Abstract
Amyloid fibrils are ordered, insoluble protein aggregates that are associated with neurodegenerative conditions such as Alzheimer’s disease1. The fibrils have a common rod-like core structure, formed from an elongated stack of β-strands, and have a rigidity similar to silk (Young’s modulus of 0.2-14 Gpa)2. They also exhibit high thermal and chemical stability3, and can be assembled in vitro from short synthetic non-disease-related peptides4,5. As a result, they are of significant interest in the development of self-assembled materials for bionanotechnology applications6. Synthetic DNA molecules have previously been used to form intricate structures and organize other materials such as metal nanoparticles7,8, and could in principle be used to nucleate and organize amyloid fibrils. Here we show that DNA origami nanotubes can sheathe amyloid fibrils formed within them. The fibrils are built by modifying the synthetic peptide fragment corresponding to residues 105-115 of the amyloidogenic protein transthyretin (TTR)9, and a DNA origami10 construct is used to form 20-helix DNA nanotubes with sufficient space for the fibrils inside. Once formed, the fibril-filled nanotubes can be organized onto predefined two-dimensional platforms via DNA-DNA hybridization interactions.
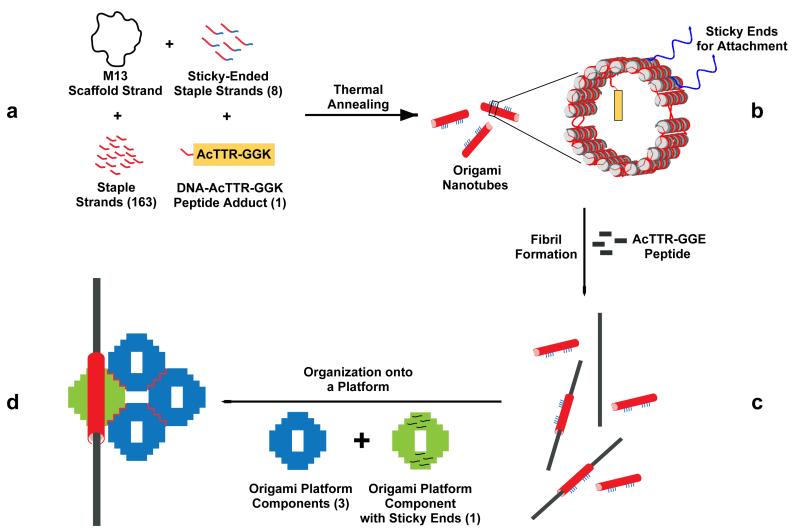
The process of [1] forming fibril-containing DNA nanotubes and [2] organizing them onto DNA origami platforms is summarized in Figure 1. To [1] build the fibril-containing DNA nanotubes, a DNA-peptide conjugate was synthesized by forming a covalent connection between an amyloid fibril peptide and a DNA molecule (Figure 1a). The DNA part of this adduct (green) was designed to behave as a staple strand in the DNA origami nanotube construct while the peptide part (gold rectangle) was designed to nucleate the formation of the amyloid fibril. DNA nanotubes were annealed containing the DNA-peptide conjugate, which protrudes from the inside of the nanotubes (Figure 1b), before mixing with the peptide solution to form amyloid fibrils (Figure 1c). The DNA platform [2] was constructed by joining four planar origami tiles containing sticky ends that form a 2 × 2 planar tile structure. The staple strands at specific sites of the platform and the nanotube (Figure 1b) were designed to hybridize with each other via DNA segments drawn in blue, thereby defining the orientations and locations of the nanotubes on the platform. We have four different designs for fibril organization on the DNA platform: three of them place one fibril on the platform and one of them puts two fibrils on the platform.
Figure 1. The process of organizing amyloid fibrils using DNA origami.
(a) The components are shown: The M13 scaffold strand, 20 sticky-ended staple strands, 151 conventional staple strands and the DNA-peptide adduct are hybridized through a thermal annealing step to yield (b) a 20-helix 2-part closed DNA nanotube containing the peptide on the inside (yellow), the sticky ends (for attachment to the platform (blue) and tube-closure sticky ends (violet). The nanotubes are mixed with AcTTR1-GGE peptide solution to form fibrils sheathed by the nanotubes (c). Platform components with and without sticky ends are added to the sample. Following another annealing step, the final assembly with the fibril organized on the platform is obtained (d).
