Abstract
Transgenic mice that overexpress the nuclear form of sterol regulatory element binding protein-1a (SREBP-1a) in liver (TgBP-1a mice) were shown previously to overproduce cholesterol and fatty acids and to accumulate massive amounts of cholesterol and triglycerides in hepatocytes. Despite the hepatic overproduction of lipids, the plasma levels of cholesterol (∼45 mg/dl) and triglycerides (∼55 mg/dl) were not elevated, perhaps owing to degradation of lipid-enriched particles by low-density lipoprotein (LDL) receptors. To test this hypothesis, in the current studies we bred TgBP-1a mice with LDL receptor knockout mice. As reported previously, LDLR–/– mice manifested a moderate elevation in plasma cholesterol (∼215 mg/dl) and triglycerides (∼155 mg/dl). In contrast, the doubly mutant TgBP-1a;LDLR–/– mice exhibited marked increases in plasma cholesterol (∼1,050 mg/dl) and triglycerides (∼900 mg/dl). These lipids were contained predominantly within large very-low-density lipoprotein (VLDL) particles that were relatively enriched in cholesterol and apolipoprotein E. Freshly isolated hepatocytes from TgBP-1a and TgBP-1a;LDLR–/– mice overproduced cholesterol and fatty acids and secreted increased amounts of these lipids into the medium. Electron micrographs of livers from TgBP-1a mice showed large amounts of enlarged lipoproteins within the secretory pathway. We conclude that the TgBP-1a mice produce large lipid-rich lipoproteins, but these particles do not accumulate in plasma because they are degraded through the action of LDL receptors.
J. Clin. Invest. 103:1067–1076 (1999).
Introduction
The synthesis of cholesterol and unsaturated fatty acids is regulated by a family of membrane-bound transcription factors called sterol regulatory element binding proteins (SREBPs) (1). The tripartite SREBPs consist of an NH2-terminal domain of ∼480 amino acids that is a transcription factor of the basic-helix-loop-helix-leucine zipper family, a central membrane attachment domain of ∼80 amino acids containing two membrane-spanning segments and a COOH-terminal regulatory domain of ∼590 amino acids. The SREBPs are bound to membranes of the endoplasmic reticulum (ER) and outer nuclear envelope in a hairpin fashion, with the NH2- and COOH-terminal domains projecting into the cytosol (2). In sterol-deprived cells, a two-step proteolytic process releases the NH2-terminal domains, allowing them to enter the nucleus where they enhance transcription of multiple genes involved in the synthesis of cholesterol and fatty acids and in their receptor-mediated uptake from plasma lipoproteins. When sterols build up in cells, the proteolytic release process is inhibited, the NH2-terminal domains of the SREBPs remain membrane bound, and transcription of the target genes declines (1).
The three known isoforms of SREBP are produced by two genes (1). SREBP-1a and SREBP-1c (also known as ADD1) (3) are encoded by a single gene by using alternate promoters that give rise to alternate first exons (4, 5). The proteins differ in the length of an acidic sequence at the extreme NH2-terminus that mediates the interaction with the coactivator CBP (6). SREBP-1a has a long acidic sequence and is a potent activator; SREBP-1c has a shorter acidic sequence and is only weakly active (7, 8). The third isoform, SREBP-2, has a long acidic sequence like that of SREBP-1a, and it too is a potent activator of transcription (8). SREBP-1a and -2 can each activate transcription of the gene encoding the low-density lipoprotein (LDL) receptor as well as genes in the cholesterol and fatty acid biosynthetic pathways, but they have different relative efficiencies. SREBP-1 isoforms tend to favor activation of genes of fatty acid biosynthesis, whereas SREBP-2 is more potent in activating genes of cholesterol biosynthesis (7–9).
The biochemistry of the SREBPs was elucidated through experiments in cultured cells. The relevance to hepatic physiology was established by two types of experiments: regulatory and genetic. The regulatory experiments demonstrated that the NH2-terminal fragment of SREBP-2 increases in hepatic nuclei of hamsters and mice that are rendered cholesterol-deficient by treatment with cholesterol synthesis inhibitors and bile acid–binding resins (4, 10, 11). Conversely, the amounts of nuclear SREBP-1 and SREBP-2 are reduced in hepatic nuclei of hamsters that are fed a high-cholesterol diet (11). These changes correlate with changes in the amounts of mRNA for the LDL receptor and enzymes of the cholesterol biosynthetic pathway.
The genetic experiments involved the production of transgenic mice that express truncated nuclear forms of SREBP-1a, -1c, or -2 in the liver (7, 9, 12). The transgene-encoded SREBPs terminate after the basic-helix-loop-helix-leucine zipper domain, but before the membrane attachment domain. These proteins enter the nucleus without proteolysis, and hence they are not susceptible to feedback regulation. The transgenic animals expressing truncated SREBP-1a and -2 had enlarged livers, owing to the accumulation of large amounts of triglycerides and cholesteryl esters. Despite the accumulation of fat, the rate of synthesis of cholesterol and fatty acids remained high, owing to large increases in hepatic mRNAs for multiple enzymes in the cholesterol and fatty acid biosynthetic pathways (9, 12). These experiments demonstrated that forced overexpression of nuclear SREBPs can overcome the normal system of feedback regulation and lead to massive overproduction of lipids in the liver.
The aforementioned studies with the transgenic mice produced one puzzling result. Despite the massive accumulation of cholesteryl esters and fatty acids in the liver, the plasma levels of these lipids were not elevated; in fact, plasma triglycerides were significantly reduced (9, 12). We hypothesized that the livers of these transgenic mice were producing lipid-rich lipoproteins, but the plasma level did not rise because of the action of hepatic LDL receptors, whose mRNA increased approximately sixfold under the influence of the SREBP transgenes (9, 12). The current experiments were designed to test this hypothesis. For this purpose, we mated SREBP-1a transgenic mice with a previously described line of mice in which the LDL receptor gene was disrupted by gene targeting (13). The LDLR–/– mice were shown previously to have moderate elevations in plasma LDL (total plasma cholesterol levels ∼240 mg/dl in LDLR–/– mice versus ∼110 mg/dl in wild-type littermates). Plasma cholesterol rose markedly only when the LDLR–/– mice were placed on a 1.25% cholesterol diet (plasma cholesterol ∼2,000 mg/dl in LDLR–/– mice versus ∼250 mg/dl in wild-type mice on the same high-cholesterol diet) (14). If low plasma lipids in the SREBP-1a transgenic mice are attributable to clearance by LDL receptors, we predicted that the plasma lipid levels would rise markedly when the SREBP-1a transgenic mice were crossed with LDLR–/– mice, and indeed this is what we observed.
