Abstract
Nitrogen catabolite repression (NCR) is the regulatory pathway through which Saccharomyces cerevisiae responds to the available nitrogen status and selectively utilizes rich nitrogen sources in preference to poor ones. Expression of NCR-sensitive genes is mediated by two transcription activators, Gln3 and Gat1, in response to provision of a poorly used nitrogen source or following treatment with the TORC1 inhibitor, rapamycin. During nitrogen excess, the transcription activators are sequestered in the cytoplasm in a Ure2-dependent fashion. Here, we show that Vps components are required for Gln3 localization and function in response to rapamycin treatment when cells are grown in defined yeast nitrogen base but not in complex yeast peptone dextrose medium. On the other hand, Gat1 function was altered in vps mutants in all conditions tested. A significant fraction of Gat1, like Gln3, is associated with light intracellular membranes. Further, our results are consistent with the possibility that Ure2 might function downstream of the Vps components during the control of GATA factor-mediated gene expression. These observations demonstrate distinct media-dependent requirements of vesicular trafficking components for wild-type responses of GATA factor localization and function. As a result, the current model describing participation of Vps system components in events associated with translocation of Gln3 into the nucleus following rapamycin treatment or growth in nitrogen-poor medium requires modification.
Keywords: GATA factor, nitrogen availability, rapamycin, vesicular trafficking, yeast
Introduction
Nitrogen is a naturally occurring element that is essential for the growth of all living cells. Many microorganisms have the ability to sense and utilize a broad range of nitrogen sources. When Saccharomyces cerevisiae cells are exposed to preferred nitrogen sources (ammonia and glutamine), the expression of genes encoding proteins required for the uptake and utilization of nonpreferred sources (proline, urea, and allantoin) are downregulated. Conversely, when provided with poorly utilized nitrogen sources, expression of these genes is derepressed. The regulatory pathway responsible for this behavior is known as nitrogen catabolite repression (NCR) (Cooper 1982; Wiame et al. 1985). Expression of NCR-sensitive genes is mediated by two DNA-binding GATA transcription activators, Gln3 and Gat1/Nil1 (Mitchell and Magasanik 1984; Coffman et al. 1995, 1996; Stanbrough et al. 1995), and is inhibited by the preprion protein Ure2, which acts as a negative regulator of Gln3 and Gat1 (Drillien and Lacroute 1972; Grenson et al. 1974; Courchesne and Magasanik 1988; Blinder et al. 1996; Coffman et al. 1996). In the presence of preferred nitrogen sources, Gln3 and Gat1 are sequestered in the cytoplasm in a Ure2-dependent manner, whereas upon growth in a poor nitrogen supply or upon transferring to nonpreferred nitrogen conditions, the GATA activators relocate to the nucleus and mediate the transcription of NCR-sensitive genes (Cox et al. 2000; Cunningham et al. 2000; Cooper 2002).
The nutrient-responsive TOR complex 1 (TORC1) was also found to participate in coordinating the regulation of GATA factor-mediated expression. Indeed, addition of the immunosuppressant drug rapamycin, inhibiting TORC1, to cells growing in the presence of a good nitrogen source transiently mimics the effects observed with a poor one, that is, nuclear localization and activation of NCR gene expression by Gln3 and Gat1 (Beck and Hall 1999; Cardenas et al. 1999; Hardwick et al. 1999). However, a growing literature demonstrates that multiple distinct regulatory mechanisms are involved in the control of the GATA activators and hence results observed in response to rapamycin treatment cannot be extrapolated to explain responses to nitrogen limitation (Cox et al. 2004; Tate et al. 2005, 2006, 2010; Puria et al. 2008; Georis et al. 2011a; Feller et al. 2013; Rai et al. 2013b; Tate and Cooper 2013). Moreover, although Gln3 and Gat1 are both required for the transcription of most NCR-sensitive genes, the two transcription factors are not always similarly regulated: (1) Gln3 and Gat1 localizations are not similarly sensitive to nitrogen limitation, rapamycin and methionine sulfoximine (Msx, an inhibitor of glutamine synthetase) treatment (Tate et al. 2010; Georis et al. 2011a). (2) In contrast to Gln3, Gat1 nuclear localization is largely Ure2-independent (Georis et al. 2008). (3) Nuclear localization of Gln3 and Gat1 exhibits distinct requirements for the TORC1-regulated phosphatases (Tate et al. 2010; Georis et al. 2011b).
Early investigations of Gln3 regulation established that under steady state, nitrogen-rich conditions, cytoplasmic Gln3 was situated within or tightly associated with a cytoplasmic membrane system (Cox et al. 2002). This membrane association was additionally visualized as Gln3 translocated into and out of the nucleus (Cox et al. 2004). Puria et al. (2008) subsequently confirmed the Gln3 membrane association by demonstrating colocalization of Gln3-Myc13 with the Golgi-to-endosome trafficking component Vps10-HA. Endogenous membranes of the protein secretory pathway have also emerged as important platforms for Tor signaling. First, all components of the TORC1 complex (Tor1, Tor2, Kog1, Lst8, and Tco89) (Cardenas and Heitman 1995; Kunz et al. 2000; Chen and Kaiser 2003; Wedaman et al. 2003; Reinke et al. 2004; Aronova et al. 2007) as well as TORC1 regulators (EGO complex) (Dubouloz et al. 2005; Gao and Kaiser 2006; Kim et al. 2008; Binda et al. 2009; Bonfils et al. 2012) and downstream effectors, such as the Tap42–Sit4 phosphatase complex and the AGC kinase Sch9 (Yan et al. 2006; Urban et al. 2007) localize to the late endosome and vacuole membranes. Moreover, the Golgi Ca2+/Mn2+ATPase Pmr1 has been described to negatively regulate TORC1 function (Devasahayam et al. 2006). Finally, genetic interactions between TORC1 and representative Class C Vps proteins that mediate docking and fusion of vesicles with vacuoles have been described (Aronova et al. 2007; Zurita-Martinez et al. 2007).
Class C and D Vps proteins regulate Golgi-to-vacuole protein transport (Peterson and Emr 2001; Bowers and Stevens 2005): the Class C Vps complex, made up of Vps11/Pep5, Vps18/Pep3, Vps16, and Vps33, is required for docking and fusion at the vacuole (Rieder and Emr 1997; Srivastava et al. 2000; Peterson and Emr 2001; Bowers and Stevens 2005), whereas the Class D proteins (including Vps3, Vps6/Pep12, Vps34 and Vps45) are thought to control vesicular trafficking between the late Golgi and the late endosome (Prescianotto-Baschong and Riezman 2002; Bowers and Stevens 2005). Puria et al. (2008) have previously observed a requirement of Class C and D Vps proteins for Gln3 nuclear translocation after transferring yeast peptone dextrose (YPD) grown cells to proline medium but not after treating them with rapamycin. Aware that Gln3 and Gat1 are sometimes regulated differently, these observations prompted two obvious but important questions: (1) Did Gat1 exhibit the same responses and requirements as Gln3? (2) Would the same responses be observed if a defined, nitrogen-rich rather than complex YPD medium was employed throughout the experiments, thereby eliminating a significant variable from their interpretation?