The DNA origami10 nanotube itself contains 20 DNA double helices; its inner diameter is 16 nm and its outer diameter is ~20 nm. Tilted side views of one section of a 20-helix DNA nanotube are illustrated in Figure 2a. The folding path of one M13 plasmid was designed to form a full tube containing two curved motifs linked on one side; each half-tube unit consists of 10 double helices (See Supplementary Figure 1). Our goal was to approximate a DNA tube with a regular 20-gon cross-section, which should have 162° dihedral angles between faces. Typically, DNA helices are connected to each other via crossover points so the number of nucleotide pairs between crossovers can be used to determine the dihedral angles. Our tube had 9 domains with 15 nucleotides between crossovers, 10 domains with 26 nucleotides between crossovers and 1 domain with 25 nucleotides between crossovers. Assuming 10.5 nucleotide pairs/turn, this generates angles near 154°, 171° and 137° respectively, with an average of exactly 162°. The full tube results from combining these angles to obtain a slightly strained DNA nanotube11. One of the linkages between the two half-tubes has 3 nucleotides of single-stranded DNA to help accommodate the strain in the system, and to allow the tube to unfold and lie flat when it is not sealed shut.
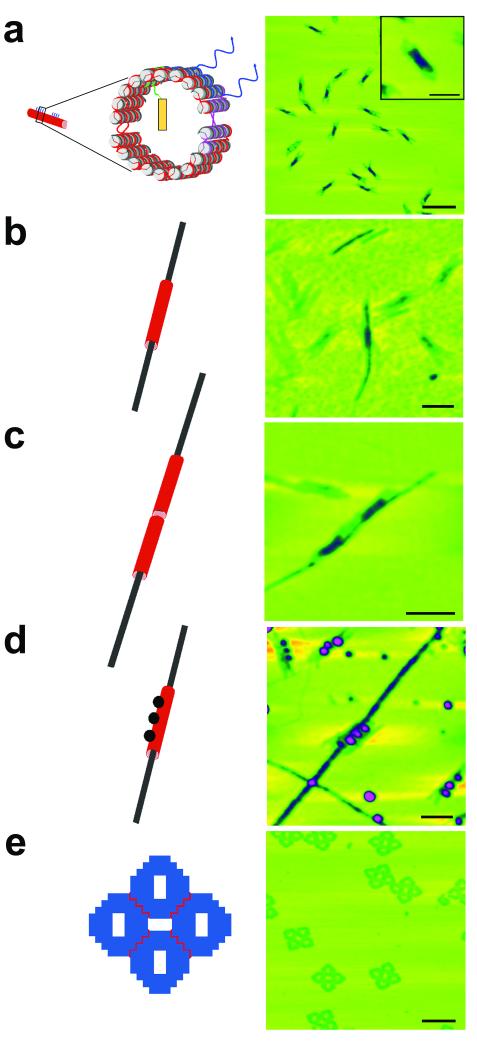
Figure 2. Components of the system.
AFM images on the right correspond to the schematics at left. Maximum height scales are 10 nm. (a) The DNA origami tube is shown. Wavy red lines between halves represent T3 linkers, but there is no gap in the purple segment, held together by sticky ends; see Supplementary Figure 1. The scale bar is 250 nm and 100 nm in the inset. The schematic has been drawn with NanoEngineer-131. (b) A DNA nanotube sheathes an amyloid fibril. The scale bar is 100 nm. Note that the nanotube is much thicker in the color coding. (c) A DNA nanotube pair sheathes a fibril. The pair of tubes surrounds the fibril, as can be seen from the height color coding. The scale bar is 125 nm. (d) A DNA nanotube decorated by a triplet of gold nanoparticles sheathes a fibril. The presence of the gold, which attaches as designed to the nanotube demonstrates that the sheath is DNA. The heights of the particles are prominent in this image. The scale bar is 100 nm. (e) The DNA origami platform. Several four-tile platforms are seen in the image. Their shapes are quite distinct. The scale bar is 250 nm.
Following the mixture and annealing of the components, formation of the closed tube was confirmed by AFM, as shown in Figure 2a. The closed tube formed as designed with uniform dimensions: 163.5 ± 10.6 nm in length, 28.8 ± 3.6 nm (~10 double helices) in width, and 3.1 ± 0.2 nm in height (two helices thick), as estimated by AFM; the helix-to-helix spacing is typically about 3 nm in DNA origami constructs10. The height of the closed tube is double the height of the open tube, while the width of the closed tube is half that of the open tube (Supplementary Figure 2).
