SUMMARY
Skeletal muscle contains Pax7-expressing muscle stem or satellite cells, enabling muscle regeneration throughout most of adult life. Here, we demonstrate that induced inactivation of Pax7 in Pax7-expressing cells of adult mice leads to loss of muscle stem cells and reduced heterochromatin condensation in rare surviving satellite cells. Inactivation of Pax7 in Myf5-expressing cells revealed that the majority of adult muscle stem cells originate from myogenic lineages, which express the myogenic regulators Myf5 or MyoD. Likewise, the majority of muscle stem cells are replenished from Myf5-expressing myogenic cells during adult life, and inactivation of Pax7 in Myf5-expressing cells after muscle damage leads to a complete arrest of muscle regeneration. Finally, we demonstrate that a relatively small number of muscle stem cells are sufficient for efficient repair of skeletal muscles. We conclude that Pax7 acts at different levels in a nonhierarchical regulatory network controlling muscle-satellite-cell-mediated muscle regeneration.
INTRODUCTION
Adult skeletal muscle is subject to continuous regeneration, especially after injury or excessive physical training. Muscle regeneration is primarily mediated by a distinct population of muscle-specific adult stem cells known as satellite cells (SCs), which are located between the basal lamina and sarcolemma of muscle fibers (Shi and Garry, 2006). Stem cells express the paired box transcription factor Pax7 (Seale et al., 2000) and are thought to originate from mesodermal cells expressing Pax7 and its paralogue Pax3 during embryogenesis (Kassar-Duchossoy et al., 2005; Relaix et al., 2005). Deletion of Pax7 in mice leads to normal numbers of stem cells at birth followed by excessive wasting of stem cells during the first weeks of postnatal development (Oustanina et al., 2004; Relaix et al., 2006). Emerging evidence indicates that heterogeneity exists within the Pax7-expressing stem cell niche and it was recently postulated that adult stem cells, unlike neonatal muscle progenitor cells, do not require Pax7 either for stem cell maintenance or for regeneration of acutely injured skeletal muscle over a short period (Lepper et al., 2009).
At present the position of Pax7 in the genetic network that directs myogenesis is disputed (Braun and Gautel, 2011). Concomitant genetic inactivation of Pax7 and Pax3 disrupts somite development and myogenesis after E10.5 in mice, although initial formation of the myotome and expression of the myogenic regulatory factor Myf5 is present in Pax3/Pax7 double mutants (Relaix et al., 2005), indicating that early Myf5 activation occurs independently of Pax3/Pax7. Conversely, Myf5/MyoD/Mrf4 compound mutants do not form the myogenic lineage (Rudnicki et al., 1993) and hence fail to express Pax7. On the other hand, there is ample evidence for direct and indirect activation of Myf5 by Pax3 during embryogenesis (reviewed by Braun and Gautel, 2011). Moreover, Pax7 seems to directly regulate Myf5 expression in satellite-cell-derived myoblasts by recruitment of a histone methyltransferase (HMT) complex (McKinnell et al., 2008). Yet, ~10% of Pax7-expressing SCs, which have never expressed Myf5, show a privileged contribution to the SC compartment compared to Pax7-positive, Myf5-expressing cells indicating heterogeneity within the SC population (Kuang et al., 2007).
Critically missing from previous studies is the role of Pax7 in long-term maintenance and expansion of these heterogeneous populations of muscle stem cells defined by Myf5 expression. Here, we show that inactivation of Pax7 during SC proliferation dramatically reduced the number of SCs and prevented muscle regeneration. Our results revealed an essential function of Pax7 in maintenance of heterochromatin and expansion of SCs, giving rise to a new model of the regulatory network between myogenic genes and Pax7 that drives muscle regeneration.
RESULTS
Conditional Deletion of the Pax7 Gene in Adult Skeletal Muscle Leads to Delayed Loss of SCs
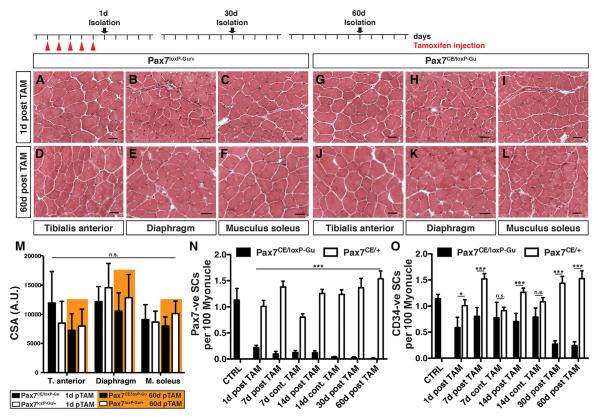
To analyze the role of Pax7 in adult muscle stem cells, we inactivated the Pax7 gene in adult mice by treating 3-month-old Pax7CE/loxP-Gu mice (n = 3) and Pax7loxP-Gu/+ controls (n = 3) with 3 mg tamoxifen (TAM) per 40 g body weight for 5 days (Lepper et al., 2009). The novel conditional Pax7 allele (Pax7loxP-Gu/loxP-Gu), which we used for this experiment, allows Cre-recombinase-mediated deletion of the transcriptional start site and the first three exons, preventing generation of an mRNA from the Pax7 locus (Figures S1A and S1B available online). Notably, generation of CMV-Cre/Pax7loxP-Gu/loxP-Gu mice, in which the Pax7 gene is deleted early during development, fully phenocopied the Pax7 germline knockout (Figure S2) (Oustanina et al., 2004), confirming that Cre-recombinase-mediated recombination generates a null allele. We observed a massive reduction, but not complete loss, of Pax7-positive SCs on isolated myofibers of Pax7CE/loxP-Gu mice already 1 day after the end of the TAM treatment (Figure 1N). Concomitant with the loss of Pax7-positive SCs, we also noted a rapid decline of Pax7 mRNA concentrations in TAM-treated Pax7CE/loxP-Gu mice (Figures S3A and S3F) and a rapid decline of SCs as assessed by the expression of calcitonin receptor (Calcr), a marker of quiescent SCs (Figure S3B) (Fukada et al., 2007). Surprisingly, when we used CD34 expression to assess SCs in these mice, virtually unchanged numbers of CD34-positive SCs were found 1 day (1d), 7d, and 14d after the end of the TAM treatment (Figure 1O). Thus, it appeared that inactivation of Pax7 led to differential changes of the SC signature. Importantly, the number of these Pax7-negative, CD34-positive SCs declined dramatically at 30d and 60d, suggesting that SCs after loss of Pax7 expression maintain some SC features for several weeks, but not indefinitely (Figure 1O; Figure S3F). At all time points TAM treatment did not affect morphological characteristics of skeletal muscles since no centrally localized nuclei indicative of regenerated muscle (Figures 1A–1L) or differences in fiber diameter indicative of muscle wasting (Figure 1M) were observed.
