Abstract
It is well established that insulin-like growth factor 1 (IGF-1) circulating levels correlate with age and that heritability and influence of IGF-1 gene variation on IGF-1 levels also well-known. However, the influence of age on the genetic factors determining IGF-1 levels is not clear. In this study, we compared heritability estimates between younger (<52 years) and older (>52 years) twins and tested: (a) whether single nucleotide polymorphisms (SNPs) lying within 100 kbp of the IGF-1 gene are also associated with IGF-1 variation and (b) whether associated SNPs show interaction with age on IGF-1 levels. To achieve these aims, we measured plasma levels of IGF-1 and genotyped 18 SNPs with minor allele frequency >0.1 in a large sample, 4,471 UK female twins. Heritability explained 42 % of IGF-1 variation adjusted for age and in unadjusted sample was independent of age. Ten SNPs in four haploblocks showed significant association with IGF-1 levels, with p = 0.01–0.0005. The most distal SNP was located up to 90 kbp from the IGF-1 gene. When their age-dependent effects were examined, one SNP, rs855203, showed significant (p = 0.0009) age-dependent interaction effect on IGF-1 levels variation. This is the first study to test the age × genotype interaction in IGF-1 levels. The genomic region marked by rs855203 may consequently be of significance for further molecular and pharmacogenetic research, in particular in advanced age.
Keywords: Genetics, Heritability, SNP, Interaction, IGF-1
Introduction
Insulin-like growth factor 1 (IGF-1) is a polypeptide product of the IGF-1 gene which is mapped to 12q23.2 human genomic region (HapMap project, release #28). It is most well-known because of its crucial role in childhood development and involvement in a variety of anabolic processes in adults. In adulthood, the decreased levels of IGF-1 have important health and life expectancy consequences. The regulation of IGF-1 circulating levels includes genetic and metabolic factors (e.g., growth hormone (GH), insulin) nutritional status, and disease-related physiological conditions (Caregaro et al. 2001; Puche and Castilla-Cortazar 2012).
It is well established that IGF-1 circulating levels correlate with age: They are maximal during puberty and progressively decline in the course of adult life (Brabant and Wallaschofski 2007; Friedrich et al. 2010). Age-dependent decreased IGF-1 levels are significantly associated and probably serve as casual factors in quite diverse illnesses. For example, one of the largest studies (>5,500 participants) of diabetes in young US adults reported that low levels were significantly associated with the disease even after adjustment for a variety of other risk factors such as smoking, alcohol intake, body mass index (BMI), hypertension, glomerular filtration rate, and serum cholesterol (Teppala and Shankar 2010). In agreement with this are studies concerning the relationship between IGF-1 levels and liver damage (Aksu et al. 2013) and IGF-1-induced atheroprotective effect, including reduction in oxidative stress and proinflammatory signaling, deteriorating with age and decreased IGF-1 levels (Higashi et al. 2012). The former study even suggested that circulating IGF-1 levels may have predictive value for determining hepatic damage that results from diabetes. On the other hand, it has been shown repeatedly that this is a significant positive association between circulating IGF-1 levels and risk of advanced colorectal adenoma, suggesting that IGF1 is associated with a pivotal precursor to colorectal cancer (Gao et al. 2012). IGF-1 is associated also with other malignancies, including breast cancer (Lann and LeRoith 2008; Endogenous Hormones and Breast Cancer Collaborative Group 2010). It is therefore important to clarify to what extent blood levels of IGF-1 depend on intrinsic factors, in particular genetic factors, and to what extent they are influenced by environmental conditions.
The heritability estimates of IGF-1 level variation are consistently statistically significant and range from 40 to 60 % depending on the sample (Lam et al. 2010; Pantsulaia et al. 2005). However, the previous studies estimated heritability of IGF-1 in samples comprising a wide range of ages (from young to old). It is therefore not known whether genetic/environmental effects on IGF-1 variation differ across age groups—as has been observed for some other biochemical factors. For example, heritability estimates for circulating insulin, which share both structural and functional similarities with IGF-1, with high sequence identity and common receptor kinases (Werner et al. 2008), have been shown to be age dependent. In one study, heritability of insulin was statistically significantly higher (p < 0.05) among older twins [h2 = 0.85 (0.69–0.92)] when compared to younger twins [h2 = 0.75 (0.55–0.86)] (Poulsen et al. 2005). Sas et al. (2012) recently detected a significant additive genetic component in the regulation of baseline serum levels of four key cytokines involved in the human inflammatory response, IL-1β, IL-6, and IL-10, and found that age acts as a moderator of the genetic influences in regulation of baseline IL-1β and TNF-α serum levels. However, such data in general are limited and we are not aware of similar estimates for IGF-1. Consequently, the primary aim of this study was to determine potential age dependence on the contribution of genetic and environmental factors to IGF-1 level.
In addition to known significant genetic effects (heritability) on IGF-1 variation, several single nucleotide polymorphisms (SNPs) have been found to associate with IGF-1 circulating levels in both genome-wide (Kaplan et al. 2011) and candidate gene association studies (Patel et al. 2008; Fletcher et al. 2005; Canzian et al. 2006; Al-Zahrani et al. 2006; Verheus et al. 2008). However, all but one study (Rietveld et al. 2003) were performed across a range of ages without taking into account possible age-dependent effects. Furthermore, the majority of such studies selected SNPs that lie inside the IGF-1 gene or relatively proximal to the gene, not extending beyond 40 kbp 5′ or 3′ from the IGF-1 locus (Canzian et al. 2006; Al-Zahrani et al. 2006; Wong et al. 2005; Schumacher et al. 2010). Interestingly, a recent study reported a significant association between circulating IGF-1 and SNPs mapped to evolutionarily conserved regions of the genome, extending as far as 70 kbp from the IGF-1 gene (Palles et al. 2008). Since SNP association studies at present explain only <5 % of IGF-1 variation (Patel et al. 2008; Al-Zahrani et al. 2006), we expanded our search for more distal SNPs relatively to the 5′ and 3′ of the IGF-1 gene. We refined our search to 100 kbp from the IGF-1 gene. In addition, we tested whether the DNA polymorphisms located at the IGF-1 locus and in its vicinity are associated with IGF-1 levels and whether these associations have an age-dependent trend.
