Abstract
To determine the specific role lipids play in membrane protein topogenesis in vivo, the orientation with respect to the membrane bilayer of Escherichia coli lactose permease (LacY) transmembrane (TM) domains and their flanking extramembrane domains was compared after assembly in native membranes and membranes with genetically modified lipid content using the substituted cysteine accessibility method for determining TM domain mapping. LacY assembled in the absence of the major membrane lipid phosphatidylethanolamine (PE) does not carry out uphill transport of substrate and displays an inverted orientation for the N-terminal six-TM domain helical bundle (Bogdanov, M., Heacock, P. N., and Dowhan, W. (2002) EMBO J. 21, 2107–2116). Strikingly, the replacement of PE in vivo by the foreign lipid monoglucosyldiacylglycerol (MGlcDAG), synthesized by the Acholeplasma laidlawii MGlcDAG synthase, restored uphill transport and supported the wild type TM topology of the N-terminal helical bundle of LacY. An interchangeable role in defining membrane protein TM domain orientation and supporting function is played by the two most abundant lipids, PE and MGlcDAG, in Gram-negative and Gram-positive bacteria, respectively. Therefore, these structurally diverse lipids endow the membrane with similar properties necessary for the proper organization of protein domains in LacY that are highly sensitive to lipids as topological determinants.
Due to the complex amphipathic nature of membrane proteins, the mechanisms and factors that determine their folding within the membrane and topological organization across the membrane are not completely understood. The majority of research in this area has focused on the information written within the amino acid sequence of membrane proteins as the primary structural, topological, and functional determinants. However, a growing body of evidence suggests that specific membrane lipids, rather than simply providing a passive hydrophobic solvent within which membrane proteins fold and organize themselves, actively determine the final structural, topological, and functional properties of membrane proteins (1).
The novel role of specific lipids acting as topological determinants for membrane proteins was established through studies of the assembly of lactose permease (LacY)2 in Escherichia coli mutants in which the membrane phospholipid composition was genetically manipulated (2). LacY belongs to the major facilitator superfamily of secondary transporters. Despite differences in substrate specificity, members of this family share significant structural and functional homology (3). They all have twelve (I–XII) transmembrane (TM) domains, which are divided into two six-TM helical bundles connected by a long hydrophilic cytoplasmic domain (Fig. 1A). The N-terminal and C-terminal extramembrane domains are both exposed to the cytoplasm. Due to extensive structural and functional information, LacY has become a paradigm for the study of this group of membrane proteins (4). LacY catalyzes the transport of substrate through two different modes: the energy-independent equilibration of substrate across the membrane (downhill transport) and the harnessing of energy released from downhill movement of protons to drive uphill transport of substrate against a concentration gradient (uphill transport).
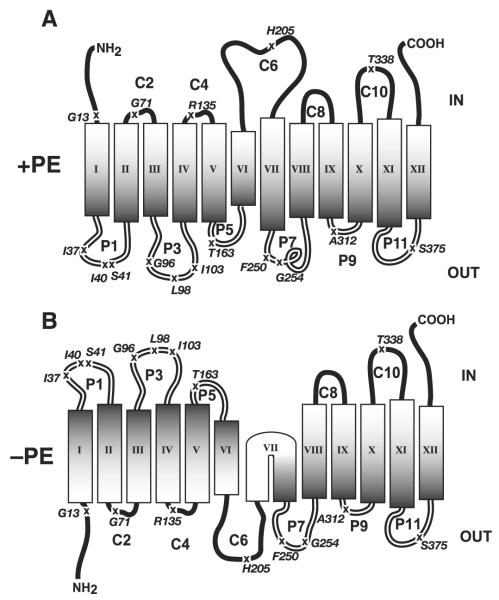
FIGURE 1. Topology models of single cysteine replacement derivatives of Cys-less LacY in +PE (A) and −PE (B) E. coli membranes (adapted from Ref. 2).
TM domains are represented as rectangular boxes and numbered from N terminus to C terminus. Extramembrane loops connecting TM domains are numbered sequentially, where P and C indicate their periplasmic (out) or cytoplasmic (in) location with respect to the bilayer plane in wild type (+PE) E. coli. X indicates the position where an amino acid residue, as indicated, is replaced by a single cysteine residue. The organization of TM VII in −PE cells remains unknown (2).
E. coli mutants lacking the major phospholipid, phosphatidylethanolamine (PE), cannot support uphill transport by LacY, but its downhill transport function remains unaffected both in vivo (5) and in reconstituted proteoliposomes (6, 7) in the absence of PE. More surprisingly, topology assays of LacY assembled in vivo in membranes with or without PE revealed profound differences in topological orientation of the TM domains as a function of membrane lipid composition (2). In the absence of PE, the N-terminal six-TM helical bundle adopts an inverted topology with respect to the plane of the membrane bilayer with the retention of correct topology for most of the C-terminal helical bundle (five of the six TM domains, Fig. 1B). The topology of LacY is dynamic with respect to membrane lipid composition since restoring PE to cells after the assembly of LacY triggered the topological reorganization of the misoriented protein accompanied by the restoration of uphill transport function. Furthermore, LacY displays similar lipid dependence in vitro when reconstituted into proteoliposomes (7). Regardless of the source of LacY (synthesized in membranes with or without PE), its topology and function are solely dependent on the presence of PE in liposomes independent of other cellular components. Interestingly, another zwitterionic lipid, phosphatidylcholine (PC), can also support the native topology of LacY in proteoliposomes, although it failed to support the uphill transport function of the permease (7).
