Abstract.
Optical mapping (OM) of cardiac electrical activity conventionally collects information from a three-dimensional (3-D) surface as a two-dimensional (2-D) projection map. When applied to measurements of the embryonic heart, this method ignores the substantial and complex curvature of the heart surface, resulting in significant errors when calculating conduction velocity, an important electrophysiological parameter. Optical coherence tomography (OCT) is capable of imaging the 3-D structure of the embryonic heart and accurately characterizing the surface topology. We demonstrate an integrated OCT/OM imaging system capable of simultaneous conduction mapping and 3-D structural imaging. From these multimodal data, we obtained 3-D activation maps and corrected conduction velocity maps of early embryonic quail hearts. 3-D correction eliminates underestimation bias in 2-D conduction velocity measurements, therefore enabling more accurate measurements with less experimental variability. The integrated system will also open the door to correlate the structure and electrophysiology, thereby improving our understanding of heart development.
Keywords: optical mapping, optical coherence tomography, cardiac electrophysiology, 3-D imaging, cardiac development, multimodel imaging
1. Introduction
Coordinated electrical activation and propagation play an important role as a heart develops from a linear tube to a complicated four-chambered form, not only for initiating rhythmic contractions of cardiac myocytes for efficient blood pumping, but also for maintaining normal cardiac development.1–3 The developmental status of the heterogeneous conduction system in the embryonic heart can be characterized by parameters such as action potential (AP) morphology, activation sequence, and conduction velocity. However, due to technology limitations, there has been a very limited number of studies of embryonic heart development that directly measure these electrophysiological parameters.4–14 Among the reported studies, the more quantitative parameter, conduction velocity, has seldom been reported.7–11 Studies of murine and zebrafish knockout models have shown that the absence of or reduced protein expression of certain connexins in the developing heart can both modify conduction velocity and lead to congenital heart defects (CHDs), such as ventricular septal defects and conotruncal heart defects.3,15,16 These types of CHDs are relatively common and are sometimes life-threatening in humans. As a result of limitations in technology to accurately measure conduction velocity, the mechanisms and the interplay between electrophysiology and heart structure are still poorly understood.
Measuring the electrophysiology of the early embryonic heart is very challenging compared to the adult heart due to the much smaller size. Yet, optical mapping (OM) has shown a great potential for studying the electrophysiology of the embryonic heart. OM uses a voltage-sensitive fluorescent dye, an excitation light source, an appropriate filter set, and a fast camera to image transmembrane potentials of the heart with high-spatial resolution over a large field of view. Compared to electrode recording, OM enables the simultaneous collection of signals from hundreds to thousands of sites, and can be used to measure the same parameters (e.g., AP morphology, activation sequence, and conduction velocity), while avoiding direct contact with delicate tissues. OM has made an enormous impact on the study of both adult and developmental cardiac electrophysiology and has advanced the understanding of mechanisms of electrical propagation.4,6,13,17–20
One limitation of OM is that the technique collects information from a three-dimensional (3-D) surface as a two-dimensional (2-D) projection map. While AP morphology analysis and general activation sequence analysis are not affected by this projection, ignoring the curvature of the heart surface results in errors when calculating conduction velocity. Curvature is especially pronounced in early stage tubular hearts and can vary greatly at different developmental stages and between individual embryos at similar stages. More importantly, in some disease models, affected embryonic hearts may differ from healthy hearts in size and curvature (e.g., abnormal looping). Efforts have been made to address this problem in adult heart models. One method is to immobilize and flatten the surface of the heart against a glass plate to reduce curvature.21 This is not applicable to embryo models because embryonic hearts are fragile and very small (). Another example is panoramic OM, which uses a rotating geometry camera to capture the surface shape of the heart.22–24 However, because embryonic hearts are very small and the heart curvature is much more complicated than the cone-shaped adult heart, this solution is also not well suited. Therefore, at present, estimation of conduction velocity in embryonic hearts based on projected 2-D mapping is difficult and error-prone. This problem complicates any comparisons made between different hearts and even between different regions of the same heart. As a result of these difficulties, measured conduction velocities of early embryonic hearts utilizing OM have been seldom reported, and the reported measurements are estimations without considering the 3-D curvature of the heart.7–11 Thus, an improved method, which can take the 3-D curvature into consideration, is needed to obtain accurate and reproducible conduction velocity measurements.
Optical coherence tomography (OCT) is a powerful noninvasive imaging modality capable of characterizing the 3-D structure of very small biological specimens. Since OCT imaging can provide high resolution (2–30 μm), sufficient tissue penetration (1–3 mm), adequate field of view, and extremely high speed, it is well suited for structural and functional imaging of models of cardiac development both in vivo and ex vivo.25–33 OCT is particularly suitable for accurately measuring the 3-D morphology of the tubular early embryonic heart.25,26,32 Furthermore, OCT uses infrared light, which does not interfere with the visible light used for OM, and it does not require special preparation or orientation of the sample. In combination with OM, OCT can provide the 3-D structural data needed to accurately measure conduction velocity in the presence of complicated curvature.
Here, we present an integrated OCT and OM imaging system capable of simultaneously imaging structure and conduction signals. Registering these two datasets enables display of the activation map overlaid on the 3-D surface of the heart, providing a more accurate representation. Furthermore, we demonstrate corrected conduction velocity measurements in looping embryonic quail hearts based on 3-D surfaces provided by OCT. Looping embryonic hearts are undergoing important structural and electrophysiological transitions, such as the initiation and development of the atrioventricular junction (AVJ) and atrioventricular (AV) delay.2,34 Also, the structure of the heart is becoming more convoluted as it loops from a C- to an S-shape.27,32,35 3-D-corrected conduction velocity calculations will eliminate the underestimation of conduction velocity inherent to 2-D OM and therefore provide more accurate measurements. Applying this new technology in future cardiac electrophysiological studies will greatly benefit our understanding of cardiac conduction system development.
