Abstract
Long-lived individuals delay aging and age-related diseases like diabetes, hypertension, and cardiovascular disease. The exact underlying mechanisms are largely unknown, but enhanced mitochondrial biogenesis and preservation of mitochondrial function have been suggested to explain healthy ageing. We investigated whether individuals belonging to long-lived families have altered mitochondrial DNA (mtDNA) content, as a biomarker of mitochondrial biogenesis and measured expression of genes regulating mitochondrial biogenesis. mtDNA and nuclear DNA (nDNA) levels were measured in blood samples from 2,734 participants from the Leiden Longevity Study: 704 nonagenarian siblings, 1,388 of their middle-aged offspring and 642 controls. We confirmed a negative correlation of mtDNA content in blood with age and a higher content in females. The middle-aged offspring had, on average, lower levels of mtDNA than controls and the nonagenarian siblings had an even lower mtDNA content (mtDNA/nDNA ratio = 0.744 ± 0.065, 0.767 ± 0.058 and 0.698 ± 0.074, respectively; pcontrols-offspring = 3.4 × 10−12, pcontrols-nonagenarians = 6.5 × 10−6), which was independent of the confounding effects of age and gender. Subsequently, we examined in a subset of the study the expression in blood of two genes regulating mitochondrial biogenesis, YY1 and PGC-1α. We found a positive association of YY1 expression and mtDNA content in controls. The observed absence of such an association in the offspring suggests an altered regulation of mitochondrial biogenesis in the members of long-lived families. In conclusion, in this study, we show that mtDNA content decreases with age and that low mtDNA content is associated with familial longevity. Our data suggest that preservation of mitochondrial function rather than enhancing mitochondrial biogenesis is a characteristic of long-lived families.
Keywords: Mitochondria, mtDNA, Healthy aging, Familial longevity, Mitochondrial biogenesis
Introduction
Mitochondria are the major source of energy in eukaryotic cells. Due to this important role in energy supply, their involvement in the aging process has already been postulated decades ago (Wallace 1992). Many more recent studies showed indeed reduced mitochondrial function in aging, e.g., a decrease in transcription and translation of mitochondrial proteins, enzyme activity, mitochondrial ATP synthesis, and overall organelle function has been observed (reviewed by Hebert et al. 2010). Mitochondria have their own circular DNA (mitochondrial DNA, mtDNA) which encodes several genes important for correct mitochondrial function (Wallace 1992) (www.mitomap.org). The amount of mtDNA differs between cell types depending on their energy consumption and is referred to as mtDNA content. mtDNA content is often used as a surrogate biomarker for the amount of mitochondria in a cell which, in itself, is the result of the balance between removal of damaged mitochondria, a process called mitophagy, and mitochondrial biogenesis, which is under control of the mTOR pathway (Scarpulla et al. 2012). In humans, associations of mtDNA content with chronological age and age-related diseases such as cancer, diabetes, obesity, and depression have been reported (reviewed by Malik and Czajka 2013). However, the relation of mtDNA content with many diseases is inconclusive since studies report a decrease, increase, or no difference, even for the same disease (Malik and Czajka 2013). Nevertheless, the general assumption is that mtDNA content decreases with age and that an altered, mostly lower, mtDNA content is associated with age-related diseases.
In various animal species, life span extension due to caloric restriction was ascribed to preservation of or enhanced mitochondrial function (Gouspillou and Hepple 2013). Given the age-related decline in mitochondrial function in humans, one could envision that long-lived individuals also preserve mitochondrial function. This could be either through increased mitochondrial biogenesis or via protection of the integrity and function of existing mitochondria. Recently, we showed familial longevity to be associated with reduced expression of genes in the mTOR pathway in blood samples of individuals belonging to long-lived families from the Leiden Longevity Study (Passtoors et al. 2013). The mTOR pathway is a known positive regulator of genes involved in mitochondrial biogenesis like PGC-1α and YY1 (Scarpulla et al. 2012). These studies suggest that decreased mitochondrial biogenesis and preservation of mitochondrial function are characteristics of longevity, rather than increased mitochondrial biogenesis. We now investigated this hypothesis in further detail in long-lived families from the Leiden Longevity Study (LLS).
