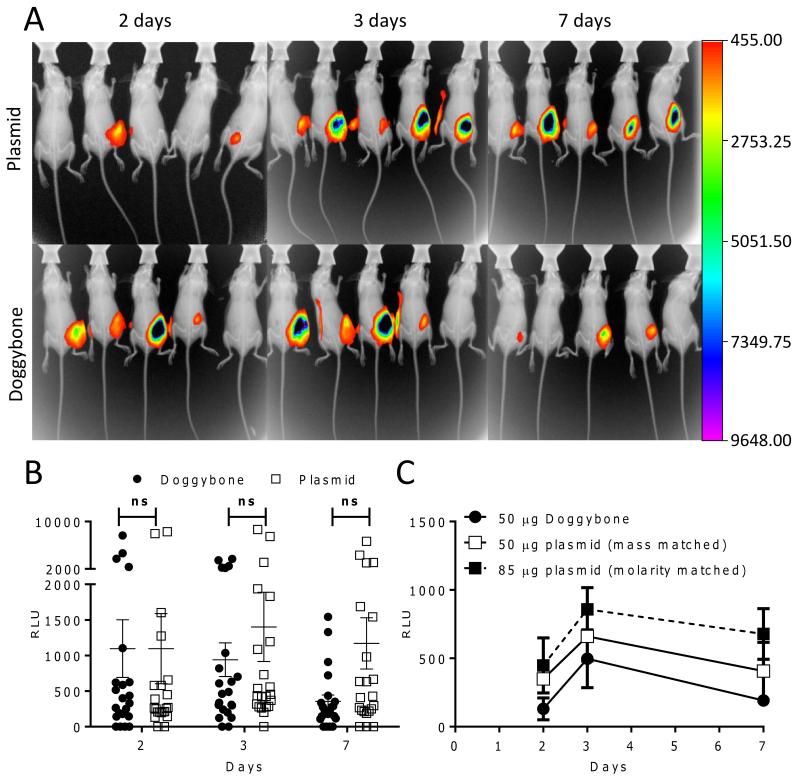
Having validated gene expression from Doggybones in vitro we sought to confirm these observations in vivo. To this end mice were immunized with luciferase-expressing constructs intramuscularly (IM) with electroporation. Light emitted after intraperitoneal luciferin injection was measured by in vivo imaging and images overlaid with X-ray images of the same animals to indicate the position of expression, background photon detection was eliminated by the concurrent imaging of control, untransfected animals (Supp. Fig 2). Luciferase gene expression was detectable in vivo from both the plasmid and the Doggybone constructs (Fig. 3A). Relative expression levels were quantified and there were no significant differences between Doggybone and plasmid (Fig. 3B), however there was a trend for reduced expression from the Doggybone constructs on day 7. Since Doggybones are smaller than plasmid DNA, in a mass-matched study, more gene copies are introduced, therefore we compared mass and molarity matched in vivo luciferase expression from the two constructs (Fig. 3C). There was a trend towards reduced expression in expression after Doggybone transfection compared to the molarity matched plasmid group. In these studies we show that genes were expressed from Doggybones and plasmid at comparable levels in vitro and in vivo.
Figure 3. In vivo Assessment of DNA construct expression levels.
BALB/c mice (n=5) were injected IM with 25 μg μg Doggybone (●) or plasmid (□) encoding luciferase with electroporation. At days 2, 3 and 7 after inoculation expression was visualized using a Carestream in vivo imaging system after intraperitoneal injection of Rediject luciferin, one representative image shown per time point (A). Luciferase levels from n=22 animals were quantified as relative light units using the Carestream analysis software (B), points represent individual mice, line represents mean +/− SEM. Mice were injected with 50μg Doggybone and compared with mass-matched plasmid (50μg) or concentration matched plasmid (85μg) expressing luciferase and expression measured by in vivo imaging over time (C), points represent mean of n=4 mice +/− SEM. ns denotes p>0.05 analysed by ANOVA and Bonferroni’s post test.