This dependence of assembly and function on PE is not limited to LacY. Phenylalanine permease (PheP) and γ-aminobutyric acid permease (GabP), both belonging to the amino acid/polyamine/organocation superfamily of secondary transporters (8), also require PE (9, 10). Both permeases show defects in uphill transport function and an inverted topology of the N-terminal two-TM helical hairpin when assembled in the absence of PE; lipid-dependent reversibility of this misorientation was demonstrated for PheP after reintroduction of PE into membranes (9).
Ongoing studies of the similarities and differences among these permeases will define the common topogenic signals written in amino acid sequences that govern TM domain orientation as a function of membrane lipid physical and chemical properties. However, in this study, we focused on the properties of lipids that determine TM domain topogenesis based on the observation that E. coli LacY, when expressed in Corynebacterium glutamicum, which lacks all amine-containing lipids including PE (11), carries out uphill transport of substrate (12). Monoglucosyldiacylglycerol (MGlcDAG) is a major membrane lipid in this and other Gram-positive bacteria (13, 14) with related glycolipids found in photosynthetic membranes of plants (15). It is the major non-bilayer prone lipid in these organisms and responsible for regulating the ratio between bilayer lipids and non-bilayer lipids in the membrane to maintain a constant radius of curvature and lipid packing stress in response to changes in the environment (16). Despite their chemical and physical differences, MGlcDAG and PE share a number of common features. They promote non-bilayer structures and thus are able to alter the lateral pressure profile of the membrane; they carry no net charge, with PE being zwitterionic and MGlcDAG being neutral; and their respective headgroups are able to form hydrogen bonds. If MGlcDAG and PE are lipids with different structures but overlapping chemical, physical, and collective phase properties, they would provide a similar lipid environment, and this interchangeability in bacterial membranes might be expected.
Introduction of MGlcDAG into PE-lacking E. coli suppresses many of the phenotypes of cells deficient in PE (17). In the absence of PE, cells grown in medium supplemented with divalent metal ions are viable, but they display a complex mixture of phenotypes. Their growth rate is greatly reduced; the filamentous morphology of these cells indicates a defect in cell division; their outer membrane integrity is compromised; and they are defective in assembly of some membrane proteins, as well as in sugar and amino acid transport (1). Introduction of MGlcDAG into PE-lacking mutants reduces the dependence for viability on divalent metal ions, greatly improves the growth rate, nearly restores normal cell division, alleviates protein-mediated osmotic stress adaptation to salts and sucrose, and partially restores uphill transport function by LacY (17). In this study, we demonstrate that MGlcDAG supports the wild type topological organization of LacY in the absence of PE, thereby demonstrating a commonality between these two lipids in their role in supporting native structure and function of LacY in their respective hosts. Defining the similarities among lipids with similar functions in the membrane will provide the foundation for understanding the specific properties of lipids required to support different aspects of cell function.
EXPERIMENTAL PROCEDURES
Materials
Chemicals and enzymes were purchased from Sigma. [14C]Acetic acid and [14C]methyl-β-d-galactopyrano-side came from American Radiolabeled Chemical Inc. Polyclonal antibody directed against the C terminus of LacY was made by ProSci Inc. Avidin-horseradish peroxidase and Super-Signal West Pico chemiluminescent substrate were from Pierce. Pansorbin cells were from Calbiochem, and Lubrol (type-PX) was from Nacalai Tesque, Inc. (Kyoto, Japan). Anti-LacZpolyclonalantibodywaspurchasedfromRocklandImmuno-chemicals. 3-(N-maleimidylpropionyl) biocytin (MPB) was purchased from Molecular Probes.
Plasmids, Bacterial Strains, and Culture Medium
Plasmids (pLacY*) encoding LacY derivatives lacking native cysteines but containing single cysteine replacements in the extramembrane domains (Fig. 1) were obtained from H. R. Kaback (UCLA). OPtac-lacY* (LacY expressed under tac operator-promoter control) was amplified from each plasmid using PCR primers 5′-ATTTGCGGCCGCGCTTATCGAAATTAATACGACTC-3′ and 5′-GGAAGATCTAAGCGACTTCATTCACCTGACG-3′ containing a BglII and NotI site, respectively. The amplified genes were positioned between the BglII and NotI sites of plasmid pTMG3, which contains the MGlcDAG synthase gene from Acholeplasma laidlawii and encodes resistance to ampicillin (17), to generate a series of plasmids pTMG3-LacY*. All strains were grown in Luria-Bertani (LB) medium containing 50 mm MgCl2. PE-lacking (−PE) strain AL95 (pss93∷kanRlacY∷Tn9) made PE-containing (+PE) by transformation with plasmid pDD72 (pssA+ camR) (+PE) was grown at 30 °C since pDD72 contains a temperature-sensitive replicon. AL95 (−PE) and AL95/pTMG3 (−PE+MGlcDAG) were grown at 37 °C. To express LacY* in strains with different lipid compositions, plasmids pLacY* were transformed into AL95/pDD72 or AL95, and plasmids pTMG-LacY* were transformed into AL95 (10). Ampicillin at 100 μg/ml was present to maintain selection for plasmids. Expression of lacY* genes was induced by growing cells for at least two generations after the addition of 1 mm isopropyl-β-d-thiogalactoside.