In this study, mtDNA content in the blood of nonagenarians and their offspring from the LLS was compared with that of controls, who are the similarly aged partners of the offspring (Schoenmaker et al. 2006). Previously, we showed that the middle-aged LLS offspring are healthier and have less age-related diseases compared to controls, which is independent from BMI or lifestyle (Westendorp et al. 2009). In addition, we investigated the relation between mtDNA content and prospective survival during ~7.5 years of follow-up in these families. Finally, we examine the relation between mtDNA content and levels of PGC-1α and YY1 gene expression in blood in a subset of the study.
Thus, by investigating the association of mtDNA content with familial longevity, prospective survival, and expression of genes involved in mitochondrial biogenesis in long-lived families, we aim to further elucidate the role of mitochondria in healthy aging.
Materials and methods
The Leiden Longevity Study
Nonagenarian siblings of European descent were recruited when aged older than 89 years for men and 91 years for women and having at least one sister or a brother fulfilling these age criteria. Offspring of the nonagenarian siblings and their spouses (as population controls) were also asked to participate in the study. In total, 3,359 individuals were included: 944 long-lived proband siblings with a mean age of 94 years (range 89–104 years), 1,671 offspring with a mean age of 60 years (range 39–81 years), and 744 controls with a mean age of 60 years (range 36–79 years) at baseline. The LLS was approved by the Medical Ethical Committee of Leiden University Medical Centre, and all participants gave written informed consent.
Anthropometry, blood sampling, blood measurements, and DNA extraction
Blood samples were taken at baseline, and blood cell counts were performed using standard procedures. DNA was isolated from whole EDTA blood using standard techniques in two different isolation centers: one in the Netherlands and one in Finland. In a small number of participants (n = 101), DNA was isolated from buccal cells after a mouth swab. In this study (data not shown) and in our previous study (Reiling et al. 2010), we observed a significant difference in mtDNA content in buccal cells compared to DNA isolated from blood. Therefore, we excluded these individuals from the study. In addition, we excluded DNA isolated from citrate blood (n = 208). Weight, height, and other anthropometric and biochemical measurements were determined according to standard procedures on the day of the research visit as described previously (Westendorp et al. 2009). BMI was not available in the nonagenarian siblings. Of the participants, 316 were excluded because of incomplete data, failed mtDNA content measurements, or outliers in any of the characteristics/measurements, defined as a deviation from the mean >3 standard deviations.
Measurement of mtDNA content
mtDNA content was measured using quantitative PCR and normalized against the amount of nuclear DNA as described previously (Reiling et al. 2010). In short, for the determination of the amount of mtDNA, a 62-bp mtDNA fragment in the MT-ND1 gene was amplified using primers 5′TCATATTATGGCCAAGGGTC and 5′CTCCTTTAACCTCTCCACCC, while for nuclear DNA, a 198-bp fragment in the beta-globin (HBB) gene was amplified using primers 5′TTTTCCCACCCTTAGGCTG and 5′CTCACTCAGTGTGGCAAAG. Each sample was measured in triplicate using Absolute qPCR SYBR Green ROX (Thermo Fisher Scientific Inc., Waltham, MA, USA) on the Applied Biosystems 7900HT system (Applied Biosystems, Foster City, CA, USA). A reference curve consisting of a serial dilution of a standard DNA was used for the quantification of mtDNA and nuclear DNA (nDNA), and the mtDNA content was calculated as the mtDNA/nDNA ratio in arbitrary units (a.u.).
Measurement of gene expression in blood
Gene expression of PGC-1α and YY1 was measured in whole blood by reverse transcriptase quantitative PCR as described previously (Passtoors et al. 2013). In short, we randomly selected offspring and controls with the largest possible age range while balancing gender between the groups (noffspring = 257, ncontrols = 262). RNA was isolated from whole blood using the PAXgene Blood RNA Kit (Qiagen, Venlo, The Netherlands), and cDNA was prepared using the First Strand cDNA Synthesis Kit (Roche Applied Science, Almere, The Netherlands). Gene expression of the two genes was measured using Taqman gene expression assays (Applied Biosystems) on the BioMark™ 48.48 and 96.96 Dynamic Arrays (Fluidigm, Amsterdam, The Netherlands). Relative gene expression was calculated using the 2−∆∆Ct method, and YKT6 was used as a reference gene.