2. Material and Methods
2.1. Embryonic Heart Preparation
Fertilized quail eggs (Coturnix coturnix, Boyd’s Bird Company, Inc., Pullman, Washington) were incubated in a humidified, forced draft incubator (G.Q.F. Manufacturing Co., Savannah, Georgia) at 38°C for 50 h. Three Hamburger–Hamilton (HH) stage 15 quail embryos36,37 were removed from their eggs. The embryonic hearts were excised and stained with 10 μM of di-4-ANEPPS (Life Technologies, Carlsbad, California) in 300 μL Tyrode’s solution (Sigma-Aldrich, St. Louis, Missouri) for 12 min at room temperature. The embryonic hearts were then placed in the imaging chamber filled with 1 mL of Tyrode’s solution containing 10 μM of cytochalasin D (Sigma-Aldrich, St. Louis, Missouri) to eliminate myocardial contraction. The experimental environment was maintained at 38°C during the subsequent imaging steps.
2.2. Integrated Optical Coherence Tomography/Optical Mapping System
To image both the structure and electrical activity, we developed an integrated OCT/OM system (the schematic diagram is in Fig. 1). The custom-built OM system14 utilized a broadband light-emitting diode light source (SOLA Light Engine, Lumencor, Beaverton, Oregon), which provides high intensity, low noise, and broadband illumination. A filter and dichroic mirror set (Chroma Technology, Bellows Falls, Vermont) was used with an excitation wavelength passband of , an emission wavelength passband of , and a dichroic mirror with cutoff wavelength of 560 nm. A low noise, high dynamic range EMCCD camera (iXon3 860, Andor Technology, South Windsor, Connecticut), and an upright microscope (Axio Scope.A1, Carl Zeiss AG, Oberkochen, Germany) were used for signal recording. Demagnifying optics (0.33X, 0.5X, or 0.63X) were utilized to control the field of view. The OM system records pixels at a maximum frame rate of 500 Hz. Depending on the field of view required by the sample, either a 10X objective lens (, C-Apochromat 10X/0.45 M27, Carl Zeiss AG, Oberkochen, Germany) or a 5X objective lens (, Fluar 5X/0.25 M27, Carl Zeiss AG) was employed. The OM imaging system was placed above an imaging chamber, which uses a coverslip as the bottom to hold the sample.
Fig. 1.
Schematic diagram of the integrated OCT/OM system. SLD: superluminescent diode. LED: light-emitting diode.
The OCT portion of the integrated system was a custom-built spectral domain OCT system38 with a compact scanner located under the sample stage. The OCT light source has a center wavelength at 1310 nm and a full-width at half-maximum bandwidth of 75 nm. The axial and lateral resolutions of the system were both in air. This system utilizes a linear-in-wavenumber spectrometer as previously described.38 The line scan camera (Sensors Unlimited, Princeton, New Jersey) in the OCT spectrometer reads out at 47 kHz. The OCT system images through the transparent coverslip bottom of the imaging chamber and the tissue to acquire full 3-D structures. The OCT scanner was tilted to reduce specular reflections.
The entire system was enclosed in a box to avoid ambient light exposure and to maintain the temperature during experiments. Two 50 W heaters (McMaster-Carr, Aurora, Ohio) were used for heating and the temperature was monitored by a temperature controller (TC-344B, Warner Instruments, Hamden, Connecticut) with the sensing tip in the solution near the sample. An OCT volume (, 1.5 mm; , 1.5 mm) and 4 s of continuous OM images were recorded at the same time from each sample heart.
2.3. Image Processing and Data Analysis
Image processing and data analysis were performed with custom software written in MATLAB® (MathWorks, Natick, Massachusetts). A cumulative normal distribution function was fit to the upstroke of each AP trace. Activation time for each pixel was determined as the inflection point of the cumulative normal distribution function. This corresponds to the middle point of the AP upstroke, which is conventionally used as the activation time. An electrical activation map was obtained based on the activation time at each pixel.
OCT images were corrected for aspect ratio and then registered with OM images. Because the OCT scanner was tilted to reduce specular reflections, the orientation of the sample in the OCT images and the OM images was not identical. The sample stage glass bottom is parallel to the OM imaging plane and is clearly visible in the OCT images; therefore, it was used as a reference to rotate the OCT image volume to match the sample orientation in the OM images as the first step in the 3-D–2-D registration process. Next, the OCT volume images of the heart were projected parallel to the OM image plane. This OCT en face projection image was registered to one of the OM images using a fully automatic partial affine transformation to match the size, orientation, and position.39 The transformation parameters were two-scale parameters (x and y directions), two translation parameters (x and y directions), and one in-plane rotation parameter. The OCT volume images were then transformed accordingly based on the transformation matrix. The OCT volume images were autosegmented using Amira (Visualization Sciences Group, FEI) to generate binary masks for surface renderings. The activation map was then projected to the 3-D volume mask for geometrically accurate visualization.
2.4. Three-Dimensional Conduction Velocity Correction Algorithm
The binary masks were also used to create a heart surface height map from the top surface, which was used to correct conduction velocity calculations. We followed conventional methods for calculating the 2-D ( and ) conduction velocity component vectors,40 then used the OCT data to determine the third () velocity component vector (Fig. 2). From the OM activation map, 2-D conduction velocities were calculated based on the activation time () gradient vectors in the and directions which are defined as
| (1) |
Fig. 2.