Lipid Analysis
Cells were cultured overnight as described above in medium containing 0.2 μCi/ml [14C]acetic acid and then diluted into the same medium, grown until mid-log phase, and harvested by centrifugation. Lipids were extracted from cell pellets by vigorously vortexing in chloroform/methanol (2:1, v/v). After centrifugation, the pellet was resuspended for a second extraction in methanol containing 0.1 n acetic acid. Supernatants from the two extractions were pooled and concentrated under vacuum using a SpeedVac concentrator (Savant). Lipids were dissolved in chloroform/methanol (1:2, v/v) and separated by silica gel thin-layer chromatography using chloroform/methanol/acetic acid (65:25:10, v/v/v) as solvent. Radiolabeled lipids were visualized, quantified using a Fluor-S™ MultiImager (Bio-Rad), and expressed as mole percent.
Chemical Labeling of Cysteines, Immunoprecipitation, and Western blot Analysis
After induction of LacY synthesis by isopropyl-β-d-thiogalactoside, cells were grown to mid-log phase and harvested by centrifugation. Cell pellets from 50-ml cultures were suspended in 1.5 ml of buffer A (100 mm KOH/HEPES buffer, 250 mm sucrose, 25 mm MgCl2, 0.1 mm KCl, adjusted to pH 7 or 9 as indicated) and divided into 0.75-ml aliquots. Samples were treated with MPB at a final concentration of 100 μm for 5 min at room temperature to label cysteines exposed to the periplasmic side of the inner membrane. The reaction was quenched by the addition of β-mercaptoethanol to 20 mm. After labeling, cells were sonicated for 1 min. To expose cysteines located on the cytoplasmic side of the cell membrane, samples treated with MPB were subjected to sonication for 1 min followed by 4 min of incubation at room temperature before quenching. In this study, sonication was used to disintegrate cell membranes, making both periplasmic and cytoplasmic cysteines accessible to MPB, whereas cysteines located within a TM domain were still protected from labeling (18). All sonicated samples were centrifuged at 4 °C and 40,000 rpm (Beckman Coulter, TLA-100) for 10 min followed by solubilization, immunoprecipitation (with anti-LacY or anti-LacZ polyclonal antibody), and Western blot analysis as described previously (2). Finally, SuperSignal West Pico chemiluminescent substrate (Pierce) was used to visualize biotinylated proteins.
LacY Transport
Transport of [14C]methyl-β-d-galactopyranoside (TMG, 0.5 μCi/ml) by LacY was measured in E. coli strains with altered lipid composition as described previously (5).
RESULTS
Lipid Analysis of Strains
Three E. coli strains that display different membrane lipid compositions were used in this work. AL95 (pss93∷kanRlacY∷Tn9 (2)) is PE-deficient (−PE) due to interruption of the pssA gene that encodes phosphatidylserine synthase, which catalyzes the committed step to PE synthesis. In the absence of PE, only anionic lipids, which are primarily phosphatidylglycerol (PG, ~40%) and cardiolipin (CL, ~50%) (Fig. 2, lane 2), are present. Complementation with the pssA gene on plasmid pDD72 (pssA+ camR (19)) results in AL95/pDD72 (+PE) displaying a wild type E. coli lipid composition in which PE accounts for 80% of the total lipid with the remainder being primarily 10% in PG and 5% in CL (Fig. 2, lane 1). Introduction of plasmid pTMG3 (ALmgs ampR (17)) into strain AL95 results in a PE-lacking but MGlcDAG-containing strain (−PE+MGlcDAG) due to the expression of the A. laidlawii MGlcDAG synthase gene. Functional expression of this enzyme resulted in significant accumulation (~35%) of the foreign glycolipid MGlcDAG in a PE-lacking background, whereas the anionic lipids PG and CL account for 35 and 25%, respectively (Fig. 2, lane 3).
FIGURE 2. Lipid content of different E. coli strains.
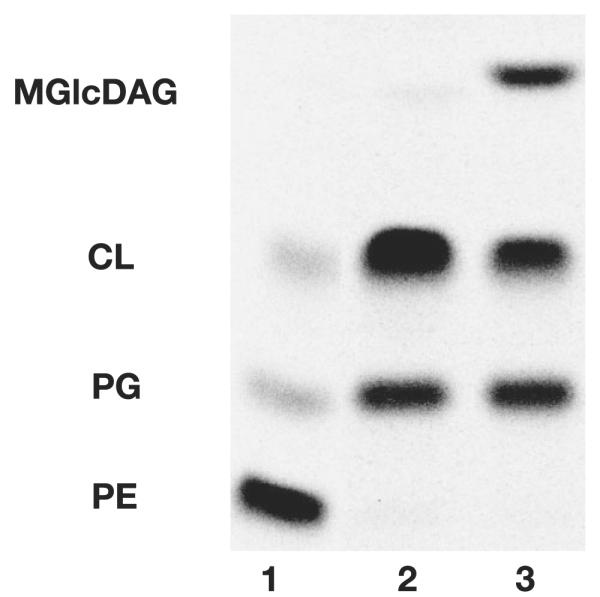
Cells were labeled with [14C]acetic acid at a final concentration of 0.2 μCi/ml and grown until mid-log phase. Lipids were extracted, separated, and quantified as described under “Experimental Procedures.” AL95/pDD72 (+PE, lane 1) carries a plasmid-borne copy of pssA gene. AL95 (−PE, lane 2) is a PE-lacking mutant. AL95/pTMG3 (−PE+MGlcDAG, lane 3) harbors a plasmid-borne copy of the gene encoding MGlcDAG synthase.