Material and methods
Sample
All subjects included for the present study were from the TwinsUK adult registry (http://www.twinsuk.ac.uk), described in detail elsewhere (Moayyeri et al. 2013a, b). In short, the registry includes 12,000 twins, 83 % female, equal number of monozygotic (MZ) and dizygotic (DZ) twins, predominantly middle-aged and older. Each volunteer was recruited regardless of any preexisting diseases and gave informed consent. Investigating a twin sample allows for an efficient way to explore the shared genetic and environmental factors, therefore estimating the proportion of variance in IGF-1 attributable to genetic variation versus the proportion that is due to shared environment or unshared environment. The current study included 4,471 individuals, all female twins, consisting of 1,935 MZ twins (789 pairs, 357 unpaired singletons) and 2,536 DZ twins (1,149 pairs, 238 unpaired singletons) with age varying from 16 to 85 years. As accurate menopause status was not available for the time when the sample for IGF-1 was drawn, menopause status was defined according to the average menopause age of 52 years old (http://www.nhs.uk/Conditions/Menopause/Pages/Introduction.aspx).
Biochemical assays
Venous blood samples were taken after an overnight fast. Within 1 h of collection, samples were centrifuged to obtain plasma, frozen in aliquots, and stored at −80 °C until analyzed. IGF-1 levels were measured by sandwich enzyme-linked immunosorbent assay (ELISA) using DuoSet ELISA development kit (R&D Systems, Minneapolis, MN, USA) and following manufacturer’s instructions. All the observed measurements were above the minimal detection sensitivity, 31.25 ng/ml. The inter- and intra-assay coefficients of variation in our analyses were 3.4 and 1.5 %, respectively.
SNP selection
The genotype data were based on genome-wide association scans performed in the TwinsUK cohort previously using the Illumina (San Diego, CA, USA) 317K and 610K SNP arrays, with a call rate of genotype ≥98 %. In total, 18 SNPs were selected for this study, located within the IGF-1 gene and up to 100 kbp upstream and downstream from the gene. SNPs were tested for association with inter-individual variation in IGF-1 levels. The criteria for SNP selection included minor allele frequency and Hardy–Weinberg equilibrium (HW), i.e., SNP with MAF < 0.1 and/or significantly deviating from HW expectation (p ≤ 0.01) was excluded from the association analysis. Table 1 shows the summary of SNP selection, including their location (HapMap project release #28).
Table 1.
Study SNP position, genotype distribution, and main results of association analysis
| Marker | Position (bp)a | Alleleb | MAF | Genotype frequency | χ 2 | p value | ||
|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | ||||||
| rs10860862 | 101310202 | T | 0.16 | 0.69 | 0.27 | 0.02 | 1.48 | 0.22 |
| rs6214 | 101317699 | T | 0.40 | 0.34 | 0.48 | 0.16 | 0.31 | 0.51 |
| rs978458 | 101326369 | T | 0.26 | 0.54 | 0.37 | 0.07 | 11.31 | 0.0007 |
| rs2288378 | 101354138 | T | 0.25 | 0.55 | 0.37 | 0.06 | 11.98 | 0.0005 |
| rs10735380 | 101368366 | G | 0.28 | 0.52 | 0.39 | 0.08 | 9.36 | 0.002 |
| rs5742629 | 101381393 | C | 0.27 | 0.52 | 0.40 | 0.07 | 1.70 | 0.19 |
| rs10778176 | 101387109 | T | 0.27 | 0.52 | 0.39 | 0.07 | 1.61 | 0.20 |
| rs1019731 | 101388555 | A | 0.14 | 0.74 | 0.23 | 0.02 | 1.09 | 0.29 |
| rs2162679 | 101395389 | C | 0.15 | 0.72 | 0.25 | 0.02 | 6.37 | 0.01 |
| rs35766 | 101404603 | C | 0.15 | 0.71 | 0.26 | 0.02 | 6.99 | 0.008 |
| rs35765 | 101405826 | T | 0.11 | 0.78 | 0.19 | 0.01 | 10.48 | 0.001 |
| rs855211 | 101434940 | A | 0.15 | 0.70 | 0.26 | 0.02 | 6.95 | 0.008 |
| rs4764702 | 101461236 | G | 0.14 | 0.73 | 0.24 | 0.02 | 1.49 | 0.22 |
| rs35760 | 101474348 | T | 0.17 | 0.68 | 0.28 | 0.03 | 1.34 | 0.24 |
| rs855205 | 101481996 | T | 0.15 | 0.71 | 0.25 | 0.02 | 2.72 | 0.09 |
| rs855203 | 101482203 | C | 0.10 | 0.81 | 0.17 | 0.01 | 11.10 | 0.0008 |
| rs1457596 | 101486637 | A | 0.11 | 0.78 | 0.19 | 0.01 | 9.16 | 0.002 |
| rs7964748 | 101489459 | G | 0.18 | 0.67 | 0.29 | 0.03 | 8.07 | 0.004 |
MAF minor allele frequency
aSNP genomic position in base pairs according to HapMap project
bMinor allele, MAF, and χ 2 test for association between the selected SNP and IGF-1 circulating levels, with accompanying p values
Statistical and quantitative genetic analysis
Preliminary statistical analysis of IGF-1 and covariates was performed using SPSS statistical package, version 19 (SPSS, Chicago, IL, USA). In this analysis, we implemented first a series of invariable and then multivariable regression analyses with IGF-1 levels as dependent variable. The independent covariates included age, BMI, and body composition, namely fat and lean body mass. Contribution of potential genetic factors and covariates to circulating IGF-1 levels in the entire sample and within age ranges was done in two steps using MAN package (MAN, Tel Aviv University, Tel Aviv, Israel). MAN takes into account family structure of the sample The program allows simultaneous estimation of the effect of covariates, e.g., age, BMI, etc, as well as contribution of the major components of variation such as additive genetic and environment factors.