Figure 1. Loss of Muscle Satellite Cells after Postnatal Inactivation of Pax7.
Pax7CE/loxP-Gu and Pax7loxP-Gu/+ littermates (n = 3, in each group) were treated with TAM for 5 days. Muscle tissues were analyzed at different time points as indicated.
(A–L) Hematoxylin/Eosin (HE) staining of muscle tissue sections from TAM-treated Pax7CE/loxP-Gu and Pax7loxP-Gu/+ mice at 1 day (A–C and G–I) and 2 months (D–F and J–L).
(M) Statistical evaluation of cross-sectional areas (CSA) of muscle fibers of TAM-treated Pax7CE/loxP-Gu and Pax7loxP-Gu/+ mice at different time points.
(N and O) Analysis of remaining SC numbers. SCs situated on isolated myotubes (n > 150) were identified by being stained with Pax7 (N) and CD34 (O) antibodies. SC numbers were calculated relative to the number of myonuclei (DAPI).
Error bars represent standard deviation of the mean (t test: n.s. = not significant; ***p < 0.001; *p < 0.05). Scale bar: (A)–(L) = 20 mm. See also Figures S1 and S2.
Pax7 Is Required during SC Expansion to Achieve Efficient Skeletal Muscle Regeneration
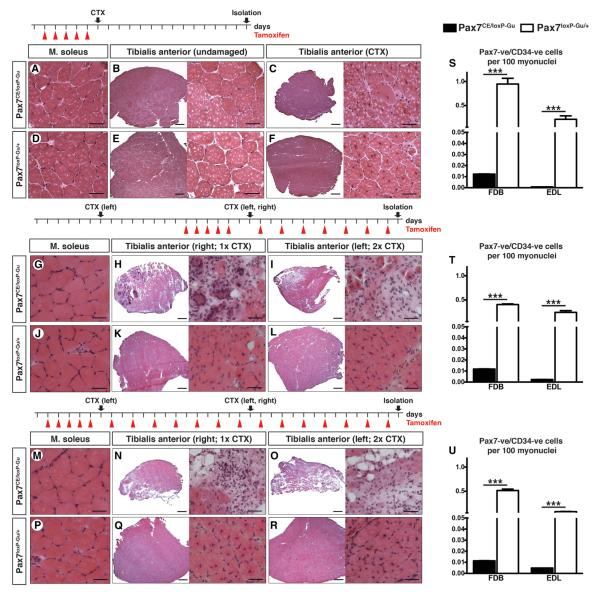
Next, we induced muscle regeneration 1 day after completion of the 5-day TAM regimen by injection of Cardiotoxin (CTX) into the Tibialis anterior (T.A.) muscle. Surprisingly, we detected a significant regenerative response resulting in the formation of multiple regenerated muscle fibers with centrally located nuclei (Figures 2C and 2F) despite a reduction of Pax7-positive SCs in TAM-treated Pax7CE/loxP-Gu mice by 97.80% ± 0.47% (n = 3) 15 days after the end of the regimen (Figure 2S). However, regenerated T.A. of TAM-treated Pax7CE/loxP-Gu mice (Figure 2C) showed a reduction of the total muscle size and of diameters of individual myofibers compared to TAM-treated Pax7loxP-Gu/+ mice and undamaged controls (Figures S3D and S3E).
Figure 2. Impaired Muscle Regeneration in Adult Mice after Enhanced Inactivation of Pax7.
(A–F, G–L, and M–R) Morphological analysis of T.A. muscles of TAM-treated Pax7CE/loxP-Gu and Pax7loxP-Gu/+ mice injected with CTX to induce muscle regeneration. TAM treatment regimens and time points of analysis are indicated at the top of each panel. (A–F) HE staining of T.A. muscles of 12-week-old mice (n = 3) 14 days after CTX injection. Cre-recombinase-mediated inactivation of Pax7 was induced by five consecutive TAM injections. (G–R) HE staining of T.A. muscles of 20-week-old mice (n = 3), which received additional administration of TAM after one (H and K) or two (I and L) rounds of muscle regeneration. TAM was administered before and during the first round of muscle regeneration (G and L) or before and during two rounds of muscle regeneration (M–R). (S–U) Number of Pax7-positive and CD34-positive SC populations on isolated myofibers relative to the number of myonuclei (DAPI). Error bars represent standard deviation of the mean (t test: ***p < 0.001). Scale bar: overview = 200 μm; magnification = 25 μm. See also Figure S3.
To answer the question of whether loss of Pax7 expression during the repair process would further compromise regeneration of skeletal muscle, we modified our experimental conditions as follows: (1) we constantly administered TAM before and during initiation of skeletal muscle injury to stimulate and sustain TAM-dependent recombination and hence deletion of the conditional Pax7 allele (Figures 2G–2L); and (2) we introduced an additional round of CTX injection either before or during TAM administration to further challenge the regenerative response (Figures 2M–2R). Sustained stimulation of Pax7 deletion in 3-month-old mice during muscle regeneration compromised the regenerative capacity more severely compared to mice in which induction of Pax7 deletion was terminated before onset of regeneration (Figures 2A–2F, 2G–2L, and 2M–2R), although no further decline of Pax7-positive SCs occurred (94.70% ± 2.24% reduction after continuous TAM treatment [n = 6] compared to 97.80% ± 0.47% [n = 3] reduction after initial TAM treatment) (Figures 2S, 2T, and 2U and Figures S3C and S4A). We observed a massive increase of both necrotic fibers and fibrotic tissue resulting in a reduction of size of the regenerating T.A. in contrast to control mice (Figures 2H, 2I, 2N, and 2O and Figures S4K–S4P). Similar results were obtained using 6-week-old mice (n = 3) (Figures S4B–S4J). Control experiments without TAM using Pax7loxP-Gu/+ and Pax7CE/loxP-Gu mice revealed that repeated rounds of muscle damage did not affect the number of Pax7-positive SCs without induction of Pax7 deletion (Figure S4Q). Our results demonstrate that continuous expression of Pax7 in SCs during muscle regeneration is essential for efficient muscle repair.