Orientation of TM Domains of LacY
We reported that the introduction of MGlcDAG into PE-lacking E. coli cells restored uphill transport activity of LacY, but the steady-state level of substrate accumulation was significantly lower than that observed in PE-containing cells (17). Previous studies (2) had revealed a dramatic change in LacY topological organization in the absence of PE accompanied by a complete loss of uphill transport function. The N-terminal six-TM helical bundle of LacY (N terminus to C6) adopts an inverted TM topology, whereas the C-terminal half (P7 to C terminus) retains its native topology (Fig. 1B). Since PC appears to restore the native topology of LacY without restoring uphill transport (7), the effects of lipids on topology and function are not necessarily coupled. Therefore, the question naturally arises as to how LacY TM domains are organized in the membrane of −PE+MGlcDAG cells. Based on the observation of only partial recovery of uphill steady-state accumulation of substrate, LacY may adopt an alternative but uniformly oriented isoform, exist in multiple or dual topological organizations within the same membrane, or display native topology.
Using the known TM orientation of LacY in +PE and −PE cells, cysteine replacements were introduced into otherwise cysteine-less LacY (Fig. 1) to map the water accessibility of the extramembrane surface of LacY using the substituted cysteine accessibility mapping for determining TM domain (SCAM™) (18). By examining the accessibility of these cysteine substitutions to the membrane-impermeable sulfhydryl reagent MPB, the orientation of the hydrophilic extramembrane domains was determined with respect to the plane of the membrane bilayer.
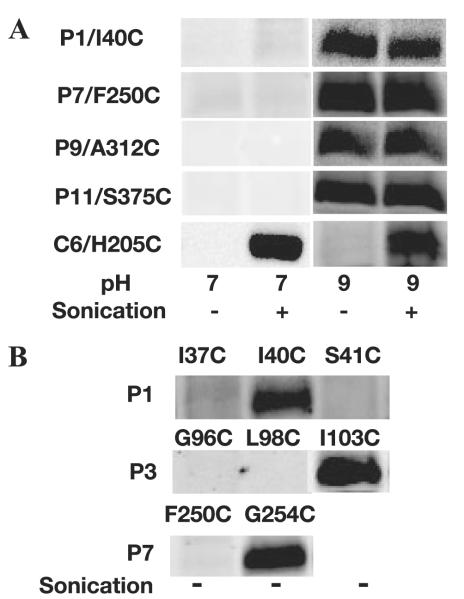
SCAM™ is dependent on the impermeability of the membrane toward labeling reagents. Cysteines located on the periplasmic side should be readily labeled by MPB, whereas cysteines located on the cytoplasmic side should not be accessible to MPB unless the cell membrane is disrupted by sonication. β-galactosidase (LacZ) is a cysteine-rich cytosolic protein; thus, it can serve as an internal control to test the impermeability of membranes, particularly those containing MGlcDAG. +PE, −PE, and −PE+MGlcDAG cells expressing LacZ were exposed to MPB. In all three strains, LacZ was only labeled by MPB if cells were first disrupted by sonication (Fig. 3), which established that the membranes of the above intact cells are not permeable to MPB under the experimental labeling conditions.
FIGURE 3. Accessibility of LacZ to MPB as a function of membrane lipid composition.

Intact whole cells (without sonication, −) or disrupted cells (with sonication, +) were treated with MPB as described under “Experimental Procedures” followed by immunoprecipitation with anti-LacZ polyclonal antibody. The biotin moiety linked to cysteines in LacZ was detected by Western blotting using avidin-horseradish peroxidase. Lanes 1 and 2, −PE cells; lanes 3 and 4, −PE+MGlcDAG cells; lanes 5 and 6, +PE cells.
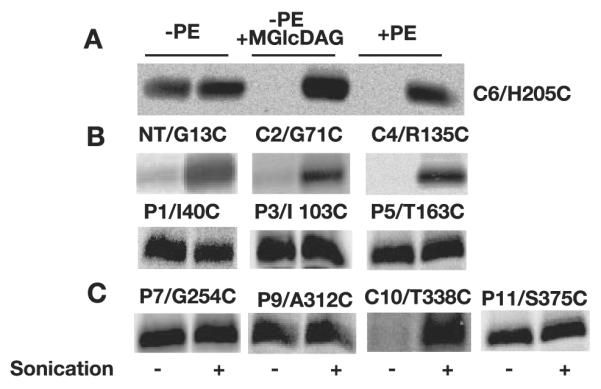
Consistent with previous results (2), labeling of LacY H205C (single cysteine in extramembrane domain C6) expressed in +PE cells was observed only after disruption of the cell membrane by sonication, whereas in −PE cells, equal labeling was observed with or without sonication, which is consistent with a cytoplasmic location of this loop in +PE membranes and a periplasmic location in −PE membranes (Fig. 4A). In the absence of PE but in the presence of the foreign glycolipid MGlcDAG, H205C showed the same labeling pattern as wild type cells, i.e. a cytoplasmic location for C6.