First, we decomposed the total observed phenotypic variation into two major family components of variation: VAD—contribution of the additive genetic factors and VTW—common familial environment shared by twins. Unexplained by familial factors and covariates, residual variation was defined as VRS. Next, we examined the contribution of each SNP on IGF-1 variation. The SNP-specific genotypes were scored by the number of minor alleles, as 0, 1 and 2, and the corresponding regression coefficients (additive model) were estimated in variance component analysis, using the previously established best fitting and most parsimonious model. That is, we tested independent linear increase (or diminution) of IGF-1 levels in relation to dose of minor allele. In addition, for exploratory purposes, we generated an allele score from relatively independent (by linkage disequilibrium (LD)) SNPs, combining the number of effect alleles, as suggested by Gao et al. (2013). We used four tag SNPs (rs2288378, rs35765, rs855203, and rs7964748) each mapping to a different haploblock. The scores were standardized by dividing them by the total number of effect allele copies (eight in this case). Thus, we created a new variable, SNP_score, ranging from 0 to 1 and tested if the combined SNPs showed stronger association with IGF-1 than each individual SNP. The comparison of the models was made using a likelihood ratio test (LRT).
Finally, we tested heritability × age and SNP × age interaction effect on IGF-1 inter-individual variation to establish whether the genetic effects are age dependent. Heritability × age effect was tested by stratifying into two age groups of approximately equal size, taking age 52 (mean age of menopause in UK women) as a dividing point. Putative genetic factors were then compared between these two groups. In order to determine potential age × genotype interaction, an interaction term was introduced as an additional parameter into the model, already containing age and genotype parameters. Under the null hypothesis, assuming no interaction, the genotype-specific regression coefficients (reflecting the pattern of the IGF-1 age dependence) and/or corresponding correlation coefficients (reflecting the strength of dependence) were expected to be the same. The hypothesis was tested by LRT, comparing the free parameter estimates to the restricted ones.
Results
The basic descriptive statistics of the sample is given in Table 2 and includes information on IGF-1 levels, anthropometric and dual-energy X-ray absorptiometry (DXA) measurements of body mass, and composition by zygosity. The sample included 4,471 individuals with mean IGF-1 levels of 48.70 ± 20.26 ng/ml and with age range 18 to 84 (average 51.2) years. There was no significant difference between the MZ and DZ twins. For example, mean levels of IGF-1 were 49.14 ± 21.45 ng/ml in MZ twins versus 48.36 ± 19.29 ng/ml in DZ twins, with p > 0.05. Additionally, age and BMI of MZ twins (52.00 ± 14.34 years and 25.61 ± 4.72 kg/m2) were very similar to DZ twins (50.71 ± 13.19 years and 25.88 ± 4.84 kg/m2, respectively), p > 0.05 for both.
Table 2.
Basic descriptive statistics of the study sample from TwinsUK
| Trait | MZ (N = 1,935) | DZ (N = 2,536) | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | SD | Mean | Min | Max | SD | |
| Age (years) | 52.00 | 17.11 | 84.54 | 14.34 | 50.71 | 18.41 | 84.44 | 13.19 |
| IGF-1 (ng/ml) | 49.14 | 2.11 | 196.278 | 21.45 | 48.36 | 3.845 | 149.25 | 19.29 |
| Weight (kg) | 66.88 | 37.90 | 128.100 | 12.31 | 68.127 | 35.10 | 140.89 | 13.12 |
| BMI (kg/m2) | 25.61 | 15.23 | 52.39 | 4.72 | 25.88 | 15.55 | 52.39 | 4.84 |
| Fat body mass (kg) | 24.33 | 5.67 | 67.51 | 8.55 | 24.60 | 6.00 | 65.03 | 8.85 |
| Lean body mass (kg) | 39.63 | 25.98 | 65.17 | 5.21 | 39.69 | 25.98 | 65.17 | 5.25 |
Comparison of MZ and DZ twins by Student's t test for quantitative continuous traits or by Mann–Whitney U test revealed no statistically significant differences
MZ monozygotic, DZ dizygotic
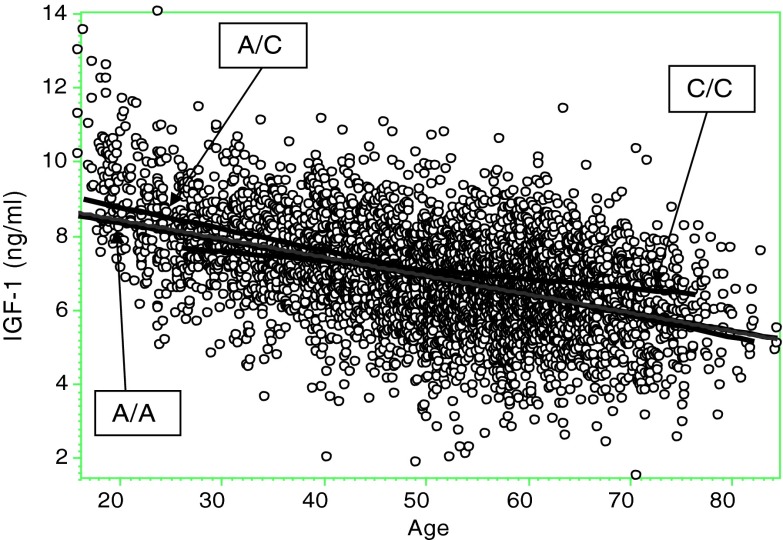
The distribution of IGF-1 showed significant deviation from normality assumptions and was transformed (square root transformation) to improve its distribution. IGF-1 levels correlated significantly with age (r =−0.47, p < 0.001, Fig. 1), but not with BMI or its components assessed by DXA, namely lean and fat body mass. Consequently, the quantitative genetic analysis was performed adjusting the IGF-1 variation for age. The intraclass correlations in IGF-1 levels in both MZ and DZ twins were statistically significant, and variance component analysis gave a heritability estimate of 42 ± 6 % adjusted for age (Table 3). In addition, common twin environment explained 10 ± 5 % of the variation, which was just marginally significant by LRT (p = 0.048). Next we divided the sample into two age groups according to the average menopause status: younger (≤52 years) and older (>52). By LRT, the heritability estimates in the best fitting and most parsimonious models in both age groups were similar and ranged between 0.53 and 0.56, while common twin environment made no significant contribution to IGF-1 variation (Table 3).