Methods
Animals.
Transgenic mice overexpressing amino acids 1–460 of human SREBP-1a under the control of the phosphoenolpyruvate carboxykinase (PEPCK) promoter were described previously (12). Males hemizygous for the SREBP-1a transgene (TgBP-1a) were bred to SJL × C57BL6/J F1 females to produce TgBP-1a and wild-type controls. TgBP-1a;LDLR–/– mice were generated by crossing TgBP-1a hemizygous males with LDLR–/– females (13). The TgBP-1a;LDLR+/– mice were bred to produce TgBP-1a;LDLR–/– offspring. The current studies were performed with offspring from TgBP-1a;LDLR–/– males bred to LDLR–/– females. All mice were housed in colony cages with a 14-h light/10-h dark cycle and were maintained on Teklad 6% (wt/wt) Mouse/Rat Diet 7002 from Harlan Teklad Premier Laboratory Diets (Madison, Wisconsin, USA).
Blot hybridization of RNA.
Total RNA was prepared from the livers of mice described in experiment A of Table 1 using the RNeasy Total RNA kit (QIAGEN Inc., Valencia, California, USA). Equal aliquots of total RNA from mice of each genotype were pooled (total of 15 μg) and subjected to Northern blot analysis with the indicated cDNA probes as described previously (12). cDNA probes for mouse apo AII, apo CI, apo CII, and apo CIII were prepared with reverse transcriptase–PCR using mouse liver poly(A)+ RNA as a template (12). The PCR primers used were as follows: apo AII: 5′ primer, 5′-GAATTCCATAGAATCGCAGCACTGTT-3′, and 3′ primer, 5′-GAATTCAACTCCTTCCGCATTTATT-3′ (15); apo CI: 5′ primer, 5′-ATGGGCCTCCTGAGAGATCCTTAGATCCAG-3′, and 3′ primer, 5′-TCTGTGATGAAGAGGGACCTGGCACATTAC-3′ (16); apo CII: 5′ primer, 5′-TGGAGTGAGCCAGGATAGTCCTTCCTGCCA-3′, and 3′ primer, 5′-GAACCTGGACTCATTTATTCTTTCACTGAA-3′ (17); apo CIII: 5′ primer, 5′-CCTGCTCAGTTTTATCCCTAGAAGCAGCTA-3′, and 3′ primer, 5′-CATACTTTCCCCTTAAAGCAACCTTCAGG-3′ (18). Northern blot filters were exposed to a Fuji PhosphorImager and quantified using a Bio-Imaging Analyzer with BAS1000 MacBAS software (Fuji Medical Systems, Stamford, Conneticut, USA).
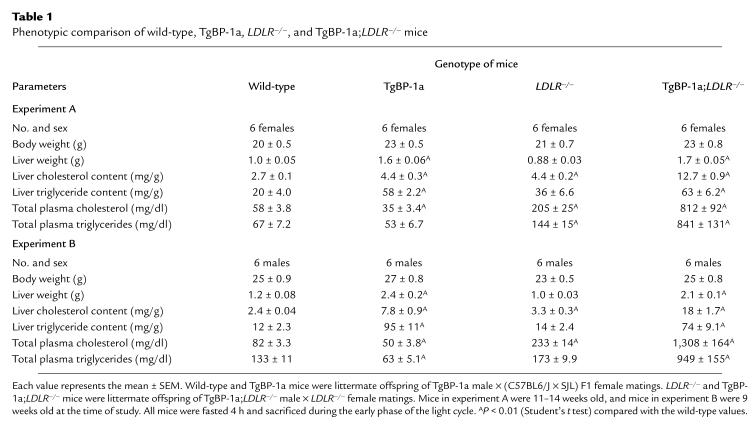
Table 1.
Phenotypic comparison of wild-type, TgBP-1a, LDLR–/–, and TgBP-1a;LDLR–/– mice
Fast performance liquid chromatography analysis.
Pooled plasma (3 ml) from six male mice of each genotype (experiment B in Table 1) was subjected to ultracentrifugation at a density (d) of 1.215 g/ml. The resulting lipoprotein fractions (d < 1.215) were adjusted to a final volume of 3 ml with a buffer containing 0.15 M sodium chloride, 0.01% (wt/vol) sodium EDTA, and 0.02% (wt/vol) sodium azide at pH 7.2, after which 2 ml was subjected to gel filtration with fast performance liquid chromatography (FPLC) (19). Fractions (2 ml) were collected, and 0.1 ml from each fraction was used to determine the content of total cholesterol and triglycerides (12, 19).
For apolipoprotein analysis, equal aliquots (0.75 ml) from each of three consecutive FPLC fractions were pooled (total of 1.5 ml) and delipidated with 15 ml of ethanol/ether (1:1), and the precipitates were collected by centrifugation at 1,000 g for 15 min. The precipitated apolipoproteins were dissolved in SDS loading buffer containing 50 mM Tris-chloride (pH 6.8), 5.5% (vol/vol) β- mercaptoethanol, 7% (wt/vol) SDS, 2.5% (wt/vol) sodium myristyl sulfate, 0.5% (wt/vol) sodium cetyl sulfate, 0.1% (vol/vol) bromophenol blue, and 10% (vol/vol) glycerol (20). The samples were boiled for 10 min and subjected to electrophoresis on 3–15% SDS gradient gels. The gels were stained with Coomassie blue and calibrated with Rainbow high-molecular-weight markers (Amersham Corp., Burlington, Massachusetts, USA).
Plasma lipoprotein isolation.
For the experiments in Table 2 and Figure 4, mice 10–14 weeks old were fasted 4 h, and plasma was collected in sodium EDTA (1 mg/ml) at pH 8 and aprotinin (2 mg/ml). Equal aliquots of plasma (0.4 ml) from 10 mice of each genotype (five males and five females) were pooled (total of 4 ml), and lipoproteins were separated by sequential ultracentrifugation at densities of 1.006, 1.019, 1.063, and 1.215 to obtain very-low-density lipoprotein (VLDL), intermediate-density lipoprotein (IDL), LDL, and high-density lipoprotein (HDL), respectively (21). Each lipoprotein fraction was dialyzed against a buffer containing 150 mM NaCl, 10 mM sodium phosphate (pH 7.4), and 0.01% sodium EDTA. The triglyceride content in each lipoprotein fraction was determined as described above for plasma samples. Phosphatidylcholine was measured with the enzyme phospholipase D according to the manufacturer’s instructions (Phospholipids B; Wako Chemicals USA Inc., Richmond, Virginia, USA). Total and unesterified cholesterol were measured fluorometrically as described (22), and esterified cholesterol was calculated by subtraction.
