Abstract
Advanced age is characterized by increased incidence of many chronic, noninfectious diseases that impair the quality of living of the elderly and pose a major burden on the healthcare systems of developed countries. These diseases are characterized by impaired or altered function at the tissue and cellular level, which is a hallmark of the aging process. Age-related impairments are likely due to loss of homeostasis at the cellular level, which leads to the accumulation of dysfunctional organelles and damaged macromolecules, such as proteins, lipids, and nucleic acids. Intriguingly, aging and age-related diseases can be delayed by modulating nutrient signaling pathways converging on the target of rapamycin (TOR) kinase, either by genetic or dietary intervention. TOR signaling influences aging through several potential mechanisms, such as autophagy, a degradation pathway that clears the dysfunctional organelles and damaged macromolecules that accumulate with aging. Autophagy substrates are targeted for degradation by associating with p62/SQSTM1, a multidomain protein that interacts with the autophagy machinery. p62/SQSTM1 is involved in several cellular processes, and its loss has been linked to accelerated aging and to age-related pathologies. In this review, we describe p62/SQSTM1, its role in autophagy and in signaling pathways, and its emerging role in aging and age-associated pathologies. Finally, we propose p62/SQSTM1 as a novel target for aging studies and age-extending interventions.
Keywords: Aging, Autophagy, Mitochondria, Senescence, p62
Introduction: impaired cellular homeostasis—a hallmark of aging
Aging tissues present increasing loss of function that is associated with structural damage and alterations. For example, the liver of aged mammals has reduced xenobiotic metabolism, hepatobiliary function, and regeneration in association with structural alterations such as increased volume, loss of functional mass, thickening of blood vessels and loss of fenestrae, loss of smooth endoplasmic reticulum in the hepatocytes, reduced phagocytic activity of the Kupffer cells, and reduced proliferative capacity (Schmucker 2005). In skeletal muscle, loss of mass and reduced contractile capacity correlate with reduced plasticity and loss of neuromuscular junctions, causing atrophy of muscle fibers (Jang and Van Remmen 2011), and in the heart, reduced function arises with increased fat deposits, fibrosis, and changes in conductivity (Kitzman and Edwards 1990). Similar changes are seen in the lungs, where reduced function is affected primarily by structural changes in the alveoli and capillaries (Lalley 2013), and in virtually all tissues.
These age-related changes are often the consequence of loss to homeostasis at the cellular level; a process common to many species across the evolutionary spectrum. For example, the yeast Saccharomyces cerevisiae accumulates extrachromosomal ribosomal DNA circles (ECR) and depolarized mitochondria with age (Sinclair and Guarente 1997; Hughes and Gottschling 2012), while old Caenorhabditis elegans nematodes accumulate lipofuscin (Klass 1977), an insoluble aggregate of oxidized proteins and lipids (Toth 1968; Porta 1991), in their intestinal cells. In higher eukaryotes, damaged organelles and proteins accumulate in long-lived postmitotic tissues, such as skeletal muscles, heart, liver, and brain (Schmucker 2005; Chaudhary et al. 2011; Szweda et al. 2003). Cells in these aging organs contain increased levels of damaged and dysfunctional cellular components with age: for example, neurons, astrocytes, microglia, hepatocytes, and cardiomyocytes accumulate lipofuscin deposits and other protein aggregates (Landfield et al. 1981; Salminen et al. 2011; Vaughan and Peters 1974; Frenzel and Feimann 1984; Schmucker and Sachs 2002); rhabdomyocytes accumulate mitochondria with oxidized bases in their DNA, reduced activity, and increased production of reactive oxygen species (Mansouri et al. 2006); and cardiac myocytes accumulate impaired mitochondria with increased fragmentation of mitochondrial DNA (Frenzel and Feimann 1984; Ozawa 1998).
Together, this evidence suggests that accumulating protein aggregates, dysfunctional mitochondria, and DNA damage contribute to the aging process and the onset of age-related conditions. Continuous, irreparable damage to DNA, proteins, and organelles can be caused by oxidants, radiations, and other insults through free radical reactions (Harman 2003), progressively impairing cellular homeostasis. Living organisms are well equipped with detoxifying, antioxidant, and repair enzymes to cope with this kind of damage (Robida-Stubbs et al. 2012; Longo 1999; Steinbaugh et al. 2012; Leiser and Miller 2010; Salmon et al. 2005; Wei et al. 2008): this suggests that raising the cellular defenses against stressors would mimic the effects of age-extending interventions. In addition to stress resistance, cellular homeostasis is also influenced by quality control mechanisms that regulate function, structure, and degradation of cellular components, such as the protein-folding machinery, mitochondrial fission and fusion dynamics, the ubiquitin-proteasome system, and autophagy (Koga et al. 2011; Seo et al. 2010). Autophagy in particular can degrade damaged proteins, protein aggregates, and damaged organelles that accumulate during aging and in age-related pathologies (Mizushima et al. 2008) and is elicited by age-extending interventions and in long-lived mutants (Hansen et al. 2008; Tóth et al. 2008; Wang and Miller 2012; Alvers et al. 2009).
Three mechanisms of autophagy have been described: macroautophagy, microautophagy, and chaperone-mediated autophagy (Cuervo 2008). Chaperone-mediated autophagy is a selective degradation pathway that recognizes soluble cytosolic proteins and translocates them directly into the lysosome for degradation (Kaushik and Cuervo 2008). Conversely, micro- and macroautophagy can degrade cellular components indiscriminately without well-defined targeting systems, even though selective mechanisms of macroautophagy have been described (Johansen and Lamark 2011; Nezis and Stenmark 2012). Microautophagy is the direct invagination of the lysosomal membrane in order to bring organelles and fractions of the cytosol into the lysosome for degradation. Microautophagy is under control of the target of rapamycin (TOR) pathway and has been most fully defined in yeast (Li et al. 2012). Conversely, macroautophagy (hereafter referred to as autophagy) is the main autophagic pathway in higher eukaryotes, and is involved both in bulk and selective substrate degradation (He and Klionsky 2009). Macroautophagy substrates are enveloped into a double membrane vesicle, called autophagosomes, which then fuses with the lysosome and releases the substrates for degradation. Autophagosomes form upon activation of the Atg1/ULK1-2 complex, formed by ULK1 or ULK2 (Atg1 in yeast), Atg13, and FIP200 (Atg17 in yeast) (Cheong et al. 2008; Jung et al. 2009). This complex recruits the protein complexes required to nucleate the autophagosomes into a structure known as phagophore assembly site (PAS). The class-III PI3K, also known as vacuolar protein sorting 34 (Vps34), Atg14, and Beclin 1 (Atg6 in yeast) form another complex that is also required for this process (Itakura et al. 2008). The Vps34 and the Atg1 complexes recruit proteins involved in two ubiquitin-like conjugation systems that elongate and shape the autophagosome membrane around the substrate: the E1-like protein Atg7 catalyzes the covalent linkage of Atg12 to the E2-like Atg10, which then transfers Atg12 to Atg5, forming the Atg5-12 complex (Geng and Klionsky 2008). The Atg5-12 complex then associates with Atg16 and is recruited to the phagophore (Mizushima et al. 2003). Similarly, Atg7 catalyzes the covalent linkage of LC3B (Atg8 in yeast) to Atg3, and Atg3 mediates the conjugation of LC3B to phosphatidyl-ethanolamine (PE). This linkage allows LC3-PE (or LC3-II) to associate with the membrane of the nascent autophagosome and regulate its size and shape (Xie et al. 2008). Once the autophagosome has formed around its substrate, it is delivered to the lysosome, where the cargo is degraded and its components recycled (He and Klionsky 2009).