Inactivation of Pax7 in Adult Skeletal Muscle Gives Rise to Rare Atypical SCs with Reduced Heterochromatin Condensation
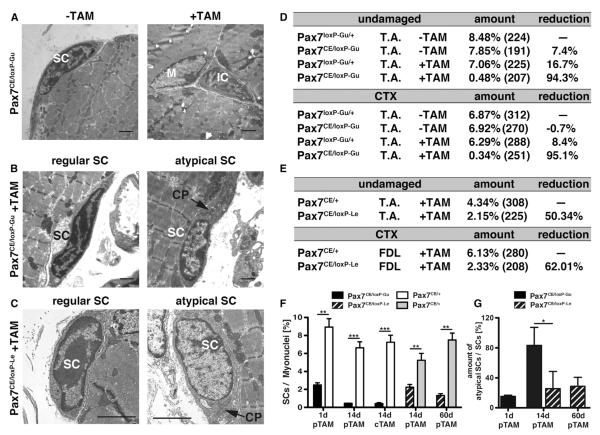
The differential results obtained by using Calcr versus CD34 to assess mutant cells in a short-term assay and the long-term survival of CD34-positive mutant cells suggested that Pax7 maintains an unknown aspect of SC properties. We therefore investigated the ultrastructure of SCs by electron microscopy (EM), which also represents the most reliable method for determining SC numbers. Under physiological conditions SCs can be readily identified as small, heterochromatin-rich cells that reside between the basal lamina and sarcolemma (Figure 3A) (Oustanina et al., 2004; Shi and Garry, 2006). By this criterion, Pax7CE/loxP-Gu mice treated with the extended TAM regimen showed a dramatic reduction of SCs in the T.A. muscle (0.48% out of 207 nuclei) compared to nontreated or TAM-treated WT controls (7.85% out of 191 nuclei and 8.48% out of 224 nuclei, respectively) 14 days after completion of TAM administration (Figures 3D and 3F). Similarly, we found only a very low number of SCs in TAM-treated Pax7CE/loxP-Gu mice in regenerated muscles 14 days after CTX-induced damage (0.34% out of 251 nuclei) (Figures 3D and 3F). We were intrigued to see that the reduction of SCs after Pax7 inactivation as determined by EM corresponded to the remaining number of Pax7-expressing SCs (Figure 1N) (94.3% reduction by EM versus 97.80% reduction by Pax7 immunofluorescence) but not to the number of CD34-positive SCs (Figure 1O). Of note, SCs detected in continuously TAM-treated Pax7CE/loxP-Gu mice lacked characteristic heterochromatin condensations and contained an abnormal amount of cytoplasm and organelles (Figure 3B). Such atypical SCs were not found in WT control mice.
Figure 3. Loss of SCs in Adult Mice Differs between Pax7 Conditional Alleles.
(A–C) EM microphotographs of ultrathin muscle sections used for quantification of SCs in mice in Pax7CE/loxP-Gu allele (A) and Pax7CE/loxP-Le mice (C). (B and C)
The majority of remaining SCs in Pax7CE/loxP-Le mutant mice are localized under the basal lamina as regular SCs but show a strong reduction of heterochromatin
(C). Similar atypical stem cells were also present in Pax7CE/loxP-Gu mice but only shortly after Pax7 inactivation (B).
(D) Number of SCs relative to myonuclei in undamaged and regenerated T.A. muscles of Pax7CE/loxP-Gu and Pax7loxP-Gu/+ mice 14 days after CTX injection. TAM was administered for 5 days before regeneration in the +TAM group. Numbers in brackets indicate the number of counted myonuclei, e.g., only a single SC was detected in TAM-treated Pax7CE/loxP-Gu mutants.
(E) Comparison of the number of SCs 14 days after Pax7 deletion reveals differences in remaining SCs between the Pax7CE/loxP-Gu and the Pax7CE/loxP-Le strains.
(F) Number of SCs identified by EM analysis in Pax7CE/loxP-Gu and Pax7CE/loxP-Le strains at different time points. Numbers include atypical SCs.
(G) Number of atypical SCs identified by reduced heterochromatin content and abnormal amounts of cytoplasm and organelles.
Abbreviations: M, myocyte; SC, satellite cell; IC, interstitial cell; CP, cytoplasm; pTAM, post TAM injection; cTAM, continuous TAM administration. Error bars represent standard deviation of the mean (t test: **p < 0.01; ***p < 0.001). Scale bar: (A)–(C) = 2 μm. See also Figure S4.
Previous studies suggested that inactivation of Pax7 in adult mice does not reduce SC number and has no aberrant effect on muscle regeneration in temporary assays (Lepper et al., 2009). The previously reported conditional Pax7loxP-Le allele (Figure S1C) theoretically allows generation of a truncated transcript by the splicing of exon I to exon III, since only exon II is deleted by Cre recombinase (Lepper et al., 2009). In fact, RT-PCR analysis of the TAM-treated Pax7CE/loxP-Le mutant strain revealed the presence of a truncated Pax7 mRNA in skeletal muscles (Figures S1C and S1D) (Lepper et al., 2009). Furthermore, overexpression of a C- and N-terminal-tagged Pax7 cDNA construct lacking exon II in HEK293T cells yielded a shortened protein, presumably initiated at a downstream ATG and predicted to contain the functional homeodomain, suggesting that the Pax7loxP-Le allele may retain residual biological activity after exon II deletion (Figures S1C–S1E) (Soleimani et al., 2012).
We next investigated the number and morphology of SCs in the Pax7CE/loxP-Le strain by EM instead of using markers. EM analysis unraveled a 50.34% to 62.01% reduction of SCs 14d after TAM administration in TAM-treated Pax7CE/loxP-Le mice (Figures 3E and 3F). Intriguingly, we also detected the same atypical SCs characterized by loss of heterochromatin condensation found in Pax7CE/loxP-Gu mice (Figure 3C). Atypical SCs represent a significant share of remaining SCs (25% of all SCs in Pax7CE/loxP-Le and 83% in Pax7CE/loxP-Gu mice 14d after initial 5d TAM treatment), although absolute numbers are low (0.53% in Pax7CE/loxP-Le and 0.57% in Pax7CE/loxP-Gu mice related to the number of myonuclei; Figure S3G). Therefore, these atypical SCs cannot fully account for the Pax7-negative, CD34-positive SC population, which exists early after Pax7 deletion (Figures 1N and 1O). Sixty days after TAM treatment, the number of SCs had declined further in Pax7CE/loxP-Le mice, resulting in an 82.41% loss of SCs. Interestingly, direct comparison of EM-counted SCs in Pax7CE/loxP-Gu and Pax7CE/loxP-Le strains at 14d (including atypical SCs) revealed significant differences of SC numbers between both strains with a more pronounced loss of SCs in the Pax7CE/loxP-Gu compared to the Pax7loxP-Le strain (94.3% versus 50.34% in undamaged muscle) (Figures 3D–3F). Hence, regardless of which conditional allele was used, EM revealed a consistent and profound change in SC number and property not found by conventional marker analysis.
