Abstract
In the nematode C. elegans, both males and self-fertile hermaphrodites produce sperm. As a result, researchers have been able to use a broad range of genetic and genomic techniques to dissect all aspects of sperm development and function. Their results show that the early stages of spermatogenesis are controlled by transcriptional and translational processes, but later stages are dominated by protein kinases and phosphatases. Once spermatids are produced, they participate in many interactions with other cells — signals from the somatic gonad determine when sperm activate and begin to crawl, signals from the female reproductive tissues guide the sperm, and signals from sperm stimulate oocytes to mature and be ovulated. The sperm also show strong competitive interactions with other sperm and oocytes. Some of the molecules that mediate these processes have conserved functions in animal sperm, others are conserved proteins that have been adapted for new roles in nematode sperm, and some are novel proteins that provide insights into evolutionary change. The advent of new techniques should keep this system on the cutting edge of research in cellular and reproductive biology.
1. Introduction to nematode sperm biology
For most eukaryotes, sexual reproduction is a central part of life. Early in the evolution of animals, this arrangement led to a specialization in gametes — females began producing low numbers of eggs, which are large and require extensive resources, whereas males began producing enormous numbers of sperm [reviewed by 1]. As a consequence, sperm are usually small (so their production requires few resources), motile (so they can find eggs), and prepared for fusion to produce a fertilized embryo [reviewed by 2]. These requirements have shaped two processes — spermatogenesis, which generates spermatids, and spermiogenesis, which activates them to become motile sperm.
In most species, using genetics to dissect these processes is difficult, because mutations that disrupt either one produce sterile males. Fortunately, one of the two sexes in the nematode C. elegans is hermaphroditic [3]. In this species, XX animals make sperm as well as oocytes, and usually reproduce by self-fertilization (Fig. 1A,B). Ward and Miwa showed that this trait simplified the genetic analysis of sperm, since hermaphrodites with defective sperm are self-sterile, but can reproduce when crossed with normal males [Fig. 1A, ref. 4]. This insight has blossomed into an international effort to use nematodes to study all aspects of sperm development and function [reviewed by 5, 6, 7]. Since many features of regulation in the germ line are conserved throughout the animal kingdom [8], these studies illuminate general mechanisms, as well as the evolutionary dynamics of sperm development and function.
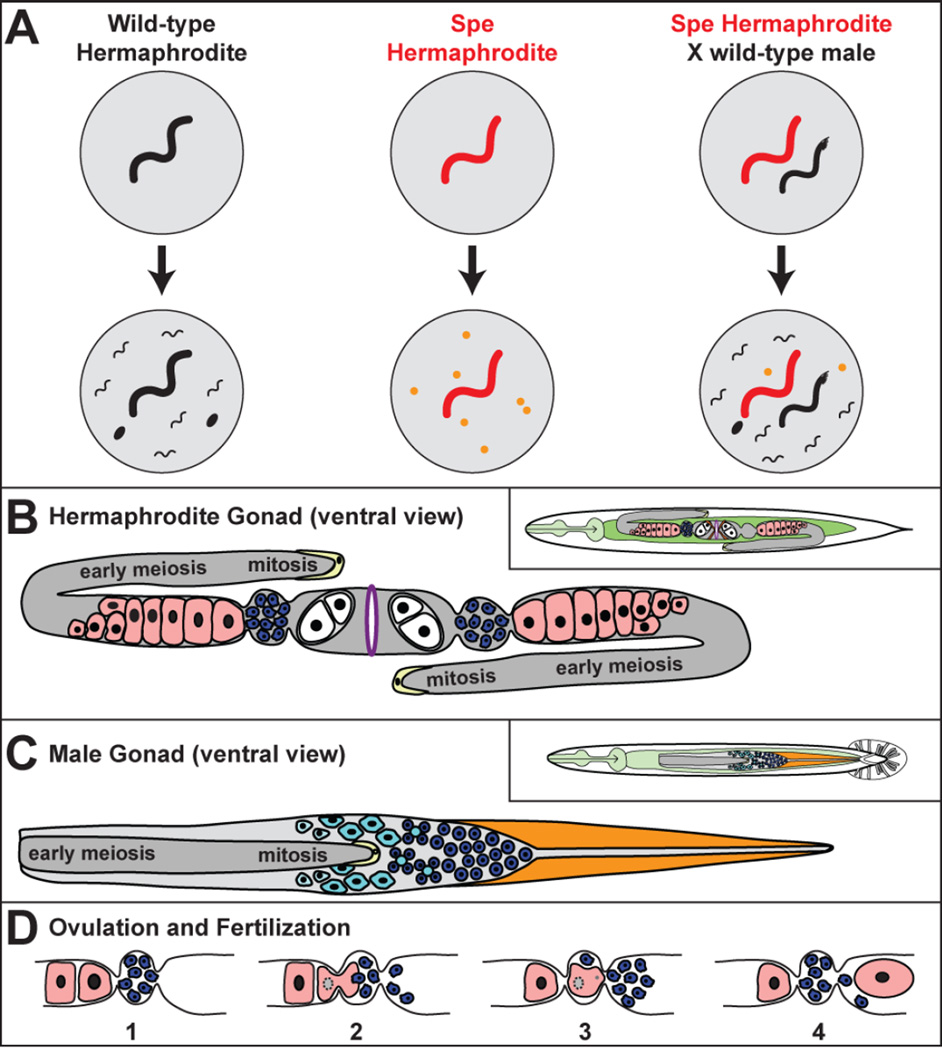
Figure 1. Reproduction in C. elegans.
A. Examples of self and cross fertilization in wild-type and mutant C. elegans. The male has an angular tail. Eggs are black ovals, and unfertilized oocytes are orange circles. B. The hermaphrodite gonad. Oocytes are pink, sperm blue, eggs white, the distal tip cells yellow and the vulva purple. The inset shows the position of the gonad in the body. Cells in mitosis or early meiosis are part of a syncytium. C. The male gonad. Cells in mitosis or early meiosis are part of a syncytium. Primary spermatocytes are light blue cells, residual bodies are light blue circles, and spermatids are dark blue circles. Distal tip cells are yellow, and the vas deferens orange. The inset shows its position in the male body. D. Ovulation and fertilization. (1) The animal is preparing to ovulate a mature oocyte (located on the right). (2) The mature oocyte nucleus undergoes nuclear envelope breakdown. During ovulation, the oocyte enters the spermatheca and is fertilized by a sperm. (3) The resulting zygote pushes many other sperm into the uterus. (4) The zygote commences embryogenesis as it passes into the uterus and sperm crawl back into the spermatheca for the next cycle.
1.1. Studying spermatogenesis in nematodes
In C. elegans males, all germ cells differentiate as sperm. However, in hermaphrodites, the first germ cells to enter meiosis produce sperm, but later ones become oocytes. This transition occurs near the beginning of adulthood. In both sexes, all germ cells in mitosis or the early stages of meiosis are part of a large syncytium (Fig. 1B,C). These immature germ cells look similar in both sexes, and there are no visible dimorphisms at the end of the pachytene stage of meiosis I. However, analyses of gene expression show that the sexual fates of these germ cells were decided earlier, around the time that they entered meiosis [9, 10].
Male germ cells will remain part of the syncytium until shortly before the first meiotic division, when they bud off the central ‘rachis’. Thus, the early stages of spermatogenesis might be coordinately regulated by factors shared in the cytoplasm. Once begun, the meiotic divisions are rapid, and the second one produces a residual body and four round spermatids (Fig 2A). In males, these spermatids remain inactive until ejaculation, whereas in hermaphrodites they remain inactive until they are pushed into the spermatheca during ovulation (Fig. 1D).
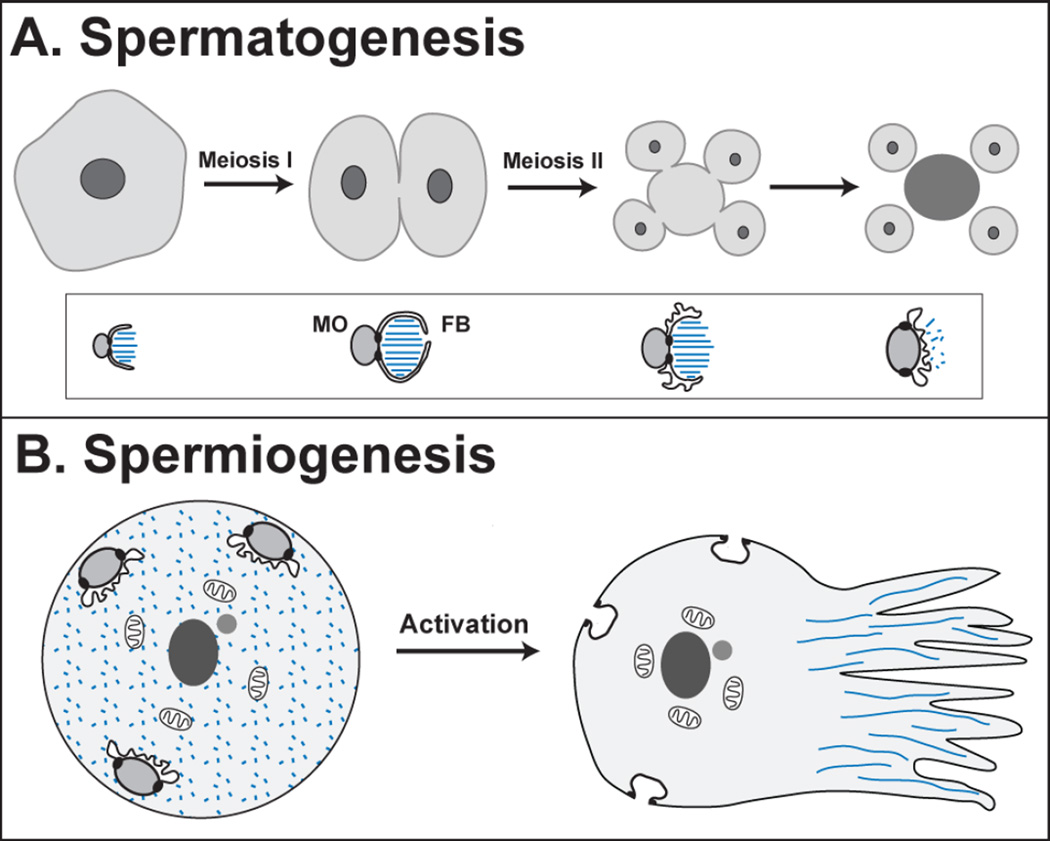
Figure 2. Spermatogenesis and Spermiogenesis.