Autophagy is under direct control of the TOR pathway: TOR complex 1 (TORC1) phosphorylates Atg1/ULK1-2 and inhibits the formation of the Atg1 complex, which is the initial step of the pathway. More specifically, autophagy is directly regulated by the nutrient and energy status of the cell and by genes involved in lifespan extension: insulin/IGF-1 and amino acids signaling activate TORC1 and repress autophagy, while the energy sensor AMP-dependent protein kinase (AMPK) can directly activate the Atg1 complex, bypassing TORC1 activity (Jung et al. 2009; Kim et al. 2011), and the FoxO transcription factor regulates the expression of several autophagy genes in skeletal muscle (Zhao et al. 2007; Mammucari et al. 2007). Autophagy can also be induced by several cellular stresses, such as ER overload, hypoxia, oxidative stress, and pathogens (He and Klionsky 2009). Because it is induced by virtually all longevity pathways, it is thought that autophagy is one of the beneficial effects of pro-longevity interventions. Indeed, autophagic degradation declines with age (Cuervo 2008), and dietary restriction (DR) prevents this age-related decline in rat liver (Del Roso et al. 2003). In addition, autophagy is required for chronological lifespan extension in yeast (Alvers et al. 2009), and for the longevity response to DR and reduced TOR and insulin signaling in C. elegans, even though its induction alone is not sufficient to extend lifespan (Hansen et al. 2008). Inhibiting autophagy triggers premature cellular senescence in human fibroblasts (Kang et al. 2011), but it is also required for the senescence transition in response to oncogenic signaling (Young et al. 2009), suggesting that autophagy could have pleiotropic effects. Indeed, excessive activation of autophagy can be deleterious and cause cell death independently of apoptosis (Pattingre et al. 2005).
Activating autophagy has beneficial effects in many models of age-related pathologies, especially metabolic syndrome, cardiovascular dysfunction (Mizushima et al. 2008; Choi et al. 2013), and neurodegenerative diseases such as Alzheimer’s (Spilman et al. 2010), Huntington’s (Ravikumar et al. 2004), and Parkinson’s disease (Narendra et al. 2010a; Geisler et al. 2010a; Burman et al. 2012). In particular, substrate-specific autophagy plays an important role in removing protein aggregates and damaged organelles that associate with these pathologies; substrate-specific autophagy is mediated by tagging targets with ubiquitin, and substrates are delivered to the autophagosomes through adaptor proteins, prominently the multifunctional protein scaffold p62/SQSTM1.
p62/SQSTM1 in autophagy, signaling, and aging
p62/SQSTM1 structure and functions
p62/SQSTM1 is a multifunctional protein involved in multiple cellular functions: the protein plays a role in signal transduction and in the degradation of both proteins and organelles (Johansen and Lamark 2011; Nezis and Stenmark 2012). Although p62/SQSTM1 partakes in a wide variety of cellular processes, mutations in the sqstm1 gene have been strongly associated with only one disease, Paget’s disease of the bone (Goode and Layfield 2010). Nevertheless, recent findings suggest that p62/SQSTM1 may also have an important role in neurodegenerative diseases, cancer, obesity, and other age-associated pathologies, and could become an interesting new target for healthy aging interventions.
Structure
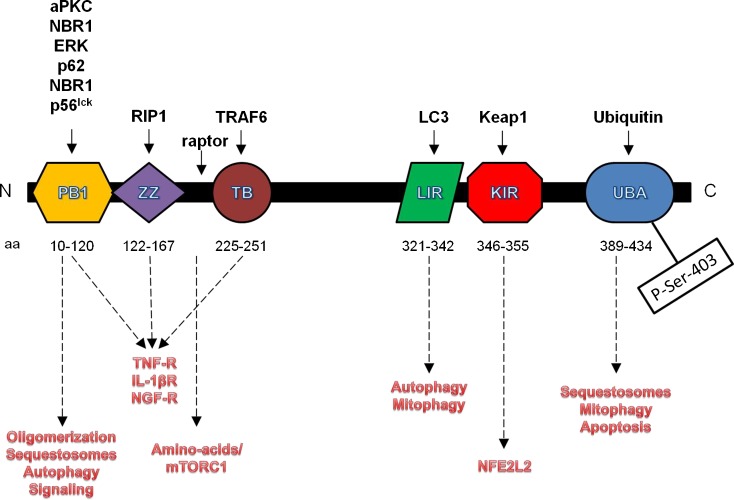
The sqstm1 gene is highly conserved among vertebrates: phylogenomic analysis in the UniProt database (entry: http://www.uniprot.org/uniprot/Q13501) reveals that p62/SQSTM1 has homologues in vertebrates and many other animal phyla, including arthropods (arthropoda), echinoderms (echinodermata), nematodes (nematoda), and mollusks (gastropoda). The human gene resides on chromosome 5g35 and encompasses a 16-kb segment of genomic DNA (Laurin et al. 2002). sqstm1 contains eight exons and is driven by a TATA-less promoter with three consensus sequences for SP1, three AP-1 sites, one NFκB site, and an antioxidant response element (ARE) (Vadlamudi and Shin 1998; Jain et al. 2010). The human sqstm1 gene encodes for a 440 amino acids protein, although a splice variant lacking the first 84 amino-acids has also been reported. The full-length protein contains several conserved domains that mediate multiple cellular functions (Fig. 1).
Fig. 1.
Architecture, interaction domain, and functions of the p62/SQSTM1 protein. The interacting partners of each domain are listed in black, while the functions each domain mediates are depicted in red
The N-terminal Phox and Bem1 (PB1) domain (aa 20–102) is necessary for p62/SQSTM1 to oligomerize and to interact with other PB1-containing proteins (Lamark et al. 2003). Through the PB1 domain, p62/SQSTM1 associates with several kinases, like atypical protein kinase C (aPKC) and p56lck, and participates in signal transduction cascades (Joung et al. 1996; Sanchez et al. 1998). Furthermore, the PB1 domain allows p62/SQSTM1 to oligomerize, a process that helps clearing selective autophagy substrates (Kirkin et al. 2009; Itakura and Mizushima 2011; Pankiv et al. 2007). PB1 domains have been described in other proteins mostly involved in signal transduction pathways, but no association of these proteins with p62/SQSTM1 has been described yet. Nevertheless, it is a possibility that p62/SQSTM1 has many interaction partners yet to be discovered, and that the protein may participate in additional pathways.
The C-terminal Ubiquitin Association (UBA) domain (aa 389–434) allows p62/SQSTM1 to associate with ubiquitin-tagged proteins and organelles (Seibenhener et al. 2004; Vadlamudi et al. 1996). Interestingly, p62/SQSTM1 seems to have increased affinity for lysine 63 (K63) poly-ubiquitin chains (Seibenhener et al. 2004; Tan et al. 2008a), a posttranslational modification that promotes the formation of protein inclusions and their clearance via autophagy (Tan et al. 2008b). The UBA domain is phosphorylated on Ser-403 by Casein Kinase II. This modification increases the domain’s affinity for poly-ubiquitin chains and the tendency of p62/SQSTM1 to form p62 bodies/sequestosomes, small protein inclusions that can function as signaling complexes and are cleared by autophagy (Matsumoto et al. 2011). The p62/SQSTM1 protein may associate with different ubiquitin-tagged cargos and direct them to the proteasome or the lysosome depending on its posttranslational modifications: in fact, knockdown of p62/SQSTM1 impairs the degradation of some proteasome substrates, and p62/SQSTM1 can associate with the proteasome through its PB1 domain (Seibenhener et al. 2004). Furthermore, p62/SQSTM1 contains two PEST sequences and is itself a proteasome substrate (Lee et al. 2012).
p62/SQSTM1 contains a zinc finger (ZZ) domain (aa 122–167), and has been found in the nucleus (Geetha and Wooten 2002), where it associates with nuclear transcription factors, such as the chicken ovalbumin upstream promoter transcription factor II (COUP-TFII), and may also act as a regulator of transcription itself (Marcus et al. 1996; Rachubinski et al. 1999). The primary sequence of p62/SQSTM1 contains two nuclear localization signals (NLS) and one Nuclear Export Signal (NES), and the protein shuttles between the cytoplasm and the nucleus, where it seems to help in sequestering and degrading ubiquitin-conjugated nuclear proteins (Pankiv et al. 2010). It has also been proposed that p62/SQSTM1 enters the nucleus in association with binding partners containing NLS, like aPKC, and that it regulates transcription in cooperation with these binding partners (Geetha and Wooten 2002). Although p62/SQSTM1 may mediate transcription in the nucleus through the ZZ domain, this domain is primarily involved in cytoplasmic signaling cascades and is in fact necessary to bind the receptor-interacting protein 1 (RIP1) kinase in the TNF receptor (TNF-R) complex. Binding of p62/SQSTM1 to RIP1 allows signaling of the TNF-R through protein kinase C (PKC) and activation of nuclear factor-kappaB (NFkB) (Sanz et al. 1999).