The first outcome of our study was that Puria et al.'s (2008) observations are medium-specific and hence cannot be generalized. Indeed, although Gln3 nuclear localization is impaired in response to nitrogen limitation but not rapamycin treatment in YPD-pregrown vps mutants, we show that in yeast nitrogen base (YNB) ammonia, Vps proteins are required for Gln3 nuclear localization even in response to rapamycin. Therefore, the model describing the requirement of Vps system components for Gln3 trafficking to the nucleus (Puria and Cardenas 2008) requires modification. Second, we show that components of Golgi-to-vacuole trafficking are required for Gat1 function either in YNB-ammonia- or YPD-grown cells treated with rapamycin or transferred to proline medium. Indeed, vps mutations reduced the ability of Gat1 to (1) translocate to the nucleus, (2) bind to the Gat1-activated DAL5 (encoding allantoate permease) promoter, and (3) elicit DAL5 expression. Additionally, we show that Gat1, like Gln3, fractionates with light intracellular membranes, raising the possibility that its regulation might occur at these locations too. Finally, our observations suggest that Ure2 might function downstream the Vps proteins during GATA factor-mediated signaling.
Experimental Procedures
Strains and culture conditions
S. cerevisiae strains used in this work are listed in Table 1. Deletion strains involving insertion of kanMX or natMX cassettes were constructed using the short and long flanking homology strategy of Wach (1996) using primers described in Table 1. Gat1-Myc13 and Gln3-Myc13 protein production was controlled in each vps mutant and their levels were comparable to the isogenic wild types (WT). Cultures were grown to midlog phase (A600nm = 0.5–0.55) in YPD medium or YNB minimal medium containing ammonia at a 1% final concentration. Appropriate supplements (100 μg mL−1 leucine, 20 μg mL−1 uracil, histidine, tryptophan) were added to the medium as necessary to cover auxotrophic requirements. Where indicated, cells were treated for 20 min with 200 ng mL−1 rapamycin or transferred to YNB minimal medium containing 0.1% proline for 60 min prior to assay.
Table 1.
Strains used in this work
| Strain | Pertinent genotype | Parent | Complete genotype | Reference | Primer/Reference |
|---|---|---|---|---|---|
| TB50 | WT | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa | Beck and Hall (1999) | ||
| TB123 | W.T. Gln3-Myc13 | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, GLN3-MYC13[KanMX] | Beck and Hall (1999) | ||
| JK9-3d | W.T. | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa | Beck and Hall (1999) | ||
| MK23 | vps3Δ | TB50 | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, vps3::kanMX | This work | vps3: 5′, −42 to −1 and 3′, 3037 to 3076 |
| MK24 | pep5Δ | TB50 | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, pep5::kanMX | This work | pep5: 5′, −41 to −1 and 3′, 3091 to 3133 |
| MK27 | vps3Δ Gln3-Myc13 | FV250 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps3::kanMX GLN3-MYC13[HIS3] | This work | vps3: 5′, −42 to −1 and 3′, 3037 to 3076 |
| MK30 | pep5Δ Gln3-Myc13 | FV250 | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, pep5::kanMX GLN3-MYC13[HIS3] | This work | pep5: 5′, −41 to −1 and 3′, 3091 to 3133 |
| MK46 | pep3Δ | TB50 | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, pep3::natMX | This work | pep3: 5′, −42 to −1 and 3′, 2758 to 2801 |
| 08047c | ure2Δvps3Δ | OK01 X MK23 | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, ure2::natMX, vps3::kanMX | This work | |
| OK01 | ure2Δ | JK9-3d | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, ure2::natMX | This work | Ure2-L1, Ure2-L2, Ure2-L3 and Ure2-L4 (Georis et al. 2008) |
| FV005 | gln3Δ | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gln3::kanMX | Georis et al. (2008) | |
| FV006 | gat1Δ | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gat1::kanMX | Georis et al. (2008) | |
| FV018 | gat1Δ Gln3-Myc13 | TB123 | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, gat1::natMX GLN3-MYC13[KanMX] | Georis et al. (2008) | |
| FV063 | W.T. Gat1-Myc13 | TB50 | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, GAT1-MYC13[HIS3] | Georis et al. (2008) | |
| FV064 | gln3Δ Gat1-Myc13 | FV005 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, gln3::kanMX GAT1-MYC13[HIS3] | Georis et al. (2008) | |
| FV250 | W.T. Gln3-Myc13 | TB50 | MATa, leu2-3, 112, ura3-52, trp1, his4, rme1, HMLa, GLN3-MYC13[HIS3] | Georis et al. (2011b) | |
| FV390 | vps34Δ Gln3-Myc13 | TB123 | MATa, leu2-3, 112, ura3-52, trp1, his3, rme1, HMLa, vps34::natMX, GLN3-MYC13[KanMX] | This work | vps34: 5′, −430 to −407 and −1 to −33; 3′, 2629 to 2664 and 3152 to 3171 |
| FV391 | vps34Δ | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps34::kanMX | This work | vps34: 5′, −430 to −407 and −1 to −33; 3′, 2629 to 2664 and 3152 to 3171 |
| FV392 | vps34Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps34::kanMX GAT1-MYC13[HIS3] | This work | vps34: 5′, −430 to −407 and −1 to −33; 3′, 2629 to 2664 and 3152 to 3171 |
| FV640 | pep5Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, pep5::kanMX GAT1-MYC13[HIS3] | This work | pep5: 5′, −41 to −1 and 3′, 3091 to 3133 |
| FV641 | vps3Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps3::kanMX GAT1-MYC13[HIS3] | This work | vps3: 5′, −42 to −1 and 3′, 3037 to 3076 |
| FV642 | vps45Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps45::kanMX GAT1-MYC13[HIS3] | This work | vps45: 5′, −42 to −1 and 3′, 1735 to 1778 |
| FV643 | vps45Δ | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps45::kanMX | This work | vps45: 5′, −42 to −1 and 3′, 1735 to 1778 |
| FV644 | vps45Δ Gln3-Myc13 | FV250 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps45::kanMX GLN3-MYC13[HIS3] | This work | vps45: 5′, −42 to −1 and 3′, 1735 to 1778 |
| FV732 | pep3Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, pep3::kanMX GAT1-MYC13[HIS3] | This work | pep3: 5′, −42 to −1 and 3′, 2758 to 2801 |
| FV733 | pep3Δ Gln3-Myc13 | FV250 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, pep3::kanMX GLN3-MYC13[HIS3] | This work | pep3: 5′, −42 to −1 and 3′, 2758 to 2801 |
| FV734 | vps16Δ Gat1-Myc13 | FV063 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps16::kanMX GAT1-MYC13[HIS3] | This work | vps16: 5′, −42 to −1 and 3′, 2397 to 2439 |
| FV735 | vps16Δ | TB50 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps16::kanMX | This work | vps16: 5′, −42 to −1 and 3′, 2397 to 2439 |
| FV736 | vps16Δ Gln3-Myc13 | FV250 | MATa, leu2-3,112, ura3-52, trp1, his3, rme1, HMLa, vps16::kanMX GLN3-MYC13[HIS3] | This work | vps16: 5′, −42 to −1 and 3′, 2397 to 2439 |
Quantitative real-time polymerase chain reaction
RNA isolation and cDNA synthesis were conducted as described in (Georis et al. 2009). Quantification of specific cDNA targets was measured by real-time polymerase chain reaction (PCR) performed on a StepOnePlus device (Applied Biosystems, Foster City, CA) using DAL5 and GAP1 primers that have been described previously (Georis et al. 2008, 2009). MEP2-specific primers are MEP2-O9 (5′-ACGAGGAATCCACTGCTTAC-3′) and MEP2-O10 (5′-TTTCTGCGTCTGTGTTACCC-3′). The values reported represent the averages of at least two experiments from independent cultures; error bars indicate standard errors.
