Abstract
The Escherichia coli cytoplasmic membrane contains the enzyme complexes of oxidative phosphorylation (OXPHOS). Not much is known about their supramolecular organization and their dynamics within the membrane in this model organism. In mitochondria and other bacteria, it was demonstrated by nondenaturing electrophoretic methods and electron microscopy that the OXPHOS complexes are organized in so-called supercomplexes, stable assemblies with a defined number of the individual enzyme complexes. To investigate the organization of the E. coli enzyme complexes of aerobic OXPHOS in vivo, we established fluorescent protein fusions of the NADH:ubiquinone oxidoreductase, the succinate:ubiquinone oxidoreductase, the cytochrome bd-I, and the cytochrome bo3 terminal oxidases, and the FoF1 ATP-synthase. The fusions were integrated in the chromosome to prevent artifacts caused by protein overproduction. Biochemical analysis revealed that all modified complexes were fully assembled, active, and stable. The distribution of the OXPHOS complexes in living cells was determined using total internal reflection fluorescence microscopy. The dynamics within the membrane were detected by fluorescence recovery after photobleaching. All aerobic OXPHOS complexes showed an uneven distribution in large mobile patches within the E. coli cytoplasmic membrane. It is discussed whether the individual OXPHOS complexes are organized as clustered individual complexes, here called “segrazones.”
Keywords: Escherichia coli, FRAP, in vivo fluorescence microsocopy, membrane protein organization, oxidative phosphorylation, TIRF microscopy
Introduction
The aerobic oxidative phosphorylation (OXPHOS) in Escherichia coli is mainly catalyzed by six enzyme complexes located in the cytoplasmic membrane. Five oxidoreductases transfer electrons from NADH and succinate to oxygen. In doing so, a proton gradient across the membrane is generated that is needed for energy-consuming processes such as ATP synthesis catalyzed by the sixth enzyme complex, the FoF1 ATP-synthase (Ingledew and Poole 1984; Senior et al. 2002; Price and Driessen 2010). The NADH:ubiquinone oxidoreductase (complex I), the alternative NADH dehydrogenase and the succinate:ubiquinone oxidoreductase (complex II) are the primary dehydrogenases acting as entry points for electrons from NADH and succinate into the respiratory chain (Friedrich 2001; Cecchini et al. 2002; Feng et al. 2012). In contrast to complex I, the reactions of the alternative NADH dehydrogenase and of complex II are not coupled with the translocation of protons across the membrane. However, the terminal cytochrome bd-I (Borisov et al. 2011) and cytochrome bo oxidases (Abramson et al. 2000) couple the reduction of oxygen to water with the generation of a proton gradient across the membrane. The primary dehydrogenases and the terminal oxidases are connected by the mobile carrier ubiquinone. Under microaerophilic and anoxic conditions menaquinone and demethylmenaquinone are used as electron carriers (Unden and Bongaerts 1997). The complexes investigated in this study are listed in Table 1.
Table 1.
The Escherichia coli aerobic OXPHOS enzyme complexes investigated in this study
| Enzyme complex | Mol. mass (kDa) | Number of different subunits | Genes | FP fusions in E. coli BW25113 (localization) |
|---|---|---|---|---|
| NADH:ubiquinone oxidoreductase (complex I) | 535 | 13 | nuoA-nuoN | nuoF-mcerulean mcherry-nuoF egfp-nuoF (all cytoplasm) |
| Succinate:ubiquinone oxidoreductase (complex II) | 120 | 4 | sdhCDAB | mcherry-sdhC (cytoplasm) |
| Cytochrome bd-I complex | 100 | 2 | cydA-cydB | cydB-egfp (cytoplasm) |
| Cytochrome bo3 complex | 140 | 2 | cyoA-cyoD | cyoA-mcherry (periplasm) |
| FoF1 ATP-synthase | 528 | 8 | atpA-atpH | atpB-egfp (cytoplasm) |
An early description of the organization of membrane proteins within the biological membrane is the “fluidic mosaic model” depicting the membrane as a two-dimensional phase in which the membrane proteins and hydrophobic electron carriers freely diffuse (Singer and Nicolson 1972). The concept of proteins freely diffusing in the membrane was challenged by fluorescence microscopy experiments (Jacobson et al. 1995; Mika and Poolman 2011) and the detection of membrane lipid domains (Groves 2006; Matsumoto et al. 2006). Furthermore, the model was questioned by experiments demonstrating a higher order organization of membrane proteins in so-called supercomplexes, stable assemblies containing a defined stoichiometry of individual complexes. The existence of supercomplexes was shown in mitochondria from several species (Schägger and Pfeiffer 2000; Zhang et al. 2005; Nübel et al. 2009; Lenaz et al. 2010) and in bacteria such as Paracoccus denitrificans (Stroh et al. 2004) by nondenaturing PAGE (polyacrylamide gel electrophoresis) techniques, by electron microscopy, (Dudkina et al. 2010, 2011; Davies et al. 2012) and the biochemical preparation of supercomplexes (Niebisch and Bott 2003). The formation of supercomplexes might help to enhance the stability of the individual complexes and might accelerate the reaction rates by substrate channeling (Matsumoto et al. 2006; Romantsov et al. 2010).
Here, we investigate the localization and dynamics of the individual OXPHOS complexes in the E. coli cytoplasmic membrane in vivo by fluorescence microscopy. To avoid artifacts possibly caused by overproduction, chromosomally encoded fusions of the OXPHOS complexes with various fluorescent proteins (FP) were established by λ-red-mediated recombination (Datsenko and Wanner 2000; Pohl et al. 2007). We integrated the genes encoding eGFP (enhanced GFP), mCerulean, and mCherry (Ito et al. 1999; Rizzo et al. 2004; Shu et al. 2006) into the nuo-, sdh-, cyd-, cyo-, and atp-operon in the E. coli chromosome. In order not to disturb the assembly of the complexes and not to influence the enzymatic activity, proper positions for the FP fusions had to be identified. In vivo fluorescence microscopy with the strains revealed that the enzyme complexes in question are unevenly distributed in the E. coli cytoplasmic membrane in large patches. Their distribution patterns and mobility in the membrane were similar in TIRF (Groves et al. 2008) and FRAP (Mullineaux 2004) experiments.