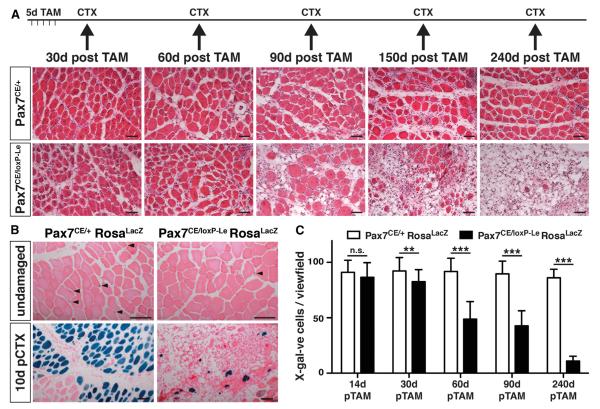
The differential decline of SC numbers in Pax7CE/loxP-Gu versus Pax7CE/loxP-Le suggested that the truncated Pax7 mRNA in Pax7CE/loxP-Le mice exerts a residual biological activity, which suffices to delay loss of SCs. In contrast, the complete loss of Pax7 mRNA expression in Pax7CE/loxP-Gu mice resulted in a more rapid loss of SCs. To determine the critical threshold after which the residual biological activity of the Pax7CE/loxP-Le becomes unable to support efficient generation of new myofibers, we determined the number of Pax7-lineage-traced SCs by X-gal staining in Pax7CE/loxP-Le/Rosa26lacZ mice 14d, 30d, 60d, 90d, and 240d after completion of TAM administration. Interestingly, we detected an increasing loss of SCs, which reached a maximum at 240d (Figure 4C). The decline of SCs at 90d and 240d corresponded to impaired skeletal muscle regeneration at 90d, 150d, and 240d post TAM administration (Figure 4A). Next, we determined whether residual SCs at 240d post TAM administration were still able to contribute to the formation of myofibers, which occasionally formed 10d after CTX-induced muscle injury. LacZ staining revealed very few, thin myotubes in regenerating muscle, which were derived from marked SCs (Figure 4B), reinforcing the concept of an absolute requirement of SCs for skeletal muscle regeneration (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). We concluded that the degree of Pax7 activity critically affects SC function and muscle regeneration and that delayed decline of SCs and/or loss of Pax7 promotes formation of low numbers of atypical SCs with irregular chromatin organization.
Figure 4. Loss of SCs and Impaired Skeletal Muscle Regeneration in Pax7CE/loxP-Le Mice Occurs Late after Cre Recombinase Activation.
(A) HE staining of T.A. muscles at different time points after TAM treatment. CTX injections were performed as indicated following 5 days of TAM administration.
(B) Representative images of HE and X-gal-stained muscles of mice 240 days after Cre recombinase activation and 10 days after CTX injection. The number of remaining lineage-traced SCs (arrowheads in upper panel) and their contribution to muscle regeneration (lower panel) is massively reduced after Pax7 inactivation.
(C) Loss of SCs in Pax7CE/loxP-Le mice becomes apparent only late (60d) after Pax7 inactivation and reached a maximum at 240d.
Error bars represent standard deviation of the mean (t test: n.s. = not significant; **p < 0.01; ***p < 0.001). Scale bar: 50 μm.
Pax7 Stimulates Proliferation of SCs in Culture
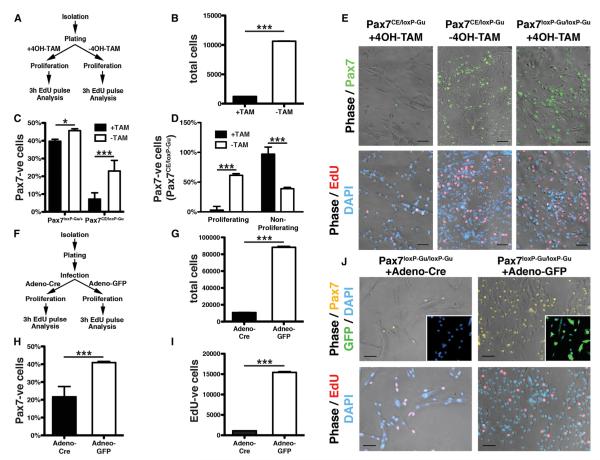
Since the Pax7loxP-Gu allele represented the most efficient means to fully neutralize Pax7 activity, we employed this strain to gain further insight into the role of Pax7 for self-renewal and expansion of SCs. We found that inactivation of Pax7 in isolated SCs (Cerletti et al., 2008) obtained from Pax7CE/loxP-Gu mice by activation of the Cre recombinase via 4OH-TAM or by infection of Pax7loxP-Gu/loxP-Gu cells with Adeno-Cre mimicked the incomplete recombination of Pax7CE/loxP-Gu mice subjected to the 5-day TAM regimen in vivo (Figure 5). Nevertheless, we observed a significant decline of Pax7-positive cells, which went along with a reduction of 5-ethynyl-2′-deoxyuridine (EdU)-incorporating proliferating cells and a corresponding increase of non-proliferating cells (Figures 5B–5D). Consistently, infection of SCs derived from Pax7loxP-Gu/loxP-Gu mice with the Adeno-Cre virus resulted in incomplete deletion of the Pax7 gene within a 5-day period, indicating a relatively sluggish recombination of the Pax7 locus (Figures 5F–5K). Similar to the 4OH-TAM treatment, we found a massive reduction of EdU-incorporating Pax7loxP-Gu/loxP-Gu SCs after their infection with Adeno-Cre compared to an Adeno-eGFP control virus (Figure 5I). Finally, we knocked down Pax7 by RNAi or overexpressed Pax7 in purified wild-type SCs via lentiviral transduction (Figure S5). Expression of the Pax7 shRNA resulted in a rapid and efficient inhibition of Pax7 expression within the 5-day cultivation period, bypassing the delay of Cre-recombinase-mediated deletion. Again, we observed significant reduction of cell cycle entry as indicated by reduced EdU incorporation and increased differentiation of SCs both in the presence and absence of fetal calf serum (Figures S5D–S5G). In contrast, overexpression of Pax7 caused an increase in the number of EdU-incorporating Pax7-positive SCs and a reduction of nonproliferating cells, as well as an impairment of SC differentiation with and without mitogen deprivation (Figures S5D–S5G), which is in line with previous results (Zammit et al., 2006).
Figure 5. Impaired Proliferation of Adult SCs after Inactivation of Pax7 in Culture.