The TNFα receptor-associated factor 6 (TRAF6) binding (TB) domain mediates the association with TRAF6. Association of p62/SQSTM1 with TRAF6 is necessary for NFkB signaling from the interleukin 1 receptor (IL-1R) and the nerve growth factor receptor (NGF-R) (Sanz et al. 2000; Wooten et al. 2001). Upon stimulation of these receptors, p62/SQSTM1 binds to TRAF6 through the TB domain and to aPKC through the PB1, bringing the kinase into the complex for activation.
p62/SQSTM1 binds the autophagosome-coating protein LC3 with a LC3 interacting region (LIR) (aa 321–342) (Pankiv et al. 2007). The LIR sequence binds to a conserved region of all mammalian homologues of S. cerevisiae Atg8 (LC3B, GABARAP,GABARAPL1, GATE16, GABARAPL3) and is necessary for the autophagic degradation of the sequestosomes (Pankiv et al. 2007). Nevertheless, the LIR is dispensable for localization of p62/SQSTM1 to the nucleation sites of the autophagosomes or to recruit the autophagy machinery to sequestosomes, a functions that appears to require oligomerization through the PB1 domain (Itakura and Mizushima 2011).
Finally, p62/SQSTM1 binds to Kelch-like ECH-associated protein 1 (Keap1) through a Keap1 interacting region (KIR) (aa 346–355) (Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010). The KIR binds to Keap1 on a site essential for Keap1 to interact with and repress nuclear factor erythroid 2-like 2 (NFE2L2)’s activation, albeit with much lower affinity (Komatsu et al. 2010). Nevertheless, p62/SQSTM1 can activate NFE2L2 by sequestering Keap1 into p62 bodies when its levels are increased, either due to autophagy impairment or direct p62/SQSTM1 overexpression (Jain et al. 2010; Komatsu et al. 2010). Interestingly, p62/SQSTM1 ability to bind Keap1 seems to be dependent on its ability to form oligomers through the PB1 domain (Jain et al. 2010).
Biological functions
Signal transduction
p62/SQSTM1 is involved in several signal transduction cascades (Sanz et al. 1999; Jin et al. 2009). p62/SQSTM1 serves as a scaffold to bring together two or more components of a signaling pathway, but it is not phosphorylated or otherwise modified by these signaling cascades, nor it has any catalytic activity of its own. For example, in TNF-R signaling, the ZZ domain of p62/SQSTM1 binds the active RIP1 kinase and the PB1 domain binds protein kinase C λ/ι (PKC λ/ι), bringing them together into a signaling complex (Sanz et al. 1999). This complex forms upon stimulation of the TNF-R and is necessary to activate NFkB, since disrupting the ZZ domain or reducing the expression of p62/SQSTM1 impairs TNFα-mediated NFkB activation (Sanz et al. 1999). In addition to TNF-R, the interleukin-1β receptor (IL-1βR) and the nerve growth factor receptors TrkA and p75NTR recruit p62/SQSTM1 to facilitate signaling to NFkB (Sanz et al. 2000; Wooten et al. 2001). These receptors signal downstream by recruiting the ubiquitin-ligase TRAF6, which subsequently binds to p62/SQSTM1 on the TB domain and to interleukin-1 receptor-associated kinase (IRAK). This complex then recruits PKC λ/ι via the PB1 domain of p62/SQSTM1 and activates it through phosphorylation by IRAK (Sanz et al. 2000; Mamidipudi et al. 2002). p62/SQSTM1 plays additional roles in IL-1β and NGF signaling: the UBA and PB1 domain are necessary to allow TRAF6 to oligomerize in the receptor complex and poly-ubiquitylate itself in response to NGF (Wooten et al. 2005). Furthermore, p62/SQSMT1 acts as a bridge between the two NGF receptors, TrkA and p75NTR, and facilitate TRAF6-mediated ubiquitin conjugation of TrkA, which is necessary for receptor internalization, signaling (Geetha et al. 2005), and turnover (Geetha et al. 2008). In IL-1 signaling, the cellular levels of p62/SQSTM1 can modulate the intensity of the response through IL-1βR (Lee et al. 2012): in fact, increased autophagic and proteasomal degradation of p62/SQSTM1 dampens IL-1βR signaling and have anti-inflammatory effects. These effects are largely mediated by the autophagy-related gene Atg16L and the E3 ligase Cullin-3 (Lee et al. 2012). Interplay between p62/SQSTM1 and Cullin-3 extends beyond IL-1 signaling: synergistic activity of Cullin-3 and p62/SQSTM1 stimulates the extrinsic apoptosis pathway (Jin et al. 2009): upon ligand binding to death receptors 3 and 4, active Cullin-3 is recruited to the death-inducing signaling complex (DISC) and catalyzes the addition of ubiquitin chains to caspase 8. Subsequently, p62/SQSTM1 facilitates the formation of Ub-tagged caspase 8 aggregates which facilitate autocatalytic cleavage of caspase 8 and thus its activation (Jin et al. 2009). These findings suggest that p62/SQSTM1 is a signaling node for multiple pathways and can potentially foster interaction between such pathways in order to maintain cellular homeostasis.
Amino acid sensing: mTOR
Amino acid levels are linked to mTOR through a mechanism that is dependent, at least to some extent, on p62/SQSTM1 (Duran et al. 2011): mTORC1 associates with the amino acid sensing Rag family of small GTPases (Kim et al. 2008; Sancak et al. 2008) on the surface of lysosomes (Sancak et al. 2010). p62/SQSTM1 mediates this process by associating with Raptor, a subunit of the mTORC1 complex, and the Rag GTPases, thereby forming a high molecular weight complex that relays the signal from amino acids to the mTORC1 pathway (Duran et al. 2011). Although the physiological role of this interaction has not been defined, it is possible that the p62/SQSTM1-mTORC1 complex responds locally to amino acids produced by the autophagy-lysosome system to prevent excessive autophagy. Alternatively, p62/SQSTM1 levels may serve as a positive feedback for both autophagy and cell growth: low p62/SQSTM1 would reduce mTORC1 activity, increase autophagy, and further lower p62/SQSTM1 levels, thereby sustaining autophagic degradation processes (Komatsu et al. 2012). Conversely, high p62/SQSTM1 would increase mTORC1 activity, reduce autophagy, and further increase its own levels, thereby sustaining cell growth (Moscat and Diaz-Meco 2011). It must be noted that, while mTORC1 is activated by many different inputs, the regulation through amino acid levels is currently the only input known to require p62/SQSTM1. Insulin, for example, can activate mTORC1 in the absence of p62/SQSTM1 (Duran et al. 2011). Furthermore, p62/SQSTM1 knockdown elicits autophagy in ways that are reminiscent of amino acid starvation (Duran et al. 2011). Thus, p62/SQSTM1 levels may serve as a rheostat to sense free amino acid availability in the cell, turning on anabolic pathways through mTORC1 when amino acids are abundant, and shutting them down when there are insufficient nutrients for protein synthesis.