Experimental Procedures
Strains, plasmids and oligonucleotides
All E. coli strains, plasmids, and oligonucleotides used in this work are listed in Tables S1, S2, and S3, respectively.
Cell growth
Cells were grown either in LB medium or in M9 minimal medium containing succinate as sole carbon source. For fluorescence microscopy 4 mL S750-medium was inoculated in a 1:100 ratio (v:v) with a 4 mL overnight culture in LB medium and grown for 3–5 h at 30°C. To select the desired mutations, cells were grown in the presence of 100 μg mL−1 ampicillin, 170 μg mL−1 (for liquid media) or 20 μg mL−1 (for agar plates) chloramphenicol or 50 μg mL−1 kanamycin.
Activity assays
The NADH and succinate oxidase activity of cytoplasmic membranes was measured with a Clarke-type oxygen electrode at 30°C in a volume of 2 mL. The assay contained 2–3 mg cytoplasmic membranes and the reaction was initiated by the addition of the corresponding substrate after obtaining a constant baseline. The “substrate”/ferricyanide oxidoreductase activities were measured as the decrease in the absorbance of ferricyanide at 410 nm with a Ultraspec spectrophotometer (Amersham Pharmacia Biotech, Munich, Germany) in a volume of 1 mL. The assay contained 10 μL membrane suspension or 30 μL of a fraction after sucrose gradient centrifugation. Detailed conditions are provided in the Data S1.
Sucrose gradient centrifugation
Membrane proteins were solubilized by an addition of 3% (w/v) n–dodecyl-β-D-maltopyranoside (DDM; AppliChem, Darmstadt, Germany) at 4°C. The extract was centrifuged for 20 min at 48,000g and 4°C. 0.9 mL of the supernatant were loaded onto 12 mL gradients of 5–30% (w/v) sucrose and centrifuged for 18 h at 160,000g. The gradients were fractionated into 0.7 mL portions and the “substrate”/ferricyanide oxidoreductase activities and the fluorescence emissions of the particular FP were measured at its specific wavelength. Detailed conditions are provided in the Data S1.
Microscopy
All strains for fluorescence microscopy were grown in S750 minimal media at 30°C. Fluorescence microscopy was performed using a Zeiss Observer Z1 equipped with a 1.45 NA objective (Jena, Germany) and a Photometrix Cascade CCD camera (Munich, Germany). The fluorophores were exited by exposition to a laser beam of 488 nm, 561 nm, or 445 nm wavelengths coupled in by a visitron VisiTIRF system (Puchheim, Germany). FRAP experiments were performed using a 50 mW argon laser of 405 nm wavelength. The 50 μm size of the laser beam was generated by a pinhole inserted into the laser beam within the module that incorporates the optical wire into the side port. For epifluorescence images, we used a xenon mercury burner with appropriate filter sets. Images were processed with the Metamorph 7.5.5 software (Sunnyvale, CA). For the TIRF- and FRAP-streams, every picture was optimized for the best signal to noise ratio.
Results
Integration of the FP-gene fusions in the E. coli BW25113 chromosome
For in vivo localization studies, it is essential to ascertain whether the proteins in question are produced at the physiological level. Therefore, chromosomally encoded FP fusions of the aerobic OXPHOS complexes were established in E. coli strain BW25113 (Datsenko and Wanner 2000; Baba et al. 2006; Table 1). Individual genes comprising the FP fusion were integrated in the chromosome via λ-red-mediated recombination (see Supporting Information). The FP fusions were established either on subclones and introduced in the chromosome in a second step or by direct recombination of the FP sequence with the chromosome (see Supporting Information).
Escherichia coli complex I is made up of 13 different subunits (Friedrich 1998). It was previously shown that a His-tag fused to the N-terminus of the cytosolic subunit NuoF containing the NADH-binding site neither prevents the assembly nor the activity of complex I making NuoF a promising candidate for a FP fusion (Pohl et al. 2007). It turned out that eGFP, mCerulean, and mCherry, respectively, can be fused to both termini of NuoF. FP fusions to other subunits either prevented the assembly of the complex or led to the production of an inactive enzyme complex. Complex II consists of four subunits (Cecchini et al. 2002). The N-terminus of the membranous subunit SdhC was decorated with mCherry. FP fusions to all other subunits lead either to the formation of inclusion bodies or to the occurrence of fluorescence in the cytoplasm. The fusion of the C-terminus of the membranous subunit CydII of the cytochrome bd-I oxidase with an eGFP has been recently described (Lenn et al. 2008). A fluorescent variant of the cytochrome bo3 complex was obtained by the fusion of mCherry to the periplasmic C-terminus of CyoII. The FoF1 ATP-synthase was decorated with an eGFP at the C-terminus of subunit Foa encoded by atpB as described (Düser et al. 2008). Further experiments were performed with the E. coli strains BW25113 nuoF-mcerulean, BW25113 mcherry-nuoF, BW25113 egfp-nuoF, BW25113 mcherry-sdhC, BW25113 cydB-egfp, BW25113 cyoA-mcherry, and BW25113 atpB-egfp (Table 1). The parental strain BW25113 was used as control.
The FP-decorated enzyme complexes are active and fully assembled
Strains containing the chromosomally encoded FP fusions showed an identical growth rate as the parental strain in LB medium and in M9-minimal medium containing succinate as sole carbon source (data not shown). The activity of the enzyme complexes decorated with the FP fusion was detected by various assays (Spehr et al. 1999). The NADH oxidase activity, a measure of the catalytic competence of complex I, and the NADH/ferricyanide oxidoreductase activity, a measure of the amount of the complex in the membrane of cytoplasmic membranes from strains BW25113 nuoF-mcerulean, BW25113 mcherry-nuoF, and BW25113 egfp-nuoF containing an FP fusion on complex I and from strain BW25113 was determined. The NADH oxidase activity was not decreased in the mutant with the mCherry fusion and only slightly decreased by 5% and 12% in the mutants carrying the mCerulean and the eGFP fusion, respectively (Table 2). The NADH oxidase activity was inhibited to 50% by piericidin A, a specific complex I inhibitor, in all strains in accordance with the literature (Pohl et al. 2007). The residual activity is due the presence of the alternative NADH dehydrogenase. The NADH/ferricyanide oxidoreductase activity did not vary significantly within the experimental error indicating that the amount of the complex decorated with the FP in the membrane did not change in comparison to the parental strain (Table 2).