FIGURE 4. Mapping the location of single cysteine replacements in Cysless LacY by SCAM™.
As described under “Experimental Procedures,” all samples were treated by MPB without sonication (−) or during sonication (+). Western blotting using avidin-horseradish peroxidase was used to detect the biotin moiety linked to cysteines that were accessible to MPB during labeling. MBP labeling was performed at pH 7 except for P1/I40C, P9/A312C, and P11/S375C, which were labeled at pH 9. A, biotinylation pattern of LacY synthesized in −PE, −PE+MGlcDAG, and +PE cells, with a single cysteine replacement at H205 in extramembrane loop C6 (C6/H205C). B, biotinylation pattern of the N-terminal half of LacY assembled in −PE+MGlcDAG membranes. NT refers to the N terminus. C, labeling pattern of the C-terminal half of LacY assembled in −PE+MGlcDAG membranes.
Cysteine replacements in N terminus (G13C), C2 (G71C), and C4 (R135C) of LacY expressed in −PE+MGlcDAG cells were examined using SCAM™. As with H205C in C6, all the above cysteines were protected from labeling in intact cells (Fig. 4B). Biotinylation of single cysteine replacements only occurred after cell disruption, which demonstrated the wild type cytoplasmic exposure of these domains, as was previously observed in +PE cells. Cysteine replacements in P1 (I40C), P3 (I103C), and P5 (T163C) of LacY were readily labeled with or without sonication, again consistent with the labeling pattern for assembly in +PE membranes; the pH dependence for reactivity of some of the P domain cysteines is discussed below. The above labeling pattern demonstrates that the N-terminal six-TM helical bundle of LacY assembled in −PE+MGlcDAG cells adopts a native TM topology.
The topological inversion of LacY in −PE membranes is limited to the N-terminal helical bundle, whereas the orientation of the C-terminal half starting from domain P7 remains the same as in +PE cells. The same is true for the C terminus of LacY in −PE+MGlcDAG-containing membranes. Periplasmic extramembrane domains P7 (G254C), P9 (A312C), and P11 (S375C) were all accessible without sonication, whereas cytoplasmic domain C10 (T338C) was only accessible after sonication (Fig. 4C).
Differences in Cysteine Accessibility
Although the overall topology of LacY assembled in the absence of PE was corrected in the presence of MGlcDAG, differences in the accessibility of some cysteine replacements were observed when compared with the data obtained from +PE and −PE cells. Previously and in experiments reported here, labeling of LacY in +PE and −PE cells was done at pH 7. However, at pH 7, cysteine replacements at Ile-40 (P1), Phe-250 (P7), Ala-312 (P9), and Ser-375 (P11) were not accessible to MPB in −PE+MGlcDAG cells even after sonication unless labeling was performed at the elevated pH of 9 (Fig. 5A). H205C in C6 remained inaccessible to MPB at pH 9 without sonication, indicating that the permeability of the −PE+MGlcDAG-containing membranes was not compromised at pH 9. Moreover, cysteines within the same extramembrane loop also displayed different labeling patterns. F250C in P7 was easily labeled in both +PE and −PE cells, whereas in −PE+MGlcDAG cells, it was not accessible at pH 7, but the cysteine replacement 3 amino acid residues away, G254C, was readily labeled at pH 7 (Fig. 5B). Similar data were also obtained for the P1 and P3 domains. Only I40C and I103C but not their neighbors were labeled under the same conditions.
FIGURE 5. Altered reactivity of cysteine replacements of LacY in –PE + MGlcDAG membranes.
A, different reactivity of cysteine replacements toward MPB at pH 7 and pH 9. B, the different accessibility of cysteine replacements to MPB within the same extramembrane loop at pH 9 (P1) and pH 7 (P3 and P7), respectively.
Transport Function of LacY
LacY assembled in cell membranes without PE is unable to catalyze uphill transport of substrate but does carry out energy-independent downhill transport (5). The non-metabolizable substrate analog TMG was used to measure the ability of LacY to accumulate substrate by uphill transport. When compared with LacY assembled in +PE cell membranes, the apparent Vmax for uphill transport of LacY synthesized in membranes without PE is 10-fold less (4 nmol/min/mg of protein), whereas the apparent Km is almost the same (0.8 mm) (5). Previously, we reported that introduction of the foreign glycolipid MGlcDAG can qualitatively restore the uphill transport activity of LacY in the absence of PE (17). Using TMG as the substrate, the kinetic parameters of a single cysteine replacement derivative of Cys-less LacY, H205C, was examined in −PE+MGlcDAG and +PE cells. The Vmax of LacY in +PE cells was 41 nmol/min/mg of protein, whereas in the presence of MGlcDAG, the Vmax for uphill transport was 35 nmol/min/mg of protein, which is also close to that previously determined for +PE cells (40 nmol/min/mg of protein) (5). Western blot analysis using LacY-specific antibody showed comparable levels of LacY in both cell types. There was no significant difference in Km (~0.8 mm) between the two cell types. Uphill transport of substrate was confirmed by the abolition of substrate uptake by the addition of the protonophore carbonyl cyanide p-trifluoromethoxyphenylhydrazone.