Diagram illustrating velocity calculations with the conventional 2-D projection method (top) and 3-D corrected method (bottom). Fluorescent signals were collected from a 3-D heart surface and projected to a 2-D plane. This diagram uses a small region on the heart surface to illustrate the process. In the conventional 2-D calculation, only velocity components in the projected plane () were calculated. In the 3-D corrected calculation, 3-D structure was used to find the normal vector () by Newell’s method. Then, the velocity component in the height dimension () was calculated and finally 3-D conduction velocity () was computed.
The default window size for calculating gradients was pixels and the window size was adaptively enlarged for increasing conduction velocity. A high conduction velocity means a low-activation time gradient, which generally requires a larger window size for accurate measurement. The velocity vector has the same direction as the gradient vector and its magnitude is the reciprocal of the gradient magnitude . As described previously,40 magnitudes and were derived as
| (2) |
3-D correction of the activation map was performed by projecting the 2-D activation map from OM onto the matched 3-D surface height map acquired from OCT, so that each OM pixel was assigned an associated -position value. For each pixel of the height map, its four neighboring pixels were used to estimate the vector normal to the heart surface, , by Newell’s method.41 The magnitude of the velocity in the z direction () was derived from and the known velocity component vector magnitudes and using the assumption that the direction of the 3-D conduction velocity vector is perpendicular to the surface normal direction. This assumption is valid because the myocardium of HH stage 15 embryonic hearts is only two cell layers thick;1 therefore, the conduction must travel along the heart surface. Therefore,
| (3) |
It follows that the 3-D-corrected conduction velocity at each pixel is . At each pixel in the region of interest, the 3-D-corrected conduction velocity was calculated, and the magnitude was mapped to the corresponding voxel on the 3-D OCT surface. Figure 2 uses a pixels area on the 3-D heart surface to illustrate the correction process for 3-D conduction velocity.
2.5. Validation and Evaluation
The ability to accurately measure sample orientation from OCT volumes was validated by imaging a plastic slide at various known angles. The plastic slide was oriented at preset angles (0 deg to 25 deg at 5 deg increments) and imaged with OCT. At each angle, a volume was recorded by OCT, and the orientation was determined from the normal that was computed using Newell’s method, as described above. Each angle was measured 15 times to estimate the error.
Quantitative comparisons were made between 2-D (uncorrected) and 3-D (corrected) conduction velocity maps. To evaluate the amount of correction from 2-D to 3-D-corrected conduction velocity, the relative difference (between 2-D and 3-D) at each pixel was calculated as . The result represents the necessary percent correction from 2-D to 3-D. A conduction velocity correction map was generated for each embryonic heart.
Direct validation of accurate conduction velocity is impossible because there is no gold standard method, and none of the existing methods take 3-D distances into consideration. Therefore, 2-D and 3-D calculation results were used as a self-validation. 2-D projection can cause nonuniform underestimations because of local structure differences. Therefore, the 2-D method can potentially produce artifacts that are not physiologically valid and the 3-D-corrected method should be able to mitigate these artifacts. Two regions that were expected to have the same conduction velocities but different slopes were selected for comparison. Each region was 109 pixels in size, and their uncorrected and corrected conduction velocities () were compared using Student’s -test. Statistical significance was set at a -value less than 0.05.
3. Results
The OCT en face projections were registered to OM image sets to accurately match the position, orientation, and scale, as described in Sec. 2.3. As shown in Fig. 3, the automatic partial affine transformation adequately registers the two image sets.
Fig. 3.
OCT/OM registration. Panel (a) shows an OCT en face projection grayscale image of a HH stage 15 embryonic heart before being registered to an OM image. The green contour represents the boundary segmentation of the OCT en face projection and the magenta contour represents the boundary segmentation of the corresponding OM image overlaid on the OCT en face projection. Panel (b) shows an OM image which served as the reference for image registration. After registration, boundary contours of OCT (green) and OM (magenta) images can be seen to correspond well.
Figure 4 shows the OCT measured angles versus actual angles from the plastic slide experiment to verify OCT measurement of surface orientation. Angles were accurately measured and the errors were all within .
Fig. 4.
Results of OCT measuring various orientations of a flat surface using Newell’s method. Error bars inside each circle represent standard deviation from 15 repeated measurements.
An AP trace was recorded at each pixel in an OM image. Representative AP traces from three different regions of the heart are shown in Fig. 5(a). Typically, there are 6–8 heartbeats during each recorded dataset. Two heartbeats in an embryonic heart were selected for illustration in Fig. 5(a). The activation time (defined as the midpoint of the upstroke) was determined at each pixel (red marks on the traces in Fig. 5(a). An activation isochrone map which displays the activation time at every pixel is shown in Fig. 5(b). The AVJ and the outflow tract exhibit denser isochrones, meaning slower propagation of APs, compared to the ventricular region, which displays sparser isochrones and higher conduction velocity. The same activation isochrone map is projected and overlaid onto the 3-D OCT surface rendering of the same embryonic heart in Fig. 5(c). The geometrically correct activation map can now be visualized in the context of 3-D features, such as the surface curvature and the looping of the heart.
Fig. 5.
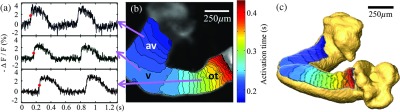
Panel (a) shows raw AP traces of three different pixels from three different regions of the heart tube, indicated by arrows from panel (b). represents the percent change in raw fluorescence signals. The red dot in each trace represents the activation time. The 0 s on the time axis is arbitrary, but chosen to show two complete cycles in each trace. The time scale is the same in each trace; therefore, the activation time delay between traces is preserved. Panel (b) shows a 2-D electrical activation map overlaid on a grayscale OM image. Color map blue to red represents the sequence of activation from early to late. Each isochrone represents 10 ms. Panel (c) shows the activation map overlaid on the 3-D OCT volume surface rendering. This panel shares the same color map as Panel (b). av- atrioventricular junction; v-ventricle; and ot-outflow tract.