Table 2.
Complex I-mediated activities in the membranes of various Escherichia coli strains
| NADH oxidase activity | NADH/ferricyanide oxidoreductase activity | |
|---|---|---|
| Strain | (μmol min−1 mg−1) | |
| BW25513 | 0.43 ± 0.02 | 3.3 ± 0.2 |
| BW25113 nuoF-mcerulean | 0.41 ± 0.04 | 3.2 ± 0.1 |
| BW25113 mcherry-nuoF | 0.44 ± 0.01 | 3.0 ± 0.1 |
| BW25113 egfp-nuoF | 0.38 ± 0.02 | 3.4 ± 0.6 |
The succinate oxidase activity mediated by complex II was reduced by one-third in strain BW25113 mcherry-sdhC to 0.05 ± 0.01 μmol min−1 mg−1 compared to that of the parental strain with 0.08 ± 0.01 μmol min−1 mg−1. However, this activity was inhibited by 20 mmol/L malonate, specifically inhibiting complex II, to more than 80% in both strains. The succinate/ferricyanide oxidoreductase activity of the FP fusion complex was decreased by 20% from 0.18 ± 0.01 μmol min−1 mg−1 to 0.15 ± 0.01μmol min−1 mg−1 due to the FP fusion. Thus, the amount of complex II in the membranes was not significantly affected by the FP fusion which in addition had only a mild effect on the physiological activity.
The NADH oxidase activity of the membranes from strain BW25113 cydB-egfp was 0.41 ± 0.02μmol min−1 mg−1 and thus, 95% of that of the parental strain in accordance with the literature. This strain containing a functional cytochrome bd oxidase has been described in detail (Lenn et al. 2008). The membranes of strain BW25113 cyoA-mCherry producing the cytochrome bo3 oxidase decorated with mCherry showed an NADH oxidase activity of 0.39 ± 0.02 μmol min−1 mg−1 corresponding to 90% of the parental strain activity. This activity was inhibited by 20% upon an addition of 30 mmol/L hydroxylamine. The NADH oxidase activity of membranes from strain BW25113 was inhibited by 21%. Thus, the FP fusion led only to a slight decrease in activity and to the assembly of a functional complex. The activity of ATP-synthase in strain BW25113 atpB-egfp was screened by its ability to grow on succinate minimal agar plates. With succinate as the sole carbon source, only cells with a functional ATP-synthase will survive. Indeed, the parental strain and strain BW25113 atpB-egfp were able to grow on these plates in contrast to a strain carrying an inactivated atpB gene in its chromosome (Fig. S1). Thus, all OXPHOS enzyme complexes decorated with an FP were active in the membrane.
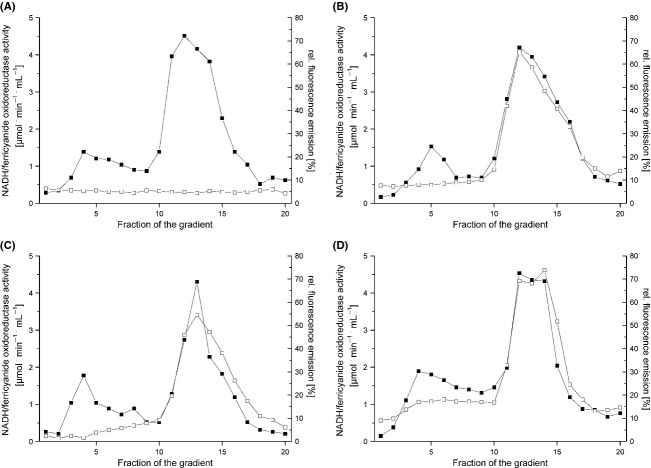
The assembly of the OXPHOS complexes containing the desired FP fusions was investigated by sucrose gradient centrifugation as described (Spehr et al. 1999). Membrane proteins were extracted from cytoplasmic membranes with dodecyl-maltoside and the detergent extract was loaded on a sucrose gradient (Fig. 1). After centrifugation, the gradient was fractionated and calibrated by measuring the NADH/ferricyanide and the succinate/ferricyanide oxidoreductase activity, respectively. In addition, all fractions of the gradients were characterized by the fluorescence emission of the respective FP. The NADH/ferricyanide oxidoreductase activity is caused by the fully assembled complex I with a molecular mass of 535 kDa sedimenting in fractions 11–13 under the chosen conditions. The sedimentation profiles of the NADH/ferricyanide oxidoreductase activity of extracts obtained from strains BW25113 nuoF-mcerulean, BW25113 mcherry-nuoF, and BW25113 egfp-nuoF were identical to the sedimentation profile of the fluorescence of the corresponding FPs (Fig. 1). This indicated that all complex I variants are fully assembled and decorated with the desired FP. The same sedimentation profile was measured with the eGFP fluorescence of the membrane extract from strain BW25113 atpB-egfp (Fig. 3). This is in line with the molecular mass of ATP-synthase of 528 kDa that cosediments with complex I under the chosen conditions (Spehr et al. 1999; Stolpe and Friedrich 2004).
Figure 1.
Sucrose gradient centrifugation of detergent-solubilized membranes from E. coli strains containing FP fusions on nuoF. The NADH/ferricyanide oxidoreductase activity (▪) and the fluorescence of the corresponding FP fusion (□) were measured in extracts from strains BW25113 (A), BW25113 nuoF-mcerulean (B), BW25113 mcherry-nuoF (C), and BW25113 egfp-nuoF (D). All data were normalized to 10 mg membrane protein extract applied per gradient. The fluorescence of mCerulean was measured at an excitation wavelength of 430 nm and an emission wavelength of 475 nm, that of mCherry at an excitation wavelength of 587 nm and an emission wavelength of 610 nm and that of eGFP at an excitation wavelength of 480 nm and an emission wavelength of 510 nm. The fluorescence shown in (A) was measured at an excitation wavelength of 430 nm and an emission wavelength of 475 nm. The fluorescence of the fractions obtained from strain BW25113 were measured at all three pairs of wavelength leading to similar profiles.