Fig. 1.
Age-dependent variation of IGF-1 circulating levels in the entire sample and stratified by SNP_rs855203 genotype. C/C homozygote at minor allele (N = 57), A/A homozygote at major allele (N = 3,630), AC heterozygote (N = 770)
Table 3.
Summary of variance component analysis of IGF-1 levels across age groups
| Agea | N | r (MZ)* | r (DZ)* | χ 2_corr | V AD | V TW | V RS |
|---|---|---|---|---|---|---|---|
| All ages | 4,471 | 0.55 | 0.31 | 88.8*** | 0.42 ± 0.06 | 0.10 ± 0.05 | 0.47 ± 0.02 |
| 16–52 years | 2,086 | 0.52 | 0.28 | 18.8** | 0.53 ± 0.04 | 0 | 0.47 ± 0.03 |
| 52–64 years | 2,385 | 0.57 | 0.34 | 88.9*** | 0.56 ± 0.03 | 0 | 0.41 ± 0.02 |
r (MZ) and r (DZ) are correlations between IGF-1 levels within twin pairs by zygosity. Parameter estimates, with their corresponding standard errors, are given for best fitting and most parsimonious models in each analysis
N available sample size, χ 2 _corr test for homogeneity of twin correlations, V AD additive genetic component, V TW common twin environment component, V RS residual variance
*All correlations were significant at p < 0.01; **p < 0.001; ***p < 0.0001
aAnalysis was conducted with simultaneous adjustment for age
Next we tested the hypothesis that IGF-1 variation is associated with one more of the selected SNPs in the IGF-1 gene including SNPs 5′ and 3′ from the IGF-1 locus. Of 18 SNPs selected for this analysis, ten SNPs showed nominally significant association, with p values in the range 0.01–0.0005. This is considerably more than what is expected (<1) from type 1 error, assuming nominal p = 0.01, if the results were obtained by chance only. The observed p values were all under 0.025, the critical p value corrected for multiple testing using the false discovery rate (FDR) method (Benjamini 2010).
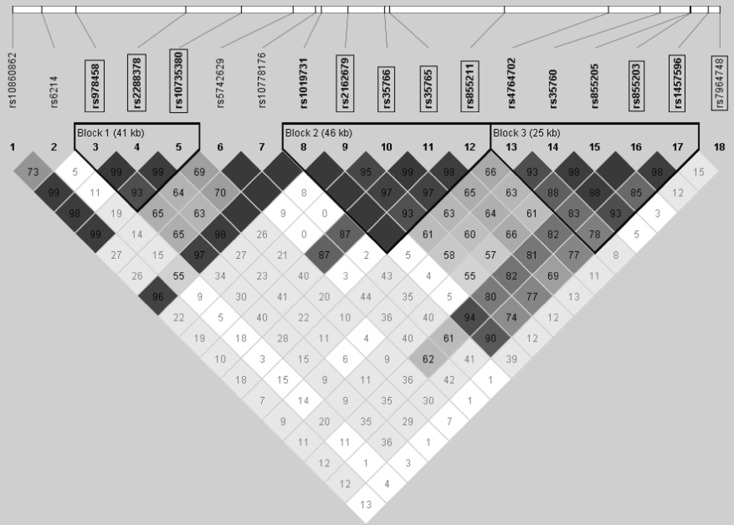
The LD pattern among the ten SNPs, except rs7964748, showed the presence of three different haploblocks (Fig. 2). Within each block, SNPs significantly associated with IGF-1 were in almost perfect LD (D′ > 0.95). We selected three tag SNPs for each haploblock: rs2288378, rs35765, rs855203, each with the most significant p value observed in association analysis with IGF-1. Additionally, rs7964748 positioned outside of the three haploblocks (virtually fourth haploblock) was also selected. The effect alleles of these four SNPs were combined to create a SNP_score (see M&M), and their association with IGF-1 levels was examined. Indeed, when included in a regression model, it showed clearly more significant association (p = 4.42 × 10−7) in comparison to the most significant individual SNP (rs2288378, p = 5 × 10−4).
Fig. 2.