Statistical analysis
Differences in mtDNA content between the dichotomous measures were analyzed using logistic regression with adjustment for age, isolation center, blood cell counts, familial relatedness, and gender (if appropriate). Continuous measures were analyzed with linear regression models.
The effect of mtDNA on prospective survival was assessed using a sex-adjusted, left-truncated Cox proportional hazards model to adjust for late entry into the data set according to age. The analyses were performed on 2,020 offspring+controls and 703 nonagenarians that have been followed for ~7.5 years.
For all statistical analyses, Stata 11.2 (StataCorp, College Station, TX, USA) was used. Results were regarded significant if p ≤ 0.05.
Results
Differences in mtDNA content between nonagenarian siblings, their offspring, and controls
In total, 2,770 participants of the LLS fulfilled the inclusion criteria and passed quality control of mtDNA content measurements. The characteristics of the participants included in the study are given in Table 1.
Table 1.
Characteristics of the Leiden Longevity Study participants included in this study
| Nonagenarian siblings | Offspring | Controls | |
|---|---|---|---|
| n = 704 | n = 1,388 | n = 642 | |
| N sibships | 369 | 607 | 0 |
| Age (years) | 93.2 ± 2.5 | 59.3 ± 6.5 | 58.7 ± 7.4 |
| Gender (% men) | 38.9 | 46.9 | 42.4 |
| BMI (kg/m2) | NA | 25.2 ± 3.2 | 25.4 ± 3.3 |
| Isolation center (% Leiden) | 44.0 | 50.9 | 54.4 |
| mtDNA/nDNA ratio | 0.698 ± 0.074 | 0.744 ± 0.065 | 0.767 ± 0.058 |
Data are means±standard deviation
NA not available
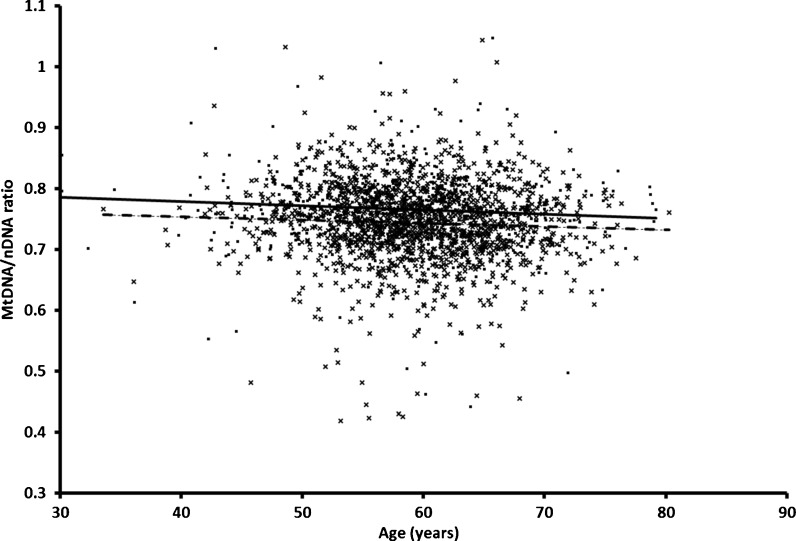
In the analysis of all groups combined, women had a significantly higher mtDNA content than men (difference 0.0167 ± 0.0025, p = 2.4 × 10−11) and there was a significant negative association with age (β = −0.0007 ± 0.0001, p = 4.7 × 10−8 (Fig. 1). The association of mtDNA content with age was similar for offspring and controls (β = −0.0006 ± 0.0003, p = 0.030 and β = −0.0005 ± 0.0003, p = 0.09, respectively). We did not observe a significant association with BMI (p = 0.24).