DISCUSSION
SCAM™ was originally used to establish that the topology of the N-terminal six-TM helical bundle of LacY and the N-terminal two-TM hairpin of PheP and GabP of E. coli are topologically inverted with respect to the membrane bilayer and that the remainder of each protein displays native topology when assembled in membranes lacking PE (2, 9, 10). Substitution of the foreign glycolipid MGlcDAG for PE partially corrects many of the lipid-related phenotypes of PE-lacking mutants of E. coli (17). Herein, we demonstrated that the replacement of PE by MGlcDAG not only supports the overall wild type TM organization but also restores the kinetic parameters of uphill transport of LacY. These new results provide an explanation for the uphill transport function of E. coli LacY when expressed in C. glutamicum, which lacks amine-containing lipids but contains MGlcDAG (12). However, E. coli membranes containing MGlcDAG displayed different properties than native membranes in that the former membranes were not rendered permeable to MPB by toluene treatment (18). Additionally, the periplasmic domains of LacY were only accessible to MPB at elevated pH, suggesting either a reduced accessibility to the reagent or an increase in the pKa of the sulfhydryl on the cysteine-substituted residues when surrounded by the hydrophilic headgroup of MGlcDAG.
Based on a combination of in vivo and in vitro results, it can be concluded that MGlcDAG, PE (2), or PC (7), but not PG or CL, support the proper topological organization of LacY, whereas MGlcDAG or PE, but not PC, PG, or CL, support uphill transport. Therefore, topogenic signals and functional determinants reside not just in proteins but also in specific lipids. What features of both lipids and proteins determine topology and function? Proteins thus far shown to respond to a change in lipid composition appear to display uncommon structural arrangements that may allow structural flexibility in the membrane bilayer so that individual domains can respond independently to the surrounding environment. LacY contains a long cytoplasmic C6 domain connecting two independently folding TM helical bundles (20), an unusually hydrophilic TM domain (VII) connected to the C6 domain, and a high content of glycine residues in the TM domains of the first half of the protein (21). PheP contains an unusually long TM domain III with a high content of glycine residues (9). This region of PheP and the C6-TM VII regions of LacY may form the molecular hinge region between independently folding domains of the respective proteins that allows for independent topological response to the lipid environment.
Generally, membrane proteins follow the “positive inside rule” in which positive charged residues are overrepresented in cytoplasmic extramembrane segments, and TM domains that traverse the lipid bilayer are most often strongly hydrophobic (22, 23). The native topology of LacY follows the positive inside rule; however, there are also several conserved negatively charged amino acid residues in cytoplasmic domains located near the membrane-protein interface of C2, C4, and C6 (21). In the case of PheP and GabP, the cytoplasmic extramembrane domains flanking the N-terminal lipid-sensitive hairpin carry a high net negative charge (24). In a PE- or PC-containing membrane, negatively charged residues in the cytoplasmic loops might be stabilized via interaction with the positively charged headgroup of PE or PC, which at neutral pH is a zwitterionic lipid like PE and was shown to support the correct topology of LacY in proteoliposomes (7). However, the fact that the uncharged glycolipid MGlcDAG supports native topology strongly suggests that the negative charge character of the membrane surface is a more important topological determinant than the positively charged headgroup of PE or PC since all three of these lipids would lower the negative charge density on the membrane surface. The pKa of negatively charged amino acids is tuned by the magnitude of negative electrostatic potential at the membrane surface, and an increase in the negative charge density would favor protonation and translocation of extramembrane domains containing acidic residues (25). Negatively charged amino acids act in a position-specific manner (26), and when located near the interface, favor translocation of hydrophilic loops to the periplasmic side of the membrane in opposition to the positive inside rule. The replacement of the zwitterionic lipid dioleoyl-PC in liposomes by dioleoyl-PE or neutral monogalactosyl diacylglycerol (MGalDAG) had no significant effect on the translocation efficiency of slightly positive domains (27). However, the stimulatory effect of anionic lipids on translocation of slightly positive domains was proportional to the net negative charge of the lipids (27). These results suggest that the net negative charge density generated at the membrane surface by the lipid headgroups is an important determinant for topological disposition of protein domains with low positive charge character. Thus, cytoplasmic domains of LacY, PheP, and GabP containing negatively charged residues may be in a metastable or “frustrated” state, making them more prone to respond to changes in the surface charge of the membrane. The increased membrane translocation frequency of these cytoplasmic domains may be of functional significance, providing for a more flexible N-terminal helical bundle, which is largely responsible for LacY substrate binding and translocation (28, 29).