Linkage disequilibrium (D′) plot of selected study SNPs. SNPs in rectangles are statistically significantly associated with circulating IGF-1 levels. All SNPs have MAF > 0.1
To test genotype × age interactive effect on IGF-1 variation, we examined their multiplicative effect (age × number of minor alleles) in a variance component model already containing age and SNP effects as covariates. Inclusion of rs855203 genotype-specific age-dependent fitting of IGF-1 variation (Fig. 1) significantly improved the model likelihood (p = 0.009), and therefore, the null hypothesis of no SNP × age interaction was rejected. Interaction terms for the three other SNPs and SNP_score did not reach statistical significance. We compared genotype-specific slopes and correlations between the IGF-1 levels and age for each of the four SNPs: rs855203, rs35765, rs2288378, and rs7964748 (Table 4). As seen, rs855203 genotype showed significantly different patterns of IGF-1/age dependence as estimated by their corresponding regression and correlation coefficients. The others confirmed the independent significant association of IGF-1 variation with age and genotypes and showed no significant interaction terms. Correlation coefficients differed significantly by rs35765 genotypes. However, regression slopes were virtually identical (Table 4).
Table 4.
Variance component analysis of IGF1, testing for independent effect of age, SNP (additive model), and their interaction
| VCAa—parameters of the model | rs855203 | rs35765 | rs2288378 | rs7964748 | |
|---|---|---|---|---|---|
| Regression parameters | β 1 (age) | −0.46 ± 0.01 | −0.46 ± 0.01 | −0.46 ± 0.01 | −0.46 ± 0.01 |
| β 2 (SNP) | −0.10 ± 0.03 | 0.09 ± 0.03 | 0.07 ± 0.02 | 0.07 ± 0.02 | |
| β 3 (age × SNP) | −0.08 ± 0.03 | 0 | 0 | 0 | |
| p 1 value (LRT) | <0.001 | 0.26 | 0.68 | 0.80 | |
| Correlationb (r) (N, p value) | AA (major) | 0.46 (3,630, <0.001) | 0.48 (3,503, <0.001) | 0.48 (2,516, <0.001) | 0.48 (3,008, <0.001) |
| AB | 0.53 (770, <0.001) | 0.48 (883, <0.001) | 0.45 ± 0.01 (1,631, <0.001) | 0.46 (1,292, <0.001) | |
| BB (minor) | 0.22 (57, 0.08) | 0.24 (70, 0.03) | 0.52 ± 0.03 (307, <0.001) | 0.49 (157, < 0.001) | |
| p 2 value (χ 2) | AA/AB | 0.009 | 0.241 | 0.20 | 0.22 |
| AA/BB | 0.020 | 0.008 | 0.18 | 0.43 | |
| AB/BB | 0.004 | 0.010 | 0.09 | 0.32 | |
| V componentsc | Given in Table 3 | ||||
VCA variance component analysis; β 1 , β 2 , β 3 regression coefficients reflecting effect of age, genotype, and age*; p 1 value (LRT) likelihood ratio test for heterogeneity of regression coefficients, showing genotype-specific age dependence of IGF-1 variation; p 2 value (χ 2 ) pairwise comparison of the corresponding genotype-specific correlations between IGF-1 level variations and age
aTesting effects of covariates on IGF-1 variation
bPresents genotype-specific correlation, by SNP, sample size, and p value for correlation coefficient estimate
cVariance component estimated in the entire sample and for “all ages” given in Table 3
Discussion
Circulating IGF-1 levels are known to be negatively correlated with age, with a gradual decline during life (Tiryakioglu et al. 2003; Brabant and Wallaschofski 2007; Friedrich et al 2010), which was also clearly observed in the present study (Fig. 1). Although the specific mechanisms affecting inter-individual IGF-1 variation are still poorly understood, it has been well documented that genetic factors contribute to circulating IGF-1 variation. While most studies have examined the genetic effect across all age groups, in this study we examined the effect of age on heritability estimations and specific genotype IGF-1 interaction.
In a variance component analysis, additive genetic factors (heritability) explained 42 % of total IGF-1 variation adjusted for age (Table 3). These estimates are in good agreement with previously published heritability estimates, ranging from 40 to 60 % (Lam et al. 2010; Pantsulaia et al. 2005; Harrela et al. 1996; Hong et al. 1996). However, dividing our sample into younger and older age groups revealed no significant trend in heritability estimates. The groups showed similar heritability estimates, of 0.53 ± 0.04 and 0.56 ± 0.03, and no contribution of common twin environment was detected.
Next, we examined individual SNP association with IGF-1 variation. Of all the selected SNPs, we found ten SNPs statistically significantly associated with IGF-1 levels, with the largest p = 0.01, <0.025, the value required for significance by FDR. The association signals may be divided in three haploblocks (Fig. 2). The first one includes three SNPs located exclusively in the intronic region. Since all three SNPs are in almost perfect LD (average D′ > 0.97), it was not surprising that all three showed significant association with IGF-1 levels. This confirms a previous study, reporting elevated premenopausal serum IGF-1 levels in women homozygous for the minor allele of rs978458 (Verheus et al. 2008). Haploblock-2 embraces rs35766 and rs35765, known to be located in the promoter region, and rs855211 mapping to 20 kbp upstream from rs35765. All three were also found to be significantly associated with IGF-1 levels (p = 0.008–0.001). Marker rs35765 has been shown to be associated with IGF-1 levels with a modest dominant effect (Canzian et al. 2006). However, some of the SNPs were previously tested, i.e., rs855211 (also rs35765 and rs2162679 from the promoter region), but reported no association with IGF-1 (Henningson et al. 2011). Note, however, they were examined in a modest sample (n = 325 versus 4,471 in our study) of young woman and that study may simply have had insufficient power to detect an association signal. Three more SNPs showed significant association with IGF-1, located in haploblock-3 (rs855203 and 1457596) and relatively far out of three blocks, rs7964748.