Chromatin immunoprecipitation
Cell extracts and immunoprecipitations were conducted as described in (Georis et al. 2008, 2009). Concentrations of specific DNA targets in immunoprecipitation (IP) and input (IN) samples were measured by real-time PCR performed on a StepOnePlus device (Applied Biosystems) using primers described in (Georis et al. 2008). The values reported represent the averages of two immunoprecipitations performed in at least two experiments from independent cultures; error bars indicate standard errors.
Intracellular Gln3 and Gat1 localization
Gln3-Myc13 and Gat1-Myc13 was visualized by indirect immunofluorescence of whole fixed cells. Immunofluorescence analysis was performed according to standard procedures (Pringle et al. 1991). Cells were fixed for 30 min in formaldehyde (3.7%). They were washed and resuspended in sorbitol buffer (1.2 mol/L sorbitol and 100 mmol/L potassium phosphate, pH 7.5). Cell walls were digested for 50 min at 30°C in sorbitol buffer supplemented with β-mercaptoethanol (40 mmol/L final) and lyticase (80 U mL−1) (92807 Fluka, St. Gallen, Switzerland). Gln3-Myc13 was detected with monoclonal antibody c-Myc (9E10) (Santa Cruz, Dallas, TX) at a working dilution of 1:30. The secondary antibody was Alexa Fluor 488 goat anti-mouse immunoglobulin G (heavy plus light chains) (A-11029; Molecular probes, Carlsbad, CA) (working dilution, 1:200). DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Cells were imaged using a fluorescence microscope (Nikon eclipse 80i; Nikon, Tokyo, Japan) equipped with digital sight DS-U3 fluorescence source. Images were captured with a digital camera (Digital sight, DS-U3) and Nikon Instrument Software elements, version 3.22 acquisition software and processed for publication with Photoshop CS (Adobe Systems, San Jose, CA).
To more representatively and completely describe images of Gln3-Myc13 and Gat1-Myc13 localization appearing in the figures of this paper, primary images were captured and saved in JPEG. We manually scored Gln3-Myc13 and Gat1-Myc13 localizations in 200 or more cells from six different microscopic fields from which these multiple primary images were taken. For scoring the unchanged images were used. For image publication, a portion of the JPEG files were processed with Photoshop CS (Adobe Systems) where changes were made only to decrease background fluorescence. Cells were classified into one of three categories: cytoplasmic (fluorescence in the cytoplasm only), nuclear–cytoplasmic (fluorescence in the cytoplasm and nucleus), and nuclear (fluorescence in the nucleus only). Although scoring limitations and reproducibility were described in Tate et al. (2006), we emphasize that the nuclear–cytoplasmic category is, of necessity, arbitrary. Placing cells in that category is based on subjective visual evaluation by the individual scoring the cells; it is not an objective instrument-based measurement. When the fluorescent signal is not restricted to a single cellular compartment, scoring depends upon repeated decisions of whether it is nuclear–cytoplasmic or a category flanking it. They will undoubtedly differ in detail from those of another observer, who sets their category dividing lines differently. Our intracellular distributions were scored as consistently as possible. Results were confirmed using independent cultures. Detailed examples of the three scoring categories for Gln3-Myc13 and Gat1-Myc13 as well as scoring precision (within and between experiments) appear in references (Tate et al. 2006, 2009, 2010; Tate and Cooper 2007, 2008; Georis et al. 2008).
Subcellular fractionation
Cells were lysed in buffer A containing 50 mmol/L Tris (pH 7.5), 0.2 mol/L sorbitol, 1 mmol/L ethylenediaminetetraacetic acid (EDTA), 1 mmol/L dithiothreitol (DTT) or buffer B containing 0.1 mol/L Tris (pH 7.5), 0.15 mol/L NaCl, 5 mmol/L EDTA. Protease and phosphatase inhibitor cocktails were added for whole-cell extract preparation. Unbroken cells were removed by centrifugation at 500g for 10 min. The cell-free extract was centrifuged at 13,000g for 15 min to yield P13 (pellet) fraction. The supernatant was then centrifuged at 100,000g for 40 min to obtain P100 (pellet) and S100 (supernatant). The different subcellular fractions were analyzed by Western blotting with monoclonal antibodies for c-Myc (Santa Cruz Biotechnology), Pep12 (Invitrogen) and Pgk1 (Invitrogen, Carlsbad, CA). The electrophoresis, blotting, and detection procedures have been described previously (Georis et al. 2009).
Results
Class C and D vps mutants are defective for activating DAL5 expression in response to rapamycin or transferring cells from ammonia to proline medium
In order to examine the requirement of proteins that participate in Golgi-to-vacuole trafficking for the control of NCR-sensitive gene expression, we assayed DAL5 expression in YNB-ammonia-grown untreated, rapamycin-treated or proline-transferred WT, gln3Δ and gat1Δ as well as Class C (pep3Δ, pep5Δ and vps16Δ) and D (vps3Δ, vps34Δ and vps45Δ) vps mutant cells (Fig. 1). The expression levels exhibited by rapamycin-treated or proline-transferred WT cells were clearly impaired in gln3Δ or gat1Δ-mutant cells, demonstrating that DAL5 transcription requires the simultaneous presence of Gln3 and Gat1 when YNB-ammonia-grown cells are transferred to proline medium, or treated with rapamycin (Georis et al. 2008). After a transfer to proline, DAL5 expression was lost in all vps mutant cells compared with a WT strain, whereas residual expression still occurred after rapamycin treatment (Fig. 1). Impaired DAL5 expression in rapamycin-treated or vps mutant cells transferred to proline could be attributed to loss of Gln3 function alone, Gat1 alone or the functions of both Gln3 and Gat1 together. Importantly, DAL5 expression decreased more when vps mutants were transferred from ammonia to proline medium than in either the gln3Δ or gat1Δ strains, suggesting that components of the Golgi-to-vacuole trafficking system may be required for both Gat1 and Gln3 function.
Figure 1.
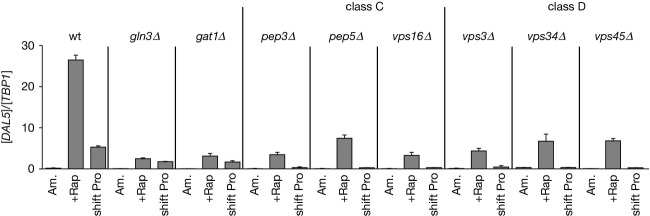
Class C and D Vps proteins requirements for efficient transcription of the Gat1-activated DAL5 gene following rapamycin treatment or transferring cells from ammonia to proline medium in YNB-grown cells. Total RNA was isolated from WT (TB50), gln3Δ (FV005), gat1Δ (FV006), the Class C pep3Δ (MK46), pep5Δ (MK24), and vps16Δ (FV735) and the Class D vps3Δ (MK23), vps34Δ (FV391), and vps45Δ (FV643) mutant cells grown in YNB-ammonia medium (Am.) and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). DAL5 mRNA levels were quantified by quantitative RT-PCR as described in “Experimental Procedures.” The values reported represent the averages of at least two experiments from independent cultures; error bars indicate standard errors.