Figure 3.
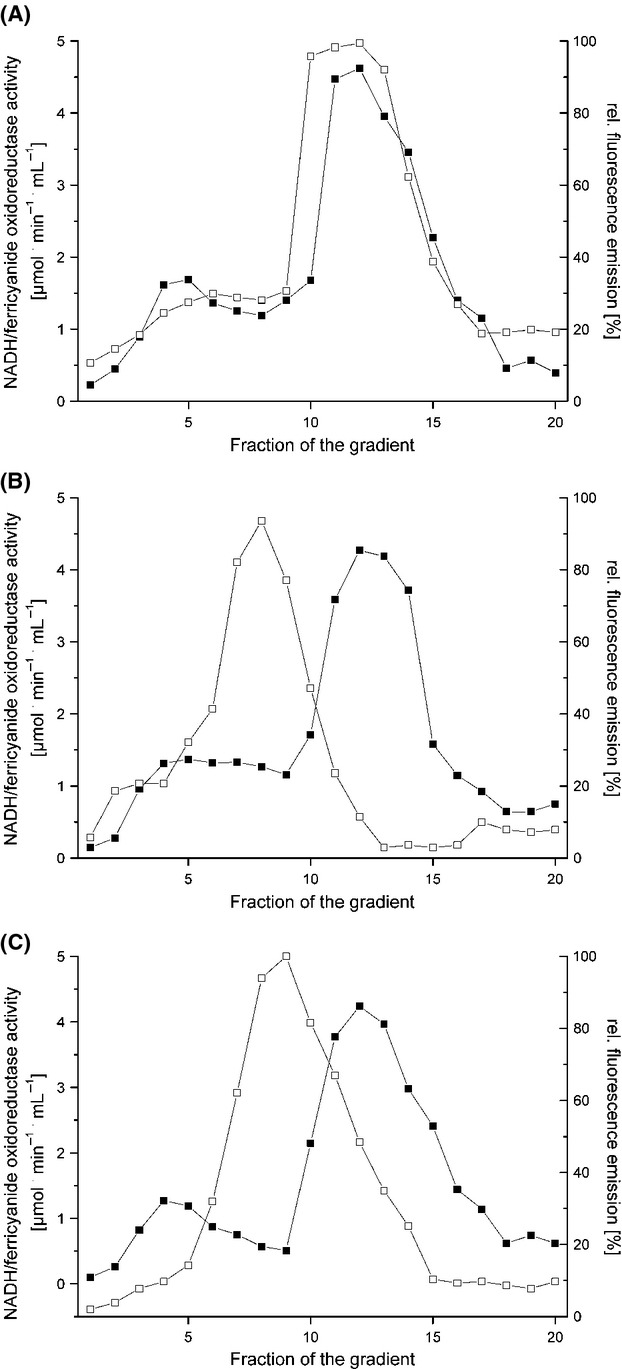
Sucrose gradient centrifugation of detergent-solubilized membranes from E. coli strains containing a FP fusion on atpB, cydB and cyoA. The NADH/ferricyanide oxidoreductase activity (▪) and the fluorescence of the corresponding FP fusion (□) were measured in extracts from strains BW25113 atpB-egfp (A), BW25113 cydB-egfp (B) and BW25113 cyoA-mCherry (C). All data were normalized to 10 mg membrane protein extract applied per gradient. The fluorescence of mCherry was measured at an excitation wavelength of 587 nm and an emission wavelength of 610 nm and that of eGFP at an excitation wavelength of 480 nm and an emission wavelength of 510 nm.
The succinate/ferricyanide oxidoreductase activity originates from the fully assembled monomeric complex II with a molecular mass of 120 kDa and from the homotrimer with a molecular mass of 360 kDa. The succinate/ferricyanide oxidoreductase activity of strain BW25113 cydB-egfp showed two activity peaks sedimenting around fraction 7 and in fractions 9–11 corresponding to the monomeric and trimeric form of complex II as described (Yankovskaya et al. 2003). The sedimentation profile of the mCherry fluorescence correlated perfectly with the activity profile exhibiting two peaks at the same positions as the activity peaks (Fig. 2). The sedimentation profile of the fluorescence emission in the extract from strains BW25113 cydB-egfp and BW25113 cyoA-mCherry showed a maximum in fractions 7–9, respectively, which is in accordance with the molecular masses of 100 and 140 kDa (Fig. 3). A fluorescence emission was not observed in any of the gradients in low molecular mass fractions corresponding to individual subunits fused with an FP excavated from a complex or to degraded proteins. Neither was fluorescence detectable in the very high molecular mass fractions corresponding to aggregates of the enzyme complexes (Figs. 1, 2 and 3). These data show that the OXPHOS complexes under investigation are fully assembled, stable, and decorated with the desired FP fusion. However, we cannot exclude the possibility that the FP fusions might have an influence on the oligomeric state of enzyme complexes.
Figure 2.
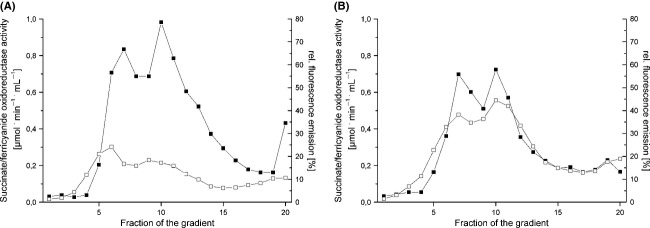
Sucrose gradient centrifugation of detergent-solubilized membranes from E. coli strains containing a FP fusion on sdhC. The succinate/ferricyanide oxidoreductase activity (▪) and the fluorescence of mCherry (□) were measured in extracts from strains BW25113 (A) and BW25113 mcherry-sdhC (B). All data were normalized to 10 mg membrane protein extract applied per gradient. The fluorescence of mCherry was measured at an excitation wavelength of 587 nm and an emission wavelength of 610 nm.