(A–E) FACS-purified SCs from Pax7CE/loxP-Gu mice were treated in culture with or without 4OH-TAM under high serum concentrations (A). Total number of SCs after TAM-induced deletion of Pax7 (B). Number of Pax7-positive cells (C) and EdU-incorporating Pax7-positive cells (D and E) after addition of TAM. Cells were analyzed after 6d of cultivation.
(F–J) Viral-mediated knockout of Pax7 in cultured SCs. FACS-sorted SCs from Pax7loxP-Gu/loxP-Gu mice were infected with Adeno-Cre and Adeno-eGFP viruses 24 hr after isolation to inactivate the Pax7 gene (F). Decreases of the total number of SC-derived cells (G), of Pax7-positive cells (H), and of EdU-incorporating Pax7-positive cells (I and J) 6d after Adeno-Cre mediated deletion of Pax7 are shown.
Error bars represent standard deviation of the mean (t test: *p < 0.05; ***p < 0.001). Scale bar: 20 mm. See also Figures S5 and S6.
To further analyze the potential of Pax7-deficient cells for proliferation and differentiation, we isolated Pax7CE-lineage-traced SCs by FACS from Pax7CE/loxP-Gu/Rosa26YFP and Pax7CE/+/Rosa26YFP mice at different time points after completion of TAM administration. Similar to the in vitro results, recombination was slow, as indicated by the gradual emergence of YFP-expressing cells (Figure S6A). We did not see a decline of permanently labeled Pax7 mutant SCs 1d and 14d after standard TAM treatment, confirming that SCs after deletion of Pax7 survive for several weeks in vivo in skeletal muscle. A decline of Pax7CE/loxP-Gu/Rosa26YFP-lineage-traced cells occurred only after 30d or after extended TAM treatment (Figures S6A and S6B). FACS-isolated Pax7-deficient cells showed a decreased rate of EdU incorporation (Figure S6C) and an enhanced propensity for differentiation (Figure S6D), corroborating the assumption that remaining Pax7 mutant SCs proliferate poorly but survive for several weeks.
Since we did not find any evidence for increased cell death of adult SCs after inactivation of Pax7 in vitro and in vivo but increased differentiation (Figures S5F and S5G and data not shown), we assume that Pax7-deficient, hypoproliferative SCs are mainly removed by differentiation into myocytes. This conclusion is also supported by lineage tracing experiments, which demonstrated differentiation of remaining Pax7 mutant SCs into myotubes in vivo (Figure 4B) and in vitro after FACS sorting (Figure S6D).
Myf5-Cre-Mediated Inactivation of Pax7 Leads to a Postnatal Decline of SC Number and Efficiently Prevents Skeletal Muscle Regeneration
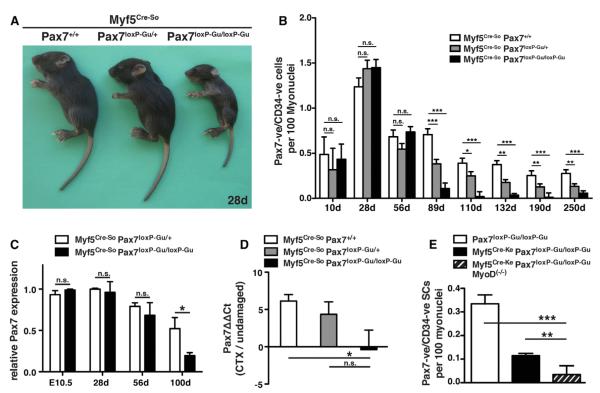
During the course of our experiments, we found that extended activation of Cre recombinase in SCs for deletion of the Pax7 gene affected muscle regeneration to a much greater extent than the short-term 5-day TAM treatment (compare Figure 2A–2F to Figure 2G–2L), although the number of remaining SCs was not reduced any further. Hence, we reasoned that Pax7 does not only control SC self-renewal and maintenance, but must also play a role in expanding muscle stem cells, in which the Myf5 locus is consistently active. To test this hypothesis we used two different Myf5Cre knockin mouse strains (Myf5Cre-So and Myf5Cre-Ke) that enable deletion of Pax7 in Myf5-expressing cells (Haldar et al., 2007; Tallquist et al., 2000). The basic helix loop helix (bHLH) transcription factor Myf5 is the earliest marker of myogenic commitment during embryonic development (Braun and Gautel, 2011). In adults, Myf5 is expressed in the majority of SCs downstream of Pax7 (Kuang et al., 2007) and in proliferating, satellite-cell-derived myoblasts (Cooper et al., 1999; Kuang et al., 2007; Ustanina et al., 2007). In addition, it has been shown that repopulation of the muscle stem cell niche is mainly achieved by a subpopulation of SCs that have never expressed Myf5, further supporting the idea that Myf5 regulates myogenic commitment, but not the self-renewal properties, of muscle stem cells (Kuang et al., 2007). Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice were viable and fertile but consistently smaller compared to Myf5Cre-So/Pax7loxP-Gu/+ littermates (Figure 6A). In situ hybridizations revealed a complete absence of Pax7 expression in somites of Myf5Cre/Pax7loxP-Gu/loxP-Gu embryos at E.10.5 while expression in the neural tube and the head mesenchyme was unaffected (Figure S7).
Figure 6. Loss of SCs and Impairment of Muscle Regeneration after Deletion of Pax7 in Myf5-Expressing Cells.
(A) Growth retardation of 28d Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice compared to Myf5Cre-So/Pax7loxP-Gu/+ and Myf5Cre-So/Pax7+/+ littermates. The growth retardation persisted during postnatal life. (B) Numbers of Pax7-positive/CD34-positive SCs in flexor digitorum brevis (FDB) muscles of Myf5Cre-So/Pax7loxP-Gu/loxP-Gu (n = 3), Myf5Cre-So/Pax7loxP-Gu/+ (n = 3), and Myf5Cre-So/Pax7+/+ (n = 3) at different postnatal stages relative to the total number of myonuclei. A reduction of SCs is evident at 89d and thereafter. (B) Analysis of Pax7 mRNA expression in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice during postnatal life in the T.A. (E). (C and D) Analysis of Pax7 mRNA expression in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice during postnatal life (D) and after CTX-induced regeneration in the T.A. (E). (E) Quantitative analysis of SC numbers in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu/MyoD(−/−) mice (striped) at an age of 100 days. Loss of MyoD results in a further loss of Pax7-positive cells. Error bars represent standard deviation of the mean (t test: n.s. = not significant; *p < 0.05; **p < 0.01; ***p < 0.001).