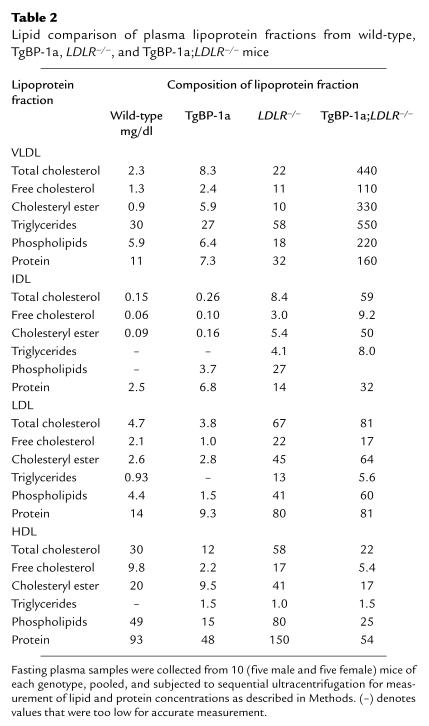
Table 2.
Lipid comparison of plasma lipoprotein fractions from wild-type, TgBP-1a, LDLR–/–, and TgBP-1a;LDLR–/– mice
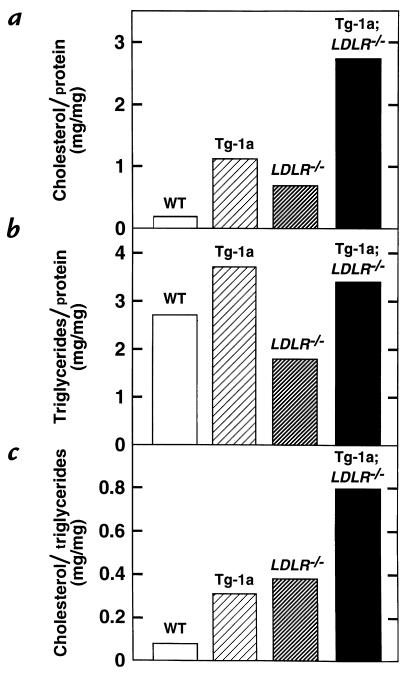
Figure 4.
Composition of plasma VLDL (d < 1.006 g/ml) from wild-type (WT), TgBP-1a, LDLR–/–, and TgBP-1a;LDLR–/– mice. The ratios of cholesterol/protein, triglycerides/protein, and cholesterol/triglycerides were calculated from the values shown in Table 2. VLDL, very-low-density lipoprotein.
Electron microscopy of plasma VLDL.
Sixteen-week-old male mice were fasted for 4 h, and plasma was collected in sodium EDTA (1 mg/ml) at pH 8 and aprotinin (2 mg/ml) from the inferior vena cava of anesthetized mice. The plasma from four mice of each genotype was pooled, and VLDL (d < 1.006) was isolated by ultracentrifugation (21). Negatively stained preparations of VLDL particles were prepared as described (23). The VLDL particles were photographed at 60,000 diameters and 80 kV in a Seimens 101 electron microscope (Seimens Corp., Iseline, New Jersey, USA).
Clearance of 125I-labeled VLDL from plasma.
125I-labeled VLDL from TgBP-1a;LDLR–/– mice was prepared as follows. Plasma was collected from five male TgBP-1a;LDLR–/– mice after a 4-h fast, pooled, and ultracentrifuged at d = 1.006. The VLDL fraction (1.3 mg protein) was radiolabeled with sodium 125I by the iodine monochloride method as described previously (21). The specific activity of the 125I-labeled VLDL was 860 cpm/ng protein. Of the total 125I-radioactivity in VLDL, ∼30% was contained in apo B, as estimated by isopropanol precipitation (24).
All recipient mice were fasted 1 h and anesthetized with sodium pentobarbital (60 mg/kg). Each mouse received an intravenous bolus of 0.1 ml of 0.15 M NaCl containing 15 μg of 125I-labeled VLDL (860 cpm/ng apo B protein) via the right external jugular vein. Blood was collected from the left jugular vein 1 min after the 125I-labeled VLDL injection and at the indicated times thereafter. Two methods were used to measure the content of 125I-labeled apo B in plasma. Method 1 measured total 125I-labeled apo B (apo B-48 plus apo B-100) by isopropanol precipitation (24). An aliquot (10 μl) of each plasma sample was adjusted to 250 μl with 0.15 M NaCl containing human LDL (100 μg) as a carrier. An equal volume of isopropanol (250 μl) was added, and the samples were placed on ice for 30 min. The samples were centrifuged at 15,000 g for 10 min to precipitate apo B. The content of 125I was determined by γ-scintillation spectrometry of samples from the original plasma and from the supernatant portion of the isopropanol-treated plasma. The amount of radiolabeled apo B was determined by subtracting the radioactivity in the supernatant from that in the original plasma. Method 2 measured 125I-labeled apo B-48 directly by subjecting 2 μl of each plasma sample to 3–15% gradient SDS-PAGE, after which the gel was dried, and the signals generated by apo B-48 were quantified by a Fuji PhosphorImager as described above.
Lipid synthesis and secretion in mouse primary hepatocytes.
Mice (14–16 weeks old) of the indicated genotype were anesthetized, and their livers were perfused via the portal vein with 30 ml of Hanks’ buffered saline, without calcium or magnesium, supplemented with 10 mM HEPES (pH 7.4), 0.5 mM sodium EGTA, and gentamycin (0.1 mg/ml). The buffer was prewarmed to 37°C and infused at a rate of ∼3 ml/min. The livers were then perfused with Liver Digest Medium containing collagenase (catalogue no. 17703-034; GIBCO-BRL, Grand Island, New York, USA) for ∼10 min at a flow rate of ∼3 ml/min. The livers were removed from the animals, the hepatic capsule was stripped, and the dissociated cells were dispersed by shaking, followed by filtration at 4°C through gauze into an equal volume of ice-cold DMEM (catalog no. 11885-084; GIBCO BRL) containing 5% (vol/vol) FCS, 10 mM HEPES (pH 7.4), and gentamycin (0.1 mg/ml). The cells were pelleted and washed twice at 4°C with the same medium. The cells were counted with an hemacytometer, and 1.5 × 106 cells were plated onto 60-mm mouse collagen IV–coated dishes (Fisher Scientific Co., Pittsburgh, Pennsylvania, USA) in the above medium and incubated at 37°C in 81% air/9% CO2 for 3 h. The cells were then washed with DMEM containing 5% human lipoprotein–deficient serum (21). They were then incubated with DMEM containing 5% human lipoprotein–deficient serum, gentamycin (0.1 mg/ml), and 0.5 mM sodium [14C]acetate (18 dpm/pmol). After incubation for the times indicated, the medium was removed and centrifuged as described above for preparation of mouse plasma VLDL. The cells were harvested by scraping into 2 ml of 0.1 N sodium hydroxide, followed by 2 ml water (25). The content of 14C-labeled cholesterol and 14C-labeled fatty acids in the cells and in the VLDL fraction of medium was quantified by thin-layer chromatography and scintillation counting as described previously (8). The cellular protein content was measured by the BCA Protein Assay Kit (Pierce Chemical Co., Rockford, Illinois, USA). The data are expressed as nanomoles of [14C]acetate incorporated into cholesterol or fatty acids per milligram of cell protein.