(A) During spermatogenesis, the primary spermatocyte undergoes two meiotic divisions, producing four spermatids (light gray) and a residual body (dark gray). The pattern of cytokinesis sometimes varies [55]. During this process, the membranous organelles (MOs) localize many sperm proteins, and their associated fibrous bodies (FBs) are made of polymerized major sperm protein (MSP, blue) until its release and depolymerization in the mature spermatid. The differentiation of these organelles is shown in the inset below each stage. (B) During spermiogenesis, a spermatid activates to become a motile sperm. The MOs fuse with the plasma membrane in the head of the sperm, and the MSP forms polymers that organize the pseudopod.
Besides the advantage of hermaphrodite genetics, several other aspects of nematode biology affect the study of sperm. First, C. elegans males have a single X chromosome but no Y. This arrangement makes males easy to produce and maintain. Furthermore, it avoids the challenge of Y-chromosome genetics, which is needed for some genes required for spermatogenesis in fruit flies [11] and humans [12]. Second, nematode sperm migrate towards and maintain their positions within the spermatheca, a part of the female gonad with numerous folds (Fig. 1B, D). In this process, nematode sperm crawl rather than swim, and several methods can induce spermiogenesis and make crawling sperm for in vitro analyses [13]. Third, some biochemical studies are simplified by the fact that the predominant component of nematode sperm is Major Sperm Protein [14], which functions in the cytoskeleton, movement and signaling.
1.2. The sperm/oocyte decision
In Caenorhabditis, germ cells of either sex are capable of undergoing spermatogenesis or oogenesis, and hermaphrodites make both types of gametes. This cell fate decision is under the ultimate control of the sex-determination pathway [reviewed by 9]. This signal transduction pathway controls the activity of tra-1, which encodes a transcription factor of the Gli family of proteins [15]. In germ cells, tra-1 regulates two genes that favor spermatogenesis over oogenesis, fog-1 [16] and fog-3 [17], by directly binding conserved elements in their promoters [18–20]. Normally, a cleaved form of TRA-1 represses these target genes, favoring oogenesis [21], and an uncleaved form works with the Tip60 HAT complex to activate the targets [22].
FOG-1 is a Cytoplasmic Polyadenylation Binding Protein, and probably works by regulating the translation of target mRNAs [19, 23, 24]. FOG-3 is a Tob protein [25], whose activity is partially controlled through phosphorylation [26]. Since both proteins are required to initiate spermatogenesis, they might act together to control key mRNAs, whose identities are unknown.
2. Regulation of spermatogenesis
In most animals, spermatogenesis operates under a major constraint — the chromatin must be reorganized into a compact form for protection. This unique chromatin structure uses special Sperm Nuclear Basic Proteins. For example, DNA in Drosophila sperm is repackaged using protamines and transition proteins [27]. In C. elegans, sperm use SPCH proteins, which resemble some invertebrate protamines, and a histone 2A variant like that found in mammals [28]. These begin to restructure the chromatin during the karyosome stage of meiosis I, just after diplotene [29]. Because this process makes DNA inaccessible, transcriptional regulation is only possible early in spermatogenesis.
2.1. Nematodes use a dedicated ‘sperm program’ to control spermatogenesis
When genes active in spermatogenesis were first cloned, Northern analyses showed that most of them are highly expressed in animals making sperm, but found at undetectable levels in animals making oocytes. Examples include the Major Sperm Protein genes [30, 30], fer-1 [31], spe-4 [32], spe-10 [33], spe-12 [34], and spe-29 [35]. There are a few exceptions to this pattern, like spe-5, which is upregulated during spermatogenesis, but also expressed during oogenesis [36] and in the soma [37].
Identifying global differences in expression was facilitated by mutants that cause XX animals to make only sperm [fem-3(gf), 38] or only oocytes [fem-1(hc17), 39]. By using microarrays to analyze gene expression in these mutants, Reinke et al. showed that transcripts from at least 1343 genes are enriched in animals that are producing sperm [40]. Furthermore, at least 468 of these genes are expressed specifically in sperm [41]. Since almost no genes are expressed during male but not hermaphrodite spermatogenesis [40], both sexes appear to share a conserved ‘sperm program’. However, only a fraction of these genes have associated mutations that disrupt spermatogenesis or sperm function [see below, reviewed by 5].
2.2. The expression of many sperm genes is controlled by transcription factors
How is this ‘sperm program’ controlled? Only 11 transcription factors are expressed specifically in sperm [40], and of these, only spe-44 has been analyzed in detail [42]. It is responsible for the expression of hundreds of other spermatogenesis genes, since mutations in spe-44 prevent the expression of these targets, resulting in defects in many aspects of spermatogenesis. Because spe-44 does not control all sperm-specific transcripts, other transcriptional regulators probably help regulate the spermatogenesis program, but have not yet been identified.
One study suggests that some of these other transcriptional regulators might be hard to find because they have pleiotropic functions. ELT-1 is required for epidermal cell fates [43], but also controls many sperm-specific genes [44]. The expression of elt-1 in the germ line is associated with spermatogenesis, ELT-1 binds a promoter sequence found in many sperm genes, and knocking down elt-1 activity affects sperm function. Thus, ELT-1 helps regulate spermatogenesis, but also has other functions that originally obscured this activity.
Gene expression might also be influenced by genomic organization. As predicted, sperm-specific genes are almost completely absent from the X chromosome, which is only present in a single copy in males [41]. More surprisingly, many sperm genes are found in three large clusters on the autosomes, whereas oocyte genes are not clustered [45]. One possible explanation for this clustering is that it allows these genes to be co-regulated across a large scale. Since some of these clusters do not exist in the related nematode C. briggsae, they must have arisen recently. During evolution, the transition to hermaphroditism favored many large-scale changes in genomic makeup and organization [46].
Although spermatogenesis in fruit flies is also under transcriptional regulation, the factors involved differ from those known to act in worms [27]. The aly class include several components of a testis-specific DREAM complex (whose orthologs regulate vulval development in nematodes), and the can class includes testis-specific paralogs of Transcription Factor IID subunits. Thus, the central role of transcriptional regulation in spermatogenesis has been conserved, but the precise factors vary.
2.3. Translational regulation also plays an important role in spermatogenesis
A large fraction of the genes that regulate germline development control the translation of messenger RNAs, rather than their transcription [reviewed by 47]. For many of these target genes, sequences in their 3’-UTRs control expression [48]. Surprisingly, many sperm genes are an exception to this rule; their 3’-UTRs do not specify sperm-specific expression, whereas their promoters are sufficient for expression during spermatogenesis [48]. These results fit nicely with the studies of spe-44 and elt-1 described above.
However, translational regulation does play a role in spermatogenesis. For example, the Cytoplasmic Polyadenylation Element Binding proteins CPB-1 and CBP-2 are translational regulators that are expressed during spermatogenesis [23]. Furthermore, RNA interference shows that CPB-1 is required for spermatocytes to progress through meiosis, although the function of CPB-2 remains unknown. Similarly, IFE-1 is an isoform of the mRNA cap-binding protein eEIF4E that is enriched in germ cells [49]. Inactivation of ife-1 blocks cytokinesis during spermatogenesis, but has only mild effects on developing oocytes [49, 50]. These effects appear to be mediated through the regulation of gsp-3 transcripts [51].
IFE-1 localizes to complex RNA/protein structures known as P granules, which are analogous to the polar granules or nuage present in the germ line of other animals. In the adult, P granules play a major role in the control of germline mRNAs. The Argonaute proteins ARG-3 and ARG-4 also localize to these P-granules, and regulate the production of 26G-RNAs, which down-regulate many mRNAs during late spermatogenesis [52, 53]. As a result, mutations in arg-3 and arg-4 cause a temperature-sensitive defect in spermatogenesis, which appears to be due to the inability to turn off some messages during differentiation.