Autophagy and protein degradation
p62/SQSTM1 has been associated with protein degradation, based upon the discovery of its UBA domain (Seibenhener et al. 2004; Vadlamudi et al. 1996). p62/SQSTM1 associates with the proteasome (Seibenhener et al. 2004) and is itself a proteasome substrate (Lee et al. 2012). Interestingly, either a reduction (Seibenhener et al. 2004) or an accumulation (Korolchuk et al. 2009) of p62/SQSTM1 leads to impaired degradation of proteasome substrates. This apparent dichotomy suggests that while single p62/SQSTM1 molecules can deliver specific substrates to the proteasome, they can also bind ubiquitin-tagged proteins indiscriminately and sequester them into p62 bodies, when the concentration of p62/SQSTM1 is increased beyond physiological levels. Interestingly, this would also suggest that when proteasome activity is impaired, p62/SQSTM1 could deliver proteasome substrates to the lysosome through the autophagy machinery, even though no evidence for such a process has been found. p62/SQSTM1 promotes selective autophagy of proteins, protein aggregates, organelles, and bacteria (Narendra et al. 2010a; Geisler et al. 2010a; Pankiv et al. 2007; Zheng et al. 2009). Autophagy substrates are usually tagged with noncanonical poly-ubiquitin chains (K63 or K27 linkages) that are brought into sequestosomes by associating with the UBA domain of p62/SQSTM1. Sequestosomes are protein/organelles aggregates formed by p62/SQSTM1oligomers and p62/SQSTM1-interacting proteins. Mature sequestosomes are targeted to the autophagosomes by the LIR sequence on their p62/SQSTM1 moieties and degraded in the lysosome (Pankiv et al. 2007). Sequestosomes can form around already existing protein inclusions, such as mutant huntingtin aggregates (Bjorkoy et al. 2005), and their formation is greatly enhanced by the phosphorylation of p62/SQSTM1 on serine 403 in the UBA domain (Matsumoto et al. 2011). Interestingly, p62/SQSTM1 must oligomerize to localize the aggregates to autophagosome-forming structures and for efficient engulfment and delivery of autophagy substrates (Itakura and Mizushima 2011). These findings suggest that p62/SQSTM1 is more than just an adaptor protein between autophagy and its substrates, but is an active component of the autophagic process (Komatsu et al. 2007; Nezis et al. 2008). Furthermore, p62/SQSTM1 can mediate autophagic clearance of non-ubiquitylated protein aggregates as well, by binding them through the PB1 domain (Watanabe and Tanaka 2011). It must be noted that p62/SQSTM1 has a functional homologue called Neighbor of BRCA 1 (NBR1), a protein with a N-terminal PB1 domain, a C-terminal UBA, and a LIR sequence. NBR1 can substitute p62/SQSTM1 in many of its autophagy-related functions (Kirkin et al. 2009), but it lacks many of its other functional domains.
Mitophagy
p62/SQSTM1 is also critical for the autophagy of mitochondria, a process called mitophagy. Mitochondria are tagged for degradation by two E3 ligases: ring finger protein 185 (Rnf185) and parkin (Narendra et al. 2010a; Geisler et al. 2010a; Tang et al. 2011). Both enzymes catalyze the formation of noncanonical ubiquitin chains (K63 and K27 linkages) on mitochondrial outer membrane proteins. Rnf185 is constitutively localized on the mitochondrial outer membrane, where it catalyzes the addition of K63-linked ubiquitin on BNIP. K63Ub-tagged BNIP is then recognized by p62/SQSTM1 that clusters the tagged mitochondria for degradation. Conversely, parkin is recruited on the mitochondria only upon loss of mitochondrial-membrane potential (Δψ) by interacting with PTEN-induced putative kinase 1 (Pink1): Pink1 is usually transported to the inner mitochondrial membrane of healthy mitochondria by a Δψ-dependent mechanism. When the Δψ is disrupted, Pink 1 is not internalized and remains on the outer mitochondrial membrane. This is recognized as a signal to recruit the E3 ligase parkin from the cytosol to the mitochondria (Narendra et al. 2010b). Here, parkin tags several mitochondrial outer membrane proteins with K63- and K27-linked poly-ubiquitin chains (Narendra et al. 2010a; Geisler et al. 2010a; Okatsu et al. 2010). Once the mitochondrial proteins are tagged with ubiquitin, p62/SQSTM1 associates with them and drives the whole organelle into sequestosomes that are then cleared by autophagy (Narendra et al. 2010a; Geisler et al. 2010a). Interestingly, p62/SQSTM1 may be dispensable for parkin mediated mitophagy, but its presence is necessary to cluster the damaged mitochondria away from the remainder of the mitochondrial network (Narendra et al. 2010a) and protect it from further damage. This function becomes critical for cellular homeostasis, especially considering that constitutive autophagy is not particularly efficient in clearing damaged mitochondria and inhibiting mTOR is required for fast removal of depolarized mitochondria (Gilkerson et al. 2011).
Antioxidant response
p62/SQSTM1 can activate the phase 2 antioxidant response by interacting with the Keap1-NFE2L2 pathway. The Keap1-NFE2L2 antioxidant response pathway is one of the major cellular defenses against oxidative insults. It is composed of NFE2L2 (also known as Nrf2), a transcription factor, and Keap1, an inhibitory protein that modulates NFE2L2 activity. In physiological conditions, NFE2L2 is kept inactive in the cytoplasm by a strong noncovalent interaction with a Keap1 homodimer. This interaction is mediated by two domains on NFE2L2, the ETGE domain, and the DLG domain. When bound to NFE2L2, Keap1 recruits the Cullin3 E3 ligase, which tags NFE2L2 with poly-ubiquitin chains and targets it for degradation by the proteasome. Conversely, when the cell is under oxidative stress, Keap1 is oxidized and changes conformation. This disrupts binding to the DLG domain on NFE2L2, thereby inhibiting NFE2L2 degradation. Subsequent phosphorylation or additional interactions facilitate complete dissociation of NFE2L2 which can migrate into the nucleus and activate transcription of several genes involved in reactive oxygen species (ROS) scavenging, detoxification and repair, and mitochondrial function (Stępkowski and Kruszewski 2011).
NFE2L2 can also be activated by p62/SQSTM1 in conditions where oxidative stress is not present: p62/SQSTM1 binds to Keap1 with the KIR on the same sequence that binds to the ETGE domain of NFE2L2 (Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010; Copple et al. 2010). The KIR has a much weaker affinity for Keap1 than NFE2L2 (Komatsu et al. 2010), hence it has been proposed that p62/SQSTM1 can activate the NFE2L2 pathway only in pathological conditions, for example, when autophagy is impaired. Nevertheless, a fraction of Keap1 associates constitutively with p62/SQSMT1 (Copple et al. 2010). Thus, it is possible that the affinity of the full-length p62/SQSTM1 protein for Keap1 is higher than that of the KIR domain alone. Alternatively, posttranslational modifications of either Keap1 or p62/SQSTM1 itself may affect binding between the two proteins and provide a physiological mechanism for this alternative NFE2L2 activation. p62/SQSTM1 brings Keap1 into sequestosomes, effectively impairing its ability to bind NFE2L2 (Lau et al. 2010), and Keap1 presence into sequestosomes seems to facilitate their clearance through autophagy (Fan et al. 2010), which would further reduce Keap1 levels and its ability to bind NFE2L2.
p62/SQSTM1 relevance in aging and age-related pathologies
p62/SQSTM1 is involved in a variety of cellular processes that are relevant for aging, including protein degradation (Pankiv et al. 2007; Seibenhener et al. 2004), mitophagy (Narendra et al. 2010a; Geisler et al. 2010a; Tang et al. 2011), oxidative stress response (Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010), metabolism regulation (Moscat and Diaz-Meco 2011), and inflammation (Lee et al. 2012). The role of p62/SQSTM1 in aging and age-related diseases is not fully understood yet, although experimental evidence suggests it is likely a critical factor: expression of p62/SQSTM1 declines with age in mice (Kwon et al. 2012), loss of p62/SQSTM1 reduces lifespan and shows several age-related effects in animal models, and reduced p62/SQSTM1 correlates with age-related pathologies in human tissues.
Neurodegenerative diseases
Alzheimer’s (AD), Parkinson’s (PD), Huntington’s (HD), and other neurodegenerative diseases are characterized by the accumulation of protein aggregates in the brain. A different aggregation-prone protein characterizes the pathology of each of these diseases, but virtually all these protein aggregates associate with p62/SQSTM1 (Kuusisto et al. 2001; Mizuno et al. 2006). The presence of p62/SQSTM1 in pathological protein inclusions initially suggested a causal role in their formation (Kuusisto et al. 2002), but recent evidence shows that p62/SQSTM1 actually mediates the degradation of at least some of these aggregate-prone proteins (Bjorkoy et al. 2005; Babu et al. 2005): in fact, p62/SQSTM1 shuttles the AD-associated protein tau to the proteasome for degradation (Babu et al. 2005). This suggests that p62/SQSTM1 has a protective role in AD. In line with this hypothesis, Sqstm1−/− mice show many characteristics of AD, including age-related accumulation of K63-Ub tau and paired helical filaments, neuronal death, impaired short-term memory, and increased anxiety (Ramesh Babu et al. 2008). Furthermore, the expression of p62/SQSTM1 is markedly reduced in the brains of AD patients, because of oxidative damage in its promoter region (Du et al. 2009a). Interestingly, the promoter of p62/SQSTM1 is increasingly damaged with age (Du et al. 2009a), and promoter damage and loss of p62/SQSTM1 expression is a common feature of several neurodegenerative diseases (Du et al. 2009b), suggesting a major involvement of the protein in the onset and the progression of most aggregate-presenting neurodegenerations.