Components of Golgi-to-vacuole trafficking are required for efficient GATA factor binding to the Gat1-activated DAL5 promoter
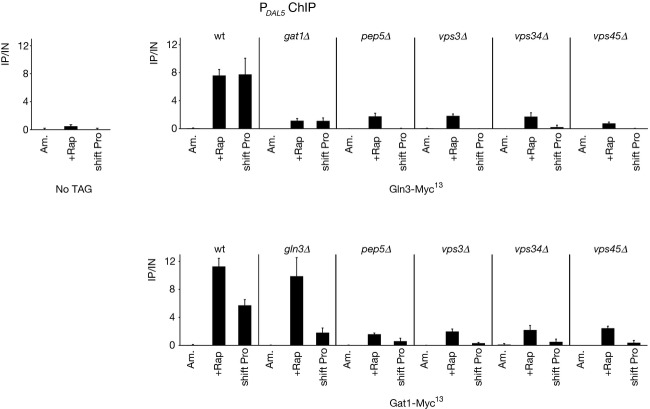
To test whether the observed requirement of representative Class C and D Vps proteins for DAL5 expression following rapamycin treatment or transfer to proline medium affected GATA factor binding to the DAL5 promoter, chromatin immunoprecipitation experiments were performed in the strains described above. Gat1-dependent Gln3-Myc13 binding to the DAL5 promoter was elicited in rapamycin-treated or proline-transferred WT cells (Fig. 2) (Georis et al. 2008). Cells lacking Gat1 or any of the Vps proteins tested displayed an impaired Gln3-Myc13 binding in both rapamycin-treated and proline-transferred cultures (Fig. 2). However, some residual binding did remain in the rapamycin-treated cells. Gat1-Myc13 was efficiently recruited to the DAL5 promoter in rapamycin-treated WT cells (Georis et al. 2008), but only about half as efficiently following transfer to proline medium (Fig. 2). Gat1 binding in cells transferred to proline medium required Gln3 and all of the Vps components tested. In contrast, high level Gat1-Myc13 binding to the DAL5 promoter following rapamycin treatment required only the Vps proteins assayed but not Gln3. This observation demonstrates that Gat1 binding in rapamycin-treated cells largely requires functional Golgi-to-vacuole trafficking components, independent of Gln3 function. On the other hand, impaired Gln3 binding observed in rapamycin-treated or proline-transferred vps mutants, could be either due to a direct requirement of the Golgi-to-vacuole trafficking component proteins we assayed or as a consequence of impaired Gat1 binding.
Figure 2.
Gln3-Myc13 and Gat1-Myc13 binding to the DAL5 promoter in WT, gln3Δ and gat1Δ cells as well as in the Class C pep5Δ mutant and the Class D vps3Δ, vps34Δ, and vps45Δ mutant cells in response to rapamycin treatment or transferring cells from YNB-ammonia to YNB-proline medium. Untagged WT (TB50), GLN3-MYC13 WT (FV250), GLN3-MYC13 gat1Δ (FV018), GLN3-MYC13 pep5Δ (MK30), GLN3-MYC13 vps3Δ (MK27), GLN3-MYC13 vps34Δ (FV390), GLN3-MYC13 vps45Δ (FV644), GAT1-MYC13 WT (FV063), GAT1-MYC13 gln3Δ (FV064), GAT1-MYC13 pep5Δ (FV640), GAT1-MYC13 vps3Δ (FV641), GAT1-MYC13 vps34Δ (FV392), and GAT1-MYC13 vps45Δ (FV642) cells were grown in YNB-ammonia medium (Am.) and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). ChIP was performed using antibodies against c-myc as described in “Experimental Procedures.” qPCR of IP and IN fractions was performed with primers specific for the DAL5 promoter (DAL5P) and for a region 2.5 kb upstream of the DAL5 open reading frame as a control (DAL5U). For each immunoprecipitation, IP/IN values were calculated as follows: ([DAL5P]IP/[DAL5P]IN − [DAL5U]IP/[DAL5U]IN). The values reported represent the averages of two immunoprecipitations performed in at least two experiments from independent cultures; error bars indicate standard errors.
Efficient nuclear GATA factor localization in rapamycin-treated or proline-transferred cells requires Class C and D Vps proteins
To determine whether impaired GATA factor binding to DNA derived from impaired nuclear localization in the vps mutants, we characterized Gat1-Myc13 and Gln3-Myc13 localization using indirect immunofluorescence. Gat1-Myc13 nuclear localization in rapamycin-treated or proline-transferred cells pregrown in YNB-ammonia was reduced in the Class C and D vps mutants relative to WT (Fig. 3). Following rapamycin treatment, Gat1-Myc13 was mainly nuclear–cytoplasmic in all tested vps mutant cells rather than fully nuclear as occurred in WT cells. In cells transferred to proline medium, the fraction of pep3Δ, pep5Δ, vps16Δ, vps3Δ, vps34Δ, vps45Δ cells where Gat1-Myc13 was nuclear or nuclear–cytoplasmic also decreased, accompanied by increased cytoplasmic Gat1-Myc13 localization. In none of the cases, however, were the Vps protein requirements absolute. These observations indicate that Class C and D Vps proteins are partially required for efficient Gat1 translocation to the nucleus. In contrast, Class C and D vps mutations totally abolished Gln3-Myc13 nuclear localization not only in cells transferred to proline medium but also, surprisingly, after rapamycin treatment (Fig. 4). These observations, obtained with YNB medium, contrast with a previous report showing that Golgi-to-vacuole trafficking components were dispensable for rapamycin-elicited Gln3 nuclear translocation (Puria et al. 2008).
Figure 3.

Requirements of Class C and D Vps proteins for intracellular Gat1-Myc13 localization in response to rapamycin or transferring cells from YNB-ammonia to YNB-proline medium. GAT1-MYC13 WT (FV063), Class C pep3Δ (FV732), pep5Δ (FV640), and vps16Δ (FV734) and Class D vps3Δ (FV641), vps34Δ (FV392) ,and vps45Δ (FV642) mutant cells were grown in YNB-ammonia medium (Am.). Cells were treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). The cultures were sampled for indirect immunofluorescence assay of Gat1-Myc13 localization. Indirect immunofluorescence assays were performed and imaged as described in Experimental Procedures. Images from which the histograms were derived are displayed on the left hand side of the figure. The upper member of each pair depicts green Gat1-Myc13-derived fluorescence and the lower one shows DAPI-positive material fluorescence. For each histogram, displayed at the right hand side of the figure, cells were scored for intracellular Gat1-Myc13 localization (cytoplasmic red bars, nuclear–cytoplasmic, yellow bars; nuclear, green bars) using criteria described in Experimental Procedures. When no histogram bar is visible on the graph, that is because there were no cells found in scoring category considered. The values reported represent the averages of at least two experiments from independent cultures.