The significant associations of rs35766 and rs35765, mapped to the IGF-1 gene promoter, are of a particular interest. It will be important to clarify whether they influence the promoter consensus sequence, thereby alternating IGF-1 transcription, or are merely markers in LD with other SNPs located in the promoter or nearby. For example, rs35766 and rs35765 are in high LD with rs35767 (D′ = 1, r2 = 0.85 and D′ = 0.97, r2 = 0.71, respectively), which was not examined in our study, but which has also been reported to be significantly associated with IGF-1 levels in a large sample by the Breast and Prostate Cancer Cohort Consortium (Patel et al. 2008).
The most remarkable findings of this study are the detection of significant associations between IGF-1 levels and several SNPs, rs855203, rs1457596, and rs7964748, located as far as 90 kbp from the IGF-1 structural locus and the significant contribution of rs855203 × age interaction to IGF-1 variation, not reported previously in the literature. It is well-known that regulatory sequences may be distant from the genes they regulate. A previous study (Palles et al. 2008) observed a significant association with a few SNPs distant from the IGF-1 gene (>70 kbp) that lie in evolutionally conserved regions. While they did not test the three SNPs mentioned above, they did report a negative association between rs10778177 and IGF-1 level variation. The polymorphism rs10778177 is in perfect LD with SNP rs4764702, which is located in the same haploblock as SNPs rs855203 and rs1457596 and are relatively in high LD (D′ ∼ 0.8). Therefore, a replication study will be needed to strengthen our result of positive association between rs855203 and rs1457596 and IGF-1 levels. Other studies identified an enhancer element for the STAT5 binding protein, a GH-activated IGF-1 gene transcription factor, mapped ∼75 kbp from the IGF-1 gene (Chia et al. 2006; Wang and Jiang 2005). In our study, the most proximal SNPs (including rs855203) lie outside the evolutionally conserved region, defined by the study of Palles et al. (2008) and the consensus STAT5-binding site. We therefore suggest that regulatory regions for the IGF-1 gene maybe positioned as far as 90 kbp from the IGF-1 gene. However, the functional role of SNPs rs855203, rs1457596, and rs7964748 is not known; we are not aware of any positive association studies involving these SNPs.
In addition to the independent effect of all four SNPs rs2288378, rs35765, rs855203, and rs7964748 with IGF-1, we report an even stronger association with IGF-1 levels when all four SNPs were combined as a SNP score variable in comparison with each independent SNP. Thus, each haploblock appears to contribute individually to the genetic variation leading to a common genetic pathway, causing an IGF-1 level variation among different individuals. However, the precise effect each SNP contributes individually or in concert with the other SNPs requires further investigation.
As mentioned, possible age × genotype interaction in determination of IGF-1 variation was poorly documented in the literature. We are aware of only one study, using a small sample, reporting that only homozygous carriers of the 192-bp alleles of the IGF-1 gene displayed an age-related decline in circulating total IGF-1 levels, but not in heterozygotes and non-carriers of the 192-bp allele (Rietveld et al. 2003). While we did not genotype the 192-bp polymorphism, we selected several tag-SNPs, covering the entire gene. Through them we obtained clear evidence of possible rs855203-genotype × age interaction. The variance component model neglecting this interaction was significantly worse (p = 0.009) than its counterpart allowing for interaction. Interestingly, the interaction concerned both genotype-specific regression slope (beta) and correlation.
Our study has some limitations and certain advantages. As mentioned above, IGF-1 levels may be influenced by nutrition status, food supplements, preexisting disease, and activity levels (Caregaro et al. 2001; Puche and Castilla-Cortazar 2012) and hormone replacement therapy (Figueroa et al. 2002). This information was not available in our study and may have thrown up the associations reported in this study. Since the study sample contains only female twins, a main limitation of this study is differences in environments between twins studied in our study and singletons which represent the more general population in addition to potential sex-related effects (Vink et al. 2012). These environmental differences may be due to shared uterine environment or the fact that MZ twins reared together share many of the same experiences. However, a study on lifestyle characteristics (Andrew et al. 2001) showed no difference between twins and singletons, suggesting that twin studies can be generalized to the general population. Gender influence on IGF-1 levels still needs further investigation. It is possible that IGF-1 levels are influenced by gender-specific factors, such as estrogen levels (Ho et al. 2006). Yet, current data suggest that sex differences in IGF-1 levels if they exist are small in healthy individuals (Aimaretti et al. 2008; Friedrich et al 2010). Moreover, using only females may not necessarily be a limitation, but rather an advantage as it is not prone to potential genotype × sex interactions, which are likely the rule rather than exception (Vink et al. 2012). Similarly we note that recent study of a large sample of European women (Friedrich et al 2010) showed that the effect of HRT on IGF-1 reference levels is negligible. Additional important advantages of this study are that we used a well-characterized cohort, consisting of a large sample of 4,471 subjects. This gives a substantial statistical power to determine small genetic effects in the present sample, as well as wide range of ages to test our major hypothesis. Previous studies performed on the TwinsUK adult registry (http://www.twinsuk.ac.uk) contributed to many collaborative GWAS showing their genetic data are in keeping with others (Moayyeri et al. 2013a, b.
In conclusion, as in previous studies, we have identified a strong age effect in IGF-1 levels. We also confirmed a significant heritability of this growth factor. This study is the first, however, to test whether the genetic determination of IGF-1 variation is also age dependent. We have identified several new polymorphisms located in a considerable distance from the IGF-1 gene that appear to function as regulatory sequences for IGF-1 transcription factors. We found no age effect on heritability estimates of IGF-1 variation, and several tested SNPs although showed significant association with IGF-1 variation also did not display genotype × age effect. However, rs855203 marker exerted clear genotype-dependent effect on pattern and strength of IGF-1 variation associated with age. The influence of the genomic region marked by rs855203 may be of significance for further molecular and pharmacogenetic research in particular, in advanced age.