Surprisingly, we found a normal number of Pax7-positive SCs during the first 10 weeks of postnatal development in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice (n = 3, SCs/100 nuclei, Myf5Cre-So/Pax7loxP-Gu/loxP-Gu 0.433 ± 0.067; Myf5Cre-So/Pax7loxP-Gu/+ 0.319 ± 0.095; Myf5Cre-So/Pax7+/+ 0.487 ± 0.077) (Figure 6B) indicating that these cells were derived from a Myf5-independent myogenic lineage (Gensch et al., 2008; Haldar et al., 2008). Similar results were obtained using the Myf5Cre-Ke strain (Figure 6E). Obviously, the lack of Pax7 deletion in the population of Myf5-negative myogenic progenitor cells allowed their survival and expansion after birth, thereby replacing myogenic cells in which Myf5Cre-So or Myf5Cre-Ke has been activated and hence in which the Pax7 gene was deleted. Such Pax7-positive/Myf5-negative cells will activate the Myf5 locus during later postnatal life to generate Pax7-positive/Myf5-positive SCs (Kuang et al., 2007).
Along this line, Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice older than 56d displayed a decreased number of SCs compared to controls and reduced expression levels of Pax7 mRNA in skeletal muscles (Figures 6B and 6C). Myotubes of Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice only rarely contained Pax7-positive SCs (n = 15, SCs/100 nuclei, Myf5Cre-So/Pax7loxP-Gu/loxP-Gu 0.047 ± 0.017; Myf5Cre-So/Pax7loxP-Gu/+ 0.213 ± 0.048; Myf5Cre-So/Pax7+/+ 0.400 ± 0.08) or CD34-positive SCs (SCs/100 nuclei, Myf5Cre-So/Pax7loxP-Gu/loxP-Gu 0.1 ± 0.03, n = 15; Myf5Cre-So/Pax7loxP-Gu/+ 0.311 ± 0.032, n = 15; Myf5Cre-So/Pax7+/+ 0.474 ± 0.063, n = 13) (Figure 6B), clearly indicating that SCs are replenished by committed, Myf5-expressing cells during later stages of postnatal development. To analyze whether Myf5-negative, MyoD-positive myogenic progenitor cells give rise to Pax7-positive SCs, we generated Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu/MyoD(−/−) mice. Interestingly, we observed a further decline, but not a complete loss, of Pax7-positive SCs in 110d Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu/MyoD(−/−) mice compared to the Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu strain (Figure 6E). From these data we concluded that an embryonic myogenic cell population that depends on MyoD but does not express Myf5 was able to partially compensate for SCs that had expressed Myf5. The presence of Pax7-positive SCs in Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu/MyoD(−/−) mice suggests that a small amount of SCs have never expressed Myf5 or MyoD during development, although both genes seem to be activated in virtually all cells that populate or repopulate the SC niche.
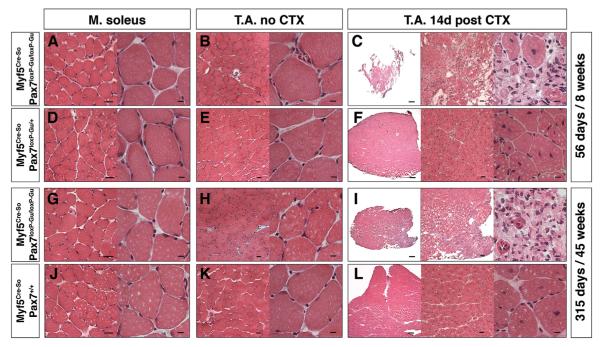
To further investigate the role of Pax7 in expanding, Myf5-expressing SCs, we induced muscle regeneration by CTX injection in 56d, 90d, and 315d Myf5Cre-So/Pax7loxP/loxP-Gu and Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu mice before and after decline of SC numbers. Most significantly, we observed a severe impairment of skeletal muscle regeneration at all stages irrespective of the number of remaining SCs (Figure 7 and data not shown). Only a few thin myotubes emerged in damaged muscles, which were mainly characterized by accumulation of connective tissue, while regeneration occurred normally in Myf5Cre-So/Pax7loxP-Gu/+ mice (Figures 7C, 7F, 7I, and 7J). Expression of Pax7 mRNA was virtually completely lost in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice after induction of muscle injury, corroborating the efficient deletion of the Pax7 gene during amplification of myogenic cells (Figure 6D). Importantly, these data correspond to the results of continuous stimulation of Cre recombinase activity for inactivation of Pax7 using Pax7CE-mice. Taken together, our data reveal that aside from its function in the maintenance of SCs, Pax7 plays an essential role for the expansion of Myf5-expressing myogenic cells, thereby enabling tissue regeneration.
Figure 7. Age-Independent Impairment of Muscle Regeneration after Deletion of Pax7 in Myf5-Expressing Cells.
Morphological analysis of T.A. muscles of Myf5Cre-So/Pax7loxP-Gu/loxP-Gu and Myf5Cre-So/Pax7loxP-Gu/+ mice of different ages with and without CTX injections to induce muscle regeneration.
(A–F) HE staining of T.A. muscles of 8-week-old mice (n = 3) 14 days after CTX injection.
(G–L) HE staining of T.A. muscles of 45-week-old mice (n = 3) 14 days after CTX injection. Myf5Cre-So-mediated deletion of Pax7 resulted in massive impairment of myofiber formation and increased fibrosis in regenerating muscles while no morphological abnormalities are apparent in nondamaged muscles. Cross-sections through nondamaged soleus muscles are shown as control (A, D, G, and J). Scale bar: overview = 20 μm; magnification = 5 μm. See also Figure S7.
DISCUSSION
Here, we describe that Pax7 is necessary for the maintenance of muscle SCs in adult mice and define a crucial function of Pax7 for expansion of SCs after injury. We also demonstrate that replenishment of muscle stem cells from Myf5-expressing myogenic cells requires Pax7, which is compromised but not completely blocked by the absence of MyoD, suggesting that muscle stem cells are partially derived from cells not expressing Myf5 or MyoD during development. Furthermore, we found that a relatively small number of muscle stem cells efficiently regenerated skeletal muscles after inactivation of the Pax7 gene in the majority of SCs.
The efficient regeneration of skeletal muscle after 94.3% reduction of SCs following inactivation of Pax7 was surprising since several previous studies demonstrated that toxin-mediated ablation of the majority of SCs leads to complete failure of muscle regeneration (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). Importantly and in contrast to the studies describing toxin-mediated ablation of Pax7-expressing SCs, here we applied a gene-targeting approach to delete the Pax7 gene. It is conceivable that toxin-expressing SCs are not killed immediately but fuse to other myoblasts, which might potentiate effects in a kind of modified bystander effect that becomes relevant when cells form syncytia (Massuda et al., 1997). Alternatively, the diphtheria-toxin-based approach, but not the gene deletion strategy, might activate processes in muscles that interfere with regeneration. Furthermore, we found that deletion of the Pax7 gene resulted in a gradual loss of SCs while activation of the diphtheria toxin will lead to cell death within a relatively narrow time window, which might prevent adaptive processes in skeletal muscles (Gensch et al., 2008). Finally, it cannot be ruled out that Pax7-negative, CD34-positive SCs initially contribute to the regeneration process, although our finding that continuous stimulation of Pax7 gene deletion before and after injury prevented regeneration argues against such a possibility.