Comparative immunogenicity of Plasmid and Doggybone expressed antigens
To test the immunogenicity of Doggybone delivered genes, HIV clade C MWS2 gp140 (the envelope protein gp120 and the transmembrane protein gp41(18)) and haemagluttinin (HA) from H1N1 influenza (A/England/195/2009) were chosen as model antigens. For the HIV gp140 studies we used a prime boost regime of four intramuscular DNA immunizations with electroporation two weeks apart followed by a single intramuscular protein boost. Electroporation was used as it gives a significantly enhanced response to in vivo DNA delivery(19, 20). All immunizations were well tolerated and no adverse reactions were observed.
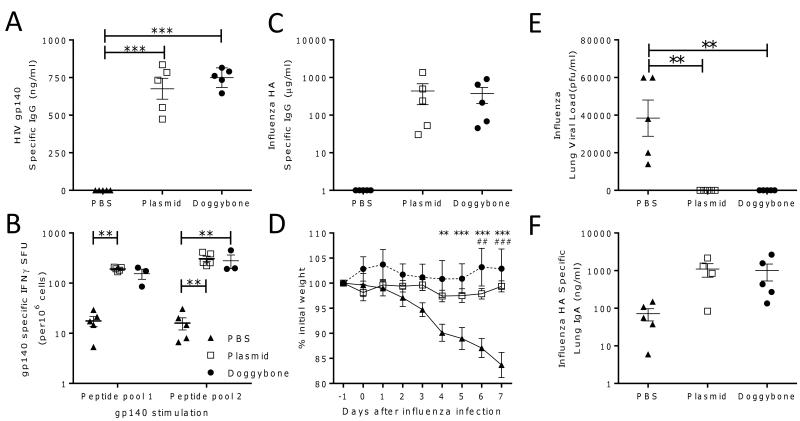
Both Doggybone and plasmid groups developed comparable gp140 specific IgG levels with the group mean at 734 and 675 ng/ml respectively; these were significantly greater than the group receiving protein alone but not significantly different from each other (Fig. 4A). Interestingly, it was only after the protein boost that an antibody response could be detected. Cellular responses against peptide pools comprising peptides derived from the N terminus (pool 1) or C terminus (pool 2) of CN54 gp140 were measured using Interferon γ (IFNγ) secretion as a readout. As with humoral responses, we observed a comparable cellular response to both the Doggybone and the plasmid. The plasmid group had an average of 191 and 306 IFNγ spot forming units (SFU) for peptide pools 1 and 2 respectively whilst the Doggybone group had an average of 154 and 280 SFU (Fig. 4B), there were negligible amounts of cells secreting IFNγ in the PBS primed group, two animals were excluded from the Doggybone group because the ELISPOT wells were too numerous to count.
Figure 4. In vivo immunogenicity assessment of DNA constructs expressing HIV gp140.
BALB/c mice (n=5) were inoculated four times IM with electroporation with 25 μg gp140 encoding plasmid (□), Doggybone (●) or PBS (▲) as a negative control. At day 56 all groups received 10 μg of recombinant gp140 IM. Serum was collected on day 70 and gp140 specific IgG levels were tested using ELISA (A). On day 70 splenocytes were taken and restimulated with synthetic peptides representing the N (pool 1) or C (pool 2) terminus of the gp140 protein, IFNγ was measured using ELISpot (B). BALB/c mice (n≥4) were inoculated IM with electroporation three times with 25 μg influenza HA encoding plasmid (□), Doggybone (●) or PBS (▲) as a negative control, at day 42 mice were infected with 105 PFU H1N1 influenza intranasally. Antibody titre was calculated on d41 prior to infection (C). Weight loss was monitored following infection (D), lung viral load (E), and lung IgA (F) were measured on day 7 after infection. Points represent individual animals, lines represent mean +/− SEM, *p<0.05, **p<0.01, ***p<0.001, analysed by ANOVA and Bonferroni’s post test. For panel D points represent mean of n=5 animals +/− SEM, **p<0.01, ***p<0.001 Doggybone compared to PBS group, ##p<0.01, ###p<0.001, Plasmid compared to PBS group.