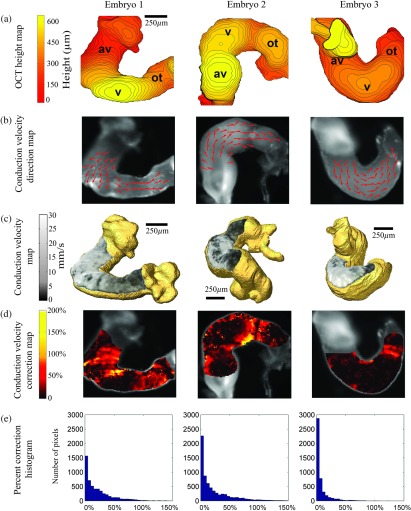
In order to compute 3-D conduction velocities from 2-D projections, the position of each OM pixel was determined from a co-registered height map generated from the corresponding OCT surface rendering. A total of three embryonic hearts were imaged, and their height maps, conduction direction quiver maps, 3-D-corrected conduction velocity maps, conduction velocity correction maps, and percent correction histograms are shown in Fig. 6. Height maps [Fig. 6(a)] were computed from 3-D OCT volumes after being registered to OM images. From the OCT height maps, the orientation of each heart can be clearly distingushed. Heart 2 was imaged from the ventral side, while hearts 1 and 3 were imaged from the dorsal side but from different angles. Because of orientation differences, the curvatures apparent in the three hearts are not identical. For example, heart 1 shows steep slopes at the AVJ and moderate slopes at the outflow tract. Heart 2 has mostly low slopes at the AVJ, the ventricle and the outflow tract, while slopes are steep between these regions. In heart 3, the AVJ is mostly obscured due to the orientation of the heart, while the ventricular region is flat and the outflow tract slopes moderately. Quiver maps were generated to indicate the propagation direction of electrical conduction [Fig. 6(b)]. The overall conduction direction was parallel to the heart tube, as expected.
Fig. 6.
Three embryonic hearts were imaged and processed. Panel (a) shows the surface height maps acquired from volumetric OCT images. The relative height of each pixel in microns is represented by the color map and each contour line represents 15 μm. Panel (b) shows quiver maps of conduction direction through the hearts. Normalized and binned arrow heads indicate the directionality only. Panel (c) shows 3-D-corrected conduction velocity maps overlaid on the surfaces of 3-D OCT volume rendering. Panel (d) shows conduction velocity magnitude correction maps indicating percent correction from the 2-D values to the 3-D-corrected values. Panel (e) shows histograms of the necessary correction amount needed for each heart with bins of 5%. av- atrioventricular junction; v-ventricle; and ot-outflow tract.
3-D-corrected conduction velocity maps of the three hearts are shown in Fig. 6(c). All hearts exhibit the lowest conduction velocity at the outflow tract (), higher conduction velocity at the AVJ (), and the highest conduction velocity at the ventricle region (). The conduction velocities at the AVJ and the outflow tract are generally uniformly distributed. The ventricle region shows more heterogeneity. The inner curvature of the ventricle region has a much lower conduction velocity than the outer curvature. Figure 6(d) shows maps of the percent change in conduction velocity resulting from the 3-D correction procedure. All correction values are non-negative because the 2-D projection error can only result in an underestimation bias, not overestimation. The correction values vary widely from 0% to more than 100%. The corrections needed for each heart are very different from one another. Heart 1 needed corrections over most of the field of views. Heart 2 mainly needed corrections at the transitions of AVJ to ventricle and ventricle to outflow tract. Heart 3 needed the least correction, mostly at the outflow tract. Histograms of the percent corrections applied to all pixels in each heart with bins of 5% are shown in Fig. 6(e). In all three hearts, most of the pixels needed small corrections (0%–20%). 41%, 37%, and 12% of the pixels from hearts 1, 2, and 3, respectively, required corrections greater than 20%.
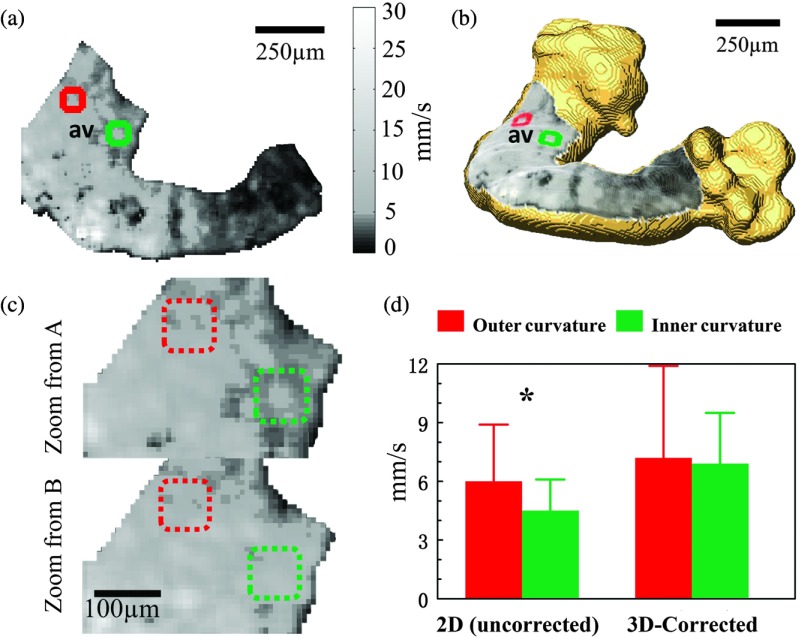
2-D calculations can produce artificial features, which may lead to misinterpretation of the data. An example is shown in Fig. 7. Because the AVJ is a straight tubular structure at HH stage 15 [this is more apparent in 3-D, Fig. 7(b), than in 2-D, Fig. 7(a)], it is expected to have a generally uniform conduction velocity around the tube. In the 2-D conduction velocity map [Fig. 7(a), magnified in 7(c)], the left (red) and right (green) sides of the AVJ appear to have a significant difference while in the 3-D-corrected conduction velocity map [Fig. 7(b), magnified in 7(c)], they are revealed to be approximately the same. Comparing the regions of interest marked in green and red, the right side showed a 25% lower conduction velocity than the left side based on the 2-D map [Fig. 7(d)]. This apparent difference is statistically significant, but it is absent in the 3-D-corrected map [Fig. 7(d)], indicating that the difference is most likely to be an artifact of the 2-D calculation.