It seems likely that the loss of SCs and the regeneration deficit of adult Pax7 mutant SCs was missed in a previous study (Lepper et al., 2009) due to various reasons: muscle regeneration was assayed within 10d after injury, addressing short-term effects of Pax7 inactivation. Since the loss of SCs was protracted for several weeks in the Pax7loxP-Le strain after completion of TAM treatment, immediate analysis of mutant mice after deletion of Pax7 might have masked the effects of Pax7 on muscle stem cell maintenance and tissue regeneration. In addition, the Pax7loxP-Le allele produces a shortened mRNA that is able, by overexpression in HEK293T cells, to generate a truncated protein lacking the paired-box domain but still containing the homeodomain, which was recently described to play a pivotal role in mediating actions of Pax7 during adult myogenesis (Soleimani et al., 2012). Such a protein might confer a residual biological activity of the Pax7loxP-Le allele responsible for the delayed decline of SCs compared to the Pax7loxP-Gu strain. Yet, Pax7loxP-Le mice eventually also suffer from a dramatic decrease of SCs and impairment of skeletal muscle regeneration, but only 150d to 240d after the initial TAM treatment.
EM-based analyses of SC numbers did not only provide unequivocal evidence for the dramatic reduction of SCs after inactivation of Pax7, but also revealed that remaining Pax7 mutant SCs show reduced heterochromatin condensation. This observation was particularly intriguing since Pax3, a transcription factor closely related to Pax7, was recently described to safeguard heterochromatin integrity in collaboration with another Pax gene family member, Pax9, in mouse embryonic fibroblasts (Bulut-Karslioglu et al., 2012). Interestingly, the authors of the study speculated that other transcription factors such as Pax7 might mediate heterochromatin silencing in muscle cells by recruitment of histone methyltransferases to reiterated transcription factor binding sites within heterochromatic repeat regions, thereby generating H3K9me3 repressive chromatin marks facilitating heterochromatin condensations. So far, mainly HMTs such as the Wdr5-Ash2L-MLL2 complex, which directs activating H3K4 histone modifications, were found to cooperate with Pax7 (McKinnell et al., 2008), but it seems likely that other HMTs such as SUV39H1, Clr4p, DIM-5, Su(var) 3-9 or SETDB1, and G9 (Karimi et al., 2011; Krauss, 2008; Shin-kai and Tachibana, 2011) also interact with Pax7. Future experiments will show how Pax7 specifically affects global chromatin organization in SCs and whether such atypical SCs possess distinct biological properties.
The loss of SCs after inactivation of the Pax7 gene in adult mice raises the question about the underlying mechanism. We and others have proposed that the progressive loss of SCs in newborn constitutive Pax7 mutants is caused by a proliferative defect in SCs (Oustanina et al., 2004; Relaix et al., 2005). This finding was later refined, demonstrating that the absence of Pax7 in newborn mice has effects on the cell cycle and leads to cell death (Relaix et al., 2005), although this conclusion does not necessarily need to hold true for adult SCs. Lastly, lineage tracing of Pax7-positive cells demonstrated that mutant cells are more readily incorporated into myofibers than control cells during the perinatal period (Lepper et al., 2009). In the current study we demonstrated that elimination of Pax7 in adult SCs in vitro and in vivo strongly reduced the number of proliferating SCs. Since we did not find evidence for increased cell death or apoptosis in cultures of Pax7-deficient SCs or in skeletal muscles but detected robust differentiation of mutant SCs into MyHC-positive myotubes, we favor the hypothesis that the loss of Pax7-deficient SCs in adult mice is mainly caused by differentiation, although we cannot rule out an enhanced rate of apoptosis in vivo over an extended time period.
The decline of SC numbers in Myf5Cre-So/Pax7loxP/loxP mice after postnatal day 56 and the massive impairment of skeletal muscle regeneration in these mice indicate that Myf5-positive SCs contribute to Pax7-dependent maintenance and expansion of muscle progenitor cells in adult mice. Most likely, lack of Pax7 in activated SCs and/or satellite-derived myogenic cells leads to cessation of cell proliferation, thereby interfering with SC renewal and expansion, which prevents muscle regeneration. Alternatively, it is possible that the lack of Pax7 expression in Myf5-expressing cells puts a considerable strain on SCs, which do not express Myf5, thereby leading to an exhaustion of this cell population and reduced contribution to SC renewal and skeletal muscle regeneration. While this argument might hold true for physiological aging, it seems less valid for regenerating muscles of Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice, which fail to restore muscle tissue before a drop of SC numbers is apparent (Figure 7). Hence, we believe that one is dealing with two different phenomena in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu mice: (1) continuous decline of SCs during physiological aging due to intermittent activation of the Myf5 locus driving Cre recombinase expression. This will result in ongoing deletion of Pax7 in SCs and continuous decline of SC numbers, which becomes apparent between postnatal days 56 and 89. (2) Massive impairment of SC expansion due to activation of Myf5-Cre expression at early stages of muscle regeneration, resulting in inactivation of the Pax7 gene. The assumption that Pax7 plays a crucial role for the expansion of SCs after injury-induced activation is in line with strong impairment of muscle regeneration after continuous stimulation of Cre recombinase activity, while temporary induction of Cre recombinase activity before muscle injury had only little effects, probably because of compensatory expansion of the remaining residual population of Pax7-positive SCs.
The relatively late decline of SCs in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu and Myf5Cre-Ke/Pax7loxP-Gu/loxP-Gu mice between postnatal days 56 and 89 suggests that Pax7-positive SCs are initially derived from myogenic progenitor cells, which do not express Myf5. Alternatively, it seems possible that a Pax7-positive myogenic progenitor cell population depending on MyoD amplifies and compensates for cells that are derived from Myf5-dependent muscle progenitor cells. In fact, we observed a further reduction of Pax7-positive SCs in Myf5Cre-So/Pax7loxP-Gu/loxP-Gu/MyoD(−/−) compound mutant mice. The lack of the MyoD gene in this strain prevents generation of MyoD-dependent muscle progenitor cells. Since we did not observe a complete loss of Pax7-positive SCs, we concluded that a part of Pax7-positive SCs is derived from myogenic cells, which require neither Myf5 nor MyoD but nevertheless give rise to Pax7-positive SCs, albeit at a lower rate. This conclusion corroborates a previous report describing a cell population that expresses the transcription factors Pax3 and Pax7 and adopts a SC position characteristic of progenitor cells in postnatal muscle (Relaix et al., 2005).