We used a short synthetic peptide fragment (105-115) from the TTR sequence to nucleate the formation of amyloid fibrils in vitro9, as the structure of fibrils formed by this peptide has been determined at atomic resolution (0.5 Å) and the packing interactions that promote assembly of protofilaments, filaments and mature fibrils described.12-14 Assembly by the TTR sequence is also robust, amenable to C-terminal modifications that do not change the core structure15,16 and similar TTR-based sequences are able to co-aggregate.15,17,18 Since DNA is a negatively charged molecule, the fibril forming sequence was designed to give negatively charged fibrils (surface zeta potential of −42.8 ± 14 mV (mean ± SD) in ultrapure water) to avoid non-specific interactions between DNA and fibrils. The TTR105-115 peptide was modified by extending it at the C-terminus with a glycine-glycine-glutamic acid (GGE) segment and an acetyl group was added at the N-terminus to yield an AcTTR1-GGE peptide, which forms negatively charged fibrils that have a typical fibril appearance when observed by TEM or AFM (See Figures S3 and S4). The presence of the correctly formed fibrils is shown by X-ray fiber diffraction which indicates a cross-β core structure with an axial reflection at 4.8 Å and an equatorial reflection at 9.0 Å arising from the inter-strand and β-sheet spacing (Supplementary Figure 3). The usual acidic conditions12 in which TTR fibrils form are inappropriate for DNA. We established that the AcTTR1-GGE peptide could form long unbranched fibrils in DNA-compatible conditions at pH 7 after incubation; these fibrils displayed a height of 3.0-4.0 ± 0.4 nm and a width of 16 ± 1 nm (See Figures S3 and S4); these conditions were used in this work.
We made a DNA-peptide conjugate by adding a 5′-carboxy-modifier C10 linker (Glen Research, Sterling VA) to the 5′ end of one of the staple strands. The linker contains an activated carboxylic acid N-hydroxysuccinimide (NHS) ester suitable for conjugation with molecules containing a primary amine, thereby resulting in a stable amide linkage19. Instead of using AcTTR1-GGE peptide for the conjugate molecule, the glutamic acid was replaced in this instance with a lysine residue (GGK) so as to have a primary amine to react with the NHS ester. These sequences share over 93% sequence identity, so are expected to coaggregate20, similar to other TTR1 based fibrils21. A schematic diagram of the reaction to construct the DNA-peptide conjugate is shown in Supplementary Figure 5. After cleavage from the solid support, the product was purified using denaturing polyacrylamide gel electrophoresis and the mass of the target product was confirmed by MALDI-TOF analysis (See Supplementary Figure 5). In addition, we demonstrated that the conjugate could be part of both the nanotube structure and the amyloid fibril (See Figures S5 and S6).
To demonstrate unambiguously that the fibrils really grow inside DNA nanotubes requires several steps. AFM does not distinguish readily between nanotubes that have fibrils within them and nanotubes with fibrils bound on their outer surfaces. Therefore, we have visualized several constructs to demonstrate the growth of fibrils inside the DNA nanotube. In the first case, we have examined fibrils growing inside a single nanotube (Figure 2b). It seems clear that the fibril is growing through both ends of the tube. In a second experiment, we have joined two tubes end-to-end, only one of which contains the nucleating peptide. If the fibrils are growing inside the nanotube, they should still align in the same direction when the nanotube gets longer. This is what we see, as shown in Figure 2c. To establish that the thicker material around the fibril is indeed the DNA nanotube, we have labeled the nanotubes with three oligo-dT-coated 5 nm gold nanoparticles using nanotubes containing three regions of three staple strands each, tailed in oligo-dA on the surface. The three regions of oligo-dA are arranged linearly on the nanotube (See Supplementary Figure 7). Figure 2d illustrates a fibril passing through a nanotube labeled with this arrangement of gold nanoparticles, thereby demonstrating that the sheathing material is the DNA nanotube.
As a DNA surface for use as a DNA platform, we chose a 2D DNA origami array planar monolayer. Figure 2e illustrates AFM images of the DNA platform, which contains four flat DNA origami motifs in a planar arrangement. After the fibrils were encapsulated within the DNA nanotubes, they were directed by hybridization of single strands extending from the nanotubes to bind to specific sites on the DNA platform surface. We designed four different orientations of the nanotubes on the origami platform. Three orientations contain only one nanotube on the platform in different configurations and the last orientation contains two nanotubes on the platform. All designs were tested and confirmed with AFM (See Supplementary Figure 8).