Fig. 1.
mtDNA content associates with age in controls and offspring of nonagenarians. For each participant in the control and offspring group, mtDNA/nDNA ratios are shown at the age of DNA collection. Controls are shown as black squares and the linear regression as a solid black line. Offspring of the nonagenarians are shown as a cross and the linear regression as a dashed line
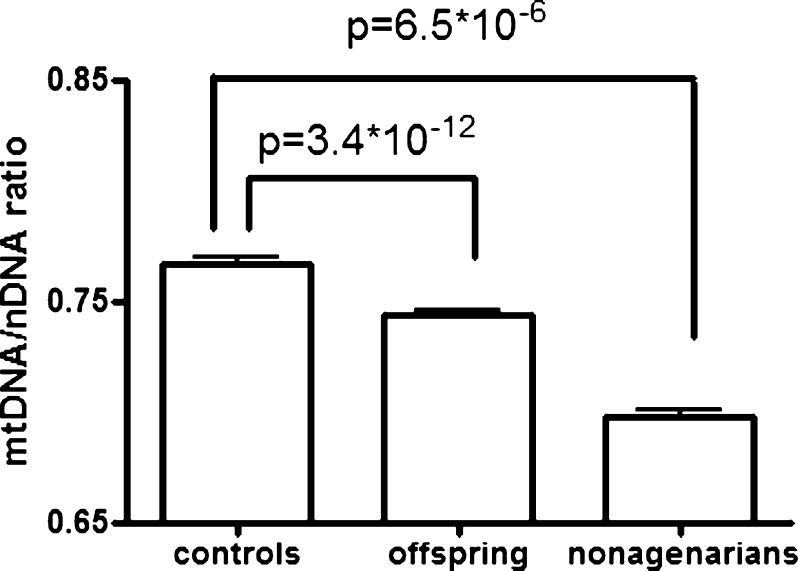
When we examined the average mtDNA content in each group and adjusted for the potential confounding effects of age and gender, we observed that the offspring of nonagenarians had lower levels of mtDNA than the similarly aged controls (pcontrols-offspring = 3.4 × 10−12, Table 1), whereas nonagenarian parents had the lowest mtDNA content (pcontrols-nonagenarians = 6.5 × 10−6; Table 1, Fig. 2). Additional adjustment for blood cell counts and BMI did not change any of the results (data not shown). Our analysis indicates that mtDNA content is negatively associated with familial longevity.
Fig. 2.
mtDNA content in the nonagenarian siblings, their offspring, and controls. MtDNA content is given as the mtDNA/nDNA ratio in arbitrary units corrected for age, gender, DNA isolation location, and family relatedness. Error bars represent the SEM
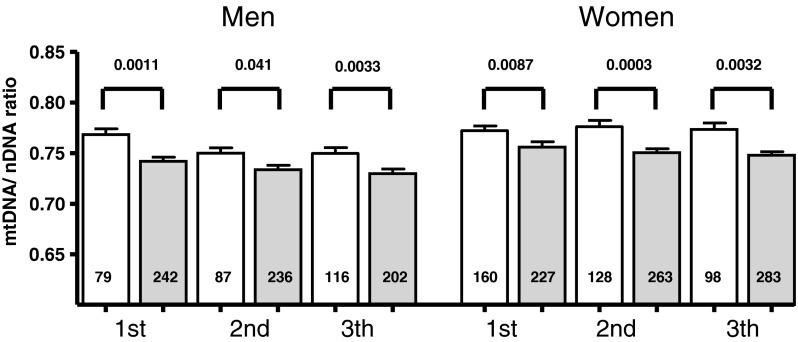
Given the association with age, gender, and familial longevity, we stratified the average mtDNA content analysis in gender-specific age tertiles for both the offspring and control groups separately to test the robustness of the observed association with familial longevity. As shown in Fig. 3, the observed lower mtDNA content in the offspring group was consistent across all age categories and in both genders.
Fig. 3.