Figure 4.
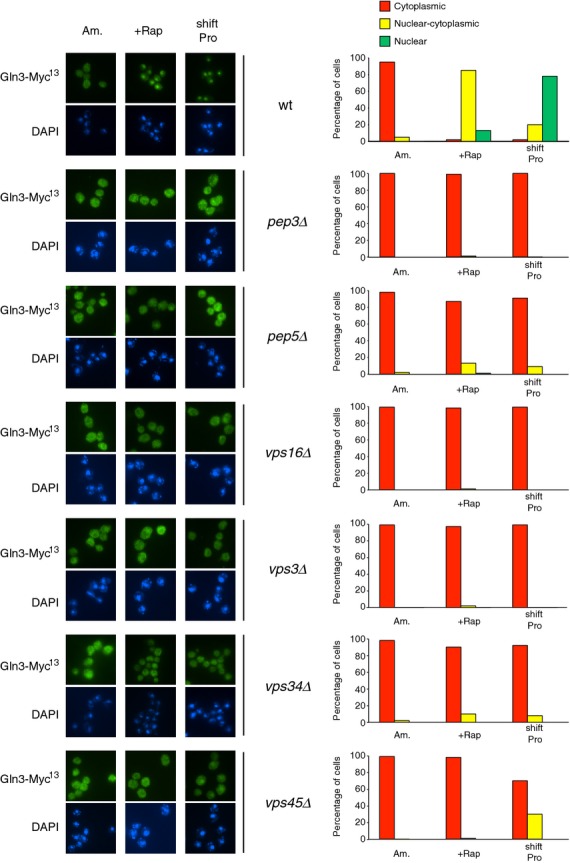
Requirements of Class C and D Vps proteins for intracellular Gln3-Myc13 localization in response to rapamycin or transferring cells from YNB-ammonia to YNB-proline medium. GLN3-MYC13 WT (FV250), Class C pep3Δ (FV733), pep5Δ (MK30), and vps16Δ (FV736) and Class D vps3Δ (MK27), vps34Δ (FV390), and vps45Δ (FV644) mutant cells were grown in YNB-ammonia medium (Am.) and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). The experimental format and data presentation are the same as those in Figure 3.
A media-dependent role of Class C and D Vps proteins for Gln3 and Gat1 nuclear translocation and DAL5 gene activation
To test if the apparent discrepancy between present and previously reported results was due to a technical problem or the different growth conditions employed, we characterized Gln3-Myc13 localization in WT, pep5Δ (chosen as Class C representative) and vps34Δ (chosen as Class D representative) mutant cells using the same growth conditions as Puria et al. (2008), that is, in YPD medium (untreated, rapamycin-treated or after transfer to proline medium). Our observations (Fig. 5A) were in agreement with previous studies showing that mutations in Class C and D VPS genes impair Gln3 nuclear localization following a transfer to proline medium but not to rapamycin treatment (Puria et al. 2008), indicating that the observed discrepancy was indeed due to the nature of the growth medium.
Figure 5.

Requirements of Class C and D Vps proteins for intracellular Gln3-Myc13 and Gat1-Myc13 localization in YPD growth conditions. GLN3-MYC13 WT (FV250), pep5Δ (MK30), vps34Δ (FV390), GAT1-MYC13 WT (FV063), pep5Δ (FV640), and vps34Δ (FV392) cells were grown in YPD medium and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). The experimental format and data presentation are the same as those in Figure 3 with one exception that all the strains were grown in YPD medium instead of YNB-ammonia.
The same procedure was followed to assess Gat1-Myc13 localization. As occurred in YNB-ammonia-grown WT cells (Fig. 3), rapamycin addition to YPD-grown cells led to a fully nuclear localization of Gat1-Myc13 (Fig. 5B). Deletion of VPS34 or PEP5 in rapamycin-treated cells reduced Gat1-Myc13 nuclear localization by increasing the fraction of cells in which Gat1-Myc13 was nuclear–cytoplasmic at the expense of cells where Gat1-Myc13 was fully nuclear. Gat1-Myc13 nuclear localization in response to transferring cells to proline medium was also partially impaired in the vps34Δ and pep5Δ mutants relative to WT. This was characterized by an increase in the fraction of cells where Gat1-Myc13 was cytoplasmic and a decrease in the fraction of cells where Gat1-Myc13 was nuclear and/or nuclear–cytoplasmic.
To test whether the observed requirement of Class C and D Vps proteins for Gat1-Myc13 nuclear localization upon rapamycin treatment or transferring cells to proline medium paralleled its ability to bind to the DAL5 promoter, chromatin immunoprecipitation experiments were performed in WT and vps34Δ cells (Fig. 6). Much more Gat1-Myc13 was recruited to the DAL5 promoter in rapamycin-treated WT cells compared with untreated YPD-grown cells. In contrast, Gat1-Myc13 binding to the DAL5 promoter in vps34Δ cells treated with rapamycin or transferred to proline medium was reduced.
Figure 6.
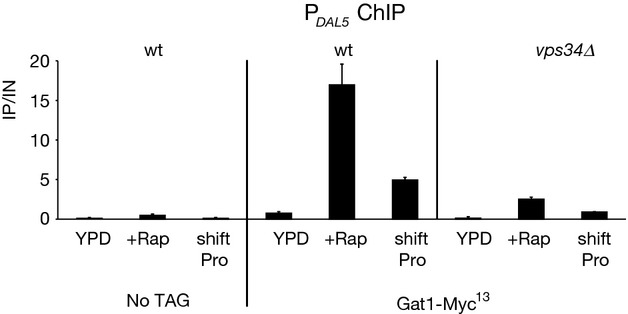
Gat1-Myc13 binding to the DAL5 promoter in WT and vps34Δ strains in response to rapamycin or transfer of YPD-grown cells to YNB-proline medium. Untagged WT (TB50), GAT1-MYC13 WT (FV063) and GAT1-MYC13 vps34Δ (FV392) cells were grown in YPD medium and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). ChIP was performed as described in Figure 2.
Finally, we determined the transcription profiles of DAL5, GAP1, and MEP2 genes in YPD-grown, untreated, rapamycin-treated, or proline-transferred WT, gln3Δ, gat1Δ and Class C and D vps mutant cells (Fig. 7). The DAL5 expression levels elicited by rapamycin-treated or proline-transferred WT cells were clearly decreased in gln3Δ or gat1Δ mutant cells, demonstrating that, as occurred in YNB medium, DAL5 transcription requires both GATA factors under these conditions (Fig. 7A). After rapamycin treatment or transfer to proline medium, DAL5 expression was clearly reduced in all vps mutants relative to WT cells (Fig. 7A). As Gln3-Myc13 nuclear localization is largely intact, impaired DAL5 expression in rapamycin-treated vps mutant cells should result from the reduced Gat1-Myc13 nuclear localization and DNA binding. On the other hand, the impaired DAL5 expression exhibited by the vps mutants in response to transferring cells into proline medium could be attributed to the impaired nuclear localization of either Gln3-Myc13, Gat1-Myc13 or of both GATA activators.