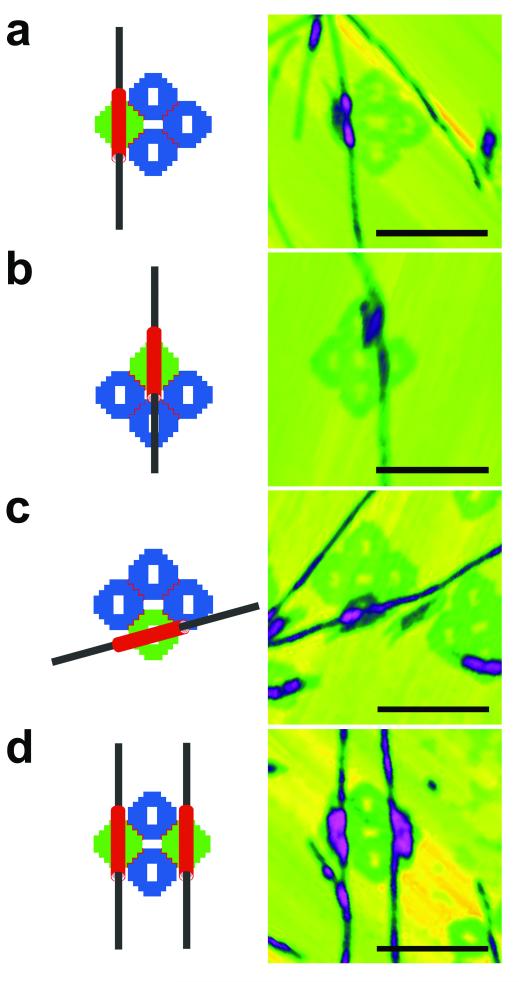
Figure 3 illustrates diagrams and AFM images of the amyloid fibrils organized onto the origami platforms: Along the main axis of one of the side origami units (Figure 3a), along the central axis of the origami platform (Figure 3b), and along a diagonal of a side origami unit (Figure 3c). To obtain these final products, DNA nanotubes with the adduct were prepared and mixed with the AcTTR1-GGE peptide solution to grow fibrils within the nanotubes. After seven days of incubation, the sample was mixed with the individual preformed DNA origami platform and annealed to yield the final assembly before AFM imaging. The images are somewhat imperfect, particularly (b), with the tubes sometimes appearing off-center, not exactly conforming to the design. Supplementary Figure 9 shows examples of centrally directed empty nanotubes that reveal the holes in the origami tile without the presence of the fibril to perturb them; this result suggests that the exact tube position is sensitive to the scanning action of the AFM. Figure 3d contains a diagram and an AFM image of two nanotubes with two amyloid fibrils arranged onto the origami platform. For this arrangement, the nanotubes with the adduct were prepared and arranged onto the platform before mixing with AcTTR1-GGE peptide solution. After seven days of incubation, the sample was analyzed with AFM.
Figure 3. Amyloid fibrils organized onto DNA origami platforms.
Schematic diagrams and AFM images of four different organizations of amyloid fibrils on DNA origami platforms. DNA nanotubes are drawn in red, amyloid fibrils are drawn in black, DNA origami platform components without sticky ends are drawn in blue, and DNA origami platform components with sticky ends are drawn in green. The scale bars are all 250 nm and the maximum heights are 10 nm. (a) The fibril is designed to pass through the left square of the platform. (b) The fibril is designed to pass through the middle of the platform, although the tube moves around a bit. (c) The fibril is designed to pass through an edge tile obliquely. (d) Two tubes are designed to place fibrils through two opposite tiles of the platform, which can be seen clearly.
The approach reported here demonstrates that it is possible to use DNA nanotubes to sheathe rod-like species and arrange them into organized patterns on a DNA surface. It illustrates a new way of organizing amyloid fibrils, which is more specific than using simple charge interactions as demonstrated in previous reports22,23. Other potential guest species include self-assembling peptide systems of similar size and carbon nanotubes, whose growth conditions are not compatible with DNA integrity, and so would have to be captured; by contrast, graphene ribbons apparently can be grown in gentle conditions24 or with the aid of self-assembling peptides,25 possibly enabling the methods used here to be emulated. Interactions between amyloid fibrils and graphene26 also provide further opportunities for hybrid materials. It is evident that yield optimization will be required before the methods reported here can be used routinely to organize peptide nanofibrils, but we have demonstrated clearly that a linear guest species can be sheathed and arranged using cylindrical DNA motifs.