Fig. 7.
Panels (a) and (b) are 2-D (uncorrected) and 3-D-corrected conduction velocity maps, respectively, for embryonic heart 1. Square regions of interest from the right (indicated in green) and left (indicated in red) sides were selected for comparison. Panel (c) top and bottom are magnified, corresponding images of the AVJ from panels (a) and (b). Panel (d) shows the conduction velocity () computed from 109 pixels of each marked region in panel (c). Conduction velocity values were compared using Student’s -test (* significant). av- atrioventricular junction.
4. Discussion
We have presented an integrated OCT and OM system. This, for the first time, provides the means to simultaneously record and directly co-register structural and electrophysiological images of embryonic hearts. We have, furthermore, demonstrated the capability of the system to recover the velocity vector in the direction and compute a 3-D conduction velocity map that is corrected for surface curvature in the embryonic heart. Imaging samples with the integrated OCT/OM system is fast (i.e., no longer than imaging the sample with one modality). Because the two imaging systems are integrated, image registration is very straightforward, which provides high accuracy and saves time in postprocessing. Fast imaging is especially important for high-throughput experiments, and therefore is advantageous for phenotyping studies.
The high-resolution 3-D structural OCT images directly correlated with OM are also useful in a qualitative way for interpretation of OM data. OM utilizes high NA objective lenses, and out-of-focus regions of the complex heart structure can be difficult to identify. Sometimes, the tissue may fold and overlap, and useful signals can only be recorded from the top surface. The tissue overlap can be difficult to interpret in 2-D fluorescence images. With correlated volumetric OCT, the 3-D morphology of the samples is clearly visualized and regions of invalid OM data can be readily excluded from analysis.
The process of 3-D correction of conduction velocity was carefully verified step by step to avoid introducing additional errors or noise to the conduction velocity measurement. First, both the OM system and the OCT system were calibrated. The results of the angle measurement verification experiment (Fig. 4) indicate that OCT correctly measures angles, with negligible errors. The OCT sample arm scanner was built based on a quasitelecentric design similar to that previously described.42 This design minimizes off-axis aberrations, which is a potential cause of inaccurate length and angle measurements. A metal mesh was imaged to verify uniformity across the field of view. Identical on-axis and off-axis grid sizes were observed (data not shown). Second, to avoid deforming the OM images from which the conduction velocity measurements were made, OCT images were registered to OM images instead of the other way around. Boundary segmentation curves on the registered OCT image and the reference OM image correspond well (Fig. 3). The registration error on the boundary is the worst case; therefore, the registration inside the heart region will be more accurate. Third, the accuracy of the height map from OCT images is also directly related to the volume segmentation. Automatic segmentation using Amira software was employed with small manual corrections applied after automatic segmentation (total time less than 10 min per heart). The performance of the automatic segmentation with correction was always satisfactory when compared to careful manual segmentation. Although we have carefully verified all steps involved in the 3-D correction process to minimize additional noise, some additional noise is unavoidable. However, some increased noise is acceptable because correcting bias due to 3-D conduction is more important. Noise can be reduced if needed by filtering or signal averaging, but the bias cannot be corrected without the additional 3-D information.
The heart surface height maps acquired from OCT provide a direct view of the slopes in depth () and an estimation of the amount of conduction velocity correction needed [Fig. 6(a)]. Steeper slopes generally result in larger errors in 2-D measurements. However, some steep regions do not necessarily require large corrections, as can be seen by comparing the height maps in Fig. 6(a) with the conduction velocity correction maps in Fig. 6(d). The reason is that only gradients in the direction of AP propagation cause errors and require correction, while the slopes perpendicular to the direction of conduction do not cause errors. The height maps in Fig. 6(a) and conduction direction maps in Fig. 6(b) together explain the patterns in the conduction velocity correction maps in Fig. 6(d).
Several other observations can be made from the conduction velocity correction maps and histograms in Fig. 6(d) and 6(e). First, all the corrections are positive, confirming that the 2-D maps only underestimate conduction velocity because 2-D projections only underestimate distances. Second, the corrections required were not uniform between hearts or even within the same heart. This is related to the morphology of the heart and the orientation of the heart when it was imaged. Third, the percent correction histograms from the three embryonic hearts show that most pixels do not need much correction because they are either at a flat region, or a region tilted perpendicular to the conduction velocity direction. Also, a wide range of correction values were needed, mainly between 0% and 100%. Corrections larger than 100% are rare, as larger corrections would only be required with a tilt angle greater than 60 deg along the conduction direction. Structures that have angles larger than 60 deg will most likely result in blurry OM pixels due to being out of focus and are likely to be excluded during OM image processing.