The OXPHOS complexes show an uneven distribution in the membrane
The localization of the aerobic OXPHOS complexes in the membrane was determined by in vivo epifluorescence microscopy. Fluorescence emission was detected only in the cytoplasmic membrane (Fig. 4). There was no enhanced fluorescence of the cytoplasm indicating the absence of degraded or not-assembled FP fusion proteins. The FP fusions of all five protein complexes under investigation showed an uneven distribution in the membrane (Fig. 4). The fluorescence was not concentrated in specific regions of the cell, such as the cell poles as described for inclusion bodies or at the division septum as reported for members of the Min-family (Szeto et al. 2002). Compared to the other mCherry-fusions (Table 1), the fluorescence in strain BW25113 mcherry-sdhC was enhanced indicating a higher concentration of this complex in the E. coli membranes. Due to its higher concentration in the membrane, the distribution of complex II is apparently more homogeneous than in the other complexes. The patches have an estimated average diameter of 300 to 500 nm in all strains. However, due to the limited resolution of 250 nm, we cannot exclude the possibility that the patches are made up of smaller, closely spaced droplets.
Figure 4.
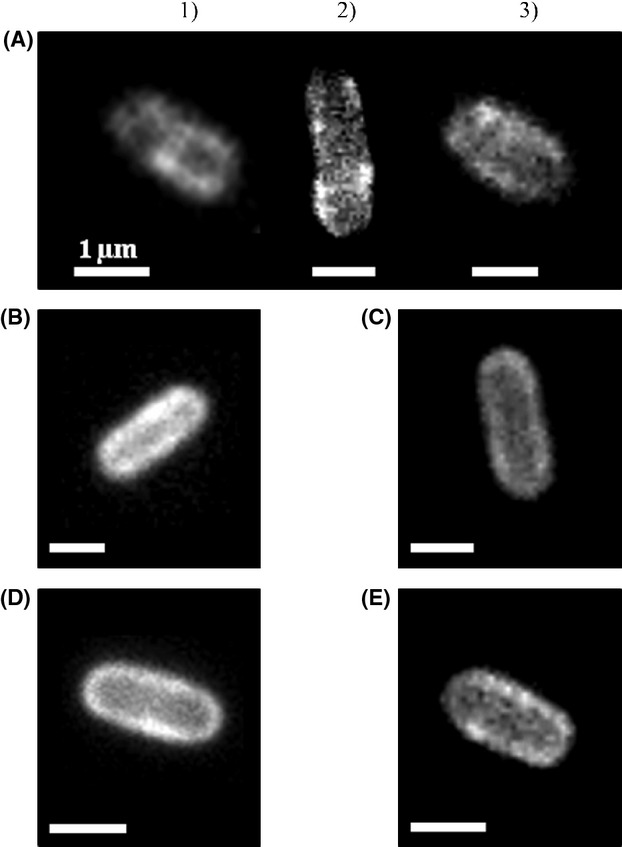
Epifluorescence images of the single cells of strains BW25113 egfp-nuoF (A1; 1 sec), BW25113 nuoF-mCerulean (A2; 1 sec), BW25113 mcherry-nuoF (A3; 1 sec), BW25113 mcherry-sdhC (B; 1 sec), BW25113 cydB-egfp (C; 0.6 sec), BW25113 cyoA-mCherry (D; 0.6 sec), and BW25113 atpB-egfp (E; 0.6 sec). The numbers in brackets indicate the exposure times in seconds. White bars 1 μm.
Patches of OXPHOS complexes are dynamic over time
The mobility of the FP-labeled OXPHOS complexes was investigated by TIRF microscopy. The images (Fig. 5) and streams showed a very similar patchy distribution pattern of the enzyme complexes to that observed by epifluorescence microscopy. The patches had the same diameter between 300 and 500 nm as determined in epifluorescence microscopy. The stream recordings revealed that the patches of the OXPHOS complexes are not static but highly dynamic (Movies S1–S4). The movement of the patches did not follow an obvious pattern and the OXPHOS complexes diffused randomly. From time to time, little droplets constricted from one patch and fused with another patch (Movies S1–S4). The dynamics of the OXPHOS complexes were investigated using FRAP experiments combined with TIRF microscopy (Fig. 6). There is a fast recovery of the fluorescence after bleaching a defined membrane area by the FRAP laser in all strains (Movies S5–S8). On average, the fluorescence recovery into the bleached area was complete after ∼10 sec. In some experiments, the bleached parts even contained the patches with the highest fluorescence emission after recovery. In most cases, the fluorescence reappeared in the bleached areas by a diffusion of an entire patch from an unbleached area. With the experimental setup, it was not possible to examine differences in the diffusion rates of the patches of fluorescence in the various strains. The observed dynamic behavior of the complexes is consistent with previous data reported for the cytochrome bd-I complex (Lenn et al. 2008).
Figure 5.
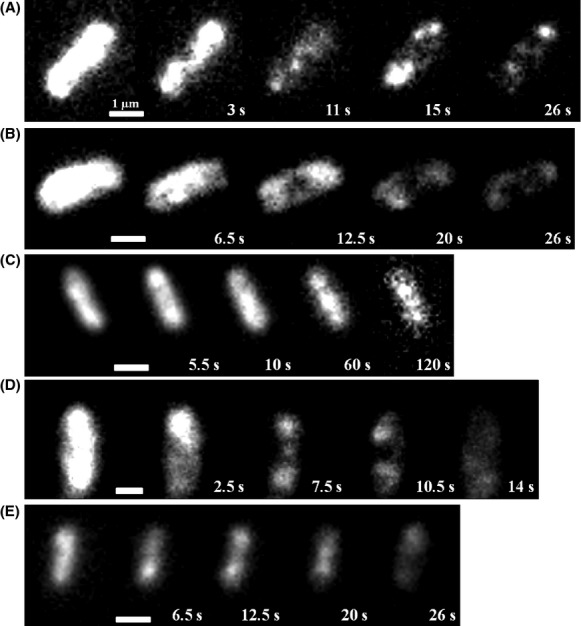
TIRF microscopy images of E. coli strains BW25113 egfp-nuoF (A), BW25113 mcherry-sdhC (B), BW25113 cydB-egfp (C), BW25113 cyoA-mCherry (D), and BW25113 atpB-egfp (E). White bars 1 μm.
Figure 6.