Ultrastructure.
The livers of one male and one female 16-week-old TgBP-1a mice, fed ad libitum, were perfused and fixed under pentobarbital anesthesia and prepared for electron microscopy as described (26). Liver lipids were stained by a modification of the Angermüller and Fahimi (27) imidazole-buffered osmium tetroxide technique as described (28). Two blocks of liver from each mouse were thin-sectioned, and ∼100 electron photomicrographs were examined in this study.
Results
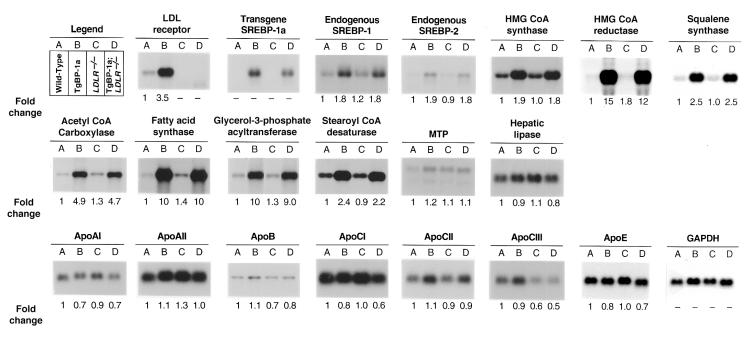
Figure 1 compares the levels of various mRNAs, as determined by Northern blotting, in pooled samples from livers of wild-type mice, transgenic SREBP-1a mice (TgBP-1a), LDLR–/– mice, and TgBP-1a;LDLR–/– mice, all studied in the same experiment. As expected, the TgBP-1a mice showed an elevation in the mRNA for the LDL receptor (3.5-fold). This mRNA was missing in the LDLR–/– and TgBP-1a;LDLR–/– mice. The level of mRNA expression for the SREBP-1a transgene in the TgBP-1a;LDLR–/– mice was ∼80 percent of that in the TgBP-1a mice. The elevations in mRNAs for the cholesterologenic enzymes (HMG CoA synthase, HMG CoA reductase, and squalene synthase) were nearly identical in the TgBP-1a and TgBP-1a;LDLR–/– mice. Similar findings pertained to the enzymes of fatty acid synthesis and esterification (acetyl CoA carboxylase, fatty acid synthase, stearoyl CoA desaturase, and glycerol-3-phosphate acyltransferase). The mRNAs for apo AI, AII, B, CI, CII, CIII, and E were not significantly or consistently altered in the gene-manipulated mice.
Figure 1.
Amounts of various mRNAs in livers of (A) wild-type, (B) TgBP-1a, (C) LDLR–/–, and (D) TgBP-1a;LDLR–/– mice, as measured by blot hybridization. Total RNA isolated from livers of mice described in experiment A of Table 1 was pooled, and 15-μg aliquots were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled probe. The amount of radioactivity in each band was quantified as described in Methods. The probe used for SCD was a mouse SCD-1 cDNA fragment that detects both SCD-1 and SCD-2 mRNAs (see ref. 35). The increase for each mRNA relative to that of wild-type mice was calculated after correction for loading differences with GAPDH as described previously (12). These values are shown below each blot. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MTP, microsomal triglyceride transfer protein; SCD, stearoyl CoA desaturase.
Consistent with the elevations in mRNAs for lipid-synthesizing enzymes, the livers of the TgBP-1a and TgBP-1a;LDLR–/– mice were enlarged, pale in color, and enriched with lipid. Table 1 summarizes two experiments in which quantitative values were obtained on livers and plasma of matched female and male mice of the four genotypes. The liver weights of the TgBP-1a and TgBP-1a;LDLR–/– mice were elevated equally when compared with the nontransgenic mice. The degree of increase was not as profound as in our previous study of TgBP-1a mice because the current animals were not placed on a low-carbohydrate/high-protein diet to induce high-level expression of the transgene, which was driven by the promoter of the gluconeogenic enzyme PEPCK (12). To avoid the possible influence of such a diet on the measured metabolic parameters, we deliberately maintained the animals on a normal chow diet, which allows a moderate, but not superabundant, expression of the transgene. The hepatic content of triglyceride was elevated equally in the TgBP-1a and TgBP-1a;LDLR–/– mice. The hepatic cholesterol content was elevated significantly in the LDLR–/– mice when compared with wild-type mice. The LDL receptor deficiency and the SREBP-1a overexpression combined to produce a higher hepatic cholesterol content in the TgBP-1a;LDLR–/– mice than in the TgBP-1a animals (Table 1).
As reported previously, total plasma cholesterol and triglyceride levels were either normal or slightly reduced in TgBP-1a mice (12). Plasma cholesterol was elevated to the range of 205–233 mg/dl in the LDLR–/– mice (Table 1). When the two genetic alterations were combined, the plasma levels of cholesterol and triglycerides rose markedly (total plasma cholesterol of 812–1308 mg/dl and triglycerides of 841–949 mg/dl in the TgBP-1a;LDLR–/– mice).
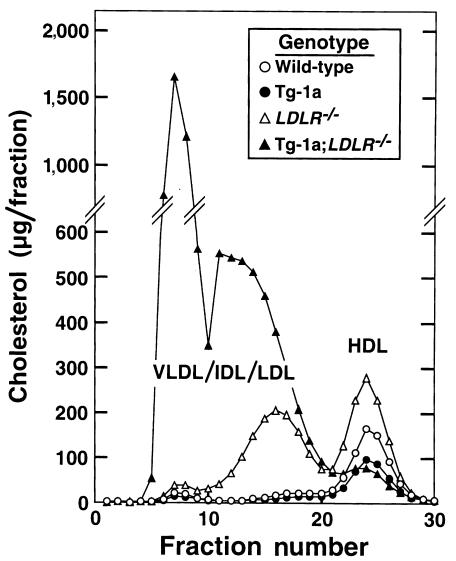
To determine the lipoprotein distribution of the excess plasma cholesterol, we fractionated the plasma lipoproteins by FPLC, which separates particles on the basis of size (Figure 2). These data repeat the earlier observation that the elevated cholesterol in LDLR–/– mice is contained within particles of the LDL size range (13). In contrast, the TgBP-1a;LDLR–/– animals exhibited a marked increase in large lipoproteins that extended from super-large VLDL to IDL and LDL.