Acknowledgments
This study was funded by the Israel Science Foundation (grant #994/10) and by the Carol and Leonora Fingrhut fund from the Sackler Faculty of Medicine, Tel University. The collection of the data for this study received support from the National Institute for Health Research (NIHR) Clinical Research Facility at Guy’s & St Thomas’ NHS Foundation Trust and NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The study was performed in partial fulfillment of the M.Sc. degree requirements of Liran Franco. We are grateful to the patients for participating in this study.
References
- Aimaretti G, Boschetti M, Corneli G, Gasco V, Valle D, Borsotti M, et al. Normal age-dependent values of serum insulin growth factor-I: results from a healthy Italian population. J Endocrinol Invest. 2008;31:445–449. doi: 10.1007/BF03346389. [DOI] [PubMed] [Google Scholar]
- Aksu I, Baykara B, Kiray M, Gurpinar T, Sisman AR, Ekerbicer N, Tas A, Gokdemir-Yazar O, Uysal N. Serum IGF-1 levels correlate negatively to liver damage in diabetic rats. Biotechnic Histochem. 2013;88:194–201. doi: 10.3109/10520295.2012.758311. [DOI] [PubMed] [Google Scholar]
- Al-Zahrani A, Sandhu MS, Luben RN, Thompson D, Baynes C, Pooley KA, et al. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Hum Mol Genet. 2006;15:1–10. doi: 10.1093/hmg/ddi398. [DOI] [PubMed] [Google Scholar]
- Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, MacGregor AJ. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–477. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- Benjamini Y. Discovering the false discovery rate. J Royal Stat Soc. 2010;72:405–416. doi: 10.1111/j.1467-9868.2010.00746.x. [DOI] [Google Scholar]
- Brabant G, Wallaschofski H. Normal levels of serum IGF-I: determinants and validity of current reference ranges. Pituitary. 2007;10:129–133. doi: 10.1007/s11102-007-0035-9. [DOI] [PubMed] [Google Scholar]
- Canzian F, McKay JD, Cleveland RJ, Dossus L, Biessy C, Rinaldi S, et al. Polymorphisms of genes coding for insulin-like growth factor 1 and its major binding proteins, circulating levels of IGF-I and IGFBP-3 and breast cancer risk: results from the EPIC study. Br J Cancer. 2006;94:299–307. doi: 10.1038/sj.bjc.6602936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caregaro L, Favaro A, Santonastaso P, Alberino F, Di Pascoli L, Nardi M, et al. Insulin-like growth factor 1 (IGF-1), a nutritional marker in patients with eating disorders. Clin Nutr. 2001;20:251–257. doi: 10.1054/clnu.2001.0397. [DOI] [PubMed] [Google Scholar]
- Chia DJ, Ono M, Woelfle J, Schlesinger-Massart M, Jiang H, Rotwein P. Characterization of distinct Stat5b binding sites that mediate growth hormone-stimulated IGF-I gene transcription. J Biol Chem. 2006;281:3190–3197. doi: 10.1074/jbc.M510204200. [DOI] [PubMed] [Google Scholar]
- Endogenous Hormones and Breast Cancer Collaborative Group. Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11:530–542. doi: 10.1016/S1470-2045(10)70095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa A, Going SB, Milliken LA, Blew R, Sharp S, Lohman TG. Body composition modulates the effects of hormone replacement therapy on growth hormone and insulin-like growth factor-I levels in postmenopausal women. Gynecol Obstet Invest. 2002;54:201–216. doi: 10.1159/000068383. [DOI] [PubMed] [Google Scholar]
- Fletcher O, Gibson L, Johnson N, Altmann DR, Holly JM, Ashworth A, et al. Polymorphisms and circulating levels in the insulin-like growth factor system and risk of breast cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:2–19. [PubMed] [Google Scholar]
- Friedrich N, Krebs A, Nauck M, Wallaschofski H. Age- and gender-specific reference ranges for serum insulin-like growth factor I (IGF-I) and IGF-binding protein-3 concentrations on the Immulite 2500: results of the Study of Health in Pomerania (SHIP) Clin Chem Lab Med. 2010;48:115–120. doi: 10.1515/CCLM.2010.009. [DOI] [PubMed] [Google Scholar]
- Gao Y, Katki H, Graubard B, Pollak M, Martin M, Tao Y, et al. Serum IGF1, IGF2 and IGFBP3 and risk of advanced colorectal adenoma. Int J Cancer. 2012;131:E105–E113. doi: 10.1002/ijc.26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Fall T, van Dam RM, Flyvbjerg A, Zethelius B, Ingelsson E, et al. Evidence of a causal relationship between adiponectin levels and insulin sensitivity: a Mendelian randomization study. Diabetes. 2013;62:1338–1344. doi: 10.2337/db12-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrela M, Koistinen H, Kaprio J, Lehtovirta M, Tuomilehto J, Eriksson J, et al. Genetic and environmental components of interindividual variation in circulating levels of IGF-I, IGF-II, IGFBP-1, and IGFBP-3. J Clin Invest. 1996;98:2612–2615. doi: 10.1172/JCI119081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningson M, Hietala M, Torngren T, Olsson H, Jernstrom H. IGF1 htSNPs in relation to IGF-1 levels in young women from high-risk breast cancer families: implications for early-onset breast cancer. Fam Cancer. 2011;10:173–185. doi: 10.1007/s10689-010-9404-z. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Sukhanov S, Anwar A, Shai S-Y, Delafontaine P. Aging, atherosclerosis, and IGF-1. J Gerontol A. 2012;67A:626–639. doi: 10.1093/gerona/gls102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KK, Gibney J, Johannsson G, Wolthers T. Regulating of growth hormone sensitivity by sex steroids: implications for therapy. Front Horm Res. 2006;35:115–128. doi: 10.1159/000094314. [DOI] [PubMed] [Google Scholar]