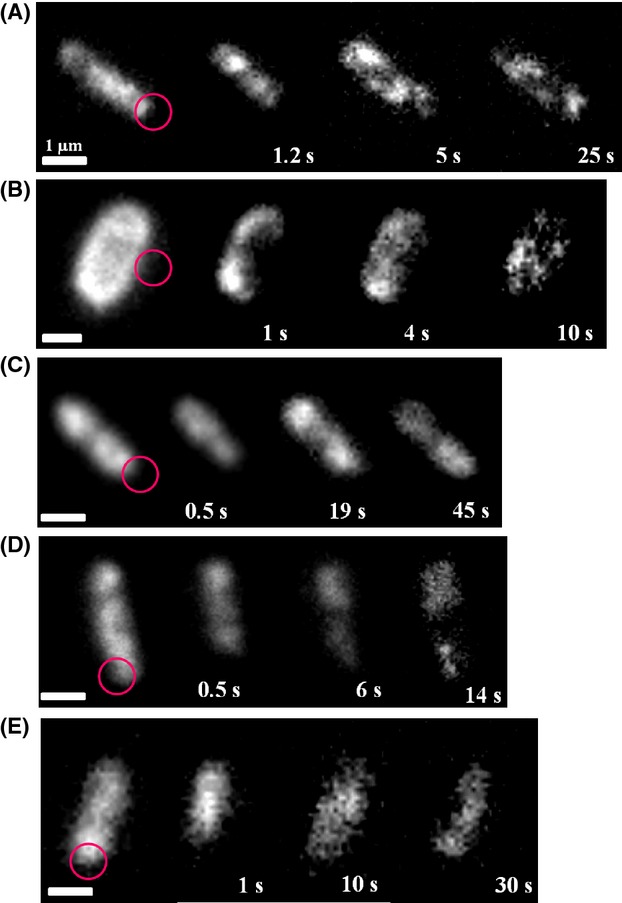
FRAP experiments using TIRF microscopy, images of E. coli strains BW25113 egfp-nuoF (A), BW25113 mcherry-sdhC (B); BW25113 cydB-egfp (C), BW25113 cyoA-mCherry (D), BW25113 atpB-egfp (E). The circle indicates the area of bleaching, numbers indicate time after bleaching. White bars 1 μm.
Discussion
In this study, the enzyme complexes of aerobic OXPHOS were localized in the cytoplasmic membrane of living E. coli cells. This was achieved by fusing the genes of either eGFP, mCerulean, or mCherry, to structural genes of respiratory complex I, complex II, cytochrome bd-I oxidase, cytochrome bo oxidase, and FoF1 ATP-synthase in the corresponding chromosomal operon. Thus, the genes were expressed and the FP-labeled proteins produced at a physiological level. This implies that the OXPHOS complexes were present in differing amounts in the membrane impeding a quantitative analysis. Considering the different fluorescence lifetime and the bleaching stability of the individual fluorophores, it was estimated that, for example, complex II is present in a more than 100-fold higher concentration in the membrane than complex I by comparison of the total fluorescence estimated for the same FP-fusion. This is most likely due to the fact that complex II is not only part of the respiratory chains but also plays a distinct role in the citric acid cycle. Due to its much higher abundance, complex II shows a more homogeneous distribution within the membrane than the other OXPHOS complexes. Accordingly, OXPHOS complexes that were present in a higher amount were observed at longer timescales than minority enzyme complexes. We were not able to determine the cellular localization of the alternative NADH dehydrogenase encoded by ndh, because fusions with any of the three fluorophores led to an increased fluorescence in the cytoplasm (data not shown). This is most likely due to the fact that the alternative NADH dehydrogenase is a peripheral membrane protein that forms an amphiphilic membrane-anchor by dimerization (Feng et al. 2012; Iwata et al. 2012). Attachment of a fluorophore enlarges the hydrophilic part so that the dimer might be pulled away from the membrane.
The fusions had no effect on the growth rate and led to a negligible decrease in enzymatic activity of complex I, complex II, cytochrome bd-I oxidase, cytochrome bo3 oxidase, and FoF1 ATP-synthase. In addition, the OXPHOS complexes were fully assembled and contained the desired FP fusions. Thus, we established a system to investigate the enzyme complexes of aerobic OXPHOS in living E. coli cells using fluorescence microscopy techniques.
Our studies show that the aerobic OXPHOS complexes are found in large patches within the E. coli cytoplasmic membrane. These patches of enzyme complexes of the same kind diffused within the membrane and fluorescence moved from one patch to another indicating the dynamic composition of these patches. There was no obvious difference detectable in the mobility of the individual enzyme complexes. Similar findings have been reported for the E. coli cytochrome bd-I oxidase (Lenn et al. 2008) and the Bacillus subtilis ATP-synthase and succinate:ubiquinone oxidoreductase (Johnson et al. 2004; Meredith et al. 2008). Therefore, the organization of the OXPHOS complexes in larger patches seems to be a general feature at least in gram-negative E. coli and gram-positive B. subtilis (see, however, discussion below). In all FRAP experiments, the bleached area recovered fluorescence much faster than possible due to de novo protein synthesis (Ogle and Ramakrishnan 2005). Thus, the FRAP is due to the diffusion of the enzyme complexes in the membrane. The difference in the molecular mass of the OXPHOS complexes should be reflected in different diffusion coefficients. However, the rates of fluorescence recovery were virtually identical in all strains, making it very likely that the OXPHOS complexes form larger supramolecular assemblies reflected in the fluorescence patches that are obscuring the determination of individual diffusion coefficients. The same patchy distribution of the OXPHOS complexes was also detected in mitochondria (Muster et al. 2010). It is proposed that the fusion and fission dynamics of mitochondria favor a dynamic reassortment of the individual OXPHOS complexes (Muster et al. 2010). The same holds true for bacteria where a static number of the OXPHOS complexes per cell would hamper cell division.