2.4. Protein kinases and phosphatases
Many developmental decisions in spermatogenesis occur after the end of transcription, and are not under translational regulation. In fact, mature spermatids lack ribosomes, which segregate to the residual body [54, 55]. Hence, one would expect protein-protein interactions to play a major role in spermatogenesis, and an even larger role in the subsequent process of sperm activation. Indeed, microarray analyses indicate that protein kinases and phosphatases are dramatically over-represented in sperm as opposed to oocytes, whereas transcripts for proteins that control chromatin structure or RNA translation are under-represented [40, 41]. Furthermore, similar patterns are seen in transcripts from the parasitic nematode Ascaris suum [56]. Thus, many aspects of the spermatogenesis and spermiogenesis programs are probably controlled by the phosphorylation state of proteins.
Molecular genetics provides concrete examples of this phenomenon. SPE-6 is a casein kinase that is essential for the segregation of cellular components early in spermatogenesis, for meiotic division, and for the control of sperm activation [57, 58]. SPE-8 is a protein tyrosine kinase that is required for sperm activation in hermaphrodites [59, 60]. The serine/threonine kinase GCK-3 is expressed ubiquitously, but plays a particularly important role in the control of meiotic divisions during spermatogenesis [61]. And the PP1 phosphatases GSP-3 and GSP-4 are required for chromosomal segregation during spermatogenesis and for Major Sperm Protein disassembly during movement [62].
3. Organizing the Sperm Cell
Mature C. elegans sperm are stable, polarized cells. Their pseudopods contain fibers of Major Sperm Protein (MSP), and their cell bodies contain a stereotypical arrangement of organelles and chromatin (Fig. 2). The chromatin and an associated pair of centrioles are located centrally, the mitochondria occupy an intermediate position, and invaginations formed by fused Membranous Organelles (MOs) line the periphery. This arrangement develops in a stepwise fashion during spermatogenesis and spermiogenesis [54, 55, 63], and genetic screens have identified at least 50 genes that define a biological pathway for sperm development [5, 59].
3.1. Meiosis and the chromosomes
A few mutants affect sperm-specific aspects of meiosis. As spermatids form, they bud off of the spermatocyte, leaving behind a residual body containing unneeded structures, such as the meiotic spindle, actin, and ribosomes. In mutants defective for the kelch gene spe-26, these divisions fail, resulting in a cell with multiple chromatin masses [64]. Because sperm contribute centrioles to the zygote, a centriole pair normally remains associated with the haploid chromatin mass after cell division; this process is controlled by the HORMA domain proteins HIM-3 and HTP-1/2 [65]. Finally, specific mutations in a Wee1 homolog, wee-1.3, cause primary spermatocytes to arrest at meiosis I but do not affect other types of cell division, in spite of a general requirement for this protein [66]. As discussed above, the sperm chromatin is eventually reorganized into a compact mass that relies on special basic proteins to pack the DNA [28].
3.2. The membranous organelles and fibrous bodies
The membranous organelles (MOs) play a central role in spermatogenesis, ensuring that specific structures are retained in spermatids and localized correctly within the cell [67]. The MOs derive from the Golgi. In spermatocytes, they localize throughout the cell and are associated with ‘fibrous bodies’ (FBs) composed of MSP arrays bounded by an extension of the MO membrane (Fig. 2A). After the completion of meiosis, the fibrous bodies disassemble, MSP disperses into the cytoplasm of the spermatid, and the MOs move to the periphery near the cell cortex, where they are ready to fuse with the plasma membrane during activation. Several membrane proteins important for fertilization localize to the MOs in spermatids and are distributed to the plasma membrane during activation. Thus, many aspects of MO fusion during activation are analogous to the acrosome reaction [67], which remodels flagellate sperm for interaction with an oocyte.
Several genes important for MO formation have been characterized. Their loss leads to structurally abnormal MOs associated with a variety of defects in the segregation of components between the spermatids and residual body, and some of these mutants arrest as terminal spermatocytes. Most genes in this class encode conserved proteins: the presenilin SPE-4, the palmitoyltransferase SPE-10, the sperm-specific V-ATPase B subunit SPE-5, and SPE-39, a protein that interacts with the HOPS complex in lysosomal trafficking [33, 36, 68–72]. In addition, spe-17 encodes a novel hydrophilic protein [73]. Thus, although the MOs have an unusual structure that is specific to nematode sperm, analysis of this class of mutants has revealed that much of their biology is actually conserved with lysosomes and lysosome-related organelles from other cell types and species.
3.3. The residual body
In the second meiotic division, the spermatids bud off from a central residual body. During this process, many components are sorted into the future spermatids, and others into the residual body to be eliminated. The sorting uses more than one molecular mechanism, since mutants in a class VI myosin fail to deliver mitochondria and fibrous body/MOs to the spermatids, but segregate chromosomes normally [74]. These mutants also fail to remove actin filaments and microtubules from the budding spermatids, so myosin VI controls traffic in both directions.
Once the residual bodies are formed, they need to be phagocytosed and their materials recycled. In the female germ line, apoptosis occurs at a high rate in developing germ cells, and cell corpses are phagocytosed by the surrounding somatic sheath cells. However, the male germ line lacks apoptosis [75]. Two semi-redundant pathways control the engulfment of somatic and germline corpses [reviewed by 76]. Both apoptotic cells and sperm residual bodies display phosphatidylserine on their cell surface [77], which triggers an ‘eat me’ response in cells of the somatic gonad. By contrast, the spermatids lack phosphatidylserine. Thus, the canonical engulfment pathways are used to clear residual bodies during hermaphrodite spermatogenesis. Much of this machinery is also used in males, although the mutations show a novel pattern of genetic interactions, which implies there are differences in the process. Interestingly, engulfment defects lead to reduced fertility in males, possibly by decreasing sperm transfer efficiency.
4. Spermiogenesis
Across species, the acquisition of motility during spermiogenesis and the modulation of motility in response to the environment are key regulatory points for sperm. In nematodes, round spermatids undergo complex morphological rearrangements that result in motile spermatozoa capable of directed migration and fusion with an oocyte. This transformative process is referred to as sperm activation or spermiogenesis. Changes that occur during activation comprise three events that are at least partially separable: fusion of MOs with the plasma membrane, formation of a pseudopod containing polymerized MSP, and sorting of other proteins and organelles to either the cell body or pseudopod (Fig 2). The activated sperm are stably polarized, though the pseudopod may be variably extended depending on the substrate and surrounding pH.
4.1. Induction of spermiogenesis
Sperm activation does not occur automatically at the end of spermatogenesis, but rather is controlled by extracellular signals (Fig. 3). Moreover, activation occurs at a specific time and place for each sex. In hermaphrodites, spermatids activate soon after they are formed, as they are pushed into the spermathecae by the forward movement of oocytes. In males, sperm activation occurs coincident with mating and ejaculation. Activation appears to be a key regulatory point in sperm development: either premature or delayed activation lowers fertility, and separable molecular pathways for activation exist in each sex to promote proper timing.
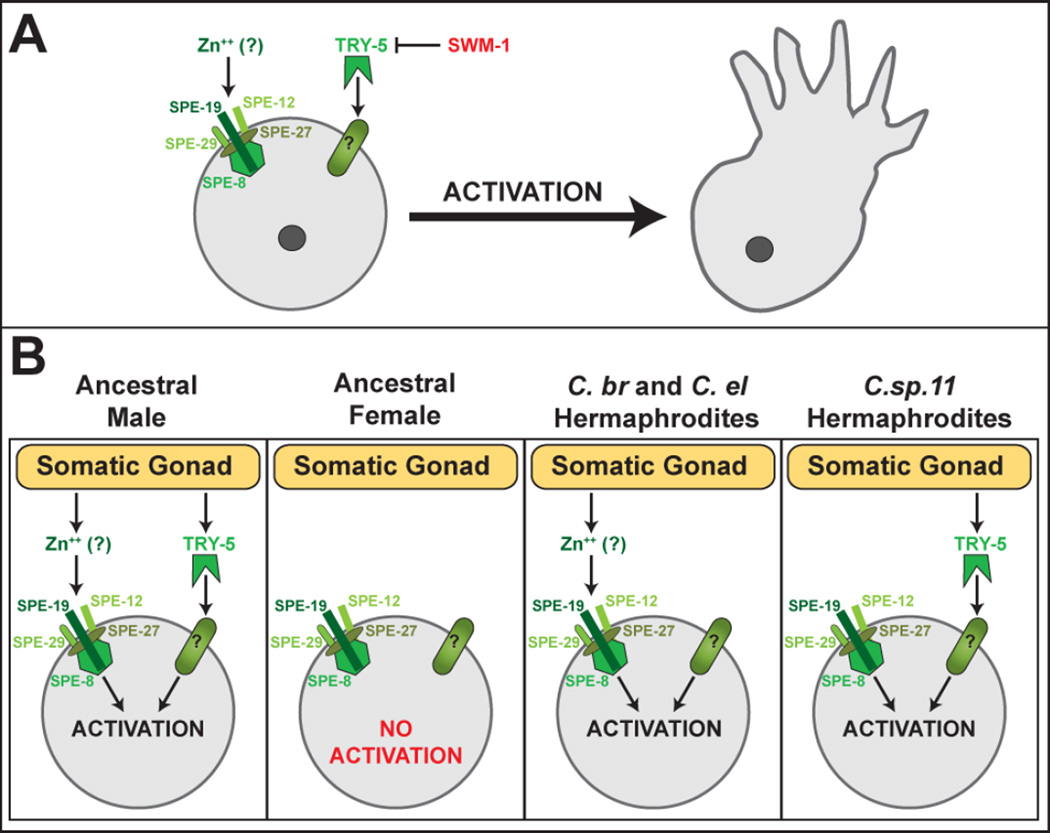
Figure 3. Regulation of Sperm Activation.