Difference in mtDNA content across different age categories in male and female controls and offspring of nonagenarians. Data represent mean mtDNA content given as the mtDNA/nDNA ratio±SEM across age tertiles in men and women separately. The number in each column represents the number of subjects in each group. The age tertiles were defined as follows: for men, first age ≤57.05, second age >57.05 and ≤62.97, and third age tertile >62.97; for women, first age ≤55.88, second age >55.88 and ≤61.53, and third age tertile >61.53. Open bars reflect mtDNA content in controls and gray bars represent the offspring
mtDNA and survival
Since we observed an association with familial longevity, we next examined whether mtDNA content is associated with survival of all individuals in the study during ~7.5 years of follow-up. We did not observe any relation between mtDNA content and prospective survival, neither in the offspring and controls combined nor in the nonagenarian siblings (offspring+controls (ndeaths = 101): hazard ratio = 2.07 (95 % CI 0.06–72.70), p = 0.69; nonagenarian siblings (ndeaths = 602): hazard ratio = 0.44 (95 % CI 0.12–1.60), p = 0.21).
Gene expression of PGC-1α and YY1 and mtDNA content
PGC-1α and YY1 are positive regulators of mitochondrial biogenesis under the influence of the mTOR pathway; therefore, we tested whether their gene expression in blood could explain the lower mtDNA content in the members of long-lived families. Offspring of nonagenarians, however, showed similar gene expression levels of PGC-1α and YY1 in blood as controls (p = 0.32 and p = 0.60, respectively). In addition, we explored whether gene expression levels of PGC-1α and YY1 were correlated with mtDNA content. Gene expression levels of PGC-1α were not associated with mtDNA content (p = 0.79), but the expression of YY1 was positively associated with mtDNA content (beta = +0.044 ± 0.017, p = 0.010). The association of mtDNA content and gene expression level was not equal for the offspring and controls, showing a significant interaction with group status (controls or offspring of nonagenarians, p = 0.021). Stratification for group status revealed a significant positive association in controls (beta = +0.017 ± 0.007, p = 0.021), but not in the offspring (beta = −0.011 ± 0.0.009, p = 0.24; Table 2), indicating that the lower mtDNA content in the members of long-lived families may be the result of altered regulation of mitochondrial biogenesis via YY1.
Table 2.
Association between mtDNA content and gene expression in blood
| Gene | Controls (n = 262) | p value | Offspring (n = 257) | p value |
|---|---|---|---|---|
| PGC-1α | +0.079 ± 0.161 | 0.63 | +0.150 ± 0.102 | 0.14 |
| YY1 | +0.017 ± 0.007 | 0.021 | −0.011 ± 0.009 | 0.24 |
Data represent beta and p values for the linear regression analysis between mtDNA content and gene expression with age, gender, and isolation center as covariates
Discussion
In two generations of the longevity families and controls, mtDNA content in blood is negatively associated with chronological age and the mtDNA content in women is higher compared to men, which is in line with our previous study (Reiling et al. 2010). However, both generations of individuals from long-lived families have lower mtDNA content compared to middle-aged population controls, irrespective of gender or age. Furthermore, the expression of a regulator of mitochondrial biogenesis, YY1, is positively associated with mtDNA content in the control group only. This relatively lower mtDNA content in middle-aged individuals from long-lived families and the lack of association between the expression of a regulator of mitochondrial biogenesis, YY1, and mtDNA content in this group support the hypothesis that the members of longevity families delay mitochondrial aging by preservation of mitochondrial function rather than by increasing mitochondrial biogenesis.