DNA constructs expressing influenza haemagluttinin (HA) were tested for their ability to protect mice from challenge with a homologous H1N1 influenza strain (A/England/195/2009). Mice received three intramuscular DNA immunizations with electroporation two weeks apart prior to intranasal infection with H1N1. Pre challenge HA specific IgG could be detected in both the DNA vaccinated groups but not in the control group (Fig. 4C). The Doggybone and plasmid protected animals from weight loss after infection whereas the PBS control group lost significantly more weight at days 6 and 7 after infection (p<0.01, Fig. 4D). Virus could be recovered from the lungs of every mouse in the group receiving PBS whereas no virus could be isolated from the lungs of animals receiving either DNA construct signifying protection (Fig. 4E). After challenge there were higher levels of HA specific lung IgA in the Doggybone and plasmid groups than the PBS control group (Fig. 4F). We show that the Doggybone and the plasmid DNA both induce an immune response to antigen and when used as a vaccine can protect from viral infection.
Discussion
The objective of this study was to compare closed linear DNA generated through enzymatic processes with circular, bacterially derived plasmids, for use as DNA vaccines. In this study we did not observe any difference between the expression levels of luciferase when delivered as a Doggybone or a plasmid. Whilst we studied the role of Doggybones as DNA vaccines, the equivalence in expression levels with plasmid suggest that these constructs could also be used for other gene delivery purposes. There are several factors that contribute to the expression levels from transfected DNA constructs, these include construct size(21, 22), DNA topology with linear DNA transfecting less well than supercoiled plasmid(23), and the number of CpG motifs affecting the time it takes for the immune system to recognize and thus clear the transfected cells(24-26). The equivalence in response between the Doggybone and plasmid reflects the balance between a number of these factors. For example whilst Doggybones contain less DNA per unit weight as plasmids, they occupy the same space in physiological environments probably due to supercoiling of the plasmids. Differences were observed between Polyfect (based upon dendrimers complexing the DNA) and the charged reagents, PEI and Lipofectamine, which may reflect the differences in charge and structure of the plasmid and Doggybone. Another linear minimal construct, the MIDGE vector, also has comparable expression in vitro and in vivo as plasmid DNA(14, 27). Future work will investigate formulating Doggybone to improve expression levels, for example formulating with PEI(28, 29) as has previously been performed with minicircles(30, 31).
We also observed an equivalent immune response to antigens expressed from either Doggybone or plasmid. Using influenza challenge we were able to demonstrate that the immune response to DNA vaccine alone was protective and that there were no differences between plasmid and Doggybone. Immunization induced both a cellular and a humoral response, though for the HIV antigen, protein boosting was required for an antibody response. In part, the poor antibody response to the gp140 vaccination may be due to the ELISA assay which, due to reagent availability, was coated with a different clade C gp140 – CN54 to the vaccine antigen MWS2, reducing the sensitivity. However, it has been previously observed that DNA vaccines induce a better cellular response than humoral response and we observed a strong cellular response in our studies. The use of DNA vaccines in humans has so far had limited success, with poor translation from pre-clinical models to clinical trials. However, the use of electroporation as in the current study has been shown to considerably improve the immunogenicity of a DNA vaccine(19). Increased immunogenicity with electroporated DNA vaccines is associated with increased expression of the delivered DNA(20) and potentially also increased inflammation due to the tissue damage after the application of the electric field(32).
Until now the most promising candidates for minimal DNA constructs were minicircles. These are circular plasmids containing only the gene of interest and the necessary regulatory elements which are excised from a larger parental plasmid grown in bacteria(13). MIDGE vectors are probably the second most prominent candidate, comprised of linear constructs sealed with hairpin loops that are derived from a plasmid with restriction endonucleases and sealed enzymatically(33). While these constructs address many of the concerns outlined by the FDA, such as integration, allergy and the presence of antibiotic resistance genes, the fact that they are still produced in bacterial represents a potential regulatory concern(34). Here we describe a third candidate minimal DNA construct for gene delivery, which does not require a bacterial production step – “Doggybone” DNA. In much the same way as PCR allowed for amplification of specific DNA through the actions of enzymes, the Doggybone process allows for the production, en masse, of GMP standard DNA constructs for use as gene delivery vectors from a template.