Figure 2.
FPLC profiles of plasma lipoproteins from wild-type (open circles), TgBP-1a (filled circles), LDLR–/– (open triangles), and TgBP-1a;LDLR–/– (filled triangles) mice. Plasma from mice described in experiment B of Table 1 was pooled (2.5 ml for each genotype) and subjected to ultracentrifugation at d = 1.215 g/ml. The lipoprotein fractions (d < 1.215 g/ml) were subjected to gel filtration by FPLC, and the cholesterol content of each fraction was measured as described in Methods. FPLC, fast performance liquid chromatography.
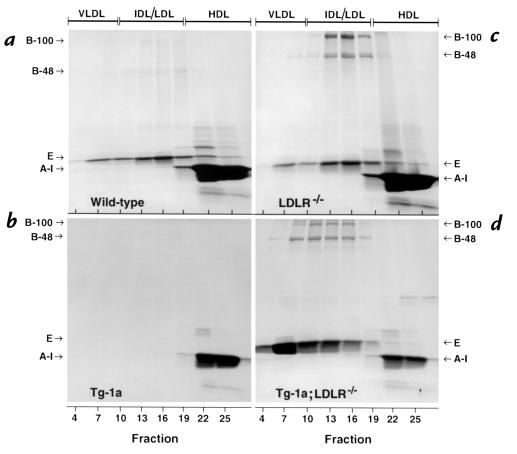
Figure 3 shows the protein composition of various lipoproteins isolated from the FPLC column. The apolipoproteins were stained with Coomassie blue and identified on the basis of size. In the plasma of wild-type mice, the major apolipoprotein seen in the VLDL/IDL/LDL fractions was apo E (Figure 3a). Apo AI was abundant in the HDL fraction. In the TgBP-1a plasma, apo E was no longer visible (Figure 3b). Apo AI was present but reduced. The LDLR–/– mice showed obvious elevations in apo B-48 and apo B-100, which were enriched in the LDL fraction (Figure 3c). The amounts of apo E and apo AI remained relatively normal. The TgBP-1a;LDLR–/– mice exhibited a major increase in apo E, most of which was found in the VLDL fraction (Figure 3d). The total amounts of apo B-48 and apo B-100 were also increased, even when compared with the elevated levels in LDLR–/– mice, and these apolipoproteins were shifted toward larger particles. The amount of apo AI was significantly reduced.
Figure 3.
SDS-PAGE of plasma apolipoproteins from (a) wild-type, (b) TgBP-1a, (c) LDLR–/–, and (d) TgBP-1a;LDLR–/– mice. Equal aliquots (0.75 ml) from three consecutive FPLC fractions shown in Figure 2 were pooled (total of 1.5 ml) and delipidated, and the apoproteins were precipitated as described in Methods. The apoproteins were subjected to 3–15% gradient SDS-PAGE and were stained with Coomassie blue. The positions of migration of apo B-100, apo B-48, apo E, and apo AI are indicated.
To study the composition of the plasma lipoproteins more quantitatively, we pooled plasma from 10 mice of each genotype and separated the lipoproteins by sequential ultracentrifugation at increasing densities as described in Methods (Table 2). The VLDL fractions from the TgBP-1a;LDLR–/– mice contained a major increase in the number of particles, as revealed by an increased content of proteins and lipids. These particles were relatively enriched in cholesterol, as determined by the marked increase in the cholesterol/protein ratio when compared with VLDL from wild-type mice (Figure 4a and Table 2). The total triglyceride content of the VLDL particles was also increased, but the triglyceride/protein ratio remained relatively normal (Figure 4b). As a result of these combined changes, the cholesterol/triglyceride ratio was elevated eightfold in the VLDL of TgBP-1a;LDLR–/– mice compared with wild-type mice (Figure 4c). A similar enrichment in cholesterol was found in the IDL fraction of TgBP-1a;LDLR–/– mice (Table 2). The content of cholesterol in the LDL fraction of the TgBP-1a;LDLR–/– mice was elevated to the same degree as in the LDLR–/– mice (Table 2). The content of both cholesterol and protein in the HDL fraction was reduced in the TgBP-1a;LDLR–/– mice (Table 2).
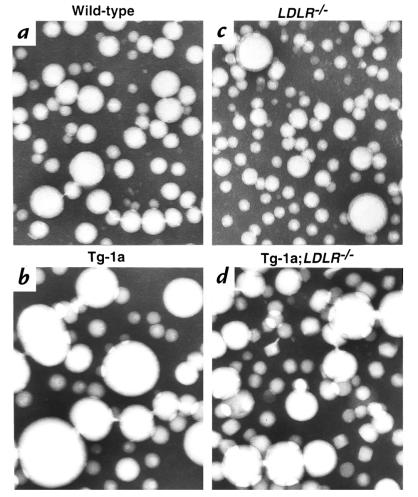
Figure 5 shows electron micrographs of negatively stained VLDL fractions from fasted animals of the four genotypes. These images confirm that the VLDL from TgBP-1a mice and TgBP-1a;LDLR–/– mice contain an increased number of extremely large particles (>80 nm in diameter) that are in the size range normally associated with chylomicrons (28). These very large particles are likely to account for the cholesterol-rich nature of the VLDL fraction in the transgenic mice, as indicated by the compositional data in Table 2.
Figure 5.
Electron microscopy of negatively stained plasma VLDL (d < 1.006 g/ml) from (a) wild-type, (b) TgBP-1a, (c) LDLR–/–, and (d) TgBP-1a;LDLR–/–mice. After a 4-h fast, plasma samples from four male mice of each genotype were pooled, and VLDL was isolated for viewing by electron microscopy as described in Methods. ×108,000.