In various animal species, it is possible to increase life span and to delay diseases related to old age by caloric restriction (Omodei and Fontana 2011). Studies in these species further showed a correlation between caloric restriction and mitochondrial function, and it has been suggested that caloric restriction promotes mitochondrial biogenesis, thereby delaying mitochondrial aging (Civitarese et al. 2007; Hepple et al. 2006; Baker et al. 2006). However, a recent study in mice and several other observations challenge this hypothesis (Lanza et al. 2012; Gouspillou and Hepple 2013). In the mouse study by Lanza and colleagues, the beneficial effects of caloric restriction on mitochondrial oxidative capacity and efficiency occur without significant effects on mitochondrial biogenesis, as also reflected by significantly reduced mtDNA content in calorie-restricted animals in their study (Lanza et al. 2012). The authors conclude: “caloric restriction preserves mitochondrial function by protecting the integrity and function of existing cellular components rather than by increasing mitochondrial biogenesis”. Based on their experiments, the authors attribute this to attenuation of oxidative damage by decreasing ROS production and increasing the endogenous antioxidant activity. In light of the findings depicted above, our results are of particular interest as they show that nonagenarian siblings and their middle-aged offspring have a low mtDNA content as compared to normally ageing controls. Based on the work by Lanza et al., we thus hypothesize that longevity families do not increase mitochondrial biogenesis to circumvent mitochondrial aging but, similar to the caloric restricted mice, are able to better preserve mitochondrial function. A better preserved mitochondrial function would slow the aging process and development of age-related diseases like diabetes and cardiovascular disease whose prevalence is indeed decreased in longevity families (Westendorp et al. 2009). With respect to the current findings, it is important to realize that the differences in health in the longevity families were not attributable to differences in BMI, food intake, or lifestyle (Westendorp et al. 2009).
Recent studies showed that an altered balance between nuclear and mitochondrial gene expression is associated with life span in various animal species (Houtkooper et al. 2013; Gomes et al. 2013). However, at present, data in humans is lacking. Our current finding of lower mtDNA content in long-lived families is seemingly in line with the proposed altered balance from these animal studies. Furthermore, we recently reported a lower mRNA expression of the mTOR pathway in whole-blood samples of long-lived participants from the LLS (Passtoors et al. 2013). mTOR is a known regulator of mitochondrial biogenesis via YY1 and PGC-1alpha (Cunningham et al. 2007). Interestingly, we observed the expected positive association between the regulator of mitochondrial biogenesis YY1 and mtDNA content in the blood of controls but not in the offspring of nonagenarians. This further suggests that the regulation of mitochondrial biogenesis is altered in these long-lived families. However, additional studies are needed to confirm this.
Whereas the members of long-lived families have lower levels of mtDNA content, we did not observe that lower mtDNA content is predictive for prospective survival. This might be due to the relatively low number of deaths in the middle-aged controls and offspring affecting power in this group. On the other hand, it might well be that survival into old age, as in our nonagenarian individuals, diminishes the effects of mtDNA content on prospective survival in that age category. Future larger prospective studies, covering a wider age range, will be needed to elucidate the role of mtDNA content in blood on survival.
Previous studies showed that exercise has beneficial effects on healthy aging, mitochondrial function, and mitochondrial biogenesis. It was also shown that exercise increases mtDNA content in muscle and that preservation of mitochondrial function in muscle is important in aging (Fiuza-Luces et al. 2013; Lanza et al. 2008; Safdar et al. 2011; Wisloff et al. 2005). Based on this, one could argue that our results could be due to differences in the level of physical activity. In the present study, we did not have information about the level of physical activity. However, the offspring and controls are partners of each other and are likely to have a similar lifestyle (Westendorp et al. 2009). Furthermore, in a previous study, we did not observe evidence for a confounding effect of self-reported level of exercise on mtDNA content in blood (Reiling et al. 2010), suggesting that mtDNA levels in blood are perhaps less influenced by the level of physical activity compared to muscle. To the best of our knowledge, there are no large controlled studies reporting on the relation between exercise and mtDNA content in blood.
Limitations of the current study include the use of DNA and RNA isolated from whole-blood samples. Therefore, results should be interpreted with caution when extrapolating them to other, perhaps more relevant tissues, such as, for instance, muscle. Another limitation of our study relates to the use of a single cohort from The Netherlands. To validate our results, replication in other cohorts with different ethnicities and age ranges is needed. This would allow to study whether our results also apply to non-familial longevity and to the oldest old, i.e., centenarians. Preferably, such studies should also take into account differences in the level of physical activity. Furthermore, mtDNA content was used as a surrogate measure of mitochondrial content and density. Direct measurements of mitochondrial proteins and mitochondrial function are necessary to confirm the associations. In addition, the gene expression studies are based on rather limited sample sizes and we thus cannot exclude false positives or small effects that were undetectable in our experimental design. Hence, further detailed studies are necessary to elucidate the full implications of our findings.