METHODS
Sequence and Structure Design, Synthesis and Purification of DNA
The conjugate and biotin-modified conjugate strands containing 5′-carboxy-modifier C10 (Glen Research) were synthesized on an Applied Biosystems 394 synthesizer without normal cleavage from the support for the conjugation reactions. The sticky-ended sequences have been designed by applying the principles of sequence symmetry minimization7, using program SEQUIN27.
All unmodified oligonucleotides were purchased from IDT (Coralville, IA). All staple strands except sticky-ended and modified strands were used without further purification. The other strands were purified by denaturing gel electrophoresis (PAGE). The bands were cut out of 15-20% denaturing gels and eluted in a solution containing 500 mM ammonium acetate, 11 mM magnesium acetate and 1 mM EDTA. The amount of DNA was estimated by OD260.
Formation of the DNA Origami Nanotube
One-unit nanotube
2.5 μL of 80 nM M13 plasmid (Bayou Biolabs) was combined with staple and sticky-ended strands (1:10 molar ratio of plasmid to staple strands) in a solution containing 40 mM Tris-HCl (pH 8.0), 20 mM acetic acid, 2 mM EDTA, and 12.5 mM magnesium acetate (TAE/Mg2+). The final volume for the system was 100 μL. The system was cooled over 120 minutes from 90 °C to 4 °C, and then kept at 16 °C overnight. The samples were purified by using an Amicon Ultra-0.5 centrifugal filter unit, 100 kD (Millipore)
Two-unit nanotube
Purified front and back-unit nanotube structures were mixed together using a 1:1 molar ratio of plasmid to staple strands with 1:1 molar ratio of plasmid to sticky-ended strands to make 2-unit tube structures. The system was cooled from 40 °C to 25 °C at a −0.6 °C/hour rate.
Formation of the DNA platform
Individual tile
5 μL of 80 nM M13 plasmid (Bayou Biolab) was combined with staple and sticky-ended strands (1:10 molar ratio of plasmid to staple strands) in a solution containing 40 mM Tris-HCl (pH 8.0), 20 mM acetic acid, 2 mM EDTA, and 12.5 mM magnesium acetate (TAE/Mg2+). The final volume for the system was 100 μL. The system was cooled from 90 °C to 4 °C on a thermo-cycling machine over 120 minutes, and then kept at 16 °C overnight. The samples were purified by using an Amicon Ultra-0.5 centrifugal filter unit, 100 kDa (Millipore).
2×2 tile
Purified DNA nanotubes and diamond tiles were mixed together using a 1:1 molar ratio of the DNA nanotubes to the diamond tiles and annealed to form a complete structure in an incubator. The system was cooled from 53 °C to 25 °C at a −0.7 °C/hour rate.
Peptide Synthesis
Peptides were synthesized on Knorr Amide resin (Novabiochem®) using standard Fmoc-Solid-phase Peptide Synthesis chemistry28 on a Liberty microwave peptide synthesizer (CEM Corporation).
Fibril Formation with DNA Nanotubes
Peptides were dissolved at 8 mM in 10% (v/v) acetonitrile in ddH2O and adjusted to pH 7. The peptide solution was mixed with 2 nM DNA nanotube in 1x TAE/Mg2+ in a 1:1 v/v ratio. The samples were incubated at 30 °C for 24 hours and then left at room temperature for 7 days.
Fibril X-ray Fiber Diffraction
An X-ray fiber diffraction pattern was collected and analyzed for a dried and aligned stalk of Ac-TTR1-GGE fibrils as described previously29 on the Macromolecular Crystallography beamline at the Australian Synchrotron, Victoria, Australia30. Diffraction patterns were acquired with a sample-to-detector distance of 300 mm and a wavelength of 0.95363 Å.
Fibril ζ-potential Measurements
The ζ-potential of AcTTR1-GGE fibrils was determined with a Malvern ZetaSizer Nano (Malvern Instruments) by taking the average of four samples of fibrils at a final concentration of 20 μg/ml in ultrapure water at pH 7.0 using a disposable zeta cell. Data were then analyzed using the Malvern Instrument Dispersion Technology Software v. 5.03.