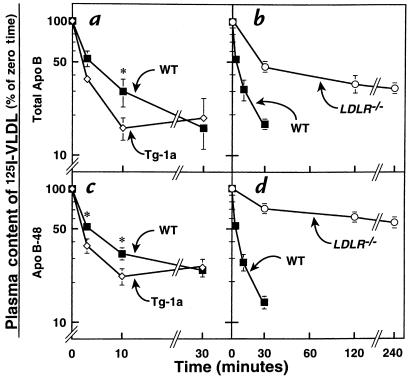
The data presented so far are consistent with the hypothesis that the production of LDL receptors in the TgBP-1a mice compensates for the oversynthesis of lipid-rich lipoproteins and that this compensation is prevented in the TgBP-1a;LDLR–/– mice, thereby unmasking a massive elevation in plasma VLDL. To test this hypothesis more directly, we measured the ability of mice from both genotypes to clear 125I-labeled VLDL from plasma. For this purpose, we isolated VLDL from fasting TgBP-1a;LDLR–/– mice and labeled the protein component with 125I. As shown in Figure 6a, the total apo B component of this 125I-labeled VLDL was cleared more rapidly from the circulation in the TgBP-1a mice compared with wild-type mice. In contrast, the LDLR–/– mice exhibited a marked delay in this clearance (Figure 6b). Similar results were obtained when we specifically followed the 125I-labeled apo B-48 component, as determined by SDS-PAGE followed by autoradiography (Figure 6, c and d). In experiments not shown, we found that the clearance of the apo B-100 and apo B-48 components of 125I-labeled VLDL were just as defective in the TgBP-1a;LDLR–/– mice as in the LDLR–/– littermates, indicating that the delayed clearance in the doubly mutant mice was due to impaired LDL receptor function.
Figure 6.
Plasma clearance of 125I-labeled VLDL in wild-type (WT), TgBP-1a, LDLR–/–, and TgBP- 1a;LDLR–/– mice. VLDL was isolated from four fasted male TgBP-1a;LDLR–/– mice and labeled with 125I as described in Methods. 125I-labeled VLDL (15 μg protein, 860 cpm/ng apo B protein) was injected intravenously into each of five 12-week-old male mice with the indicated genotype. Blood was drawn at the indicated times to quantify the content of 125I-labeled apo B in plasma. (a and b) Plasma content of 125I-labeled total apo B was measured by isopropanol precipitation followed by scintillation counting (as described in Methods). (c and d) Plasma samples were subjected to SDS-PAGE, and the signal generated by apo B-48 was quantified using a Fuji PhosphorImager as described in Methods. The data are plotted as the percent of the zero time value. *P < 0.05 (Student’s t test) compared with wild-type values. Each value represents the mean ± SEM of data from five mice.
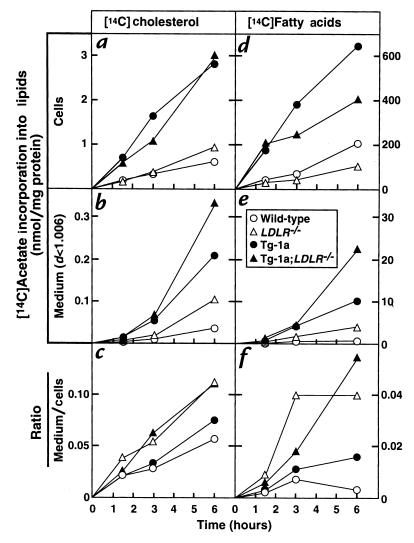
To compare the rates of lipid synthesis and secretion in the mice of the four genotypes, we turned to the use of freshly isolated hepatocytes (Figure 7). To mimic in vivo conditions as closely as possible, these cells were studied three hours after plating. The time required to prepare the hepatocytes before plating was ∼40 minutes. The cells were incubated for various intervals with [14C]acetate, after which the cells and medium were harvested separately and the content of total 14C-labeled cholesterol and 14C-labeled fatty acids (unesterified plus esterified) was measured. As expected, hepatocytes from the TgBP-1a and TgBP-1a;LDLR–/– mice synthesized much more 14C-labeled cholesterol and 14C-labeled fatty acids than did the wild-type and LDLR–/– mice (filled symbols in Figure 7, a and d). They also secreted more of these lipids into the culture medium (filled symbols in Figure 7, b and e). An interesting phenomenon emerged when we calculated the efficiency of secretion, i.e., the ratio between the amount of radioactive lipids in the medium compared with the cells. Relative hypersecretion was seen in those animals lacking LDL receptors, i.e., the LDLR–/– and TgBP-1a;LDLR–/– mice (triangles in Figure 7, c and f). Similar results were obtained when this experiment was repeated on three other occasions.
Figure 7.
Rates of lipid synthesis and secretion by primary hepatocytes from wild-type (open circles), TgBP-1a (filled circles), LDLR–/– (open triangles), and TgBP-1a;LDLR–/– (filled triangles) mice. Primary hepatocytes were prepared from one mouse of each genotype as described in Methods. The hepatocytes were incubated with DMEM containing 5% FCS for 3 h at 37°C, after which the cells were incubated with 0.5 mM sodium [14C]acetate (18 dpm/pmol) in DMEM supplemented with 5% human lipoprotein–deficient serum. At the indicated time, the medium was removed for centrifugation at d < 1.006 g/ml, the monolayers were washed, and the cells were harvested. The content of 14C-labeled cholesterol and fatty acids in cells and medium was quantified as described in Methods. Each value is the average of duplicate incubations. Similar results were obtained in three other experiments in which different groups of mice were studied at a single time point.
To further test the hypothesis that the livers of TgBP-1a mice are producing increased amounts of lipid-enriched lipoproteins, we studied the liver by electron microscopy. At low magnification (approximately ×12,000), the excess nonpolar lipids in the liver were apparently partitioned largely within two distinct compartments of the hepatocytes: large cytosolic lipid droplets (∼1–3 μm in diameter) and smaller lipid-staining particles (∼100–250 nm) within the membrane-bound compartments of the VLDL secretory pathway. Remarkably, there were very few lipid-staining droplets between these two size ranges (images not shown). The hepatocyte VLDL secretory compartments were better resolved at higher magnification (×27,000), as shown in Figure 8, a–d.
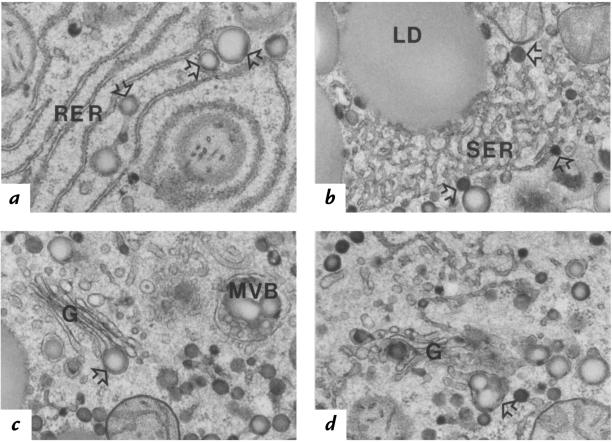
Figure 8.