(A) Two signal transduction pathways control sperm activation. In one pathway, a signal acts through five proteins of the SPE-8 group. Three of these proteins have transmembrane domains and are depicted crossing the plasma membrane. Studies in C. elegans show that labile Zn++ activates sperm in a process that depends on these genes. In the other pathway, the extracellular protease TRY-5 activates sperm, and is opposed by the protease inhibitor SWM-1. (B) Hermaphrodites use one of the two male pathways. In the ancestor of Caenorhabditis, both sperm activation pathways were used in males, but neither signal was produced by females (Wei et al, in preparation), During the evolution of self-fertility, each species co-opted only one of the two pathways for use in hermaphrodites. TRY-5 is conserved in nematodes, but it is not known if Zn++ acts in sperm activation in other species.
Activation can be elicited in vitro by incubating spermatids in any of a wide variety of compounds, including the protease mixture Pronase, the weak base triethanolamine (TEA), the ionophore monensin, the stilbene chloride channel inhibitor DIDS (4,4’-diisocyanatostilbene-2,2’-disulfonic acid), calmodulin inhibitors, wortmannin, or zinc [13, 63, 78–81]. Studies using these activators have been useful for defining the structural changes that occur and the ionic dependence of the process [63, 82, 83]. The media used for in vitro activation contain only a simple mixture of buffered salts, of which Na+ and K+ are generally required [78]. By contrast, Ca++ is needed inside the cell, but is dispensable in the extracellular medium, and calcium ionophores do not activate sperm [63]. In most cases the mechanism of action of these compounds is unknown. However, they presumably feed into the normal pathways for activation, and this has been confirmed for some compounds. Finally, a rise in intracellular pH is associated with activation by either monensin or TEA, but not Pronase, suggesting that it is not an obligatory feature of the process.
4.2. Sexually dimorphic signals control spermiogenesis
Sexually dimorphic activation was discovered through the analysis of a set of genes referred to as the ‘spe-8 group,’ based on the name of their founding member [13, 59]. These genes are required for hermaphrodite self-fertility, but sperm from mutant males function normally. Moreover, the fertility of some mutant hermaphrodites can be restored by mating with sterile males, which ‘trans-activates’ their own spermatids. Taken together, these results show that hermaphrodites use a spe-8-dependent pathway to activate sperm, and that males have a spe-8-independent pathway. Furthermore, trans-activation implies that the source of male activator is seminal fluid, and that sperm from hermaphrodites are capable of responding to the male signal. Subsequent analyses have shown that sperm from males can be activated via the hermaphrodite pathway as well, and that most if not all components are present in sperm from both sexes.
The five spe-8 group genes, spe-8, spe-12, spe-19, spe-27, and spe-29, all encode proteins expressed in sperm [34, 35, 60, 84, 85]. SPE-12, SPE-19 and SPE-29 have transmembrane domains, and SPE-8 is a non-receptor tyrosine kinase. The working model in the field is that these proteins form a complex at the sperm plasma membrane that receives and transduces a sperm activation signal. Surprisingly, it was recently reported that this signal might be labile extracellular zinc [81]. Zinc activates sperm in vitro and is concentrated within both sperm mitochondria and MOs in a pattern that changes during activation. Furthermore, labile zinc is present in the hermaphrodite and male gonads within secretory cells, placing it in the right place to promote activation. Finally, zinc has no effect on sperm from spe-8 group mutants. However, it is not clear how a zinc signal might act through the putative SPE-8 group complex.
In males, sperm activation is controlled by a protease and its inhibitor, which allows precise regulation of timing. In the gonad, spermatids are prevented from activating by SWM-1, a secreted protein with dual trypsin inhibitor-like (TIL) domains [86]. SWM-1 appears to function within the male gonad to block the activity of TRY-5, a serine protease that is transferred in seminal fluid during mating [87]. Since TRY-5 is a protease, and proteases can activate sperm in vitro, it might work by cleaving proteins on the surface of spermatids, though definitive targets have not yet been identified. One candidate is SNF-10, an SLC6-family transporter protein that is required for spermatids to respond to male activator (Fenker et al., submitted). Additional genes in the male pathway have been identified in genetic screens but remain to be analyzed (G.M.S., unpublished). The activation of nematode spermatids by proteases is conserved, since it has also been observed in the distant relative Ascaris suum [88, 89]
4.3. Downstream components of the sperm activation pathway
Some defects in spermatogenesis lead to the premature activation of sperm. This was first revealed by the analysis of suppressors that restored fertility to the spe-8 group mutant spe-27, while also causing premature sperm activation in males. These mutations turned out to be partial loss-of-function alleles of spe-6, a casein kinase 1 gene required for spermatogenesis and the localization of MSP to the Fibrous Body-MOs [57, 58, 70]. Thus, SPE-6 was suspected to have a secondary function as a ‘brake’ that prevented sperm activation. However, two additional genes, spe-4 and spe-46, were found to have dual spermatogenesis and activation phenotypes [32, 90, 91], suggesting an alternate model whereby defects in MO formation uncouple activation from signaling. MO morphogenesis defects might be associated with the mislocalization of proteins or ions (like zinc), leading to an inability to delay activation. In other words, the ‘brake’ might be a properly organized spermatid rather than a specific protein.
In C. elegans, FER-1 is associated with the MOs and is required for them to fuse with the plasma membrane [31, 55, 83]; although fer-1 mutant spermatids activate, they have short pseudopods. FER-1 is related to dysferlin, which mediates membrane fusion events in muscle development, muscle repair, and synaptic transmission in the mammalian auditory system [92].
Finally, several other proteins and pathways have been implicated in the activation process. Two groups reported that the spermatid membrane harbors cholesterol microdomains that relocalize during activation; however, cholesterol depletion experiments had conflicting results, in one case enhancing and in the other case reducing activation in vitro [93, 94]. Loss of cil-1, a phosphoinositide 5-phosphatase, leads to reduced sperm activation, as well as a reduction in pseudopod length [80]. Finally, experiments with chemical inhibitors and activators of the MAPK pathway and calcium signaling implicate both processes in sperm activation [95].
4.4. Seminal fluid
In most species, male ejaculate not only contains sperm, but also seminal fluid, which promotes reproductive success through its effects on sperm and on the female reproductive system [reviewed by 96]. Although C. elegans males produce seminal fluid, it is dispensable for basal fertility, since sperm transferred by artificial insemination are able to fertilize eggs and outcompete sperm from hermaphrodites [97]. To date, two protein components of seminal fluid have been identified: TRY-5, the putative male activator [87], and PLG-1, a mucin required for the deposition of a mating plug over the hermaphrodite vulva [98, 99].
Although plugging is a widespread phenotype among wild isolates of C. elegans, the standard laboratory strain, N2 Bristol, lacks plg-1 activity. The purpose of the plug remains controversial, since a plug only slightly deters subsequent mating in lab assays [100]. Crosses between different C. elegans strains revealed that lines containing mostly Bristol DNA, with the exception of the plg-1 region, have a weaker plugging phenotype than other wild strains, providing direct evidence that additional seminal fluid components affect this trait. Genomic data indicate that 20% of putative secreted proteins are upregulated in males relative to hermaphrodites [101], and many of these could be seminal fluid factors. Finally, the release of seminal fluid by the male gonad is surprisingly complex. TRY-5 and PLG-1 are expressed in overlapping sets of secretory cells, which release their contents during discrete steps of the copulation program, suggesting a specialization of functions within the male gonad. However, much remains to be discovered about the role of seminal fluid in C. elegans sperm biology. Furthermore, hermaphrodites might have evolved to provide some of these functions within their own gonads, as they appear to have done with the sperm activation pathways.
5. Regulation of Motility
Nematode sperm differ in their motility from the sperm of many other animals. The mature sperm lack flagella and do not swim; rather, they have a pseudopod and move by crawling. Furthermore, they have almost no actin or myosin [102]. Instead, their cytoskeleton is composed of major sperm protein (MSP), a 14 kDa molecule completely unrelated to actin [103, 104] but capable of forming nonpolar fibers and higher-order structures that can drive lamellopodium-like protrusion and cell locomotion through their assembly and disassembly [105].
Most of our information about MSP-based motility comes from studies of the parasitic nematode Ascaris suum. Ascaris is much larger than C. elegans, providing a more-tractable source of material for in vitro studies of MSP assembly, and larger sperm cells that simplify analyzing locomotion. Like actin, MSP is dynamically regulated to achieve cell movement. The polymerization of MSP at the leading edge of the cell drives protrusion, and fiber disassembly and recycling of subunits at the cell body-pseudopod junction drives retraction. Polymerization is regulated by MPOP, a protein localized to the leading edge [106]. The MSP-nucleating activity of MPOP depends on a pH-sensitive tyrosine phosphorylation, mediated by an unknown soluble protein. In addition, membrane tension also influences polymerization rates [107]. Finally, two proteins, MFP1 and MFP2, can associate directly with the MSP cytoskeleton but have opposing effects on assembly [108].