Conjugation Reaction
Resin-bound NHS-activated oligonucleotide was treated with 10 eq of AcTTR1-GGK peptide, AcYTIAALLSPYSGGK, predissolved in 1:9 triethylamine:dichloromethane19. The reaction vessel was shaken at room temperature; after for 4 hours, the resin was washed with dichloromethane and air dried. Oligonucleotides were deprotected and cleaved from the support with ammonium hydroxide at room temperature.
Preparation of Thiolated DNA
The disulfide bond in the thiolated DNA was reduced by adding 200 mM tris (2-carboxyethyl) phosphine (TCEP) solution into DNA solution using a 1:1 volume/volume ratio and incubated at room temperature overnight. The unreacted TCEP was removed using a G25 spin column (Pharmacia).
Phosphination and Concentration of the Gold Nanoparticles
Citrate ion stabilized gold nanoparticles (5 nm, Ted Pella Inc.) were stabilized with adsorption of Bis (p-sulfonatophenyl) phenylphosphine dihydrate dipotassium salt (BSPP, Strem Chemicals Inc.). BSPP (20 mg) was added to the colloidal nanoparticle solution (50 mL) and the mixture was shaken overnight at room temperature for ligand exchange. The mixture was concentrated up to the micromolar range after phosphine coating and the concentration was determined by measuring the absorbance at 520 nm.
Preparation of the Single-Stranded DNA Covered Gold Nanoparticles
Gold-DNA conjugates were prepared by mixing gold nanoparticles (5 nm, Ted Pella Inc.) with 3′ end-thiolated (-SH) ssDNA with a molar ratio of 1:100 and incubated in 0.5x TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8) containing 50 mM NaCl overnight at room temperature.
Gold Nanoparticle Binding
Preformed DNA nanotubes were mixed with ssDNA-covered gold nanoparticles using a 1:5 molar ratio of the nanotubes to ssDNA-covered gold nanoparticles. The mixture was cooled down from 40 °C to 25 °C at a rate of −0.6 °C/hour.
Atomic Force Microscopy (tapping in air)
For DNA samples
5 μL of the annealed sample was spotted on freshly cleaved mica (Ted Pella, Inc.) and left to adsorb for 2 minutes. The mica was washed with double-distilled water (ddH2O) three times. The mica was then air-dried with compressed air before being mounted onto the microscope. The imaging was performed in the tapping-in-air mode on a NanoScope IV (Digital Instruments), employing commercial cantilevers with Si3N4 tips for the air mode.
For negatively-charged fibril samples
A freshly cleaved mica surface (Ted Pella, Inc.) was treated with poly-L-lysine (1 mg/mL) before sample deposition. 5 μL of the annealed sample was spotted onto poly-L-lysine modified mica and left to adsorb for 2 minutes. The sample was washed with ddH2O 3 times. Then the sample was air-dried before being mounted on to the microscope. The imaging was performed in the tapping-in-air mode using a NanoScope IV (Digital Instruments), with commercial cantilevers with Si3N4 tips for air mode.
Supplementary Material
Acknowledgements
This research has been supported by the following grants to NCS: GM-29554 from NIGMS, grants CMMI-1120890 and CCF-1117210 from the NSF, MURI W911NF-11-1-0024 from ARO, grants N000141110729 and N000140911118 from ONR. MNB was supported by an Australian Nanotechnology Network Overseas Travel Fellowship and the Bio21 Institute. Research carried out in part at the Center for Functional Nanomaterials, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DEAC02-98CH10886.
Footnotes
Supplementary Information accompanies this paper at www.nature.com/naturenanotechnology/. Reprints and permission information is available online at http://npg.nature.com/reprintsand permissions/. Correspondence and requests for materials should be addressed to NCS and SLG.
Author Contributions. AU did experiments and wrote the paper. MNB did experiments and wrote the paper. RJS did experiments and wrote the paper. WBS did experiments and wrote the paper. TW did experiments and wrote the paper. PSA co-designed peptide conjugation experiments and wrote the paper. JWC wrote the paper. SLG conceived of and directed the project and wrote the paper. NCS conceived of and directed the project and wrote the paper.