Figure 7.
Class C and D Vps protein requirements for transcription of the GATA factor-activated genes DAL5, GAP1 and MEP2 in response to rapamycin or transfer of YPD-grown cells to YNB-proline medium. The strains, experimental format and data presentation are the same as those in Figure 1 with one exception that all the strains were grown in YPD medium instead of YNB-ammonia. (A) DAL5. (B) GAP1. (C) MEP2.
GAP1 or MEP2 expression levels in YPD-grown WT cells treated with rapamycin or transferred to proline medium were only slightly reduced in gln3Δ or gat1Δ mutant cells, demonstrating that either Gln3 or Gat1 alone could sustain GAP1 or MEP2 expression in rapamycin-treated cells or in cells transferred to proline medium (Fig. 7B and C). Consistent with previous data (Puria et al. 2008), only in cells transferred to proline medium but not those treated with rapamycin, were GAP1 and MEP2 expression levels reduced in all vps mutant cells compared with a WT strain (Fig. 7B and C). The unaffected expression observed after rapamycin treatment in the vps mutants correlated with the intact nuclear translocation of Gln3. On the other hand, impaired expression observed in vps mutant cells transferred to proline medium was in agreement with the impaired nuclear translocation of both Gln3 and Gat1.
A fraction of Gat1 associates with light intracellular membranes
Several regulators of GATA factor-activated genes, for example, TORC1, Tap42-Sit4 phosphatase, as well as Gln3 have been shown to be associated with intracellular light membranes (Cardenas and Heitman 1995; Kunz et al. 2000; Chen and Kaiser 2003; Wedaman et al. 2003; Reinke et al. 2004; Yan et al. 2006; Aronova et al. 2007; Puria et al. 2008). These findings, and our observations that vps mutant cells exhibit defects in Gat1 localization and function, raised the possibility that it too might be associated with light membranes. Therefore, Gat1-Myc13 subcellular fractionation was performed to assess this possibility (Fig. 8A). To evaluate the degree to which our procedure cleanly separated soluble from insoluble proteins, we examined the distribution of the cytosolic marker Pgk1 and the membrane-associated protein, Pep12. As expected, Pgk1 was recovered primarily in the supernatant fraction, whereas the endosomal marker Pep12 was distributed between both the P13 and P100 fractions (Fig. 8A), as described previously (Becherer et al. 1996). Gat1-Myc13 was detected as a typical double band, corresponding to two isoforms possessing different N-termini (Rai et al. 2013a). In WT cells, Gat1-Myc13, like Gln3-Myc13, was fractionated not only with heavy (P13) membranes (Fig. 8A for Gat1-Myc13 and B for Gln3-Myc13) known to contain plasma and endoplasmic reticulum membranes but also with lighter (P100) membranes containing Golgi and the endosomal marker Pep12. In contrast with membrane-associated Pep12, significant portions of Gat1-Myc13 and Gln3-Myc13 were still detected in the S13 and S100 fractions along with the cytosolic marker Pgk1, indicating that only a portion of Gat1-Myc13 and Gln3-Myc13 were associated with intracellular membranes.
Figure 8.
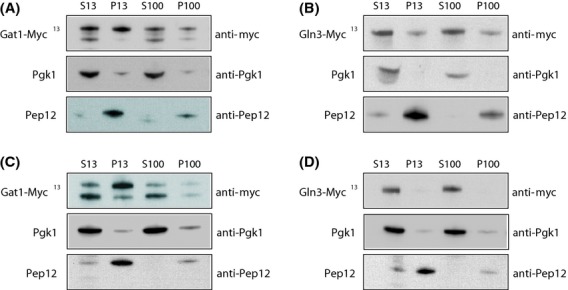
Subcellular fractionation of Gat1-Myc13 and Gln3-Myc13. Cell-free lysates from YPD-grown GAT1-MYC13 WT (FV063; A and C) and GLN3-MYC13 WT (FV250; B and D) cells were subjected to differential centrifugation to yield low-speed pellet (P13), supernatant (S13), high-speed pellet (P100), and soluble (S100) fractions. Equal cell equivalents were examined by Western blot to detect Gln3-Myc13, Gat1-Myc13, Pep12, and Pgk1. Protein extracts were prepared using a lysis buffer lacking NaCl (A and B) or containing 0.15 mol/L NaCl (C and D).
Gln3 association with intracellular membranes has already been reported as likely being peripheral as indicated by its sensitivity to extraction with NaCl (Puria et al. 2008). To test this characteristic for Gat1, protein extracts were prepared using a stringent lysis buffer (containing 0.15 mol/L NaCl). Under such stringent conditions, Gat1-Myc13 fractionated largely with heavy (P13) membranes (Fig. 8C) but only residually with lighter (P100) ones. Interestingly, the two isoforms of Gat1-Myc13 were unevenly distributed between the supernatant and the pellet fractions. The faster migrating isoform, beginning at Gat1 methionines 95 or 102 (Rai et al. 2013a), fractionated mainly in the soluble fractions, whereas the slower migrating isoform, beginning at Gat1 methionine 40 (Rai et al. 2013a), was mainly associated with the P13 fraction. As expected, Gln3-Myc13 was not detected in the pellet fraction under these stringent conditions (Fig. 8D). These findings indicate that Gat1 appears to be more stably associated with light membranes than Gln3.
Ure2 seems to act downstream of Vps3 to control DAL5 expression
Ure2 is a well-known negative regulator of GATA factor-mediated gene expression (Grenson et al. 1974; Courchesne and Magasanik 1988). Aiming at determining the epistatic relation between the ure2Δ and vps mutations, all attempts to delete URE2 in vps34Δ, pep5Δ, or vps45Δ mutant cells failed, suggesting that combining the ure2Δ with either vps34Δ, pep5Δ, or vps45Δ mutations might be resulting in synthetic lethality. To test this possibility, tetrad analysis was conducted in crosses between the vps34Δ, pep5Δ, and vps45Δ and ure2Δ strains. Haploid meiotic progeny generated by sporulation of heterozygous diploids containing the above mutations carried only one mutation. No meiotic segregants carrying a double mutation were recovered, confirming that these double mutants were synthetically lethal. Double ure2Δvps3Δ mutants, however, were recovered on YPD medium following sporulation of a VPS3ure2Δ/vps3ΔURE2 heterozygous diploid strain, but these were unable to grow in YNB-ammonia medium.
DAL5 expression was assayed in YPD-grown untreated, rapamycin-treated, or proline-transferred WT, ure2Δ, vps3Δ, and ure2Δvps3Δ cells (Fig. 9). In YPD-grown cells, high-level DAL5 expression observed in a ure2Δ was lost upon additionally deleting VPS3. However, in response to rapamycin treatment or a transfer to proline, the elevated DAL5 expression levels observed in ure2Δ were only lowered, but not lost in the double mutant. In fact, the double mutant exhibited a phenotype resembling the WT. Altogether, these observations suggest that the vps3Δ and ure2Δ mutations more likely only compensate the effects of one another.
Figure 9.