Methods
DNA constructs
The proTLx expression plasmid consisted of the CMV Immediate early promoter plus enhancer, a multiple cloning site and an SV40 late polyadenylation signal all flanked by two telRL sequences, the site of protelomerase TelN recognition and cleavage. The plasmid backbone contained an ampicillin resistance gene and the pUC origin of replication. The Luciferase gene from Photinus pyralis (firefly), the gene encoding gp140 from HIV-1 Clade C MSW2 and the haemagluttinin gene from H1N1 Influenza (A/England/195/2009) were cloned (including a GCCACC Kozak sequence) into the HindIII and EcoRI sites on the proTLx base plasmid by restriction digest. Plasmids were maintained in recombination deficient E. coli strains. For use in vivo, plasmid was prepared from E. coli using an EndoFree Giga kit (Qiagen). Recombinant plasmids were verified by restriction endonuclease digestion and sequencing.
Purification of TelN protelomerase
The gene encoding protelomerase TelN, from E. coli phage N15, was cloned into pQE-30 UA (Qiagen) under the control of the IPTG-inducible promoter from phage T5. The enzyme was overproduced in E. coli M15 pREP4 cells (Qiagen), with an N-terminal 6 X Histidine tag, and purified on a HisTrap column (GE Healthcare) using a linear gradient of imidazole (0-500mM) and standard purification techniques. The eluted protein was buffer exchanged into 10mM Tris HCl pH 7.4, 75mM NaCl, 50% (v/v) glycerol, 1mM DTT & 0.1mM EDTA) and stored at a concentration of 15μM at −20°C.
Preparation of closed linear DNA (Doggybones)
Plasmid was prepared using a miniprep kit (Qiagen) and used as a template for Rolling Circle Amplification by the method described in patent EP2391731(16). The template plasmid containing gene of interest flanked by telRL sites, was mixed with custom primers (50μM) and the template denatured by heating to 95°C. To initiate rolling circle amplification from the denatured template, the reaction was first mixed with reaction buffer (30mM Tris-HCl pH 7.4, 5mM (NH4)2SO4, 30mM KCl, 7.5mM MgCl2, 2mM DTT) and then 2mM dNTPs (Bioline) were added together with 2000 units of Phi29 DNA polymerase (Lucigen) and 4 units of thermostable pyrophosphatase (Lucigen). Upon mixing, the reaction was incubated at 30°C for 18 hours with custom primers (2μM) and 2mM dNTPs. In order to produce Doggybones by cleaving at the telRL sites and capping the double stranded ends, the protelomerase TelN (1μM) was added to the reaction and the reaction mixture was incubated at 30°C for10 minutes. The amplified concatamers were processed by adding 2X unit excess of ApaLI restriction enzyme (NEB) and 1.5 Molar excess amounts of the single turnover TelN protelomerase. This cleaved the concatamers to produce Doggybone DNA encompassing the expression cassette containing luciferase, gp140 or HA and a second Doggybone consisting of the backbone components which was also cleaved by ApaLI to produce 3 fragments. These backbone fragments were digested by the addition of 500U/mg DNA exonuclease III. The reaction was monitored until all the backbone fragments had been removed by exonuclease. The Doggybone DNA was purified using Cleanup Maxi columns (A&A Biotechnology, Gdynia, Poland), as manufacturer’s instructions, and eluted into Tris EDTA buffer at pH8 or H2O. The resulting elutions were concentrated using 50 MWCO Amicon columns to a final concentration of 1mg/ml.
DNA Hairpin assay
Two different telRL-containing plasmids were constructed to investigate the ability of TelN to cleave DNA to produce the hairpin ends. The commercially available Luciferase-encoding plasmid pGL4.13 (Promega) was modified to include 2 telRL sequences flanking the Luciferase cassette. It was hereafter named pGL DOG with the Doggybone created from this construct called DB GL. A PCR-amplified version of the pGL DOG cassette (PCR GL) incorporating the telRL-flanked expression region was created. pUC18 telRL was created by the addition of a telRL site into the HindIII and BamHI sites of pUC18 (Thermo Scientific).