Among the three hearts, heart 2 generally had a lower conduction velocity [Fig. 6(c)], which may be because heart 2 was imaged from a different side or because it was less healthy. In all the hearts, the inner curvature of the ventricle regions has a lower conduction velocity than the outer curvature because the outer curvature has a much longer distance for the conduction wave to travel than the inner curvature. In the middle of each heart, there are some small patches of very low apparent conduction velocity values that are distinct from surrounding regions. These patches have lower signal-to-noise ratio (SNR) in raw fluorescent signals, which increased the noise in the activation time estimations. This activation time noise may not be readily observed in the activation map [e.g., for heart 1, Fig. 5(b)], but the conduction velocity calculation is a gradient estimation; therefore, it is very sensitive to noise and a small error in activation time could result in a large error in conduction velocity.
Relative conduction velocity can be estimated and compared qualitatively from the activation map. However, absolute conduction velocity values have been rarely reported for embryonic hearts. Kamino et al. have measured conduction velocities for chick embryonic hearts at the stage of 7–9 somites (HH stage 9–10) using OM. From a series of photodiode readings and estimated 2-D distances along the heart tube, the conduction velocity was calculated to be .7,8,10 Argüello et al. have reported conduction velocities for 45–47 h (HH stage 13) chick embryonic heart using electrode recordings from very limited sites (two points per region: at the AVJ and at the ventricle, averaged from four hearts).9 This study used similar stage embryos to those in the demonstration presented here, and the results were comparable to our averaged results [Fig. 6(c)]. These previous measurements provide valuable comparisons. However, they all have low-spatial resolution and did not take 3-D distances into consideration. Conduction velocity increases significantly at HH stage 24–26, which was measured in 2-D by Rothenberg et al. using OM.11 These results are not comparable to the work presented here because of the stage differences, but 3-D topology was also not taken into account.
Conventional 2-D conduction velocity calculations from OM can create artifacts and be misleading when comparing different regions of the same heart. For example, in heart 1, 2-D calculations presented an apparent difference in conduction velocity between the left and right sides of the AVJ, while 3-D-corrected calculations showed no significant difference [Figs. 7(c) and 7(d)]. Both the left and right sides of the AVJ have relatively steep slopes [Fig. 6(a), heart 1]; however, the slope at the right side is mostly parallel to the conduction direction, while the slope at the left side is mostly perpendicular to the conduction direction [Figs. 6(a) and 6(b), heart 1]. Therefore, more correction was needed at the right side than the left side of the AVJ [Fig. 6(c)]. Any uneven curvatures can cause problems because their projections will result in nonuniform underestimations. Artifacts caused by nonuniform curvatures in conventional 2-D conduction velocity calculations can be mitigated by 3-D correction.
The instrument described here successfully demonstrated the conduction velocity correction method, but results could be further improved by additional technical development. The main source of conduction velocity noise is the OM recording. Compared to OM measurements of adult heart models, OM of early embryonic hearts suffers from low SNR because each pixel is integrating signals from a much smaller volume (i.e., fewer cells). Activation times determined from low-SNR AP traces are relatively noisy and, in turn, result in noisy calculated conduction velocity. Therefore, optimizing the imaging and postprocessing steps for OM will improve the accuracy of calculating the conduction velocity. Potential improvements include optimizing the imaging protocol to maximize the SNR of acquired OM data so that the activation time can be more accurately determined and developing gradient estimation methods more robust to low-SNR signals. For example, instead of using a two-point difference, a more advanced global or local fitting algorithm may be adopted to estimate the gradients.43 Another limitation of the instrument described here is that because the heart is imaged by OCT from the opposite side of the OM, the size of the hearts that may be accommodated are limited to a maximum of about HH stage 21. This is because the penetration depth of OCT in tissue is limited to about 2 mm. This limitation could be overcome by coupling both the OCT and OM lights into the same objective lens, and allowing OCT and OM imaging from the same side.
The integrated OCT/OM system and 3-D conduction velocity correction algorithm presented here will reduce the experimental variability due to inconsistent heart morphology and orientation when measuring conduction velocity using OM. This will enable improved comparisons of conduction velocity between hearts and easier identification of differences caused by diseases and defects. The ability to accurately measure the conduction velocity can benefit the study of cardiac development in multiple ways. It may aid the study of ion channels and gap junctions at early stages of development. It may also benefit the study of other animal models. For example, compared to the avian heart, the looping-stage mouse heart exhibits more significant 3-D curvature.35,44 The knockout mouse phenotyping project (KOMP) has already found of transgenic mice have CHDs.45 It is likely that a large proportion of these defective hearts have conduction abnormalities. Yet, these hearts have not been assayed for conduction defects. The methods described here could enable such electrophysiological assays. Furthermore, the integrated OCT/OM system provides high-resolution 3-D structural images of the hearts simultaneously with the conduction measurements. Important features inside the embryonic heart can be characterized by OCT imaging and their influences on conduction velocity can be studied. For example, cardiac cushions, which are valve progenitors, have been shown to modulate APs,46 and some common genes and pathways can affect both conduction at the AVJ region and the formation of cardiac cushions.47,48 The direct correlation between the morphology of the AV cushions and conduction velocity at the AVJ has never been available and may now be investigated with this newly developed integrated OCT/OM system.
In conclusion, we have presented a novel integrated OCT/OM system, which can acquire structural and electrophysiological images simultaneously. We have also developed a 3-D-corrected conduction velocity calculation method using the combined information. This can correct conduction velocity underestimations and artifacts caused by the 2-D projection and provide more accurate measurements and more reliable comparisons for studies of normal and diseased models, thus improving our understanding of the electrophysiology of developing embryonic hearts.
Acknowledgments
The authors would like to thank Dr. D. L. Wilson for valuable advice on image processing. The project was supported by the National Heart, Lung, and Blood Institute through NIH R01-HL083048, R01-HL095717 and R21-HL115373. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. P. M. It was partially supported by the Chinese Government Scholarship.