Other common features of neurodegenerative diseases are oxidative stress and mitochondrial dysfunction (Lin and Beal 2006), thus p62/SQSTM1 may also protect the brain by promoting the activity of NFE2L2 and clearing damaged mitochondria, in addition to aggregate-prone proteins. This is evident especially in PD, where impaired mitochondrial dynamics and oxidative stress are among the main characteristics of the disease (Narendra et al. 2008; Buttner et al. 2008; Geisler et al. 2010b). p62/SQSTM1 aids in clearing dysfunctional mitochondria tagged for degradation by the Pink1-Parkin system, two genes involved in familial PD (see above and (Narendra et al. 2010a; Geisler et al. 2010a)). Loss of p62/SQSTM also correlates with reduced activity of NFE2L2 in neurons and astrocytes affected by neurodegenerative diseases (Du et al. 2009a; Ramsey et al. 2007). Interestingly, a lower incidence of PD can be found in individuals carrying a Nfe2l2 haplotype that correlates with increased transcriptional activity (von Otter et al. 2010), and expression of NFE2L2 in a Drosophila melanogaster model of PD is protective against neurodegeneration (Barone et al. 2011). Loss of p62/SQSTM1 in the brain could thus lead to increased oxidative stress, possibly eliciting a stress-induced senescence response in astrocytes (Bitto et al. 2010a), which can also contribute to the progression of AD and other neurodegenerative diseases (Salminen et al. 2011; Bhat et al. 2012).
Inclusion body myositis
Inclusion body myositis is the most common age-related myopathy. This disorder has been traditionally included in the category of inflammatory disorders of the musculoskeletal system (Ringel et al. 1987; Dalakas 1991) but recently, it has been recognized that the inflammatory response in inclusion body myositis is secondary to the degenerative changes in the muscle fibers (Askanas and Engel 2002), which include the presence of characteristic rimmed vacuoles, atrophy, and the accumulation of both amyloid-β precursor protein and tau proteins (Nogalska et al. 2011; Nogalska et al. 2010). The presence of both amyloid-β and tau accumulations suggest a common mechanisms with AD and perhaps other forms of neurodegenerative disorders. Clinical studies evaluating biomarkers of the disease have identified p62/SQSTM1 as a significant component of the intracellular inclusions found in inclusion body myositis (Inamori et al. 2012; Nogalska et al. 2009; Dubourg et al. 2011).
Age-related metabolic dysfunction
Aging is an important risk factor for several metabolic disorders, including obesity and type-2 diabetes (Gong and Muzumdar 2012). Impaired metabolic homeostasis in the elderly is likely due to a variety of environmental and genetic factors. Among these factors, p62/SQSTM1 seems to play a critical role in regulating metabolism in white adipose tissue, liver, and other metabolic organs: indeed, sqstm1−/− mice show mature-onset obesity and several features of metabolic syndrome, including: increased fat accumulation in the adipose and liver, impaired glucose tolerance and insulin sensitivity, increased plasma triglycerides and cholesterol levels, and decreased energy expenditure (Rodriguez et al. 2006). These effects are due to excessive signaling from the extracellular signal-regulated kinase 1 (ERK1) in response to insulin in Sqstm−/− mice: in physiological conditions, ERK1 signaling is dampened by p62/SQSTM1, which sequesters the kinase into p62 bodies through its PB1 domain, thereby preventing excessive proliferation and differentiation of white adipocytes (Rodriguez et al. 2006). Thus, loss of p62/SQSTM1 with age may contribute to the increased fat accumulation and metabolic dysfunctions seen in the elderly. Consistent with this hypothesis, p62/SQSTM1 is necessary for PKCζ activity (Sanz et al. 1999), which counteracts glucose intolerance by reducing the production of pro-inflammatory cytokines in mouse’s white adipocytes (Lee et al. 2010). p62/SQSTM1 expression may also favor the differentiation of brown over white adipocytes: knockout of Atg7 in white adipose tissue causes the white adipocytes to acquire features of brown adipocytes, including increased mitochondrial mass, increased β-oxidation and mitochondrial uncoupling, and formation of multiple lipid droplets per cell, instead of a single big lipid vacuole (Zhang et al. 2009). Although no causal relation was established between these changes and p62/SQSTM1 accumulation, p62/SQSTM1 levels are elevated in Atg7−/− adipocytes, due to impairment of autophagy. Thus, p62/SQSTM1 may favor the acquisition of brown fat features, for example, by promoting β-oxidation through PGC-1α, NFE2L2, and NRF1 transcription factors involved in the generation of mitochondria and in the expression of genes for lipid catabolism (Komatsu et al. 2010; Adam et al. 2010; Piantadosi et al. 2008). Brown fat is progressively lost with age, and higher amounts of brown fat have been negatively correlated with obesity, insulin resistance, and neurodegenerative diseases (see (Lecoultre and Ravussin 2011) and (Mattson 2010) for review). Hence, it is conceivable that loss of p62/SQSTM1 expression contributes to age-related loss of brown fat and thus to the onset of age-related metabolic dysfunctions.
Another p62/SQSTM1 partner, mTORC1, influences the differentiation and proliferation of white adipocytes. In fact, adipose-specific ablation of raptor reduces weight gain, fat accumulation, and white adipocytes size and number, while increasing glucose tolerance and insulin sensitivity in mouse (Polak et al. 2008). The mTORC1 pathway is also responsible for peripheral resistance to insulin: increasing the activity of S6K1, a mTORC1 substrate, can trigger a negative feedback on the insulin receptor substrate 1 (IRS-1), thereby impairing insulin signaling (Um et al. 2004). mTORC1 response to insulin is independent of p62/SQSTM1 (Duran et al. 2011) and the insulin signaling pathway is not impaired in young Sqstm1−/− mice (Rodriguez et al. 2006). Nevertheless, p62/SQSTM1 may help modulating the response to insulin by sequestering mTORC1 into insulin-insensitive, amino acid-responsive complexes, thereby reducing S6K1 activation and the negative feedback on IRS-1, when the adipocytes are excessively stimulated. In support of this hypothesis, the adipocytes of S6k1−/− mice show features of adipocyte-specific Atg7−/− mice, including increased mitochondrial mass, increased β-oxidation, and multiple lipid droplet per cell (Um et al. 2004). Importantly though, protein overload can also cause insulin resistance through the S6K1/IRS-1 negative feedback (Um et al. 2006). Thus, p62/SQSTM1 may also contribute to insulin resistance by increasing S6K1 activation in amino acid-sensitive tissues, like skeletal muscle and liver, while increasing insulin sensitivity in the more lipid and glucose-sensitive adipose. Tissue specific knockout studies may help define the role of p62/SQSTM1 in insulin resistance in more detail (Moscat and Diaz-Meco 2011).