Several studies have also shown that the collective charge properties of the membrane surface can serve as a membrane protein topological determinant. For instance, lowering the positive charge density of cytoplasmic domains of leader peptidase or lowering the content of anionic phospholipids in the membrane results in increased translocation of these cytoplasmic domains to the periplasmic side of the membrane (30). The topology of the outer envelope protein of spinach chloroplasts (OEP7) is sensitive to lipid headgroup charge (31). Native orientation was observed after reconstitution into liposomes containing the high PC content of the outer leaflet of the chloroplast outer membrane, whereas an inverted orientation was attained in liposomes with the high PG content of the inner leaflet of the outer membrane. Therefore, surface charge density of the membrane surface and the charge character of extramembrane domains act in concert to determine TM domain orientation, which is further supported by our results.
Both in vivo and in vitro studies have shown that only primary amine-containing lipids such as PE or its precursor phosphatidylserine (PS), but not PG, CL, or PC, support the energy-dependent uphill transport function of LacY (5, 6). However, in the membrane of C. glutamicum, where MGlcDAG is the major lipid constituent, E. coli LacY was fully functional even in the absence of any primary amine-containing lipids (12). Uphill transport by LacY assembled in +PE and −PE+MGlcDAG membranes displayed comparable values for the apparent Vmax and Km. The ability of MGlcDAG to replace PE may stem from either or both of its similarities with PE. The non-bilayer prone property enables it to affect the lateral pressure profile across the membrane. Its headgroup has no net charge yet is able to form hydrogen bonds, which may participate in the proton wire of LacY to facilitate the coupling of proton translocation to substrate transport. In vitro data showed that although zwitterionic PC can support the proper topology of LacY in the region of C6-TMVII-P7 (7), it does not support uphill transport or the native conformation of extramembrane domain P7 (32), which is associated with uphill transport function (33). PC is a bilayer-forming phospholipid, and its amino moiety cannot form hydrogen bonds.
Although it is too early to propose a common role of these two lipids in cell function, there is a growing body of evidence suggesting a similar role for PE and MGlcDAG or its plant counterpart MGalDAG (Gal referring to galactose) in supporting the proper function of membrane proteins. The branched chain amino acid transporter of Streptococcus cremoris (contains MGlcDAG but not PE) functions as an active transporter when reconstituted into proteoliposomes containing PE, MGlcDAG, or MGalDAG but was inactive with combinations of only PG, CL, and/or PC (13). Since S. cremoris membranes are devoid of PE, it was concluded that PE and glycolipids are physiologically interchangeable and required for full function of these membrane proteins. The Ca2+-ATPase of the sarcoplasmic reticulum reconstituted with either PE or MGalDAG exhibited a high initial rate of ATP-dependent coupled Ca2+ uptake (35). Reconstitution with lipids of increasing degrees of methylation (PE, monomethyl PE, and dimethyl PE versus PC) or increasing degrees of glycosylation (MGalDAG versus diagalactosyldiacylglycerol) revealed a progressive decrease in coupled uptake. In both cases, the progressive loss of function correlates with a shift from non-bilayer to bilayer potential for the lipids. The function of these membrane proteins, therefore, seems to depend on a higher-order structure in the form of the increased radius of curvature and unique packing order provided by nonbilayer prone lipids. Moreover, a similarity in the physical properties of these lipids is supported by the fact that PE-specific polypeptide antibiotic duramycin also strongly interacts with MGalDAG (36).
The role of membrane lipids is much more diverse and profound than simply forming a permeability barrier for the cell. Lipids actively participate in various cellular processes. However, it is difficult to determine whether it is the collective physical-chemical properties of the membrane bilayer or the unique chemical and physical properties of an individual lipid by which lipids assert their effect. By taking advantage of the simple yet well characterized E. coli system, we can easily manipulate the lipid composition to isolate the function of individual lipids involved in complex processes. Through comparing the interchangeability and specificity of two lipids, PE and MGlcDAG, which have overlapping and different physical and/or chemical properties, we have begun to define the specific features of individual lipids required in different cellular processes. Since PG and PC adopt a bilayer lipid structure exclusively, only PE and MGlcDAG share the ability to simultaneously assume a nonbilayer structure and form hydrogen bonds. The lack of net charge of the latter two lipids plus PC, particularly in membranes containing anionic lipids, appears to be required for proper topological organization of LacY, whereas headgroup hydrogen-forming ability and possibly non-bilayer prone properties are required for coupling proton translocation to substrate transport during uphill transport. Generalization of these concepts beyond LacY remains tenuous since the lipid-sensitive topogenic signals within LacY, PheP, and GabP have not been defined. Given the differences among these proteins, their response to different lipid environments may not be the same.
It is tempting to speculate that during the course of evolution, both proteins and lipids co-evolved together in the context of the lipid environment of membrane systems in which both are mutually dependent on each other. Although it is surprising that E. coli is tolerant to such major changes in membrane lipid composition, our results emphasize the importance of similar physical and chemical properties of lipids in supporting cell functions rather than a more strict structural requirement, as observed for proteins. Thus far, major structural response to changes in lipid environment has only been noted in non-essential solute transport proteins. The changes in structure observed may be a function of the highly mobile and flexible nature of these proteins that allows subdomains within the proteins to coexist in multiple topological states. More rigid proteins may be forced to maintain native topological organization due to the overall stability of the native state. A recognition of the significance of specific protein-lipid interactions in determining membrane protein topology is important not only for improvement of the accuracy of topology prediction (37) using lipid profiles as one of the structural constraints but also for development of vectoral proteomics (34).