Retraction is driven by fiber rearrangements and disassembly at the rear of the pseudopod [109, 110]. In C. elegans, a PP1 phosphatase, GSP-3/4, localizes to this region and is required for MSP dynamics [62]. Taken together, the number of accessory proteins involved in MSP-based cell motility is small compared to actin, and this simplification may be an advantage for this mode of motility. MSP is intriguing from both a cell biological and an evolutionary standpoint, since MSP and actin represent convergent solutions for powering cell motility.
6. Communication with female cells
Much of the C. elegans lifestyle is geared toward reproduction. Thus, it makes sense that a robust communication network exists between sperm and the female to ensure that resources are only invested in gamete production when offspring can result (Fig. 4). In one direction, signals from the female gonad and germ line attract sperm toward mature oocytes [111]. In the other direction, signals from sperm increase the rate of oocyte maturation, ovulation, and egg laying [111, 112]. This bidirectional communication links oocyte production to the availability of sperm. Communication between oocytes and the somatic gonad further ensures that the different female processes are coupled to one another [112]. For excellent in-depth reviews of these processes, see [113, 114].
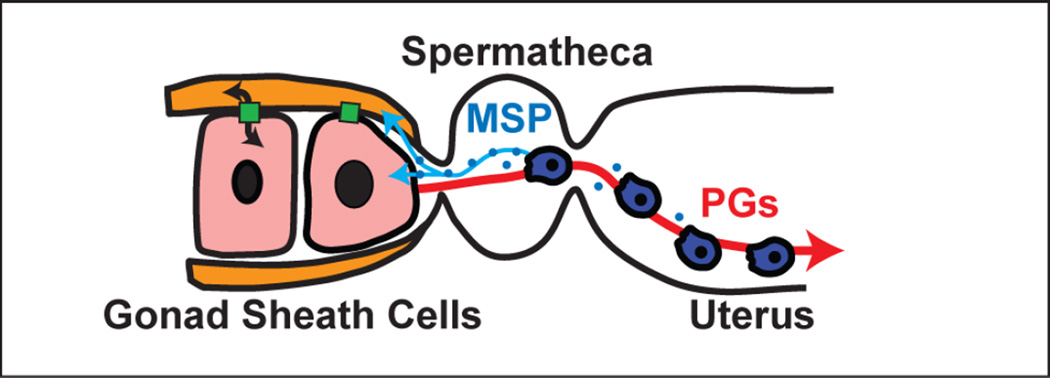
Figure 4. Sperm communication.
The sperm release small vesicles containing Major Sperm Protein (MSP, blue circles), which signals to the oocytes and the somatic gonad to promote maturation and ovulation (light blue arrows). The oocytes release F-series prostaglandins (PGs) that provide guidance cues to direct sperm migration to the spermatheca (red arrow). Oocytes and sheath cells also exchange information (black arrows) through gap junctions (green rectangles) that couples processes in the somatic gonad to those in the oocytes.
6.1. Signaling from the hermaphrodite to sperm: guidance cues
How do C. elegans sperm know which way to go? As in a number of other systems, they appear to be guided by chemoattractive molecules. In wild-type worms, sperm migrate rapidly and directionally toward the two spermathecae. However, in hermaphrodites that lack oocytes, sperm movements are slow and adirectional, which is consistent with the absence of signals that depend on the female germ line [115]. Furthermore, the production of guidance signals is dependent on the nutritional status of the worm, as determined by state of the insulin signaling pathway in intestinal cells. Mutants have been used to define the cellular source of this signal and to identify pathways required for its production, and biochemical approaches have been used to define precise components [115–117]. When food is abundant, intestinal cells produce and secrete yolk, a mixture of vitellogenins and lipids [118, 119]. Oocytes take up yolk and utilize its polyunsaturated fatty acid (PUFA) components to generate F-series prostaglandins (PGs), which serve as a guidance signal. Mutations that block PUFA synthesis or transport to the oocytes eliminate this signal, affecting sperm guidance. This phenotype can be rescued by dietary PUFA supplementation or by direct injection of the relevant PG species into the gonad.
Although this pathway provides a satisfying model for sperm guidance, some complexities exist. Gap junction-mediated interactions between oocytes and the somatic gonad also promote sperm guidance through a step downstream of PG synthesis; this process could involve transport or activity [120]. It depends on the gap-junction proteins INX-14, INX-8, and INX-9, which function in transcriptionally active, immature oocytes that are coupled to gonad sheath cells. Thus, oocytes may become competent to activate or release sperm guidance cues during their development. Similar gap junctions formed in the proximal gonad function in the sperm-sensing pathway described below.
Little is known about the machinery that responds to the prostaglandin signal in sperm. A Srb class G protein-coupled chemoreceptor, srb-13, is a candidate to act as a receptor in sperm [116]. Other sperm genes involved in guidance have not been identified, and it is not known how G protein-coupled receptors (or any other receptor types) modulate the MSP cytoskeleton to effect directional motility.
6.2. Signaling from sperm to the oocyte
During oogenesis, C. elegans oocytes arrest at prophase of meiosis I. Upon release from this arrest, they mature by activating MAPK pathways and undergoing cortical rearrangements, nuclear envelope breakdown, and other processes that prepare them for fertilization [reviewed by 114]. These oocytes are surrounded by somatic gonadal sheath cells, which signal to promote maturation and contract to move oocytes through the gonad. When no sperm are present, producing and laying unfertilized oocytes would be wasteful. Thus, hermaphrodites actively suppress ovulation and associated processes in the absence of sperm. Sperm signal their presence directly to oocytes and indirectly via the sheath cells to promote oocyte maturation and sheath cell contractions. This process does not require sperm to be active or capable of fertilization, since many spe mutants lay unfertilized oocytes [59]. The proteins involved in each of these effects overlap, leading to a complex signaling network that is outlined below [reviewed by 113].
Sperm communicate their presence by releasing a soluble, hormonal signal. Using biochemical purification coupled with an injection-based bioassay, the active molecule was shown to be MSP, the same protein that composes the sperm cytoskeleton [121]. Extracellular MSP directly interacts with both oocytes and the somatic gonad, and distinct regions of MSP are sufficient for mediating its effects on these two tissues. Interestingly, MSP lacks a secretion signal and is released by sperm in novel double-membrane vesicles; so far, the mechanism of secretion is not understood, but might involve the protrusive activity of MSP itself [122].
MSP transduces signals via an incompletely understood mechanism involving multiple cell surface receptors and intracellular pathways. In oocytes, MSP directly binds to VAB-1, an ephrin receptor [123, 124]. In the absence of sperm, VAB-1 inhibits oocyte maturation through a process involving the ephrin EFN-2 and DAF-18/PTEN [123, 125]. During this process, DAB-1/disabled and RAN-1 mediate the localization of VAB-1 to the endocytic recycling compartment [126]. However, if sperm are present, MSP binds VAB-1, which alters its trafficking and augments MAPK activity [127]. VAB-1 functionally interacts with multiple intracellular Ca++ regulators [127], but how these interactions couple to the MAPK cascade is not well understood. Components of a conventional receptor tyrosine kinase (RTK) pathway, including the Shp protein tyrosine phosphatase PTP-2, are essential for MAPK activation [128]. However, this unidentified RTK is not likely to be VAB-1. Genetic screens also identified other proteins that function with VAB-1 to block oocyte maturation, including VAV-1 (a vav-family GEF), PKC-1 (a protein kinase C), and PQN-19 (STAM) [124].
In sheath cells, a parallel pathway also regulates oocyte maturation. Here, MSP signaling counteracts an inhibitory signal that depends on the POU-class homeoprotein CEH-18, which is required for sheath cell differentiation and gap junction assembly [129]. In sheath cells, the identity of the MSP receptor is not known, but its effects are mediated through a Gs/adenylate cyclase pathway involving GSA-1 and KIN-2 [124, 130]. Finally, the sheath cell signals the oocyte to mature through gap junctions that comprise the oocyte innexins INX-14 and INX-22, which inhibit MAPK signaling in the absence of sperm [131]. An early growth response factor protein, EGRH-1, helps inhibit oocyte maturation in the absence of sperm [132], but its relationship to the CEH-18 pathway is unknown.
6.3. Signaling from sperm to the somatic gonad
Sheath cell contractions are necessary for oocytes to move through the gonad. These contractions are elicited by the oocyte itself, through the release of the EGF protein LIN-3 [112, 133], as well as by MSP binding to VAB-1 on sheath cells [123]. Furthermore, the EGL-30 Gq alpha subunit, the PLC-3 phospholipase C, and ITR-1 are all required for an increased contraction rate, but GSA-1 and ACY-4 are not [130]. In the absence of sperm, gap junctions with the oocyte, defined by INX-14, negatively regulate sheath cell contraction [131].