In addition to insulin resistance, aging correlates also with decreased production of insulin (Gong and Muzumdar 2012). Insulin is released by the pancreatic β-islets in response to glucose availability. Briefly, glucose oxidation in the islets raises ATP concentration, which in turn closes ATP-sensitive K+ channels. The ensuing depolarization triggers a Ca2+spike which then promotes the release of insulin-containing vesicles. Interestingly, the β-islets from type-2 diabetes patients show mitochondrial dysfunction, which leads to impaired ATP production, and reduced insulin release (Anello et al. 2005). Given the importance of p62/SQSTM1 in maintaining mitochondrial homeostasis (Narendra et al. 2010a; Geisler et al. 2010a), it is possible that an age-related loss of p62/SQSTM1 in β cells could contribute to this mitochondrial dysfunction and thereby reduce insulin release. Furthermore, β cells regenerative/proliferative potential decreases with age in mouse, due to accumulation of the cell cycle-dependent kinase inhibitor p16INK4A. p16INK4A is a marker of cellular senescence (Alcorta et al. 1996), a terminal proliferative arrest triggered by a series of insults, including oxidative stress. Interestingly, autophagy impairment causes cellular senescence and accumulation of p16INK4A in human fibroblasts, because of increased oxidative stress from the mitochondria (Kang et al. 2011). Thus, loss of p62/SQSTM1 in the elderly may further reduce the production of insulin by promoting cellular senescence in β cells and thus diminishing their regenerative capacity.
p62/SQSTM1: a novel aging gene
The p62/SQSTM1 protein is involved in several age-related pathologies, but perhaps the most striking evidence of its importance in longevity is the premature aging phenotype of Sqstm1−/− mice (Kwon et al. 2012). Sqstm1−/− mice have reduced lifespan, show premature signs of aging and increased oxidative stress, as well as increased mitochondrial damage and dysfunction, due to loss of a basal NFE2L2 antioxidant response (Kwon et al. 2012). These features are recapitulated by observations in human fibroblasts and by other studies showing that chronic activation of mTORC1 with IGF-1 reduces proliferative potential and causes accumulation of damaged mitochondria and ROS, leading to cellular senescence (Bitto et al. 2010b; Handayaningsih et al. 2012). Conversely, long-lived Snell/Dwarf mice show increased basal activation of the NFE2L2 pathway, suggesting that IIS and the antioxidant response cooperate to influence longevity (Leiser and Miller 2010). In fact, Snell/Dwarf mice-derived fibroblasts show lower levels of mTORC1 activity and enhanced autophagy in response to stress (Wang and Miller 2012). Similarly, we and others show that prolonged reduction of mTORC1 signaling increases autophagy and delays cellular senescence (Demidenko et al. 2009; Lerner et al. 2013; Cao et al. 2011). Since the expression of p62/SQSTM1 is critical to prevent so many age-related processes and is necessary for the beneficial effects of rapamycin on human cells, it is possible that the pro-longevity effects of reducing IGF-1/mTORC1 signaling are due, at least in part, to altered p62/SQSTM1 dynamics. Considering that p62/SQSTM1 interacts with mTORC1 through Raptor (Duran et al. 2011), disrupting the assembly of mTORC1 with rapamycin probably increases the pool of p62/SQSTM1 available for selective autophagy and other homeostatic processes. In fact, rapamycin does not have any effect on the accumulation of dysfunctional mitochondria and the activation of the NFE2L2 pathway in cells with reduced p62/SQSTM1 expression (Lerner et al. 2013): thus, p62/SQSTM1 is necessary to activate these processes in response to rapamycin. In addition, reducing the expression of p62/SQSTM1 leads to an increase in senescence-associated markers (Lerner et al. 2013). These results suggest a role for p62/SQSTM1 in homeostatic processes that prevent cellular senescence. Maintaining these processes could prevent aging independently of mTOR modulation. Indeed, Sqstm1−/− mice accumulate damaged mitochondria and exhibit reduced activation of the NFE2L2 pathway and accelerated aging (Kwon et al. 2012). It would be interesting to see whether Sqstm1−/− mice or MEFs show increased markers of cellular senescence and whether clearing senescent cells in these mice would rescue the accelerated aging phenotype, as it does in the BubR1 progeroid mouse (Baker et al. 2011).
Genetic activation of p62/SQSTM1 has not been tested yet as a therapeutic approach for aging pathologies, such as neurodegenerative diseases, but related approaches have been tried successfully. For example, chronic treatment with rapamycin reduces the accumulation of amyloid plaques and the decline in cognitive behavior in a mouse model of AD which expresses the human amyloid precursor protein (hAPP) (Spilman et al. 2010). The expression and the degradation of p62/SQSTM1 are increased in human fibroblasts by chronic treatment with the mTORC1-inhibitor rapamycin, and rapamycin reduces the accumulation of damaged mitochondria in a p62/SQSTM1-dependent manner (Lerner et al. 2013). It is thus an intriguing possibility that rapamycin protects from AD by influencing the turnover of p62/SQSTM1 in vivo, as it does in vitro, possibly by increasing its expression through NFE2L2 (Jain et al. 2010). Changes in p62/SQSTM1 have been linked to neurodegeneration in vivo: damage to the Sqstm1 promoter has been identified in the brains of patients affected by neurodegenerative diseases (Du et al. 2009a; Du et al. 2009b), and knockout of Sqstm1 recapitulates many features of AD in mouse (Ramesh Babu et al. 2008). Genetic ablation of p62/SQSTM1 in hAPP mice would reveal whether p62/SQSTM1 is necessary for rapamycin to revert the symptoms of AD-like pathology in these animals and would confirm the importance of p62/SQSTM1 in preventing neurodegenerative disorders.
As suggested elsewhere (Nezis and Stenmark 2012), artificially increasing p62/SQSTM1 expression may have the same pro-longevity effects of mTORC1 inhibition, perhaps with the advantage of maintaining cell growth and proliferation. Overexpression of p62/SQSTM1 could reduce adipogenic signaling and inflammation, activate the antioxidant response, and sequester hazardous cellular components into inert p62 bodies. However, unchecked overexpression of p62/SQSTM1 can also have some deleterious effects, especially in the liver and when autophagy is impaired (Komatsu et al. 2010). Increasing the turnover of p62/SQSTM1 could achieve the same effects as increasing its expression, but may also increase clearance of sequestosomes containing damaged mitochondria and proteins, thereby maintaining pro-survival signaling pathways, and generally improving cellular and organism homeostasis. Many questions remained unanswered about the role of p62/SQSTM1 in physiological processes and in age-related pathologies. For example, it is still unknown how the various cellular processes converging on p62/SQSTM1 interact with each other and how these interactions, or lack thereof in aging, influence cellular homeostasis. In the past few years, exciting findings have begun to uncover the physiological relevance of this scaffold protein in aging, but a comprehensive and integrative view of its functions is yet to be found. We have provided a link between p62/SQSTM1 and the IGF-1/mTOR pathways, in the hope it will provide a framework to study the involvement of this protein in aging and age-related pathologies. Future studies will help define the importance of p62/SQSTM1 in vertebrate aging and whether modulating its expression can bring to a healthy aging population.