References
- 1.Tan SY, Pepys MB. Amyloidosis. Histopathology. 1994;25:403–414. doi: 10.1111/j.1365-2559.1994.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 2.Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nature Nanotech. 2011;6:469–479. doi: 10.1038/nnano.2011.102. [DOI] [PubMed] [Google Scholar]
- 3.Mesquida P, Riener CK, MacPhee CE, McKendry RA. Morphology and mechanical stability of amyloid-like peptide fibrils. J. Mater. Sci. Mater. Med. 2007;18:1325–1331. doi: 10.1007/s10856-006-0075-0. [DOI] [PubMed] [Google Scholar]
- 4.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Amyloid fibril formation by an SH3 domain. Proc. Natl Acad. Sci. USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti F, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gras SL. Amyloid fibrils: From disease to design. New biomaterial applications for self-assembling cross-beta fibrils. Australian J. Chem. 2007;60:333–342. [Google Scholar]
- 7.Seeman NC. Nucleic acid junctions and lattice. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 8.Seeman NC. Nanomaterials based on DNA. Ann. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gustavsson A, Engstrom U, Westermark P. Normal transthyretin and synthetic transthyretin fragments form amyloid-like fibrils in vitro. Biochem. Biophys. Res. Commun. 1991;175:1159–1164. doi: 10.1016/0006-291x(91)91687-8. [DOI] [PubMed] [Google Scholar]
- 10.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 11.Sherman WB, Seeman NC. Design of minimally strained nucleic acid nanotubes. Biophys. J. 2006;90:4546–4547. doi: 10.1529/biophysj.105.080390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzpatrick AWP, et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl Acad. Sci. USA. 2013;110:5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc. Natl Acad. Sci. USA. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporini MA, et al. Accurate determination of interstrand distances and alignment in amyloid fibrils by magic angle spinning NMR. J. Phys. Chem. B. 2010;114:13555–13561. doi: 10.1021/jp106675h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bongiovanni MN, Puri D, Goldie KN, Gras SL. Noncore residues influence the kinetics of functional TTR105–115-based amyloid fibril assembly. J. Mol. Biol. 2012;421:256–269. doi: 10.1016/j.jmb.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 16.Bongiovanni MN, Caruso F, Gras SL. Lysine functionalised amyloid fibrils: the design and assembly of a TTR1-based peptide. Soft Matter. 2013;9:3315–3330. [Google Scholar]
- 17.Gras SL, et al. Functionalized amyloid fibrils for roles in cell adhesion. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 18.MacPhee CE, Dobson CM. Formation of mixed fibrils demonstrates the generic nature and potential utility of amyloid nanostructures. J. Am. Chem.Soc. 2000;122:12707–12713. [Google Scholar]
- 19.Glen Report 15.1, A novel route to activated carboxylate modified oligonucleotides.
- 20.Wright CF, Teichmann SA, Clark J, Dobson CM. The importance of sequence diversity to the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 21.Bongiovanni MN, Puri D, Goldie KN, Gras SL. Noncore residues influence the kinetics of functional TTR105-115-based amyloid fibril assembly. J. Mol. Biol. 2012;421:256–269. doi: 10.1016/j.jmb.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Mesquida P, Ammann DL, MacPhee CE, McKendry RA. Microarrays of peptide fibrils created by electrostatically controlled deposition. Adv. Mater. 2005;17:893–897. [Google Scholar]
- 23.Mesquida P, Blanco EM, McKendry RA. Patterning amyloid peptide fibrils by AFM charge writing. Langmuir. 2006;22:9089–9091. doi: 10.1021/la061485t. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, et al. Two-dimensional graphene nanoribbons. J. Am. Chem. Soc. 2008;130:4216–4217. doi: 10.1021/ja710234t. [DOI] [PubMed] [Google Scholar]
- 25.Han TH, et al. Peptide/graphene hybrid assembly into core/shell nanowires. Adv. Mater. 2010;22:2060–2064. doi: 10.1002/adma.200903221. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Adamcik J, Raffaele Mezzenga R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nature Nanotech. 2012;7:421–427. doi: 10.1038/nnano.2012.62. [DOI] [PubMed] [Google Scholar]
- 27.Seeman NC. De novo design of sequences for nucleic acid structure engineering. J. Biomol. Struct. Dyn. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 28.Merrifield RB. Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J. Am. Chem. Soc. 1963;85:2149–2154. [Google Scholar]
- 29.Gras SL, et al. Functionalized amyloid fibrils for roles in cell adhesion. Biomaterials. 2008;29:1553–1562. doi: 10.1016/j.biomaterials.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 30.McPhillips TM, et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Rad. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 31. http://www.nanoengineer-1.com/content/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.