Sperm have additional effects on the hermaphrodite germline. MSP promotes oogenesis by increasing the rate of cytoplasmic flow from the syncytial gonad into developing oocytes, a process required for oocyte growth; this activity is mediated by the Gs/adenylate cyclase pathway in sheath cells [130]. The rate of egg laying is also positively regulated by sperm, but it is not clear whether this phenomenon is also dependent on MSP, or which cells in the nervous system or musculature are responsive [134]. Thus, nothing is left to chance: nearly every process of hermaphrodite reproduction appears to be modulated by the presence of sperm inside the reproductive tract. Moreover, the predominant components of the sperm (MSP) and oocyte (yolk) were recruited for use in these signaling processes.
6.4. Broader effects of sperm signals
Hermaphrodites also interact with males in antagonistic ways that affect their physiology outside of the gonad, and may limit rather than promote reproductive fitness of one of the sexes. Although hermaphrodites largely play a passive role in copulation, they can influence the probability of mating by emitting pheromones to which males respond with positive chemotaxis and copulatory behaviors [135–137]. One component of this ‘come hither’ signal is dependent on the presence of sperm, since hermaphrodites lacking sperm are more attractive to males [138]. This effect requires activated sperm and depends on signals mediated by ceh-18, but not vab-1. In addition, sperm-depleted hermaphrodites are more receptive towards males [139].
7. Formation of the embryo
Each sperm not only delivers a haploid genome to an oocyte, but also plays a critical role in the initiation of embryonic development. These functions depend on proteins that recognize the oocyte during fertilization, proteins that initiate appropriate changes in the sperm, and factors that contribute directly to the new embryo, like the two centrioles.
7.1. Fertilization
During fertilization, proteins on the surface of the sperm must interact with others on the surface of the oocyte, triggering a response in both gametes that leads to fusion and embryogenesis. In nematodes, the sperm proteins that carry out these functions are members of the spe-9 class, which is named after its founding member [reviewed by 140]. Mutations in this group of spe genes share a specific set of phenotypes — the spermatids appear normal, they signal to oocytes and activate at the appropriate time, the mature sperm move normally and compete for positions in the spermatheca, but the sperm cannot fertilize oocytes.
One might expect these proteins would fall into two broad groups — those that interact with the oocyte, and those that mediate the response within sperm. Both SPE-9 and SPE-38 are excellent candidates for the first role. Each is a transmembrane protein, and SPE-9 has extracellular EGF repeats that are critical for fertilization [141, 142]. Furthermore, both proteins localize to the pseudopod in activated sperm [143, 144]. Since the pseudopod is the first part of the sperm to make contact with the oocyte, these proteins might interact with surface proteins on the oocyte at the beginning of fertilization.
By contrast, TRP-3, also known as SPE-41, appears to mediate the response to the oocyte. TRP-3 is a calcium-permeable cation channel that is initially found in the membranous organelles, but which is redistributed over the entire surface of the sperm during activation [145]. This redistribution depends on SPE-38, although SPE-38’s localization does not require TRP-3 [146]. During fertilization, TRP-3 controls a Ca++ influx, which is likely to trigger major changes in the sperm. Since oocytes appear to bind trp-3 sperm without fusing, one possibility is that TRP-3 controls fusion [145].
Other genes of the spe-9 class are still being investigated. Although spe-42 encodes a protein with a transmembrane domain and a C-terminal RING domain that is essential for fertilization, its localization is not yet known [147, 148]. Three additional genes are known by mutations but not yet characterized molecularly — spe-13, fer-14 [59, 149] and spe-36 (D. Shakes and A. Singson, pers. comm.). Finally, fer-7 might also belong to this group [150].
Many proteins involved in sex evolve at a rapid pace, and discerning broad patterns of conservation can be challenging. Thus, it is not surprising that SPE-9 and SPE-38 appear unique to nematodes. By contrast, there are homologs of SPE-42 in most animals [147], and one of these controls fertilization in Drosophila [151]. However, C. briggsae spe-42 cannot substitute for C. elegans spe-42 in transgenic animals [148], so its functional domains might be evolving rapidly. In addition, TRP-3 is a member of the conserved family of TRPC channels [145], but it is not known if it plays a conserved role in fertilization.
7.2. Embryogenesis
Most aspects of early development are controlled by factors present in the oocyte, such as the initiation of embryogenesis, which depends on EGG proteins in nematodes [reviewed by 152]. However, some sperm factors also play critical roles in early development.
The entry point of the sperm itself determines the future antero-posterior axis, since the most posterior point is marked by the sperm pronucleus/centrosome complex [153]. This process depends on SPD-2 [154], which plays a critical role in centrosome duplication [155] and is required for rapid assembly of the sperm aster. Thus, the centrioles that are provided by the sperm not only permit future cell division, but also help determine the embryo’s body plan. The sperm also supplies CYK-4, a rho-guanosine triphosphatase activating protein that works with ECT-2 to regulate RHO-1 and alter the actomyosin gradient in the new embryo, helping to establish the antero-posterior axis [156]. Orthologs of CYK-4 are found in the sperm of many other animals.
At least one other sperm factor is critical early in embryogenesis — spe-11 mutations cause paternal effect lethality [59, 157]. These mutant embryos have problems with meiosis, which are in part caused by a failure in cytokinesis [157, 158]. Furthermore, the spe-11 embryos form weak eggshells [157] that fail to prevent polyspermy [159]. Although spe-11 is normally required only in sperm, its ectopic expression in oocytes is sufficient for normal development, which proves that it functions in the embryo [160]. Since SPE-11 is novel and only found in Caenorhabditis nematodes, it might have evolved this function recently.
7.3. Anucleate sperm
Defects in chromatin repackaging are associated with reduced fertility in mammals and sensitivity to DNA damage in Drosophila [reviewed by 161]. However, in C. elegans, sperm chromatin is not required to initiate embryogenesis. Using temperature-sensitive mutations in genes required for chromosome segregation, Sadler et al. [162] created sperm completely lacking nuclear DNA. Not only were these anucleate sperm capable of fertilizing oocytes, they showed normal male precedence in crosses to hermaphrodites, by blocking the production of self progeny (see below). Furthermore, many oocytes fertilized by anucleate sperm were able to complete meiosis, initiate development and establish normal antero-posterior polarity. Thus, chromatin is not needed for sperm function, once gene expression ends in the spermatocyte.
7.4. Paternal toxins
In C. elegans, some strains carry an ancient pair of genes, zeel-1 and peel-1, whereas others lack both genes [163]. This polymorphism is maintained by paternal effect lethality — embryos lacking zeel-1 activity die if fertilized by sperm that contain PEEL-1 [164]. The PEEL-1 toxin is a transmembrane protein that is initially found in the membranous organelles, but relocalizes to the surface of the sperm opposite the pseudopod during activation. Surprisingly, the PEEL-1 toxin acts late in embryogenesis, and its effects depend on the dose delivered by the sperm. Thus, sperm proteins are capable of influencing late stages of development, and are not restricted to regulating the first cell divisions. Furthermore, even small doses of some sperm proteins can have a potent effect on the embryo, despite the fact that the sperm only contributes about 1/200th of the surface area of the fertilized oocyte. Perhaps other paternal effect genes that act later in embryogenesis remain to be discovered.
8. Germline warfare
Sexual competition plays a major role in evolution [165], so gametes are selected not only for their ability to fuse and form embryos, but also to compete with each other.
8.1. Sperm competition
The male/hermaphrodite reproductive strategy employed by C. elegans has implications for many aspects of sperm biology. In the absence of mating, the hermaphrodite’s own sperm are used with unusually high efficiency: essentially every self sperm produced goes on to fertilize an oocyte. If mating occurs, sperm from males compete with these self sperm for access to oocytes. As in other species with a high risk of sperm competition, this situation places selective pressure on sperm [discussed by 166], especially those of males, who produce no other types of gametes. Perhaps as a result, C. elegans exhibits a very strong male-hermaphrodite sperm precedence order: if mating occurs and sperm are transferred, sperm from males preferentially fertilize oocytes, to the extent that many crosses result in the exclusive generation of male-sired offspring.
Using a variety of methods to control when and how sperm compete with one another, several mechanisms for male precedence have been excluded [97, 167–169]. First, although males transfer more sperm than the hermaphrodite produces, their competitive advantage is largely independent of sperm number. Second, artificial insemination experiments demonstrated that neither seminal fluid nor the mode of activation is important. Third, sperm transferred from one hermaphrodite to another by artificial insemination or sperm from sequential male matings do not show precedence, so the order of introduction into the hermaphrodite reproductive tract does not matter. Finally, even fertilization is not required: motile but fertilization-incompetent sperm from males still outcompete self sperm, though this results in sterility [149].
Instead, precedence depends on an intrinsic property of sperm from males [167] and requires the ability of these sperm to migrate to the spermathecae [149], where they localize to the exclusion of the hermaphrodite’s sperm [111]. It makes sense that preferential localization within this sperm storage organ would result in preferential usage. However, how this pattern is attained is not fully understood. Sperm from males are larger than those from hermaphrodites, and size correlates with greater adhesion and faster crawling speeds in vitro [168]. Furthermore, the movement of an oocyte through the spermatheca following ovulation pushes sperm out into the uterus, and they need to migrate back to compete for future oocytes. Thus, differences in retention in the spermatheca or the speed of reentry, which could both depend on size, might contribute to male precedence. Fusion of sperm with the oocyte usually occurs on the side of the oocyte that enters the spermatheca [111, 153], suggesting that the precise position within the spermatheca could be important [113], but the existence of such a privileged ‘fertilization zone’ has not been tested directly.