The fluorescent patches in the E. coli cytoplasmic membrane are much larger than the respiratory supercomplexes characterized so far (Stroh et al. 2004; Boekema and Braun 2007; Dudkina et al. 2011; Davies et al. 2012). Thus, they cannot represent individual supercomplexes and to our knowledge a clustering of supercomplexes has not been reported. Using in vitro experiments, it was reported that supercomplexes are also formed in E. coli. The homotrimeric organization of the succinate:ubiquinone oxidoreductase detected by other groups (Yankovskaya et al. 2003; Sousa et al. 2011) fits nicely with our data obtained by sucrose gradient centrifugation. In addition, defined associations between the two NADH dehydrogenases, between the two terminal oxidases and the formate dehydrogenase as well as between cytochrome bd-I and succinate dehydrogenase have been proposed (Sousa et al. 2012; Sousa et al. 2013a,b). With the strains described here, it is not possible to decide whether these supercomplexes are formed in vivo.
The question is still open whether the assemblies clustered in patches consist of only one kind of enzyme complex, that might be called “segrazones” or whether they contain various OXPHOS complexes as proposed for the “respirazones” (Lenn et al. 2008). To answer this question, we are currently in the process of generating new E. coli strains that contain pairs of FP-labeled complexes with the FP fusions of different colors each at the same position of the enzyme complexes described here. A quantitative analysis of these strains will elucidate whether the fluorescent patches contain only one kind or several kinds of the OXPHOS enzyme complexes. However, as the cytochrome bc1 complex, the respiratory complex III, is the central and connecting part of most supercomplexes described so far and because this complex is missing in E. coli, we propose that the OXPHOS complexes in the E. coli membrane are indeed organized in “segrazones” and, thus, represent a novel type of supramolecular organization. This would also question the interpretation that the patches of ATP-synthase and succinate:ubiquinone oxidoreductase observed in B. subtilis (Johnson et al. 2004; Meredith et al. 2008) do not reflect a supramolecular structure, because this organism contains a cytochrome bc complex (Yu et al. 1995). Indeed, supercomplex assemblies consisting of bc complex, caa3 oxidase and most likely ATP-synthase and of succinate:ubiquinone oxidoreductase and cytochrome aa3 menaquinol oxidase (Garcia Montes de Oca et al. 2012) as well as of the bc and caa3 complexes in various stoichiometries (Sousa et al. 2013a,b) were reported to be present in B. subtilis. However, supercomplexes lacking bc complex such as a cytochrome bd oxidase/cytochrome bo3 oxidase/formate dehydrogenase supercomplex and a cytochrome bd/succinate dehydrogenase supercomplex have also been described (Sousa et al. 2012, 2013a,b). Noteworthy, all other bacterial supercomplexes reported in the literature so far contain a member of the cytochrome bc family (Magalon et al. 2012).
Acknowledgments
We thank Peter Gräber and Michael Börsch (Universities of Freiburg and Jena) for the gift of pKF2-6, Gary Cecchini (University of California) for pSDH15, Conrad Mullineaux (Queen Mary University London) and Tchern Lenn (University of California, Berkeley) for providing E. coli strain BW25113 cydB-egfp. We thank Conrad Mullineaux, London, and Mark Leake, York, for helpful discussions. We thank Linda Williams for her help in correcting the manuscript. This work was supported by Deutsche Forschungsgemeinschaft by grant for 929.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Movies S1-S4. The movies show streams of live cells from strains BW25113 egfp-nuoF (Movie S1), BW25113 mcherry-sdhC (Movie S2), BW25113 cyoA-mcherry (Movie S3) and BW25113 atpB-egfp (Movie S4) obtained by TIRF-microscopy. The streams show the localization of the OXPHOS complexes in clusters that are dynamic over time (Movies S1-S4).
Movies S5-S8. The movies show streams of live cells from strains BW25113 egfp-nuoF (Movie S5), BW25113 mcherry-sdhC (Movie S6), BW25113 cyoA-mcherry (Movie S7) and BW25113 atpB-egfp (Movie S8) obtained by TIRF microscopy in a FRAP experiment. A part of the cell was bleached with a short laser pulse and the recovery of fluorescence in the bleached areas is seen (Movies S5-S8). All OXPHOS complexes rapidly diffuse into the bleached areas with similar kinetics.
References
- Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema EJ, Braun HP. Supramolecular structure of the mitochondrial oxidative phosphorylation system. J. Biol. Chem. 2007;282:1–4. doi: 10.1074/jbc.R600031200. [DOI] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta. 2011;1807:1398–1413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini G, Schröder I, Gunsalus RP, Maklashina E. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta. 2002;1553:140–157. doi: 10.1016/s0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc. Natl. Acad. Sci. USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Kouril R, Peters K, Braun HP, Boekema EJ. Structure and function of mitochondrial supercomplexes. Biochim. Biophys. Acta. 2010;1797:664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düser MG, Bi Y, Zarrabi N, Dunn SD, Börsch M. The proton-translocating a subunit of F0F1-ATP synthase is allocated asymmetrically to the peripheral stalk. J. Biol. Chem. 2008;283:33602–33610. doi: 10.1074/jbc.M805170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Li W, Li J, Wang J, Ge J, Xu D, et al. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature. 2012;491:478–482. doi: 10.1038/nature11541. [DOI] [PubMed] [Google Scholar]
- Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim. Biophys. Acta. 1998;1364:134–146. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- Friedrich T. Complex I: a chimaera of a redox and conformation driven proton-pump? J. Bioenerg. Biomembr. 2001;33:169–177. doi: 10.1023/a:1010722717257. [DOI] [PubMed] [Google Scholar]
- Garcia Montes de Oca LY, Chagolla-Lopez A, Gonzales de la Vara L, Cabellos-Avelar T, Gomes-Lojero C, Gutierrez Cirlos EB. The composition of the Bacillus subtilis aerobic respiratory chain supercomplexes. J. Bioenerg. Biomembr. 2012;44:473–486. doi: 10.1007/s10863-012-9454-z. [DOI] [PubMed] [Google Scholar]
- Groves JT. Unveiling the membrane domains. Science. 2006;313:1901–1902. doi: 10.1126/science.1133760. [DOI] [PubMed] [Google Scholar]