Doggybone and linear DNA constructs were run on native and denaturing gels. Native gel 100ng of each sample mixed with 6X gel loading buffer (Sigma) was run on a 0.8% agarose in TAE gel run in 1xTAE. Denaturing gel 500ng of each sample was mixed with 6X denaturing sample buffer (300mM NaOH, 6mM EDTA, 18% Ficoll, 0.15% bromocresol green, 0.25% xylene cyanol). The samples were then run on a 1% agarose (in H2O) at 10V in denaturing running buffer (50mM NaOH, 0.1mM EDTA) overnight on ice to minimize overheating. Once sufficiently separated the gel neutralized for 30 min in 1M Tris HCl pH 7.6, 1.5M NaCl. Both gels were stained for DNA with ethidium bromide and visualized under UV light.
DNA construct characterization by dynamic light scattering
DNA size and charge were calculated using dynamic light scattering on a Zetasizer Nano (Malvern Instruments) in a disposable zeta cell. DNA or DNA complexes were first diluted to 2 μg/ml in filter sterilized degassed water. Samples were read at 25 °C with a back scatter set at 173°, the average of 20 readings in triplicate was taken in each case. Data was analyzed using Malvern instruments Zetasizer software 7.01.
In vitro transfection
CHO-K1 cells were grown to 95% confluence in compete DMEM before being transfected with 1ug of luciferase or tdTomato-expressing plasmid or Doggybone. DNA was delivered by electroporation using a Nucleofector II device with cell line transfection kit T (Lonza) or using the following transfection reagents; Lipofectamine 2000 (Invitrogen), Polyfect (Qiagen) or PEI (Polysciences). Each was used according to supplier’s specification. Luciferase expression: At the appropriate juncture, media was removed, cells were lysed using luciferase lysis buffer (Promega) and frozen for 24 hours. Expression was visualized after the addition of luciferase assay substrate (Promega) and light emission was read using a FluorStar OPTIMA multilablel plate reader (BMG Labtech) and recorded as relative light units (RLU). tdTomato red fluorescent protein expression was imaged using a Nikon Eclipse TE2000-S inverted microscope under bright light and a TxRed filter set (excitation 540-580nm and emission 600-660nm) at 24 and 48 hours after transfection.
Animals
Female BALB/c mice were obtained from Harlan Scientific (Bevil’s Hill, UK) and used at 6-8 weeks of age. All procedures undertaken had been approved by the local ethics review board and performed by personal licensees under the appropriate project license. Experiments carried out were in accordance with Animals (Scientific Procedures) Act 1986.
DNA injections
Mice were injected intramuscularly (IM) into the anterior tibialis with 25μg plasmid or Doggybone in 50μl of sterile PBS followed by electroporation (EP). Two lots of 5 pulses of 150V with switched polarity between pulses were delivered using a CUY21 EDIT system (BEX, Japan). For the HIV gp140 immunisation study, mice received a prime boost regime of four IM DNA immunizations with electroporation two weeks apart followed by a single IM protein boost of CN54 gp140 protein (10 μg). For the influenza study, mice received three IM DNA immunizations with electroporation two weeks apart prior to infection with H1N1.
In vivo imaging
After transfection with plasmid or Doggybone encoding luciferase, expression was visualized using an In vivo FX pro (Carestream, USA) following intraperitoneal injection of Rediject D-Luciferin (Caliper) in accordance with manufacturer’s specification. Light emission was measured for 4 minutes without binning and an X-ray was taken for 30 seconds, the two images were overlaid using Carestream MI SE software. Relative luminescence was quantified by using the software’s region of interest (ROI) analysis function, with background levels set using control, untransfected animals.
Antigen specific ELISA
A quantitative assay was used to determine serum antibody levels adapted from Donelly et al(35). 96-well plates were coated with 1μg/ml gp140 or HA1 (A/England/195/2009, Life Technologies) and blocked with 1% BSA. A dilution series of recombinant murine IgG was used on each plate as a standard to quantify antigen specific antibodies. Sera were diluted 1:500, 1:5,000 and 1: 50,000 to ensure the absorbance reading measured fell within the linear range of the standard curve. Bound IgG was detected by incubation for 2 hr at 37 °C with HRP-conjugated goat anti-mouse IgG (AbD Serotec). Plates were washed and developed with 50 μl TMB/E substrate and the reaction was terminated by the addition of 50 μl of 2M H2SO4 and read at A450. For IgA measurements on lung mash supernatants, a similar protocol was performed using biotinylated anti-IgA and detected using HRP-Streptavidin, compared to a standard curve of recombinant murine IgA.