References
- Adam T, Opie LH, Essop MF. AMPK activation represses the human gene promoter of the cardiac isoform of acetyl-CoA carboxylase: role of nuclear respiratory factor-1. Biochem Biophys Res Commun. 2010;398(3):495–499. doi: 10.1016/j.bbrc.2010.06.106. [DOI] [PubMed] [Google Scholar]
- Alcorta DA, et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93(24):13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers A, et al. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5(6):847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anello M, et al. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia. 2005;48(2):282–289. doi: 10.1007/s00125-004-1627-9. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel W. Inclusion-body myositis and myopathies: different etiologies, possibly similar pathogenic mechanisms. Curr Opin Neurol. 2002;15(5):525–531. doi: 10.1097/00019052-200210000-00002. [DOI] [PubMed] [Google Scholar]
- Babu J, Geetha T, Wooten M. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94(f27f4e63-a010-5926-f8b4-116f27f87842):192–395. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- Baker D, et al. Clearance of p16(Ink4a)-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone M, Sykiotis G, Bohmann D. Genetic activation of Nrf2 signaling is sufficient to ameliorate neurodegenerative phenotypes in a Drosophila model of Parkinson's disease. Dis Models Mech. 2011;4(29b98216-6e5f-44c2-693d-1f4bd0dc641e):701–708. doi: 10.1242/dmm.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R et al (2012) Astrocyte senescence as a component of Alzheimer's disease. PLoS One 7(9) [DOI] [PMC free article] [PubMed]
- Bitto A, et al. Stress-induced senescence in human and rodent astrocytes. Exp Cell Res. 2010;316:2961–2968. doi: 10.1016/j.yexcr.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Bitto A et al (2010b) Long-term IGF-I exposure decreases autophagy and cell viability. PLoS One 5(9) [DOI] [PMC free article] [PubMed]
- Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman J, et al. Analysis of neural subtypes reveals selective mitochondrial dysfunction in dopaminergic neurons from parkin mutants. Proc Natl Acad Sci U S A. 2012;109(26):10438–10443. doi: 10.1073/pnas.1120688109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner S, et al. Functional mitochondria are required for alpha-synuclein toxicity in aging yeast. J Biol Chem. 2008;283(12):7554–7560. doi: 10.1074/jbc.M708477200. [DOI] [PubMed] [Google Scholar]
- Cao K et al (2011) Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med 3(89) [DOI] [PubMed]
- Chaudhary K, El-Sikhry H, Seubert J. Mitochondria and the aging heart. JGC. 2011;8(3):159–167. doi: 10.3724/SP.J.1263.2011.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, et al. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(2):668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A, Ryter S, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Copple I, et al. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem. 2010;285(81009124-2876-05da-270c-84b2eb4bf664):16782–16790. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. Autophagy and aging: keeping that old broom working. TIG. 2008;24(12):604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakas M. Polymyositis, dermatomyositis and inclusion-body myositis. N Engl J Med. 1991;325(21):1487–1498. doi: 10.1056/NEJM199111213252107. [DOI] [PubMed] [Google Scholar]
- Del Roso A, et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol. 2003;38(5):519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- Demidenko ZN, et al. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8(12):1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- Du Y, et al. Age-associated oxidative damage to the p62 promoter: implications for Alzheimer disease. Free Radic Biol Med. 2009;46(23d068fb-0594-c642-03d0-07ddf3c86ab0):492–993. doi: 10.1016/j.freeradbiomed.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Wooten M, Wooten M. Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiol Dis. 2009;35(685e8d56-503b-eab5-c2b8-07dd602f3f7f):302–312. doi: 10.1016/j.nbd.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg O et al (2011) Diagnostic value of markers of muscle degeneration in sporadic inclusion body myositis. Acta myologica: myopathies and cardiomyopathies: official journal of the Mediterranean Society of Myology / edited by the Gaetano Conte Academy for the study of striated muscle diseases. 30(2):103-108 [PMC free article] [PubMed]
- Duran A, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44(1):134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W et al (2010) Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy 6(a2b5697d-a785-653c-42b7-84b2eb17c4d1) [DOI] [PMC free article] [PubMed]
- Frenzel H, Feimann J. Age-dependent structural changes in the myocardium of rats. A quantitative light- and electron-microscopic study on the right and left chamber wall. Mech Ageing Dev. 1984;27(1):29–41. doi: 10.1016/0047-6374(84)90080-0. [DOI] [PubMed] [Google Scholar]
- Geetha T, Wooten M. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512(24344ce0-5262-a90c-1802-fc2e2b0a525f):19–43. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- Geetha T, Jiang J, Wooten M. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell. 2005;20(1db9e0fb-5676-7dcd-89aa-116f27f72f5c):301–313. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Geetha T, et al. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem Biophys Res Commun. 2008;374(f569f091-6fb4-2905-c0a9-1b280377cbcc):33–40. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6(7):871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky D. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9(9):859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson R, et al. (2011) Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Human Mol Genet 21(5):978–990 [DOI] [PMC free article] [PubMed]
- Gong Z, Muzumdar R. Pancreatic function, type 2 diabetes, and metabolism in aging. Int J Endocrinol. 2012;2012(94bf4f10-7e61-8f0d-47cb-395213c68671):320482. doi: 10.1155/2012/320482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode A, Layfield R. Recent advances in understanding the molecular basis of Paget disease of bone. J Clin Pathol. 2010;63(e67bfe6b-8256-f35a-f589-fc0dcd60d5a1):199–402. doi: 10.1136/jcp.2009.064428. [DOI] [PubMed] [Google Scholar]
- Handayaningsih A-E et al. (2012) IGF-I enhances cellular senescence via the reactive oxygen species-p53 pathway. Biochem Biophys Res Commun 425(2):478–484 [DOI] [PubMed]
- Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4(2):e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. The free radical theory of aging. Antioxid Redox Signal. 2003;5(5):557–561. doi: 10.1089/152308603770310202. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky D. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Gottschling D. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamori Y, et al. Inclusion body myositis coexisting with hypertrophic cardiomyopathy: an autopsy study. NMD. 2012;22(8):747–754. doi: 10.1016/j.nmd.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192(35dd33bb-6bd5-d6ad-3d10-fcbe10fbbcb6):17–44. doi: 10.1083/jcb.201009067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y, Van Remmen H. Age-associated alterations of the neuromuscular junction. Exp Gerontol. 2011;46(2–3):193–198. doi: 10.1016/j.exger.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137(4):721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I, Strominger J, Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci U S A. 1996;93(529cf5e6-37c8-49cb-37e6-008f126febd7):5991–5996. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HT, et al. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One. 2011;6(8):e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo A. Chaperone-mediated autophagy. Methods Mol Biol (Clifton, N.J.) 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10(552acb39-f20a-30f5-80fb-6f46f5589ac8):935–980. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kitzman D, Edwards W. Age-related changes in the anatomy of the normal human heart. J Gerontol. 1990;45(2):9. doi: 10.1093/geronj/45.2.m33. [DOI] [PubMed] [Google Scholar]
- Klass M. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6(6):413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo A. Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res Rev. 2011;10(2):205–215. doi: 10.1016/j.arr.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kageyama S, Ichimura Y (2012) p62/SQSTM1/A170: physiology and pathology. Pharmacol Res Off J Ital Pharmacol Soc 66(6):457–462 [DOI] [PubMed]
- Korolchuk V, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(47c733bb-2560-6596-2d60-07bda3a4c8b9):517–544. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusisto E, Salminen A, Alafuzoff I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport. 2001;12(10):2085–2090. doi: 10.1097/00001756-200107200-00009. [DOI] [PubMed] [Google Scholar]
- Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer's disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28(3bceda89-d3ed-f98e-1484-07daef4daadc):228–265. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- Kwon J, et al. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep. 2012;13(e6cd8e22-20d2-22e1-d21d-e3874e265257):150–156. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalley P. The aging respiratory system—pulmonary structure, function and neural control. Respir Physiol Neurobiol. 2013;187:199–210. doi: 10.1016/j.resp.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Lamark T, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278(4a65a5a4-71cb-2ade-e9ee-fc2e2b09dbc8):34568–34649. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Landfield P, et al. Hippocampal aging in rats: a morphometric study of multiple variables in semithin sections. Neurobiol Aging. 1981;2(4):265–275. doi: 10.1016/0197-4580(81)90034-8. [DOI] [PubMed] [Google Scholar]
- Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30(13):3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin N, et al. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70(6):1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoultre V, Ravussin E. Brown adipose tissue and aging. Curr Opin Clin Nutr Metab Care. 2011;14(1):1–6. doi: 10.1097/MCO.0b013e328341221e. [DOI] [PubMed] [Google Scholar]