Biographies
Pei Ma is a PhD candidate at Case Western Reserve University, Ohio. She received her BS degree in optical engineering from Tianjin University, Tianjin, China, in 2010.
Biographies of the other authors are not available.
References
- 1.Sedmera D., “Form follows function: developmental and physiological view on ventricular myocardial architecture,” Eur. J. Cardio-Thorac. Surg. 28(4), 526–528 (2005). 10.1016/j.ejcts.2005.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikawa T., Hurtado R., “Development of the cardiac conduction system,” Semin. Cell. Dev. Biol. 18(1), 90–100 (2007). 10.1016/j.semcdb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 3.Chi N. C., et al. , “Cardiac conduction is required to preserve cardiac chamber morphology,” Proc. Natl. Acad. Sci. 107(33), 14662–14667 (2010). 10.1073/pnas.0909432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reckova M., et al. , “Hemodynamics is a key epigenetic factor in development of the cardiac conduction system,” Circ. Res. 93(1), 77–85 (2003). 10.1161/01.RES.0000079488.91342.B7 [DOI] [PubMed] [Google Scholar]
- 5.Rentschler S., et al. , “Visualization and functional characterization of the developing murine cardiac conduction system,” Development 128(10), 1785–1792 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamino K., “Optical approaches to ontogeny of electrical activity and related functional organization during early heart development,” Physiol. Rev. 71(1), 53–91 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Hirota A., et al. , “Early events in development of electrical activity and contraction in embryonic rat heart assessed by optical recording,” J. Physiol. 369(1), 209–227 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirota A., et al. , “Initial development of conduction pattern of spontaneous action potential in early embryonic precontractile chick heart,” Dev. Biol. 99(2), 517–523 (1983). 10.1016/0012-1606(83)90301-9 [DOI] [PubMed] [Google Scholar]
- 9.Argüello C., et al. , “Electrophysiological and ultrastructural study of the atrioventricular canal during the development of the chick embryo,” J. Mol. Cell. Cardiol. 18(5), 499–510 (1986). 10.1016/S0022-2828(86)80915-4 [DOI] [PubMed] [Google Scholar]
- 10.Kamino K., et al. , “Functional pacemaking area in the early embryonic chick heart assessed by simultaneous multiple-site optical recording of spontaneous action potentials,” J. Gen. Physiol. 91(4), 573–591 (1988). 10.1085/jgp.91.4.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberg F., et al. , “Emerging patterns of cardiac conduction in the chick embryo: Waveform analysis with photodiode array-based optical imaging,” Dev. Dyn. 233(2), 456–465 (2005). 10.1002/(ISSN)1097-0177 [DOI] [PubMed] [Google Scholar]
- 12.Panakova D., Werdich A. A., MacRae C. A., “Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel,” Nature 466(7308), U874–U109 (2010). 10.1038/nature09249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bressan M., Liu G., Mikawa T., “Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field,” Science 340(6133), 744–748 (2013). 10.1126/science.1232877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y. T., et al. , “Optical stimulation enables paced electrophysiological studies in embryonic hearts,” Biomed. Opt. Express 5(4), 1000–1013 (2014). 10.1364/BOE.5.001000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salameh A., Blanke K., Daehnert I., “Role of connexins in human congenital heart disease: the chicken and egg problem,” Front Pharmacol. 4(70) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchhoff S., et al. , “Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double-deficient mice,” Circ. Res. 87(5), 399–405 (2000). 10.1161/01.RES.87.5.399 [DOI] [PubMed] [Google Scholar]
- 17.Efimov I. R., Nikolski V. P., Salama G., “Optical imaging of the heart,” Cir. Res. 95(1), 21–33 (2004). 10.1161/01.RES.0000130529.18016.35 [DOI] [PubMed] [Google Scholar]
- 18.Werdich A. A., et al. , “The zebrafish as a novel animal model to study the molecular mechanisms of mechano-electrical feedback in the heart,” Prog. Biophys. Mol. Biol. 110(2–3), 154–165 (2012). 10.1016/j.pbiomolbio.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pennisi D. J., et al. , “Induction and patterning of the cardiac conduction system,” Int. J. Dev. Biol. 46(6), 765–775 (2002). [PubMed] [Google Scholar]
- 20.Rothenberg F., Efimov I. R., Watanabe M., “Functional imaging of the embryonic pacemaking and cardiac conduction system over the past 150 years: technologies to overcome the challenges,” Anat. Rec.: Discov. Mol. Cell. Evol. Biol. 280A(2), 980–989 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Salama G., Lombardi R., Elson J., “Maps of optical action potentials and NADH fluorescence in intact working hearts,” Am. J. Physiol-Heart Circ. Physiol. 252(2), H384–H394 (1987). [DOI] [PubMed] [Google Scholar]
- 22.Lin S.-F., Wikswo J. J. P., “Panoramic optical imaging of electrical propagation in isolated heart,” J. Biomed. Opt. 4(2), 200–207 (1999). 10.1117/1.429910 [DOI] [PubMed] [Google Scholar]
- 23.Kay M. W., Amison P. M., Rogers J. M., “Three-dimensional surface reconstruction and panoramic optical mapping of large hearts,” IEEE Trans. Biomed. Eng. 51(7), 1219–1229 (2004). 10.1109/TBME.2004.827261 [DOI] [PubMed] [Google Scholar]
- 24.Qu F., et al. , “Three-dimensional panoramic imaging of cardiac arrhythmias in rabbit heart,” J. Biomed. Opt. 12(4), 044019 (2007). 10.1117/1.2753748 [DOI] [PubMed] [Google Scholar]