Cellular Assays
Splenocytes were isolated by mechanical dissociation through sterile nylon mesh, followed by red blood cell lysis with ammonium chloride. The splenocytes were cultured for 72h at 2.5× 105 cells/well in the presence of HIV gp140 CN54 peptides divided into two pools as has previously been described(29) and 2μg/ml anti CD28 (BD Pharmigen). Concanavalin A (ConA, Sigma) and culture media were used as the positive and negative control respectively. ELISPOT assays were performed using a commercial kit from MABTECH (Nacka Strand, Sweden) following the manufacturer’s recommendations. The spots were counted using the AID ELISPOT reader ELR03 (Autoimmune Diagnostika).
Influenza Infection
H1N1 Influenza strain (A/England/195/2009) was grown in Madin-Darby Canine Kidney (MDCK) cells, in serum free DMEM supplemented with 1ug/ml trypsin. The virus was harvested 3 days after inoculation and stored at −80°C. Viral titre and lung viral load determined by plaque assay as previously described(36). Mice were infected intranasally with 5×105 PFU virus. Weight was measured daily to monitor disease severity. The harvesting of lung tissues was carried out as previously described(37). For the preparation of lung mash supernatants, lungs were homogenized through 100 μm cell strainers (BD Pharmingen) and washed through with a 1ml volume of RPMI five times, following centrifugation this supernatant was retained for viral load analysis. After the removal of the supernatants from all tissues, cells were treated with ACK lysing buffer for 5 minutes and they were resuspended in RPMI. Cell viability was assessed by trypan blue exclusion, and total cell numbers were counted by disposable multiwell haemocytometer.
Statistical analysis
The appropriate statistical test was performed using GraphPad prism 5.01 (GraphPad Software Inc, La Jolla, CA, USA). Based on the type of data either a 2 way ANOVA with Bonferroni’s post test or a student’s T-test was performed.
Supplementary Material
Acknowledgements
The authors would like to thank Qin Hu (St George’s University of London) for the MWS2 gp140 gene sequence, Ruth Elderfield and Wendy Barclay (Imperial College London) for the influenza haemagluttinin gene sequence and influenza virus and Paul Rothwell, Jon Extance, Karen Oliver and Kinga Karbowniczek (Touchlight Genetics Ltd) for the generation of the Doggybone material. This work was funded in part by Touchlight Genetics Ltd. and The Exgenomes project under the Seventh Framework program for funding. E.K. is funded by an MRC CASE studentship MR/J006548/1. We are indebted to Dormeur Investment Service Ltd., for funding of equipment used in this project.
Footnotes
Conflict of Interest Statement: The study was funded in part by Touchlight Genetics, who were involved in all stages of the study conduct and analysis. Dr Caproni and Dr Porter are employees of Touchlight Genetics. The corresponding author had final responsibility to submit for publication.