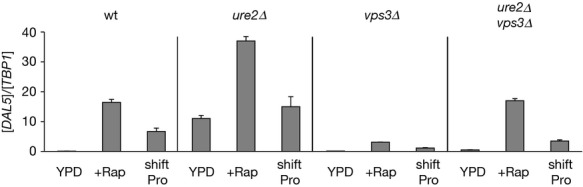
Epistatic Relation between Ure2 and Vps3. WT (TB50), ure2Δ (OK01), vps3Δ (MK23), ure2Δvps3Δ (08047c) cells were grown in YPD medium and treated with rapamycin (+Rap) or transferred to proline medium (shift Pro). DAL5 mRNA levels were quantified by quantitative RT-PCR as described in “Experimental Procedures.” The experimental format and data presentation are the same as those in Figure 1.
Discussion
In this report, we show that mutations in Class C and D Vps proteins led to defects in DAL5 expression, after treating cells with rapamycin or transferring them to YNB-proline medium. These defects correlated with altered Gat1 and Gln3 nuclear localization and DNA binding. Vps protein requirements for Gat1 localization and function were observed in cells grown either in defined, nitrogen-rich or complex YPD medium, whereas the requirements of representative Vps proteins for Gln3 function were media-specific: a requirement after rapamycin treatment was observed in YNB and not YPD medium. The results in YPD, but not YNB medium were in agreement with Puria et al.'s (2008) initial conclusions. However, the media-dependence of the Vps proteins requirements for the rapamycin control of Gln3 localization and function suggests that the previously published model describing Vps protein participation in GATA factor regulation may not be correct and hence in need of revision (Puria and Cardenas 2008).
The influence of medium composition (YPD vs. YNB) on the Vps requirement for the control of Gln3 localization may not be too surprising. A previous report has indicated that key components of the medium, such as zinc or the pH of the medium, can influence Gln3 cellular localization and GATA factor responses (Feller et al. 2006). Several differences exist between YPD and YNB media, including the pH and the quality of the nitrogen sources provided. Our YNB cultures started at a pH of 5 and were harvested at a pH of 2.7, whereas in YPD, starting and ending pHs were 6 and 4.8, respectively. Another major difference between the YPD and YNB-ammonia media is the quality of the nitrogen source, influencing the yeasts' growth rate (faster on YPD than on YNB). A third major difference is the availability of all amino acids in YPD medium, whereas YNB-ammonia-grown cells need to synthesize their own amino acids (Magasanik and Kaiser 2002; Ljungdahl and Daignan-Fornier 2012). Although it is still not obvious if and how the Vps proteins may be connected to the presence of external amino acids, we speculate that our data do not exclude the possibility that Vps proteins could be required for Gln3 relocation to the nucleus at multiple steps other than only those associated with protein trafficking to the vacuole. In mammalian cells, it has been reported that Vps34 is required for amino acids sensing and transmitting the stimulatory effects of amino acids to the TOR pathway (Byfield et al. 2005; Nobukuni et al. 2005; Backer 2008). Moreover, and in agreement with their multiple functions, two distinct Vps34 complexes function in autophagy and carboxypeptidase Y sorting in response to nitrogen starvation (Kihara et al. 2001).
In addition to the requirement of normal Golgi-to-vacuole trafficking components for Gln3 and Gat1 nuclear localization, we show that Gat1, like Gln3 (Puria et al. 2008), associates with light membranes probably derived from the Golgi apparatus, although the former seems to be more stably associated with light membranes than the latter. Interestingly, the two Gat1 isoforms appeared to behave differently: irrespective of the lysis conditions, the faster migrating form was always observed in the cytosolic fraction, whereas the slower migrating form was more associated with the membrane fraction. It is possible that the 55 N-terminal amino acids lacking in the shorter isoform (Rai et al. 2013a) are determinants of interactions leading to membrane association. In line with this hypothesis, mutants affecting the N-terminal methionines display altered cytoplasmic retention in repressing conditions although this did not affect Gat1's transactivation capacities (Rai et al. 2013a).
Our results also suggest that Ure2 most likely participates in GATA factor regulation downstream the Vps proteins. Aware that genetic interactions exist between URE2 and several VPS genes (Costanzo et al. 2010; Hoppins et al. 2011), the synthetic lethality exhibited upon deleting URE2 in combination with VPS34, VPS45, or PEP5 was not very surprising. The ure2Δvps3Δ double mutant exhibited a WT transcription profile under derepressive conditions, thus, suggesting that Vps3 may be dispensable for GATA factor function, at least when Ure2 is absent. Multiple speculative explanations are possible. Among them, Vps components could be incorporated in a membrane-based nitrogen sensing system where mutating the Vps components would impair transmission of the signal and thus leading to constitutive negative regulation of the GATA factors in a Ure2-dependent manner. However, adding a ure2 mutation to a vps mutation (i.e., ure2Δvps3Δ double mutant) will relieve the negative regulation of the GATA factors. Accordingly, Ure2 would appear to function downstream of the VPS system. Another possibility might be that Vps proteins and Ure2 might exert GATA factor regulation through separate pathways. Taking advantage of the inability of the ure2Δvps3Δ double mutant to grow on YNB-ammonia medium, it could prove useful to select suppressors or mutants able to bypass this lethality. This would enable the identification of new components involved either in GATA factor regulatory pathways or in membrane trafficking as well as other potential unrelated roles of Ure2.
In S. cerevisiae, some, but not all, amino acids are compartmentalized in the vacuoles (Wiemken and Durr 1974; Kitamoto et al. 1988; Sekito et al. 2008). Major known regulators of GATA factor-mediated gene activation (TORC1, Tap42-Sit4 and Gln3) have been localized to membranes of the secretory pathway (Cardenas and Heitman 1995; Kunz et al. 2000; Chen and Kaiser 2003; Wedaman et al. 2003; Reinke et al. 2004; Yan et al. 2006; Aronova et al. 2007; Puria et al. 2008). Moreover, the EGO complex, which is thought to couple amino acid signals to TORC1 activity, resides on the vacuolar membrane (Binda et al. 2009). Together, our results and these observations are consistent with the conclusion that Class C and D Vps proteins are required for the responses of GATA factor intracellular localization and function to changing nitrogen availability. Further, the data suggest that cytoplasmic membranes, which are associated with Golgi-to-vacuolar trafficking, appear to be additionally involved with the generation and/or implementation of these responses.
Together with our data, a recent report (Han and Emr 2011) supports the growing view that transcription factor regulation occurs not only inside the nucleus, but potentially at cytoplasmic compartments as well. In this manner, cytoplasmic signals may directly or indirectly regulate the function of transcription factors, thus strengthening the connection between gene expression and the intra- and extracellular environments to which it responds.
Acknowledgments
We thank André Feller for preparing the artwork, Anna Maria Marini for useful suggestions to improve the manuscript, and Bruno André for allowing the use of the fluorescence microscope. This work was supported by the Commission Communautaire Française (COCOF; E. D., F. V., and I. G.) and the Fonds de la Recherche Fondamentale Collective (FRFC 2.4547.11; E. D, and I. G.) and National Institute of General Medical Sciences grant GM-35642 (J. J. T. and T. G. C.). M. F. K. is an FNRS Research Fellow (Fonds de la Recherche Scientifique).
Conflict of Interest
None declared.