- 25.Yelbuz T. M., et al. , “Optical coherence tomography: a new high-resolution imaging technology to study cardiac development in chick embryos,” Circulation 106(22), 2771–2774 (2002). 10.1161/01.CIR.0000042672.51054.7B [DOI] [PubMed] [Google Scholar]
- 26.Filas B. A., Efimov I. R., Taber L. A., “Optical coherence tomography as a tool for measuring morphogenetic deformation of the looping heart,” Anat. Rec. Adva. Inte. Anat. Evol. Biol. 290(9), 1057–1068 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Männer J., et al. , “High-resolution in vivo imaging of the cross-sectional deformations of contracting embryonic heart loops using optical coherence tomography,” Dev. Dyn. 237(4), 953–961 (2008). 10.1002/dvdy.v237:4 [DOI] [PubMed] [Google Scholar]
- 28.Davis A. M., et al. , “In vivo spectral domain optical coherence tomography volumetric imaging and spectral Doppler velocimetry of early stage embryonic chicken heart development,” J. Opt. Soc. Am. A 25(12), 3134–3143 (2008). 10.1364/JOSAA.25.003134 [DOI] [PubMed] [Google Scholar]
- 29.Peterson L. M., et al. , “4D shear stress maps of the developing heart using Doppler optical coherence tomography,” Biomed. Opt. Express 3(11), 3022–3032 (2012). 10.1364/BOE.3.003022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garita B., et al. , “Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping,” Am. J. Physiol-Heart Circ. Physiol. 300(3), H879 (2011). 10.1152/ajpheart.00433.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karunamuni G., et al. , “Ethanol exposure alters early cardiac function in the looping heart: a mechanism for congenital heart defects?,” Am. J. Physiol-Heart Circ. Physiol. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins M. W., Watanabe M., Rollins A. M., “Longitudinal imaging of heart development with optical coherence tomography,” IEEE J. Sel. Topics Quantum Electron. 18(3), 1166–1175 (2012). 10.1109/JSTQE.2011.2166060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu A., et al. , “Dynamic variation of hemodynamic shear stress on the walls of developing chick hearts: computational models of the heart outflow tract,” Eng. Comput. 25(1), 73–86 (2009). 10.1007/s00366-008-0107-0 [DOI] [Google Scholar]
- 34.Sedmera D., et al. , “Developmental transitions in electrical activation patterns in chick embryonic heart,” Anat. Rec. A: Discov. Mol. Cell. Evol. Biol. 280A(2), 1001–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Männer J., “Cardiac looping in the chick embryo: a morphological review with special reference to terminological and biomechanical aspects of the looping process,” Anat. Rec. 259(3), 248–262 (2000). 10.1002/(ISSN)1097-0185 [DOI] [PubMed] [Google Scholar]
- 36.Viktor H., Howard L. H., “A series of normal stages in the development of the chick embryo,” Dev. Dyn. 195(4), 231–272 (1992). 10.1002/aja.1001950404 [DOI] [PubMed] [Google Scholar]
- 37.Ainsworth S. J., Stanley R. L., Evans D. J., “Developmental stages of the Japanese quail,” J. Anat. 216(1), 3–15 (2010). 10.1111/joa.2009.216.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Z., Rollins A. M., “Fourier domain optical coherence tomography with a linear-in-wavenumber spectrometer,” Opt. Lett. 32(24), 3525–3527 (2007). 10.1364/OL.32.003525 [DOI] [PubMed] [Google Scholar]
- 39.Rueckert D., et al. , “Nonrigid registration using free-form deformations: application to breast MR images,” IEEE Trans. Med. Imaging 18(8), 712–721 (1999). 10.1109/42.796284 [DOI] [PubMed] [Google Scholar]
- 40.Salama G., Kanai A., Efimov I. R., “Subthreshold stimulation of Purkinje fibers interrupts ventricular tachycardia in intact hearts. Experimental study with voltage-sensitive dyes and imaging techniques,” Circ. Res. 74(4), 604–619 (1994). 10.1161/01.RES.74.4.604 [DOI] [PubMed] [Google Scholar]
- 41.Sutherland I. E., Sproull R. F., Schumacker R. A., “A characterization of ten hidden-surface algorithms,” ACM Comput. Surv. 6(1), 1–55 (1974). 10.1145/356625.356626 [DOI] [Google Scholar]
- 42.Hu Z., Rollins A., “Quasi-telecentric optical design of a microscope-compatible OCT scanner,” Opt. Express 13(17), 6407–6415 (2005). 10.1364/OPEX.13.006407 [DOI] [PubMed] [Google Scholar]
- 43.Laughner J. I., et al. , “Processing and analysis of cardiac optical mapping data obtained with potentiometric dyes,” Am. J. Physiol-Heart Circ. Physiol. 303(7), H753–H765 (2012). 10.1152/ajpheart.00404.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaufman M. H., The Atlas of Mouse Development, Academic Press, London: (1992). [Google Scholar]
- 45.Adams D., et al. , “Bloomsbury report on mouse embryo phenotyping: recommendations from the IMPC workshop on embryonic lethal screening,” Dis. Model. Mech. 6(3), 571–579 (2013). 10.1242/dmm.011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polo-Parada L., Zhang X., Modgi A., “Cardiac cushions modulate action potential phenotype during heart development,” Dev. Dyn. 238(3), 611–623 (2009). 10.1002/dvdy.v238:3 [DOI] [PubMed] [Google Scholar]
- 47.Christoffels V. M., Moorman A. F. M., “Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others?,” Circ. Arrhythm. Electrophysiol. 2(2), 195–207 (2009). 10.1161/CIRCEP.108.829341 [DOI] [PubMed] [Google Scholar]
- 48.Milan D. J., et al. , “Notch1b and neuregulin are required for specification of central cardiac conduction tissue,” Development 133(6), 1125–1132 (2006). 10.1242/dev.02279 [DOI] [PubMed] [Google Scholar]