Electron micrographs of thin sections depicting lipoprotein assembly and degradation in fatty hepatocytes of TgBP-1a mouse liver. These hepatocytes contained numerous very large (∼100–250 nm) nascent VLDL particles (open arrows) in the following areas: (a) within the lumen of the unique smooth-surfaced (ribosome-free) terminal ends of rough ER (RER); (b) within the smooth ER (SER) proper; (c and d) within forming secretory vesicles of the Golgi apparatus (G), singly (c) or clustered (d). Multivesicular bodies (MVB) that contained partially degraded lipoproteins the same size as nascent VLDL were often seen (c). Larger than normal patches of tightly packed membranes of SER were common in these fatty hepatocytes and sometimes appeared to be associated with cytosolic lipid droplets (LD) (b). Differences in the intensity of lipid staining between nascent VLDL particles of different diameters may reflect differential absorbency of the imidazole mordant and/or differences in unsaturated fatty acid content of the core lipids (27). ×27,000.
The specialized compartment of ER that has been strongly implicated in nascent VLDL assembly — namely, the ribosome-free regions of the terminal ends of the rough ER (RER) (Figure 8a) — was far more evident in the hepatocytes of the TgBP-1a mice than in those of mice or of other species studied previously (29). In many hepatocytes of the TgBP-1a mice, virtually all of these putative VLDL assembly compartments appeared to contain a chylomicron-sized nascent VLDL precursor particle. This morphologic observation alone strongly suggests that VLDL core-lipid assembly in the fatty livers of TgBP-1a mice may be greatly stimulated, even though the level of apo B mRNA remains unchanged. Similarly sized lipid-staining particles were commonly found associated with the smooth ER (SER), mostly around the periphery of the large patches of tightly packed membranes of SER proper (Figure 8b). These putative nascent VLDL particles of chylomicron diameter were often observed singly within forming secretory vesicles of the Golgi apparatus (Figure 8c) or as clusters of a few particles (Figure 8d). This finding differs from hepatocytes of many species, including those of wild-type mice, in which nascent VLDL are almost always clustered in large numbers within forming Golgi secretory vesicles (29).
Interestingly, late hepatocytic endosomal compartments, which have been shown in rat livers to be prelysosomal multivesicular bodies (MVB) filled with remnants of apo B–containing VLDL and chylomicrons (30), contained lipid-staining particles that were not distinguishable from those within the secretory organelles (Figure 8c). In some cases, the large cytosolic lipid droplets (LD) appeared to be closely associated with the surface of tightly packed SER membranes (Figure 8b).
Discussion
The current data provide an explanation for the previous paradoxical observation that plasma levels of cholesterol and triglycerides are normal or slightly reduced in SREBP-1a transgenic mice despite massive overproduction and accumulation of these lipids in the liver (9, 12). The data suggest that the livers of these transgenic mice produce extremely enlarged, cholesterol-enriched VLDL but that these particles do not accumulate in plasma because they are degraded through the action of LDL receptors. When LDL receptors were eliminated by producing TgBP-1a;LDLR–/– doubly mutant mice, this degradation was prevented and massive hyperlipidemia was unmasked.
In the SREBP-1a transgenic mice, the clearance of 125I-labeled VLDL was faster than normal despite massive hepatic cholesterol accumulation, which ordinarily would have downregulated LDL receptors. This enhanced clearance was mediated by LDL receptors because it was abolished in the TgBP-1a;LDLR–/– mice.
The notion that the TgBP-1a animals overproduce large cholesterol-rich VLDL particles is supported by two observations. First, when freshly isolated hepatocytes from these animals were incubated with [14C]acetate, they secreted increased amounts of 14C-labeled cholesterol and 14C-labeled fatty acids. Second, electron micrographs of livers from TgBP-1a mice showed large lipoproteins that were formed in the ER and carried to the Golgi where they were observed within budding secretory vesicles.
Receptor-mediated degradation of the cholesterol-enriched VLDL particles could have occurred at one or all of three levels: (a) before secretion from the hepatocytes; (b) after secretion but before escape from the Dissé’s space into the hepatic sinusoids; and (c) after the particles had circulated in plasma. We believe it likely that some, but not all, of the nascent VLDL was destroyed before secretion from the cell. This is based on findings from the studies in which hepatocytes were incubated with [14C]acetate. These studies showed that the relative efficiency of secretion, as determined by the ratio between secreted lipid compared with the amount retained within the cell, was higher in LDL receptor–deficient mice. This relative hypersecretion was observed in the LDLR–/– mice as well as in the TgBP-1a;LDLR–/– mice, even though the absolute rates of lipid synthesis and secretion differed markedly between these two genotypes (Figure 7, c and f). This finding would be explained if LDL receptors, functioning within hepatocytes, mediate the degradation of some VLDL particles before their secretion. These findings would also be consistent with numerous studies of lipoprotein turnover in humans that show an apparent oversecretion of VLDL (and perhaps LDL) in homozygotes with familial hypercholesterolemia who lack LDL receptors (31). This finding is also consistent with recent data from Twisk and coworkers (32), who showed a decrease in intrahepatic degradation of apo B–containing lipoproteins in LDLR–/– mice, which also lack LDL receptors .
The electron microscopic images in Figure 8c suggest that some of the large lipoprotein particles are secreted and then taken back into the cells. This conclusion is based on the finding of large lipoprotein-like particles within multivesicular bodies that ordinarily contain lipoproteins taken up from the surface by receptor-mediated endocytosis (30). The chylomicron-sized VLDL particles are so large (many >150 nm) that they might be hindered in penetrating through the sinusoidal endothelial pores, whose diameters are limited to about 120 nm (33). They would then remain retarded in the Dissé’s space, from which they would be cleared by cell-surface hepatocyte LDL receptors and delivered to multivesicular bodies. In the absence of LDL receptors, some of these particles may more readily pass into the sinusoids from the Dissé’s space.
The large size of the secreted VLDL particles is likely attributable to an imbalance between the large amount of triglyceride and cholesteryl esters and the limiting amount of apo B, whose mRNA levels do not increase in the TgBP-1a mice. A similar phenomenon has been observed in enterocytes of apo B–deficient mice (28).
The observation of marked hyperlipidemia in the TgBP-1a;LDLR–/– mice, but not in TgBP-1a mice, provides further support for the emerging concept that LDL receptors are able to compensate for large increases in cholesterol input from either dietary sources or endogenous synthesis and that significant hypercholesterolemia occurs only when the function of the LDL receptor is impaired through one of several mechanisms — namely, feedback suppression of the SREBP pathway, mutation in the LDL receptor gene, or mutation in the genes encoding either of its ligands, apo B or apo E (14, 31, 34).