- Hong Y, Pedersen NL, Brismar K, Hall K, de Faire U. Quantitative genetic analyses of insulin-like growth factor I (IGF-I), IGF-binding protein-1, and insulin levels in middle-aged and elderly twins. J Clin Endocrinol Metab. 1996;81:1791–1797. doi: 10.1210/jcem.81.5.8626837. [DOI] [PubMed] [Google Scholar]
- Kaplan RC, Petersen AK, Chen MH, Teumer A, Glazer NL, Doring A, et al. A genome-wide association study identifies novel loci associated with circulating IGF-I and IGFBP-3. Hum Mol Genet. 2011;20:1241–1251. doi: 10.1093/hmg/ddq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, Chen MH, Lacey SM, Yang Q, Sullivan LM, Xanthakis V, et al. Circulating insulin-like growth factor-1 and its binding protein-3: metabolic and genetic correlates in the community. Arterioscler Thromb Vasc Biol. 2010;30:1479–1484. doi: 10.1161/ATVBAHA.110.203943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lann D, LeRoith D. The role of endocrine insulin-like growth factor-I and insulin in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:371–379. doi: 10.1007/s10911-008-9100-x. [DOI] [PubMed] [Google Scholar]
- Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK adult twin registry (TwinsUK resource) Twin Res Hum Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort profile: TwinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palles C, Johnson N, Coupland B, Taylor C, Carvajal J, Holly J, et al. Identification of genetic variants that influence circulating IGF1 levels: a targeted search strategy. Hum Mol Genet. 2008;17:1457–1464. doi: 10.1093/hmg/ddn034. [DOI] [PubMed] [Google Scholar]
- Pantsulaia I, Trofimov S, Kobyliansky E, Livshits G. Genetic regulation of the variation of circulating insulin-like growth factors and leptin in human pedigrees. Metabolism. 2005;54:975–981. doi: 10.1016/j.metabol.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Patel AV, Cheng I, Canzian F, Le Marchand L, Thun MJ, Berg CD, et al. IGF-1, IGFBP-1, and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort Consortium (BPC3) PLoS One. 2008;3:e2578. doi: 10.1371/journal.pone.0002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, Vaag A. Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes. 2005;54:275–283. doi: 10.2337/diabetes.54.1.275. [DOI] [PubMed] [Google Scholar]
- Puche JE, Castilla-Cortazar I. Human conditions of insulin-like growth factor-I (IGF-I) deficiency. J Transl Med. 2012;10:224. doi: 10.1186/1479-5876-10-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld I, Janssen JA, Hofman A, Pols HA, van Duijn CM, Lamberts SW. A polymorphism in the IGF-I gene influences the age-related decline in circulating total IGF-I levels. Eur J Endocrinol. 2003;148:171–175. doi: 10.1530/eje.0.1480171. [DOI] [PubMed] [Google Scholar]
- Sas AA, Jamshidi Y, Zheng D, Wu T, Korf J, Alizadeh BZ, et al. The age-dependency of genetic and environmental influences on serum cytokine levels: a twin study. Cytokine. 2012;60:108–113. doi: 10.1016/j.cyto.2012.04.047. [DOI] [PubMed] [Google Scholar]
- Schumacher FR, Cheng I, Freedman ML, Mucci L, Allen NE, Pollak MN, et al. A comprehensive analysis of common IGF1, IGFBP1 and IGFBP3 genetic variation with prospective IGF-I and IGFBP-3 blood levels and prostate cancer risk among Caucasians. Hum Mol Genet. 2010;19:3089–3101. doi: 10.1093/hmg/ddq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teppala S, Shankar A. Association between serum IGF-1 and diabetes among U.S. adults. Adults Diabetes Care. 2010;33:2257–2259. doi: 10.2337/dc10-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryakioglu O, Kadiolgu P, Canerolgu NU, Hatemi H. Age dependency of serum insulin-like growth factor (IGF)-1 in healthy Turkish adolescents and adults. Indian J Med Sci. 2003;57:543–548. [PubMed] [Google Scholar]
- Verheus M, McKay JD, Kaaks R, Canzian F, Biessy C, Johansson M, et al. Common genetic variation in the IGF-1 gene, serum IGF-I levels and breast density. Breast Cancer Res Treat. 2008;112:109–122. doi: 10.1007/s10549-007-9827-x. [DOI] [PubMed] [Google Scholar]
- Vink JM, Bartels M, van Beijsterveldt TC, van Dongen J, van Beek JH, Distel MA, et al. Sex differences in genetic architecture of complex phenotypes? PLoS One. 2012;7:e47371. doi: 10.1371/journal.pone.0047371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang H. Identification of a distal STAT5-binding DNA region that may mediate growth hormone regulation of insulin-like growth factor-I gene expression. J Biol Chem. 2005;280:10955–10963. doi: 10.1074/jbc.M412808200. [DOI] [PubMed] [Google Scholar]
- Werner H, Weinstein D, Bentov I. Similarities and differences between insulin and IGF-I: structures, receptors, and signalling pathways. Arch Physiol Biochem. 2008;114:17–22. doi: 10.1080/13813450801900694. [DOI] [PubMed] [Google Scholar]
- Wong HL, Delellis K, Probst-Hensch N, Koh WP, Van Den Berg D, Lee HP, et al. A new single nucleotide polymorphism in the insulin-like growth factor I regulatory region associates with colorectal cancer risk in Singapore Chinese. Cancer Epidemiol Biomarkers Prev. 2005;14:144–151. [PubMed] [Google Scholar]