Acknowledgment
We thank Dr. H. R. Kaback for supplying us with plasmids encoding various derivatives of lactose permease, which were essential to carry out the reported experiments.
Footnotes
This work was supported by Grant R37 GM20478 from the National Institutes of General Medical Sciences (to W. D.) and by a grant from the John S. Dunn, Sr. Research Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: LacY, lactose permease; CL, cardiolipin; GabP, γ-aminobutyric acid permease; MGalDAG, monogalactosyldiacylglycerol; MGlcDAG, monoglucosyldiacylglycerol; MPB, 3-(N-maleimidylpropionyl) biocytin; PheP, phenylalanine permease; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; TM, transmembrane; SCAM™, substituted cysteine accessibility method for determining TM domain mapping; TMG, methyl-β-d-galactopyranoside.
REFERENCES
- 1.Dowhan W, Mileykovskaya E, Bogdanov M. Biochim. Biophys. Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdanov M, Heacock PN, Dowhan W. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saier MH., Jr. Mol. Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 4.Abramson J, Iwata S, Kaback HR. Mol. Membr. Biol. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov M, Dowhan W. J. Biol. Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 6.Chen CC, Wilson TH. J. Biol. Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 7.Wang X, Bogdanov M, Dowhan W. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack DL, Paulsen IT, Saier MH. Microbiology (Reading) 2000;146:1797–1814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. J. Biol. Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Campbell HA, King SC, Dowhan W. J. Biol. Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 11.Hoischen C, Kramer R. J. Bacteriol. 1990;172:3409–3416. doi: 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brabetz W, Liebl W, Schleifer KH. J. Bacteriol. 1993;175:7488–7491. doi: 10.1128/jb.175.22.7488-7491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driessen AJ, Zheng T, In't Veld G, Op den Kamp JA, Konings WN. Biochemistry. 1988;27:865–872. doi: 10.1021/bi00403a005. [DOI] [PubMed] [Google Scholar]
- 14.Berg S, Edman M, Li L, Wikstrom M, Wieslander A. J. Biol. Chem. 2001;276:22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 15.Dormann P, Benning C. Trends Plant Sci. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 16.Osterberg F, Rilfors L, Wieslander A, Lindblom G, Gruner SM. Biochim. Biophys. Acta. 1995;1257:18–24. doi: 10.1016/0005-2760(95)00042-b. [DOI] [PubMed] [Google Scholar]
- 17.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. J. Biol. Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 18.Bogdanov M, Zhang W, Xie J, Dowhan W. Methods (Amst.) 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeChavigny A, Heacock PN, Dowhan W. J. Biol. Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 20.Sahin-Toth M, Kaback HR, Friedlander M. Biochemistry. 1996;35:2016–2021. doi: 10.1021/bi952496g. [DOI] [PubMed] [Google Scholar]
- 21.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 22.von Heijne G. J. Mol. Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson J, Persson B, von Heijne G. Proteins. 2005;60:606–616. doi: 10.1002/prot.20583. [DOI] [PubMed] [Google Scholar]
- 24.Dogovski C, Pi J, Pittard AJ. J. Bacteriol. 2003;185:6225–6232. doi: 10.1128/JB.185.21.6225-6232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishtalik LI, Cramer WA. FEBS Lett. 1995;369:140–143. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 26.Rutz C, Rosenthal W, Schulein R. J. Biol. Chem. 1999;274:33757–33763. doi: 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 27.Ridder AN, Kuhn A, Killian JA, de Kruijff B. EMBO Rep. 2001;2:403–408. doi: 10.1093/embo-reports/kve087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez-Ibar JL, Guan L, Weinglass AB, Verner G, Gordillo R, Kaback HR. J. Biol. Chem. 2004;279:49214–49221. doi: 10.1074/jbc.M407408200. [DOI] [PubMed] [Google Scholar]
- 29.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 30.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schleiff E, Tien R, Salomon M, Soll J. Mol. Biol. Cell. 2001;12:4090–4102. doi: 10.1091/mbc.12.12.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanov M, Sun J, Kaback HR, Dowhan W. J. Biol. Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 33.Sun J, Wu J, Carrasco N, Kaback HR. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 34.Vener AV, Stralfors P. IUBMB Life. 2005;57:433–440. doi: 10.1080/15216540500138360. [DOI] [PubMed] [Google Scholar]
- 35.Navarro J, Toivio-Kinnucan M, Racker E. Biochemistry. 1984;23:130–135. doi: 10.1021/bi00296a021. [DOI] [PubMed] [Google Scholar]
- 36.Navarro J, Chabot J, Sherrill K, Aneja R, Zahler SA, Racker E. Biochemistry. 1985;24:4645–4650. doi: 10.1021/bi00338a025. [DOI] [PubMed] [Google Scholar]
- 37.Adamian L, Nanda V, DeGrado WF, Liang J. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]