References
- 1.Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–4. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 2.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization. Annual review of immunology. 2000;18:927–74. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 3.Evensen O, Leong JA. DNA vaccines against viral diseases of farmed fish. Fish and shellfish immunology. 2013;35:1751–8. doi: 10.1016/j.fsi.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Liu MA. DNA vaccines: an historical perspective and view to the future. Immunological reviews. 2011;239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferraro B, Morrow MP, Hutnick NA, Shin TH, Lucke CE, Weiner DB. Clinical applications of DNA vaccines: current progress. Clinical infectious diseases. 2011;53:296–302. doi: 10.1093/cid/cir334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klinman DM, Klaschik S, Tross D, Shirota H, Steinhagen F. FDA guidance on prophylactic DNA vaccines: analysis and recommendations. Vaccine. 2010;28:2801–5. doi: 10.1016/j.vaccine.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belakova J, Horynova M, Krupka M, Weigl E, Raska M. DNA vaccines: are they still just a powerful tool for the future? Archivum immunologiae et therapiae experimentalis. 2007;55:387–98. doi: 10.1007/s00005-007-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luke JM, Vincent JM, Du SX, Gerdemann U, Leen AM, Whalen RG, et al. Improved antibiotic-free plasmid vector design by incorporation of transient expression enhancers. Gene therapy. 2011;18:334–43. doi: 10.1038/gt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luke J, Carnes AE, Hodgson CP, Williams JA. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine. 2009;27:6454–9. doi: 10.1016/j.vaccine.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mairhofer J, Cserjan-Puschmann M, Striedner G, Nöbauer K, Razzazi-Fazeli E, Grabherr R. Marker-free plasmids for gene therapeutic applications—Lack of antibiotic resistance gene substantially improves the manufacturing process. Journal of biotechnology. 2010;146:130–7. doi: 10.1016/j.jbiotec.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 11.van der Heijden I, Gomez-Eerland R, van den Berg JH, Oosterhuis K, Schumacher TN, Haanen JB, et al. Transposon leads to contamination of clinical pDNA vaccine. Vaccine. 2013;31:3274–80. doi: 10.1016/j.vaccine.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Prather KJ, Sagar S, Murphy J, Chartrain M. Industrial scale production of plasmid DNA for vaccine and gene therapy: plasmid design, production, and purification. Enzyme and Microbial Technology. 2003;33:865–83. [Google Scholar]
- 13.Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene therapy. 1997;4:1341–9. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- 14.Moreno S, Lopez-Fuertes L, Vila-Coro AJ, Sack F, Smith CA, Konig SA, et al. DNA immunisation with minimalistic expression constructs. Vaccine. 2004;22:1709–16. doi: 10.1016/j.vaccine.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 15.Kay MA, He CY, Chen ZY. A robust system for production of minicircle DNA vectors. Nature biotechnology. 2010;28:1287–9. doi: 10.1038/nbt.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill V, inventor; Touchlight Genetics Ltd, assignee , assignee. Production of Closed Linear DNA2010. 2010 Aug 05;
- 17.Heinrich J, Schultz J, Bosse M, Ziegelin G, Lanka E, Moelling K. Linear closed mini DNA generated by the prokaryotic cleaving-joining enzyme TelN is functional in mammalian cells. Journal of molecular medicine. 2002;80:648–54. doi: 10.1007/s00109-002-0362-2. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Jin W, Hu K, Luo S, Du T, Griffin GE, et al. Highly conserved HIV-1 gp120 glycans proximal to CD4-binding region affect viral infectivity and neutralizing antibody induction. Virology. 2012;423:97–106. doi: 10.1016/j.virol.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Kopycinski J, Cheeseman H, Ashraf A, Gill D, Hayes P, Hannaman D, et al. A DNA-based candidate HIV vaccine delivered via in vivo electroporation induces CD4 responses toward the alpha4beta7-binding V2 loop of HIV gp120 in healthy volunteers. Clinical and vaccine immunology: CVI. 2012;19:1557–9. doi: 10.1128/CVI.00327-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosazza C, Buntz A, Riess T, Woll D, Zumbusch A, Rols MP. Intracellular tracking of single plasmid DNA-particles after delivery by electroporation. Molecular therapy. 2013 doi: 10.1038/mt.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin W, Xiang P, Li Q. Investigations of the effect of DNA size in transient transfection assay using dual luciferase system. Analytical biochemistry. 2005;346:289–94. doi: 10.1016/j.ab.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Zhang G, Ludtke JJ, Thioudellet C, Kleinpeter P, Antoniou M, Herweijer H, et al. Intraarterial delivery of naked plasmid DNA expressing full-length mouse dystrophin in the mdx mouse model of duchenne muscular dystrophy. Human gene therapy. 2004;15:770–82. doi: 10.1089/1043034041648408. [DOI] [PubMed] [Google Scholar]