- Groves JT, Parthasarathy R, Forstner MB. Fluorescence imaging of membrane dynamics. Annu. Rev. Biomed. Eng. 2008;10:311–338. doi: 10.1146/annurev.bioeng.10.061807.160431. [DOI] [PubMed] [Google Scholar]
- Ingledew WJ, Poole RK. The respiratory chains of Escherichia coli. Microbiol. Rev. 1984;48:222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Suzuki M, Husimi Y. A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem. Biophys. Res. Commun. 1999;264:556–560. doi: 10.1006/bbrc.1999.1541. [DOI] [PubMed] [Google Scholar]
- Iwata M, Lee Y, Yamashita T, Yagi T, Iwata S, Cameron AD, et al. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc. Natl. Acad. Sci. USA. 2012;109:15247–15252. doi: 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Sheets ED, Simson R. Revisiting the fluid mosaic model of membranes. Science. 1995;268:1441–1442. doi: 10.1126/science.7770769. [DOI] [PubMed] [Google Scholar]
- Johnson AS, van Horck S, Lewis PJ. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology. 2004;150:2815–2824. doi: 10.1099/mic.0.27223-0. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Baracca A, Barbero G, Bergamini C, Dalmonte ME, Del Sole M, et al. Mitochondrial respiratory chain super-complex I-III in physiology and pathology. Biochim. Biophys. Acta. 2010;1797:633–640. doi: 10.1016/j.bbabio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Lenn T, Leake MC, Mullineaux CW. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coli plasma membrane in vivo. Mol. Microbiol. 2008;70:1397–1407. doi: 10.1111/j.1365-2958.2008.06486.x. [DOI] [PubMed] [Google Scholar]
- Magalon A, Arias-Cartin R, Walburger A. Supramolecular organization in prokaryotic respiratory systems. Adv. Microb. Physiol. 2012;61:217–266. doi: 10.1016/B978-0-12-394423-8.00006-8. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol. Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- Meredith DH, Plank M, Lewis PJ. Different patterns of integral membrane protein localization during cell division in Bacillus subtilis. Microbiology. 2008;154:64–71. doi: 10.1099/mic.0.2007/013268-0. [DOI] [PubMed] [Google Scholar]
- Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr. Opin. Biotechnol. 2011;22:117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Mullineaux CW. FRAP analysis of photosynthetic membranes. J. Exp. Bot. 2004;55:1207–1211. doi: 10.1093/jxb/erh106. [DOI] [PubMed] [Google Scholar]
- Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, et al. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One. 2010;5:e11910. doi: 10.1371/journal.pone.0011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebisch A, Bott M. Purification of a cytochrome bcaa3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum. Identification of a fourth subunity of cytochrome aa3 oxidase and mutational analysis of diheme cytochrome c1. J. Biol. Chem. 2003;278:4339–4346. doi: 10.1074/jbc.M210499200. [DOI] [PubMed] [Google Scholar]
- Nübel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9:2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Pohl T, Uhlmann M, Kaufenstein M, Friedrich T. Lambda red-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2007;46:10694–10702. doi: 10.1021/bi701057t. [DOI] [PubMed] [Google Scholar]
- Price CE, Driessen AJM. Biogenesis of membrane bound respiratory complexes in Escherichia coli. Biochim. Biophys. Acta. 2010;1803:748–766. doi: 10.1016/j.bbamcr.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM. Protein localization in Escherichia coli cells: comparison of the cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J. Bacteriol. 2010;192:912–924. doi: 10.1128/JB.00967-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta. 2002;1553:188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- Sousa PM, Silva ST, Hood BL, Charro N, Carita JN, Vaz F, et al. Supramolecular organizations in the aerobic respiratory chain of Escherichia coli. Biochimie. 2011;93:418–425. doi: 10.1016/j.biochi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Sousa PM, Videira MA, Bohn A, Hood BL, Conrads TP, Goulao LF, et al. The aerobic respiratory chain of Escherichia coli: from genes to supercomplexes. Microbiology. 2012;158:2408–2418. doi: 10.1099/mic.0.056531-0. [DOI] [PubMed] [Google Scholar]
- Sousa PM, Videira MA, Melo AM. The formate:oxygen oxidoreductase supercomplex of Escherichia coli aerobic respiratory chain. FEBS Lett. 2013a;587:2559–2564. doi: 10.1016/j.febslet.2013.06.031. [DOI] [PubMed] [Google Scholar]
- Sousa PM, Videira MA, Santos FA, Hood BL, Conrads TP, Melo AM. The bccaa3 supercomplexes from the Gram positive bacterium Bacillus subtilis respiratory chain: a megacomplex organization? Arch. Biochem. Biophys. 2013b;537:153–160. doi: 10.1016/j.abb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Spehr V, Schlitt A, Scheide D, Guénebaut V, Friedrich T. Overexpression of the Escherichia coli nuo-operon and isolation of the overproduced NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 1999;38:16261–16267. doi: 10.1021/bi9919605. [DOI] [PubMed] [Google Scholar]
- Stolpe S, Friedrich T. The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J. Biol. Chem. 2004;279:18377–18383. doi: 10.1074/jbc.M311242200. [DOI] [PubMed] [Google Scholar]
- Stroh A, Anderka O, Pfeiffer K, Yagi T, Finel M, Ludwig B, et al. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 2004;279:5000–5007. doi: 10.1074/jbc.M309505200. [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl Acad. Sci. USA. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- Yu J, Hederstedt L, Piggot PJ. The cytochrome bc complex (menaquinone:cytochrome c reductase) in Bacillus subtilis has a nontraditional subunit organization. J. Bacteriol. 1995;177:6751–6760. doi: 10.1128/jb.177.23.6751-6760.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movies S1-S4. The movies show streams of live cells from strains BW25113 egfp-nuoF (Movie S1), BW25113 mcherry-sdhC (Movie S2), BW25113 cyoA-mcherry (Movie S3) and BW25113 atpB-egfp (Movie S4) obtained by TIRF-microscopy. The streams show the localization of the OXPHOS complexes in clusters that are dynamic over time (Movies S1-S4).
Movies S5-S8. The movies show streams of live cells from strains BW25113 egfp-nuoF (Movie S5), BW25113 mcherry-sdhC (Movie S6), BW25113 cyoA-mcherry (Movie S7) and BW25113 atpB-egfp (Movie S8) obtained by TIRF microscopy in a FRAP experiment. A part of the cell was bleached with a short laser pulse and the recovery of fluorescence in the bleached areas is seen (Movies S5-S8). All OXPHOS complexes rapidly diffuse into the bleached areas with similar kinetics.