References
- Aronova S, Wedaman K, Anderson S, Yates J, III, Powers T. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2779–2794. doi: 10.1091/mbc.E07-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Blinder D, Coschigano PW, Magasanik B. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 1996;178:4734–4736. doi: 10.1128/jb.178.15.4734-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- Cardenas ME, Heitman J. FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EJ, Kaiser CA. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA, Rai R, Cooper TG. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J. Bacteriol. 1995;177:6910–6918. doi: 10.1128/jb.177.23.6910-6918.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:847–858. doi: 10.1128/mcb.16.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The molecular biology of the yeast saccharomyces: metabolism and gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 39–99. [Google Scholar]
- Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne WE, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Rai R, Distler M, Daugherty JR, Coffman JA, Cooper TG. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem. 2000;275:17611–17618. doi: 10.1074/jbc.M001648200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37559–37566. doi: 10.1074/jbc.M204879200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Tate JJ, Cooper TG. Actin cytoskeleton is required for nuclear accumulation of Gln3 in response to nitrogen limitation but not rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:19294–19301. doi: 10.1074/jbc.M309240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devasahayam G, Ritz D, Helliwell SB, Burke DJ, Sturgill TW. Pmr1, a Golgi Ca2+/Mn2+-ATPase, is a regulator of the target of rapamycin (TOR) signaling pathway in yeast. Proc. Natl. Acad. Sci. USA. 2006;103:17840–17845. doi: 10.1073/pnas.0604303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillien R, Lacroute F. Ureidosuccinic acid uptake in yeast and some aspects of its regulation. J. Bacteriol. 1972;109:203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO Protein Complexes Orchestrate Microautophagy in Yeast. Mol. Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Feller A, Boeckstaens M, Marini AM, Dubois E. Transduction of the nitrogen signal activating Gln3-mediated transcription is independent of Npr1 kinase and Rsp5-Bul1/2 ubiquitin ligase in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:28546–28554. doi: 10.1074/jbc.M605551200. [DOI] [PubMed] [Google Scholar]
- Feller A, Georis I, Tate JJ, Cooper TG, Dubois E. Alterations in the Ure2 alphaCap domain elicit different GATA factor responses to rapamycin treatment and nitrogen limitation. J. Biol. Chem. 2013;288:1841–1855. doi: 10.1074/jbc.M112.385054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J. Biol. Chem. 2008;283:8919–8929. doi: 10.1074/jbc.M708811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I, Feller A, Vierendeels F, Dubois E. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol. Cell. Biol. 2009;29:3803–3815. doi: 10.1128/MCB.00399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I, Tate JJ, Cooper TG, Dubois E. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J. Biol. Chem. 2011a;286:44897–44912. doi: 10.1074/jbc.M111.290577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georis I, Tate JJ, Feller A, Cooper TG, Dubois E. Intranuclear function for protein phosphatase 2A: Pph21 and Pph22 are required for rapamycin-induced GATA factor binding to the DAL5 promoter in yeast. Mol. Cell. Biol. 2011b;31:92–104. doi: 10.1128/MCB.00482-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M, Dubois E, Piotrowska M, Drillien R, Aigle M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol. Gen. Genet. 1974;128:73–85. doi: 10.1007/BF00267295. [DOI] [PubMed] [Google Scholar]
- Han BK, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J. Bacteriol. 1988;170:2683–2686. doi: 10.1128/jb.170.6.2683-2686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Schneider U, Howald I, Schmidt A, Hall MN. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J. Biol. Chem. 2000;275:37011–37020. doi: 10.1074/jbc.M007296200. [DOI] [PubMed] [Google Scholar]
- Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics. 2012;190:885–929. doi: 10.1534/genetics.111.133306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Mitchell AP, Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong C, Riezman H. Ordering of compartments in the yeast endocytic pathway. Traffic. 2002;3:37–49. doi: 10.1034/j.1600-0854.2002.30106.x. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Puria R, Cardenas ME. Rapamycin bypasses vesicle-mediated signaling events to activate Gln3 in Saccharomyces cerevisiae. Commun. Integr. Biol. 2008;1:23–25. doi: 10.4161/cib.1.1.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puria R, Zurita-Martinez SA, Cardenas ME. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2008;105:7194–7199. doi: 10.1073/pnas.0801087105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Tate JJ, Georis I, Dubois E, Cooper TG. Constitutive and Nitrogen Catabolite Repression-Sensitive Production of Gat1 Isoforms. J. Biol. Chem. 2013a;289:2918–2933. doi: 10.1074/jbc.M113.516740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Tate JJ, Nelson DR, Cooper TG. gln3 mutations dissociate responses to nitrogen limitation (nitrogen catabolite repression) and rapamycin inhibition of TorC1. J. Biol. Chem. 2013b;288:2789–2804. doi: 10.1074/jbc.M112.421826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, Yates J, III, Aronova S, Chu S, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Fujiki Y, Ohsumi Y, Kakinuma Y. Novel families of vacuolar amino acid transporters. IUBMB Life. 2008;60:519–525. doi: 10.1002/iub.92. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Woolford CA, Jones EW. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics. 2000;156:105–122. doi: 10.1093/genetics/156.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanbrough M, Rowen DW, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Stress-responsive Gln3 localization in Saccharomyces cerevisiae is separable from and can overwhelm nitrogen source regulation. J. Biol. Chem. 2007;282:18467–18480. doi: 10.1074/jbc.M609550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Formalin can alter the intracellular localization of some transcription factors in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8:1223–1235. doi: 10.1111/j.1567-1364.2008.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Cooper TG. Five Conditions Commonly Used to Down-regulate Tor Complex 1 Generate Different Physiological Situations Exhibiting Distinct Requirements and Outcomes. J. Biol. Chem. 2013;288:27243–27262. doi: 10.1074/jbc.M113.484386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Rai R, Cooper TG. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:27195–27204. doi: 10.1074/jbc.M504052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Feller A, Dubois E, Cooper TG. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J. Biol. Chem. 2006;281:37980–37992. doi: 10.1074/jbc.M606973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Georis I, Feller A, Dubois E, Cooper TG. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J. Biol. Chem. 2009;284:2522–2534. doi: 10.1074/jbc.M806162200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JJ, Georis I, Dubois E, Cooper TG. Distinct Phosphatase Requirements and GATA Factor Responses to Nitrogen Catabolite Repression and Rapamycin Treatment in Saccharomyces cerevisiae. J. Biol. Chem. 2010;285:17880–17895. doi: 10.1074/jbc.M109.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wedaman KP, Reinke A, Anderson S, Yates J, III, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiame JM, Grenson M, Arst HN., Jr Nitrogen catabolite repression in yeasts and filamentous fungi. Adv. Microb. Physiol. 1985;26:1–88. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- Wiemken A, Durr M. Characterization of amino acid pools in the vacuolar compartment of Saccharomyces cerevisiae. Arch. Microbiol. 1974;101:45–57. doi: 10.1007/BF00455924. [DOI] [PubMed] [Google Scholar]
- Yan G, Shen X, Jiang Y. Rapamycin activates Tap42-associated phosphatases by abrogating their association with Tor complex 1. EMBO J. 2006;25:3546–3555. doi: 10.1038/sj.emboj.7601239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]