- Lee S, et al. PKCzeta-regulated inflammation in the nonhematopoietic compartment is critical for obesity-induced glucose intolerance. Cell Metab. 2010;12(64b8228e-3e87-bfd5-890a-3e48b17b5d2c):65–142. doi: 10.1016/j.cmet.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, et al. Autophagy suppresses interleukin-1β (IL-1β) signaling by activation of p62 degradation via lysosomal and proteasomal pathways. J Biol Chem. 2012;287(21aff234-8231-4f10-7d43-1535e01980f9):4033–4073. doi: 10.1074/jbc.M111.280065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser S, Miller R. Nrf2 signaling, a mechanism for cellular stress resistance in long-lived mice. Mol Cell Biol. 2010;30(f6928770-a92a-c84b-70bf-3f502d906abb):871–955. doi: 10.1128/MCB.01145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner C et al (2013) Reduced mTOR activity facilitates mitochondrial retrograde signaling and increases lifespan in normal fibroblasts. Aging Cell (In press) [DOI] [PMC free article] [PubMed]
- Li W-W, Li J, Bao J-K. Microautophagy: lesser-known self-eating. CMLS. 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Longo V. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol Aging. 1999;20(5):479–486. doi: 10.1016/s0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- Mamidipudi V, Li X, Wooten M. Identification of interleukin 1 receptor-associated kinase as a conserved component in the p75-neurotrophin receptor activation of nuclear factor-kappa B. J Biol Chem. 2002;277(48659987-f63a-21a3-c089-152bf7a64ec3):28010–28018. doi: 10.1074/jbc.M109730200. [DOI] [PubMed] [Google Scholar]
- Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6(6):458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mansouri A, et al. Alterations in mitochondrial function, hydrogen peroxide release and oxidative damage in mouse hind-limb skeletal muscle during aging. Mech Ageing Dev. 2006;127(3):298–306. doi: 10.1016/j.mad.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Marcus S, et al. A p56(lck) ligand serves as a coactivator of an orphan nuclear hormone receptor. J Biol Chem. 1996;271(b9b0da87-fccd-43cc-0095-0174c302442e):27197–27397. doi: 10.1074/jbc.271.44.27197. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, et al. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44(75e058b6-11c7-0417-c1eb-374a0a09fa5f):279–368. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Perspective: does brown fat protect against diseases of aging? Ageing Res Rev. 2010;9(1):69–76. doi: 10.1016/j.arr.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, et al. Immunoreactivities of p62, an ubiquitin-binding protein, in the spinal anterior horn cells of patients with amyotrophic lateral sclerosis. J Neurol Sci. 2006;249(1):13–18. doi: 10.1016/j.jns.2006.05.060. [DOI] [PubMed] [Google Scholar]
- Mizushima N, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116(Pt 9):1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco M (2011) Feedback on fat: p62-mTORC1-autophagy connections. Cell 147(7fe468c7-09ee-421a-bbe5-0fa750b8a785) [DOI] [PMC free article] [PubMed]
- Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(89cd27ab-ae69-8528-c6f4-1f3f639dc970):795–1598. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6(8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis I, Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid Redox Signal. 2012;17(5):786–793. doi: 10.1089/ars.2011.4394. [DOI] [PubMed] [Google Scholar]
- Nezis I, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180(6):1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalska A, et al. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118(3):407–413. doi: 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- Nogalska A, et al. Novel demonstration of amyloid-β oligomers in sporadic inclusion-body myositis muscle fibers. Acta Neuropathol. 2010;120(5):661–666. doi: 10.1007/s00401-010-0737-3. [DOI] [PubMed] [Google Scholar]
- Nogalska A, et al. Novel demonstration of conformationally modified tau in sporadic inclusion-body myositis muscle fibers. Neurosci Lett. 2011;503(3):229–233. doi: 10.1016/j.neulet.2011.08.042. [DOI] [PubMed] [Google Scholar]
- Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T. Mitochondrial DNA mutations and age. Ann N Y Acad Sci. 1998;854:128–154. doi: 10.1111/j.1749-6632.1998.tb09898.x. [DOI] [PubMed] [Google Scholar]
- Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Pankiv S, et al. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285(3c704f44-98ab-1180-47d3-0682c131049e):5941–5994. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Piantadosi CA, et al. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res. 2008;103(11):1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8(84e93330-7322-b853-cd6e-3a6ea7481b27):399–809. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Porta EA. Advances in age pigment research. Arch Gerontol Geriatr. 1991;12(2–3):303–320. doi: 10.1016/0167-4943(91)90036-p. [DOI] [PubMed] [Google Scholar]
- Rachubinski R, Marcus S, Capone J. The p56(lck)-interacting protein p62 stimulates transcription via the SV40 enhancer. J Biol Chem. 1999;274(5ae492ce-035d-a8aa-4ee9-fc2e2b0a2e07):18278–18362. doi: 10.1074/jbc.274.26.18278. [DOI] [PubMed] [Google Scholar]
- Ramesh Babu J, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106(27a74d97-a739-d297-646d-16c8da17e322):107–127. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- Ramsey C, et al. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66(6a48f324-2d51-2aa2-8725-1b0928e05358):75–160. doi: 10.1097/nen.0b013e31802d6da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Ringel S, et al. Spectrum of inclusion body myositis. Arch Neurol. 1987;44(11):1154–1157. doi: 10.1001/archneur.1987.00520230042011. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15(5):713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(0a30ac0c-ca9e-5ee9-59ec-24cbc90092cf):211–233. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Salminen A, et al. Astrocytes in the aging brain express characteristics of senescence-associated secretory phenotype. Eur J Neurosci. 2011;34(1):3–11. doi: 10.1111/j.1460-9568.2011.07738.x. [DOI] [PubMed] [Google Scholar]
- Salmon A, et al. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289(1):9. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Sci (New York, N.Y.) 2008;320(56ca5f23-896a-b94e-de3e-0c4be356193c):1496–1997. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(01f0f45f-84d6-a5ce-e325-0c4b1f2cde36):290–593. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez P, et al. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18(6abf35eb-a291-e542-7256-008f126e35a9):3069–3149. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, et al. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999;18(28a44be7-14d0-4c74-cc5a-01ea9df668a3):3044–3097. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, et al. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19(dec466e5-a9e0-db20-d2db-01c4befea22f):1576–1662. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D. Age-related changes in liver structure and function: implications for disease ? Exp Gerontol. 2005;40(8–9):650–659. doi: 10.1016/j.exger.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Sachs H. Quantifying dense bodies and lipofuscin during aging: a morphologist's perspective. Arch Gerontol Geriatr. 2002;34(3):249–261. doi: 10.1016/s0167-4943(01)00218-7. [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo A, et al. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123(Pt 15):2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Spilman P, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbaugh M, et al. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am J Physiol Endocrinol Metab. 2012;303(4):95. doi: 10.1152/ajpendo.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stępkowski T, Kruszewski M. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic Biol Med. 2011;50(36577d97-5454-ace8-d631-84b2eb1acb05):1186–1281. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- Szweda P, et al. Aging, lipofuscin formation, and free radical-mediated inhibition of cellular proteolytic systems. Ageing Res Rev. 2003;2(4):383–405. doi: 10.1016/s1568-1637(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Tan J, et al (2008) Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy 4:251–253
- Tan JM, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17(3):431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- Tang F et al (2011) RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS One 6(2caa5a85-183c-3c9d-537c-84b2eb293c9e) [DOI] [PMC free article] [PubMed]
- Toth SE. The origin of lipofuscin age pigments. Exp Gerontol. 1968;3(1):19–30. doi: 10.1016/0531-5565(68)90052-1. [DOI] [PubMed] [Google Scholar]
- Tóth M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Um S, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431(1ae7851e-f1fe-5adc-6b8c-3a5109b98611):200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Um S, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3(fb9365a2-b3f2-6ed9-7322-3edcb615be48):393–795. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R, Shin J. Genomic structure and promoter analysis of the p62 gene encoding a non-proteasomal multiubiquitin chain binding protein. FEBS Lett. 1998;435(2–3):138–142. doi: 10.1016/s0014-5793(98)01021-7. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R, et al. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J BiolChem. 1996;271(e002a393-07b2-a95e-5354-fc56877148c3):20235–20242. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- Vaughan D, Peters A. Neuroglial cells in the cerebral cortex of rats from young adulthood to old age: an electron microscope study. J Neurocytol. 1974;3(4):405–429. doi: 10.1007/BF01098730. [DOI] [PubMed] [Google Scholar]
- von Otter M, et al. Association of Nrf2-encoding NFE2L2 haplotypes with Parkinson's disease. BMC Med Genet. 2010;11(ea342ab4-65bd-117a-1b99-1f4bd0e008b2):36. doi: 10.1186/1471-2350-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Miller R. Fibroblasts from long-lived mutant mice exhibit increased autophagy and lower TOR activity after nutrient deprivation or oxidative stress. Aging Cell. 2012;11(4):668–674. doi: 10.1111/j.1474-9726.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Tanaka M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J Cell Sci. 2011;124(Pt 16):2692–2701. doi: 10.1242/jcs.081232. [DOI] [PubMed] [Google Scholar]
- Wei M et al (2008) Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet 4(1) [DOI] [PMC free article] [PubMed]
- Wooten M, et al. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J Biol Chem. 2001;276(95729bab-6726-13b1-9482-01c430398cd6):7709–7721. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- Wooten M, et al. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem. 2005;280(2978358a-d99d-7f3a-c8da-01c23fc80840):35625–35634. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky D. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19(8):3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23(7):798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106(f900c956-7e9b-e365-2109-3a67f3160fb4):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]