In conclusion, in this study, we show that low mtDNA content in blood is associated with familial longevity and that members of long-lived families have an altered association between YY1 gene expression and mtDNA content, suggesting lower mitochondrial biogenesis. Hence, we conclude that members of long-lived families delay the aging process, not by increasing mitochondrial biogenesis, but by preservation of mitochondrial function.
Acknowledgments
We thank all the participants of the Leiden Longevity Study. The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2011) under grant agreement no. 259679. This study was supported by a grant from the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Ageing (grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), by Unilever Colworth and by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007), and a grant from ZonMW, Priority Medicines Elderly program (project no. 113102006).
References
- Baker DJ, Betik AC, Krause DJ, Hepple RT. No decline in skeletal muscle oxidative capacity with aging in long-term calorically restricted rats: effects are independent of mitochondrial DNA integrity. J Gerontol A Biol Sci Med Sci. 2006;61:675–684. doi: 10.1093/gerona/61.7.675. [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Fiuza-Luces C, Garatachea N, Berger NA, Lucia A. Exercise is the real polypill. Physiology (Bethesda) 2013;28:330–358. doi: 10.1152/physiol.00019.2013. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouspillou G, Hepple RT. Facts and controversies in our understanding of how caloric restriction impacts the mitochondrion. Exp Gerontol. 2013;48(10):1075–1084. doi: 10.1016/j.exger.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Hebert SL, Lanza IR, Nair KS. Mitochondrial DNA alterations and reduced mitochondrial function in aging. Mech Ageing Dev. 2010;131:451–462. doi: 10.1016/j.mad.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepple RT, Baker DJ, McConkey M, Murynka T, Norris R. Caloric restriction protects mitochondrial function with aging in skeletal and cardiac muscles. Rejuvenation Res. 2006;9:219–222. doi: 10.1089/rej.2006.9.219. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza IR, Zabielski P, Klaus KA, Morse DM, Heppelmann CJ, Bergen HR, III, Dasari S, Walrand S, Short KR, Johnson ML, Robinson MM, Schimke JM, Jakaitis DR, Asmann YW, Sun Z, Nair KS. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012;16:777–788. doi: 10.1016/j.cmet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;5:481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Omodei D, Fontana L. Calorie restriction and prevention of age-associated chronic disease. FEBS Lett. 2011;585:1537–1542. doi: 10.1016/j.febslet.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passtoors WM, Beekman M, Deelen J, van der Breggen R, Maier AB, Guigas B, Derhovanessian E, van Heemst D, de Craen AJ, Gunn DA, Pawelec G, Slagboom PE. Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell. 2013;12:24–31. doi: 10.1111/acel.12015. [DOI] [PubMed] [Google Scholar]
- Reiling E, Ling C, Uitterlinden AG, van 't Riet E, Welschen LM, Ladenvall C, Almgren P, Lyssenko V, Nijpels G, van Hove EC, Maassen JA, de Geus EJ, Boomsma DI, Dekker JM, Groop L, Willemsen G, 't Hart LM (2010) The association of mitochondrial content with prevalent and incident type 2 diabetes. J Clin Endocrinol Metab 95:1909–1915 [DOI] [PubMed]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci USA. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenmaker M, de Craen AJ, de Meijer PH, Beekman M, Blauw GJ, Slagboom PE, Westendorp RG. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet. 2006;14:79–84. doi: 10.1038/sj.ejhg.5201508. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- Westendorp RG, van Heemst D, Rozing MP, Frolich M, Mooijaart SP, Blauw GJ, Beekman M, Heijmans BT, de Craen AJ, Slagboom PE. Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: the Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, Fernstrom M, Rezaei K, Lee SJ, Koch LG, Britton SL. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]