C. elegans males normally show no precedence order with respect to each other — when genetically similar males sequentially mate with a hermaphrodite, each one has an equal chance of siring progeny. However, three results imply that fierce sperm competition might exist among Caenorhabditis males. First, when males from two different wild isolates of C. elegans (N2 and AB1) were each mated to the same hermaphrodite, the AB1 sperm took precedence [168]. Second, long-term competition between C. elegans males in the laboratory leads to larger sperm [170]. And third, interspecies crosses imply that sperm from male/female species are more competitive than those from male/hermaphrodite species, where fewer opportunities for sperm competition exist (Ting et al, submitted).
8.2. Sperm size
As discussed above, sperm size might play a role in competitiveness. In C. elegans, males make larger sperm than hermaphrodites [168]. Furthermore, these male sperm outcompete sperm from hermaphrodites [111, 167], and larger male-derived sperm appear to be favored over smaller ones [169]. Perhaps larger sperm are faster at repopulating the spermatheca and better at dislodging smaller competitors from the choicest positions near maturing oocytes (Fig. 1C).
This model implies that males from gonochristic species should have large sperm, since they are likely to compete with others at every mating, and a general survey of nematode species confirmed this prediction [169]. Furthermore, sexual competition between C. elegans males led to an increase in sperm size within only 60 generations [170]. Taken together, these results suggest that larger sperm are more competitive, and that sexual competition favors larger sperm in males. Because sperm from hermaphrodites only rarely compete with sperm from males in the wild [171, 172], these hermaphrodite sperm are probably not under selection for size.
Indeed, the presence of smaller sperm in hermaphrodites seems to have been originally caused by a developmental bias, rather than selection [173]. If females from a male/female species are induced to produce sperm by manipulating the sex determination pathway, these sperm are smaller than those from their male counterparts. Furthermore, the analysis of C. elegans mutants showed that several factors characteristic of XX hermaphrodites influence sperm size. Thus, it appears that newly evolving hermaphrodites initially made smaller sperm because their female gonads and germ lines were not optimized for making large sperm. Although this dimorphism was originally caused by a developmental bias, it was probably maintained by the selective forces described above.
8.3. Segregation and post-segregation distortion
Sometimes competition involving sperm occurs at higher levels than the race to fertilize an oocyte. Dramatic examples involve meiotic drive and molecular or endosymbiotic parasites that manipulate the reproductive process to further their own success. Perhaps the most widespread example of distortion involves Wolbachia bacteria, which are transmitted from mother to offspring through the oocyte [reviewed by 174]. In some species, the embryos produced when sperm from an infected male fertilize an uninfected oocyte all die, and this cytoplasmic incompatibility increases the frequency of Wolbachia-infected animals in the population. So far, there are no examples of Wolbachia infection from Caenorhabditis, but they play important roles in many other nematodes [reviewed by 175].
The zeel-1/peel-1 system highlights the importance of sperm in some postsegregation distorter systems [reviewed by 176]. As described above, sperm from males carrying the zeel-1/peel-1 genetic cassette cause embryonic lethality when they fertilize oocytes that lack these genes [163]. This effect is caused by the paternal PEEL-1 toxin, and helps increase the frequency of this cassette in the population.
In these examples, selection does not ensure that the fittest sperm fertilize oocytes; instead, sperm prevent the production of viable progeny from animals lacking either the Wolbachia endosymbiont, or the zeel-1/peel-1 cassette. Other distortion mechanisms might exist too.
8.4. Sexual conflict
Finally, sperm could be involved in sexual conflict between males and females, although proving such relationships is difficult. Mating can lead to adverse effects for the hermaphrodite, including reduction of lifespan, altered stress responses, and shrinkage in size [177–179]. Sperm, seminal fluid, and possibly other secreted compounds play specific roles in inducing these effects, which are independent of gamete or progeny production by the hermaphrodite.
One recent study shows that sperm from males can migrate to ectopic locations in the female or hermaphrodite gonad, suppressing female reproduction (Ting et al, submitted). These invasive sperm were detected in interspecies crosses, where they can easily be shown to lower fecundity, and similar events sometimes occur in crosses between males and females of the same species. The data suggest that competition between males leads to the production of highly invasive sperm that compete well with other male sperm; females respond through adaptations to their own reproductive tracts to guide and accommodate these sperm. As a consequence, this arms race leads to sperm that are not matched to the female gonads of related species, where the invasive behavior can run unchecked. Whether the sterilization of sympatric females of other species is merely a side effect or is favored by selection remains unknown.
These examples show that males and hermaphrodites exhibit both cooperative and antagonistic interactions, like those seen in male-female species [180, 181]. Thus, C. elegans has potential as a model for studying sexual conflict, in addition to other facets of reproductive and evolutionary biology.
8.5. Interspecies crosses
Interspecies crosses also demonstrate that some sperm functions change at faster rates than others during evolution. For example, sperm from most Caenorhabditis males are capable of migrating towards the spermatheca in XX animals of other species, which implies that their interactions with guidance cues are largely conserved [182]. These sperm are also capable of stimulating ovulation, so the signal from the sperm to the female germ line is mostly conserved [182]. This makes sense because the signaling molecule is MSP, and its sequence is highly conserved among nematodes. By contrast, the signals that initiate spermiogenesis are changing more rapidly, but some interspecies interactions can still be detected [182, 183]. Finally, fertilization only occurs between gametes of closely related species, suggesting that the interactions between EGG and SPE-9 class proteins are undergoing rapid change [182, 184–186].
9. Conclusion
Although we have learned an enormous amount about nematode sperm, several techniques are opening up new areas for future study. First, reverse genetics had been challenging, since many sperm genes do not respond to RNA interference. However, new techniques for gene editing that use TALENs [187–189] or CRISPRs [188, 190] are rapid and inexpensive. Thus, genes that have been identified in genomic expression studies can now be quickly assayed for their functions in sperm. Second, labeling germline proteins for in vivo studies of cell and protein dynamics had once been difficult. Now, the MosSCI [191, 192], TALEN or CRISPR systems can be used to create GFP fusions by homologous recombination. Third, a large set of wild isolates of C. elegans and several related species are now available, and the genomes of eight relatives are nearly complete. Thus, sperm biology and sperm competition can be compared in male/female and male/hermaphroditic systems.
As these studies reveal, nematode sperm provide insight into a range of biological processes. Some nematode proteins have homologs that function in mammalian sperm, so they provide a good model for human reproduction. Other nematode processes differ from those in mammals, but share some important similarities, like the processing of the nematode membranous organelle and the acrosome reaction [67]. In addition, many conserved proteins have adopted new roles in nematode sperm; the powerful set of genetic, genomic and cell-biological techniques that are available make nematodes ideal for exploring the molecular functions of these proteins. Finally, many unique aspects of nematode sperm make this system ideal for studying evolutionary change in reproductive systems.
Highlights.