Ultimately, we propose that in aging mice a significant amount of SCs is generated from a pool of amplifying, Myf5-expressing muscle stem cells in a Pax7-dependent manner, contributing to the heterogeneity of the muscle stem cell population. Similar networks that do not depend on the classical hierarchical mode of self-renewal and differentiation but on neutral competition between symmetrically dividing stem cells have recently been described for stem cell maintenance in the mouse intestine (Snippert et al., 2010) and for the self-renewal of radial glia-like neural stem cells (Bonaguidi et al., 2011). The concept of nonhierarchical maintenance and expansion of muscle stem cells revises previous ideas and offers new ways to exploit stem cells for improved regeneration of muscle tissues.
EXPERIMENTAL PROCEDURES
Animals
The strategy to generate the Pax7loxP-Gu allele is described in Figure S1. Gen eration of Pax7CE (Lepper et al., 2009), Rosa26YFP (Srinivas et al., 2001), CMV-Cre (Schwenk et al., 1995), Myf5Cre-So (Tallquist et al., 2000), and Myf5Cre-Ke (Haldar et al., 2007) mice has been described previously. TAM (Sigma) was administered intraperitoneally at 3 mg per 40 g body weight per injection. CTX (0.06 mg/ml, Sigma) was injected into T.A. muscles in a volume of 50 μl. All animal experiments were done in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and according to the regulations issued by the Committee for Animal Rights Protection of the State of Hessen (Regierungspraesidium Darmstadt).
PCR Genotyping, RT-PCR, Immunofluorescence, and Histology
Primers used for genotyping and RT-PCR are listed in the Supplemental Experimental Procedures. T.A. muscles were frozen in isopentane/liquid nitrogen and cryosectioned at 8 μm. Cryosections were counterstained with Hematoxylin/Eosin (Chroma) using established techniques. Antibodies for immunofluorescence staining of cryosections are listed in the Supplemental Experimental Procedures. EdU incorporation was visualized using the Click-iT Imaging Kit (Invitrogen).
EM
Identification of SCs in skeletal muscle by EM was based on cell size, content of heterochromatin, and position to basal lamina and sarcolemma as outlined before (Oustanina et al., 2004).
Cell Culture
FACS-based isolation of SCs was accomplished with a FACSAriaIII (BD Biosciences) using antibodies listed in the Supplemental Experimental Procedures. For FACS-based SC isolation, hind limb and trunk muscles were minced, digested with 100 CU Dispase (BD) and 0.2% type II collagenase (Worthington Biochemicals), and filtered through 100 μm, 70 μm, and 40 μm cell strainers (BD). After lysis of red blood cells by hypotonic shock, the cell suspension was loaded onto a 30% / 70% Percoll (Sigma) gradient. Mononuclear cells were located in the interphase between 30% and 70% Percoll while debris and dead cells were situated on top of the 30% phase. Next, mononuclear cells were washed with PBS, blocked with 1% FCS, and immunostained with fluorescence coupled primary antibodies to exclude CD11b PE-Cy7, CD45 PE, and CD31 PE cells and to include mCadherin APC and CD34 Alexa-450-expressing cells. Unstained cells were used to distinguish autofluorescence background from positive signals. SCs were cultured on Matrigel-coated 96- or 384-well μClear plates (BD Biosciences, Greiner) as described previously (Ustanina et al., 2007) and subjected to immunofluorescence staining. 4OH-TAM (0.4 μM, Calbiochem) was added to the medium after plating of cells and was exchanged daily. EdU (Invitrogen) was added to the culture 3 hr before fixation in a final concentration of 40 μM. Detection of EdU was done with Alexa Fluor 594 (Invitrogen) following standard protocols. Image acquisition and analysis were performed on ImageXpress micro high throughput microscope with MetaXpress software (Molecular Devices).
Muscle Fiber Size and Myotube Analysis
Digital images of Hematoxylin/Eosin-stained sections were processed by ImageJ software (NIH) measuring the area of visually localized fibers (>400 fibers per animal). Myotubes were isolated as described (Oustanina et al., 2004). SCs situated on myotubes were identified by being immunostained with Pax7, CD34, and CalcR antibodes (see Supplemental Experimental Procedures). Statistical analyses were performed using Microsoft Excel and GraphPad Prism software.
Supplementary Material
ACKNOWLEDGMENTS
The authors are indebted to Dr. Frank Constanini, Dr. Charles Keller, and Dr. Phil Soriano for the contribution of mouse strains. T.B. was supported by the DFG (Excellence Cluster Cardio-Pulmonary System [ECCPS], Br1416, and SFB TR81), the LOEWE Center for Cell and Gene Therapy, the German Center for Cardiovascular Research, and the Universities of Giessen and Marburg Lung Center (UGMLC). C.-M.F. is supported by the NIH (RO1AR060042 and R21AR063847). C.L. is supported by DP5OD009208 and R21AR063847).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information for this article includes Supplemental Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2013.07.016.
REFERENCES
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat. Struct. Mol. Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RN, Tajbakhsh S, Mouly V, Cossu G, Buckingham M, Butler-Browne GS. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 1999;112:2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gensch N, Borchardt T, Schneider A, Riethmacher D, Braun T. Different autonomous myogenic cell populations revealed by ablation of Myf5-expressing cells during mouse embryogenesis. Development. 2008;135:1597–1604. doi: 10.1242/dev.019331. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Haldar M, Karan G, Tvrdik P, Capecchi MR. Two cell lineages, myf5 and myf5-independent, participate in mouse skeletal myogenesis. Dev. Cell. 2008;14:437–445. doi: 10.1016/j.devcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes &. Development. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133:93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massuda ES, Dunphy EJ, Redman RA, Schreiber JJ, Nauta LE, Barr FG, Maxwell IH, Cripe TP. Regulated expression of the diphtheria toxin A chain by a tumor-specific chimeric transcription factor results in selective toxicity for alveolar rhabdomyosarcoma cells. Proc. Natl. Acad. Sci. USA. 1997;94:14701–14706. doi: 10.1073/pnas.94.26.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes &. Development. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, Punch VG, Kawabe Y, Jones AE, Palidwor GA, Porter CJ, Cross JW, Carvajal JJ, Kockx CE, van IJcken WF, et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev. Cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellström M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.