Acknowledgments
We thank Scott Clark, Robin Craddock, and Debra Morgan for excellent technical assistance; Richard Gibson for invaluable help with animal care; Jinny Wong for help with outstanding electron microscopy; and our colleague Shinji Ikemoto for help with primary hepatocytes. This study was supported by grants from the National Institutes of Health (HL-20948 and HL-60844), the Moss Heart Foundation, and the Perot Family Foundation. J.D. Horton was the recipient of a Postdoctoral Fellowship for Physicians from the Howard Hughes Medical Institute.
Footnotes
J.D. Horton and H. Shimano contributed equally to this work.
References
- 1.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Hua X, Sakai J, Ho YK, Goldstein JL, Brown MS. Hairpin orientation of sterol regulatory element binding protein-2 in cell membranes as determined by protease protection. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 3.Tontonoz P, Kim JB, Graves RA, Spiegelman BM. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993;13:4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimomura I, Shimano H, Horton JD, Goldstein JL, Brown MS. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua X, Wu J, Goldstein JL, Brown MS, Hobbs HH. Structure of human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 6.Näär AM, et al. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimano H, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pai J, Guryev O, Brown MS, Goldstein JL. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element binding proteins. J Biol Chem. 1998;273:26138–26148. doi: 10.1074/jbc.273.40.26138. [DOI] [PubMed] [Google Scholar]
- 9.Horton JD, et al. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing SREBP-2. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng Z, Otani H, Brown MS, Goldstein JL. Independent regulation of sterol regulatory element binding proteins 1 and 2 in hamster liver. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimomura I, et al. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc Natl Acad Sci USA. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimano H, et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishibashi S, et al. Hypercholesterolemia in LDL receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishibashi S, Goldstein JL, Brown MS, Herz J, Burns DK. Massive xanthomatosis and atherosclerosis in cholesterol-fed LDL receptor-negative mice. J Clin Invest. 1994;93:1885–1893. doi: 10.1172/JCI117179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doolittle MH, LeBoeuf RC, Warden CH, Bee LM, Lusis AJ. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J Biol Chem. 1990;265:16380–16388. [PubMed] [Google Scholar]
- 16.Hoffer MJV, van Eck MM, Havekes LM, Hofker MH, Frants RR. The mouse apolipoprotein C1 gene: structure and expression. Genomics. 1993;18:37–42. doi: 10.1006/geno.1993.1424. [DOI] [PubMed] [Google Scholar]
- 17.Hoffer MJV, Hofker MH, van Eck MM, Havekes LM, Frants RR. Evolutionary conservation of the mouse apolipoprotein e-c1-c2 gene cluster: structure and genetic variability in inbred mice. Genomics. 1993;15:62–67. doi: 10.1006/geno.1993.1010. [DOI] [PubMed] [Google Scholar]
- 18.Januzzi JL, Azrolan N, O’Connell A, Aalto-Setälä K, Breslow JL. Characterization of the mouse apolipoprotein Apoa-1/Apoc-3 gene locus: genomic, mRNA, and protein sequences with comparisons to other species. Genomics. 1992;14:1081–1088. doi: 10.1016/s0888-7543(05)80133-8. [DOI] [PubMed] [Google Scholar]
- 19.Yokode M, Hammer RE, Ishibashi S, Brown MS, Goldstein JL. Diet-induced hypercholesterolemia in mice: Prevention by overexpression of LDL receptors. Science. 1990;250:1273–1275. doi: 10.1126/science.2244210. [DOI] [PubMed] [Google Scholar]
- 20.Cruzado ID, Cockrill SL, McNeal CJ, Macfarlane RD. Characterization and quantitation of apolipoprotein B-100 by capillary electrophoresis. J Lipid Res. 1998;39:205–217. [PubMed] [Google Scholar]
- 21.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of LDL in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 22.Heider JG, Boyett RL. The picomole determination of free and total cholesterol in cells in culture. J Lipid Res. 1978;19:514–518. [PubMed] [Google Scholar]
- 23.Hamilton RL, Jr, Goerke J, Guo LSS, Williams MC, Havel RJ. Unilamellar liposomes made with the French pressure cell: a simple preparative and semiquantitative technique. J Lipid Res. 1980;21:981–992. [PubMed] [Google Scholar]
- 24.Yamada N, Havel RJ. Measurement of apolipoprotein B radioactivity in whole blood plasma by precipitation with isopropanol. J Lipid Res. 1986;27:910–912. [PubMed] [Google Scholar]
- 25.Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 26.Young SG, et al. A genetic model for absent chylomicron formation: mice producing apolipoprotein B in the liver, but not in the intestine. J Clin Invest. 1995;96:2932–2946. doi: 10.1172/JCI118365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angermüller S, Fahimi HD. Imidazole-buffered osmium tetroxide: an excellent stain for visualization of lipids in transmission electron microscopy. Histochem J. 1982;14:823–835. doi: 10.1007/BF01033631. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton RL, Wong JS, Cham CM, Nielsen LB, Young SG. Chylomicron-sized lipid particles are formed in the setting of apolipoprotein B deficiency. J Lipid Res. 1998;39:1543–1557. [PubMed] [Google Scholar]
- 29.Alexander CA, Hamilton RL, Havel RJ. Subcellular localization of B apoprotein of plasma lipoproteins in rat liver. J Cell Biol. 1976;69:241–263. doi: 10.1083/jcb.69.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havel RJ, Hamilton RL. Hepatocytic lipoprotein receptors and intracellular lipoprotein catabolism. Hepatology. 1988;8:1689–1704. doi: 10.1002/hep.1840080637. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein, J.L., Hobbs, H.H., and Brown, M.S. 1995. Familial hypercholesterolemia. In The metabolic and molecular bases of inherited disease. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill. New York, NY. 1981–2030.
- 32.Twisk J, Gillian-Daniel DL, Lin W, Attie AD. The low density lipoprotein receptor promotes post-translational degradation of apolipoprotein B. Circulation. 1998;98(Suppl. 1):I–178. [Google Scholar]
- 33.Wisse E. An electron microscopic study of the fenestrated endothelial lining of rat liver sinusoids. J Ultrastruct Res. 1970;31:125–150. doi: 10.1016/s0022-5320(70)90150-4. [DOI] [PubMed] [Google Scholar]
- 34.Mahley, R.W., Weisgraber, K.H., and Farese, R.V., Jr. 1998. Disorders of lipid metabolism. In Williams textbook of endocrinology. J.D. Wilson, D.W. Foster, H.M. Kronenberg, and P.R. Larsen, editors. W.B. Saunders. Philadelphia, PA. 1099–1153.
- 35.Shimomura I, Shimano H, Korn BS, Bashmakov Y, Horton JD. Nuclear sterol regulatory element binding proteins activate genes responsible for entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J Biol Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]