- 23.von Groll A, Levin Y, Barbosa MC, Ravazzolo AP. Linear DNA low efficiency transfection by liposome can be improved by the use of cationic lipid as charge neutralizer. Biotechnology progress. 2006;22:1220–4. doi: 10.1021/bp060029s. [DOI] [PubMed] [Google Scholar]
- 24.Darquet AM, Rangara R, Kreiss P, Schwartz B, Naimi S, Delaere P, et al. Minicircle: an improved DNA molecule for in vitro and in vivo gene transfer. Gene therapy. 1999;6:209–18. doi: 10.1038/sj.gt.3300816. [DOI] [PubMed] [Google Scholar]
- 25.Madeira C, Rodrigues CA, Reis MS, Ferreira FF, Correia RE, Diogo MM, et al. Nonviral gene delivery to neural stem cells with minicircles by microporation. Biomacromolecules. 2013;14:1379–87. doi: 10.1021/bm400015b. [DOI] [PubMed] [Google Scholar]
- 26.Chen ZY, He CY, Ehrhardt A, Kay MA. Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Molecular therapy. 2003;8:495–500. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 27.Endmann A, Baden M, Weisermann E, Kapp K, Schroff M, Kleuss C, et al. Immune response induced by a linear DNA vector: influence of dose, formulation and route of injection. Vaccine. 2010;28:3642–9. doi: 10.1016/j.vaccine.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 28.Mann JF, McKay PF, Arokiasamy S, Patel RK, Klein K, Shattock RJ. Pulmonary delivery of DNA vaccine constructs using deacylated PEI elicits immune responses and protects against viral challenge infection. Journal of controlled release. 2013 doi: 10.1016/j.jconrel.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann JF, McKay PF, Arokiasamy S, Patel RK, Tregoning JS, Shattock RJ. Mucosal Application of gp140 Encoding DNA Polyplexes to Different Tissues Results in Altered Immunological Outcomes in Mice. PloS one. 2013;8:e67412. doi: 10.1371/journal.pone.0067412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang C, Gao S, Jiang W, Lin S, Du F, Li Z, et al. Targeted minicircle DNA delivery using folate-poly(ethylene glycol)-polyethylenimine as non-viral carrier. Biomaterials. 2010;31:6075–86. doi: 10.1016/j.biomaterials.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 31.Vaysse L, Gregory LG, Harbottle RP, Perouzel E, Tolmachov O, Coutelle C. Nuclear-targeted minicircle to enhance gene transfer with non-viral vectors in vitro and in vivo. The journal of gene medicine. 2006;8:754–63. doi: 10.1002/jgm.883. [DOI] [PubMed] [Google Scholar]
- 32.Hutnick NA, Myles DJ, Bian CB, Muthumani K, Weiner DB. Selected approaches for increasing HIV DNA vaccine immunogenicity in vivo. Current opinion in virology. 2011;1:233–40. doi: 10.1016/j.coviro.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Fuertes L, Perez-Jimenez E, Vila-Coro AJ, Sack F, Moreno S, Konig SA, et al. DNA vaccination with linear minimalistic (MIDGE) vectors confers protection against Leishmania major infection in mice. Vaccine. 2002;21:247–57. doi: 10.1016/s0264-410x(02)00450-4. [DOI] [PubMed] [Google Scholar]
- 34.Smith HA. Regulatory considerations for nucleic acid vaccines. Vaccine. 1994;12:1515–9. doi: 10.1016/0264-410x(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 35.Donnelly L, Curran RM, Tregoning JS, McKay PF, Cole T, Morrow RJ, et al. Intravaginal immunization using the recombinant HIV-1 clade-C trimeric envelope glycoprotein CN54gp140 formulated within lyophilized solid dosage forms. Vaccine. 2011;29:4512–20. doi: 10.1016/j.vaccine.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elleman CJ, Barclay WS. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology. 2004;321:144–53. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Tregoning JS, Yamaguchi Y, Wang B, Mihm D, Harker JA, Bushell ES, et al. Genetic susceptibility to the delayed sequelae of neonatal respiratory syncytial virus infection is MHC dependent. Journal of immunology. 2010;185:5384–91. doi: 10.4049/jimmunol.1001594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.