Spermatogenesis in nematodes is controlled at multiple levels
Redundant pathways control nematode spermiogenesis
Movement of nematode sperm depends on the Major Sperm Protein
Nematode sperm engage in sophisticated crosstalk with female reproductive cells
Nematode sperm provide a model for studying sperm competition
Acknowledgements
RE was supported by NSF grant 1021128, and GS by NIH R01-GM087705. We thank Mike Miller and Steve L’Hernault for comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.White-Cooper H, Doggett K, Ellis RE. The evolution of spermatogenesis. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington, MA: Elsevier/Academic Press; 2009. pp. 151–183. [Google Scholar]
- 2.Lessells CM, Snook RR, Hosken D. The evolutionary origin and maintenance of sperm: selection for a small, motile gamete mating type. In: Birkhead TR, Hosken DJ, Pitnick S, editors. Sperm Biology: An Evolutionary Perspective. Burlington, MA: Elsevier/Academic Press; 2009. pp. 43–67. [Google Scholar]
- 3.Nigon V. Les modalités de la réproduction et le déterminisme de sexe chez quelques Nématodes libres. Ann Sci Nat Zool. 1949;11:1–132. [Google Scholar]
- 4.Ward S, Miwa J. Characterization of temperature-sensitive, fertilization-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1978;88:285–303. doi: 10.1093/genetics/88.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.L'Hernault SW. Spermatogenesis. In: WormBook, editor. The C. elegans Research Community. 2006. WormBook.org. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimura H, L'Hernault SW. Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev Dyn. 2010;239:1502–1514. doi: 10.1002/dvdy.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu DS, Shakes DC. Spermatogenesis. Adv Exp Med Biol. 2013;757:171–203. doi: 10.1007/978-1-4614-4015-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesch BJ, Page DC. Genetics of germ cell development. Nat Rev Genet. 2012;13:781–794. doi: 10.1038/nrg3294. [DOI] [PubMed] [Google Scholar]
- 9.Ellis RE, Schedl T. Sex-determination in the germ line. In: WormBook, editor. The C. elegans Research Community. 2006. Wormbook.org. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan CT, Noble D, Kimble J. Mitosis-meiosis and sperm-oocyte fate decisions are separable regulatory events. Proc Natl Acad Sci U S A. 2013;110:3411–3416. doi: 10.1073/pnas.1300928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bridges CB. Direct proof through non-disjunction that the sex-linked genes of Drosophila are borne by the X-chromosome. Science. 1914;40:107–109. doi: 10.1126/science.40.1020.107. [DOI] [PubMed] [Google Scholar]
- 12.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 13.Shakes DC, Ward S. Initiation of spermiogenesis in C. elegans: a pharmacological and genetic analysis. Dev Biol. 1989;134:189–200. doi: 10.1016/0012-1606(89)90088-2. [DOI] [PubMed] [Google Scholar]
- 14.Klass MR, Hirsh D. Sperm isolation and biochemical analysis of the major sperm protein from Caenorhabditis elegans. Dev Biol. 1981;84:299–312. doi: 10.1016/0012-1606(81)90398-5. [DOI] [PubMed] [Google Scholar]
- 15.Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- 16.Barton MK, Kimble J. fog-1 a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen PJ, Ellis RE. TRA-1A regulates transcription of fog-3 which controls germ cell fate in C. elegans. Development. 2000;127:3119–3129. doi: 10.1242/dev.127.14.3119. [DOI] [PubMed] [Google Scholar]
- 19.Jin SW, Kimble J, Ellis RE. Regulation of cell fate in Caenorhabditis elegans by a novel cytoplasmic polyadenylation element binding protein. Dev Biol. 2001;229:537–553. doi: 10.1006/dbio.2000.9993. [DOI] [PubMed] [Google Scholar]
- 20.Chen PJ, Cho S, Jin SW, Ellis RE. Specification of germ cell fates by FOG-3 has been conserved during nematode evolution. Genetics. 2001;158:1513–1525. doi: 10.1093/genetics/158.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006;11:733–740. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Guo Y, Chen X, Ellis RE. Evolutionary change within a bipotential switch shaped the sperm/oocyte decision in hermaphroditic nematodes. PLoS Genet. 2013;9:e1003850. doi: 10.1371/journal.pgen.1003850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin SW, Arno N, Cohen A, Shah A, Xu Q, et al. In Caenorhabditis elegans, the RNA-binding domains of the cytoplasmic polyadenylation element binding protein FOG-1 are needed to regulate germ cell fates. Genetics. 2001;159:1617–1630. doi: 10.1093/genetics/159.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PJ, Singal A, Kimble J, Ellis RE. A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev Biol. 2000;217:77–90. doi: 10.1006/dbio.1999.9521. [DOI] [PubMed] [Google Scholar]
- 26.Lee MH, Kim KW, Morgan CT, Morgan DE, Kimble J. Phosphorylation state of a Tob/BTG protein, FOG-3, regulates initiation and maintenance of the Caenorhabditis elegans sperm fate program. Proc Natl Acad Sci U S A. 2011;108:9125–9130. doi: 10.1073/pnas.1106027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White-Cooper H, Davidson I. Unique aspects of transcription regulation in male germ cells. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a002626. pii: a002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu DS, Liu H, Nix P, Wu TF, Ralston EJ, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shakes DC, Wu JC, Sadler PL, Laprade K, Moore LL, et al. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000611. doi: 10.1371/journal.pgen.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klass M, Dow B, Herndon M. Cell-specific transcriptional regulation of the major sperm protein in Caenorhabditis elegans. Dev Biol. 1982;93:152–164. doi: 10.1016/0012-1606(82)90249-4. [DOI] [PubMed] [Google Scholar]
- 31.Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. J Cell Sci. 1997;110:1073–1081. doi: 10.1242/jcs.110.9.1073. [DOI] [PubMed] [Google Scholar]
- 32.L'Hernault SW, Arduengo PM. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol. 1992;119:55–68. doi: 10.1083/jcb.119.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gleason EJ, Lindsey WC, Kroft TL, Singson AW, L'Hernault SW. spe-10 encodes a DHHC-CRD zinc-finger membrane protein required for endoplasmic reticulum/Golgi membrane morphogenesis during Caenorhabditis elegans spermatogenesis. Genetics. 2006;172:145–158. doi: 10.1534/genetics.105.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nance J, Minniti AN, Sadler C, Ward S. spe-12 encodes a sperm cell surface protein that promotes spermiogenesis in Caenorhabditis elegans. Genetics. 1999;152:209–220. doi: 10.1093/genetics/152.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nance J, Davis EB, Ward S. spe-29 encodes a small predicted membrane protein required for the initiation of sperm activation in Caenorhabditis elegans. Genetics. 2000;156:1623–1633. doi: 10.1093/genetics/156.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gleason EJ, Hartley PD, Henderson M, Hill-Harfe KL, Price PW, et al. Developmental genetics of secretory vesicle acidification during Caenorhabditis elegans spermatogenesis. Genetics. 2012;191:477–491. doi: 10.1534/genetics.112.139618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Syntichaki P, Samara C, Tavernarakis N. The vacuolar H+ -ATPase mediates intracellular acidification required for neurodegeneration in C. elegans. Curr Biol. 2005;15:1249–1254. doi: 10.1016/j.cub.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 38.Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- 40.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 41.Reinke V, Smith HE, Nance J, Wang J, Van Doren C, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni M, Shakes DC, Guevel K, Smith HE. SPE-44 implements sperm cell fate. PLoS Genet. 2012;8:e1002678. doi: 10.1371/journal.pgen.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page BD, Zhang W, Steward K, Blumenthal T, Priess JR. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 1997;11:1651–1661. doi: 10.1101/gad.11.13.1651. [DOI] [PubMed] [Google Scholar]
- 44.del Castillo-Olivares A, Kulkarni M, Smith HE. Regulation of sperm gene expression by the GATA factor ELT-1. Dev Biol. 2009;333:397–408. doi: 10.1016/j.ydbio.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller MA, Cutter AD, Yamamoto I, Ward S, Greenstein D. Clustered organization of reproductive genes in the C. elegans genome. Curr Biol. 2004;14:1284–1290. doi: 10.1016/j.cub.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 46.Thomas CG, Li R, Smith HE, Woodruff GC, Oliver B, et al. Simplification and desexualization of gene expression in self-fertile nematodes. Curr Biol. 2012;22:2167–2172. doi: 10.1016/j.cub.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinke V. Gene regulation: a tale of germline mRNA tails. Curr Biol. 2008;18:R915–R916. doi: 10.1016/j.cub.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Merritt C, Rasoloson D, Ko D, Seydoux G. 3' UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, et al. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. 2001;128:3899–3912. doi: 10.1242/dev.128.20.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson MA, Cronland E, Dunkelbarger S, Contreras V, Strome S, et al. A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J Cell Sci. 2009;122:1529–1539. doi: 10.1242/jcs.046771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawasaki I, Jeong MH, Shim YH. Regulation of sperm-specific proteins by IFE-1, a germline-specific homolog of eIF4E, in C. elegans. Mol Cells. 2011;31:191–197. doi: 10.1007/s10059-011-0021-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu W, Shirayama M, Conte DJ, Vasale J, Batista PJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, et al. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf N, Hirsh D, McIntosh JR. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastruct Res. 1978;63:155–169. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- 55.Ward S, Argon Y, Nelson GA. Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol. 1981;91:26–44. doi: 10.1083/jcb.91.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma X, Zhu Y, Li C, Shang Y, Meng F, et al. Comparative transcriptome sequencing of germline and somatic tissues of the Ascaris suum gonad. BMC Genomics. 2011;12:481. doi: 10.1186/1471-2164-12-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varkey JP, Jansma PL, Minniti AN, Ward S. The Caenorhabditis elegans spe-6 gene is required for major sperm protein assembly and shows second site non-complementation with an unlinked deficiency. Genetics. 1993;133:79–86. doi: 10.1093/genetics/133.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muhlrad PJ, Ward S. Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics. 2002;161:143–155. doi: 10.1093/genetics/161.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L'Hernault SW, Shakes DC, Ward S. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muhlrad PJ. A genetic and molecular analysis of spermiogenesis initiation in Caenorhabditis elegans. Mol Cell Biol PhD. 2001:192. [Google Scholar]
- 61.Kupinski AP, Muller-Reichert T, Eckmann CR. The Caenorhabditis elegans Ste20 kinase, GCK-3, is essential for postembryonic developmental timing and regulates meiotic chromosome segregation. Dev Biol. 2010;344:758–771. doi: 10.1016/j.ydbio.2010.05.505. [DOI] [PubMed] [Google Scholar]
- 62.Wu JC, Go AC, Samson M, Cintra T, Mirsoian S, et al. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics. 2012;190:143–157. doi: 10.1534/genetics.111.135376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson GA, Ward S. Vesicle fusion, pseudopod extension and amoeboid motility are induced in nematode spermatids by the ionophore monensin. Cell. 1980;19:457–464. doi: 10.1016/0092-8674(80)90520-6. [DOI] [PubMed] [Google Scholar]
- 64.Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26 gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- 65.Schvarzstein M, Pattabiraman D, Bembenek JN, Villeneuve AM. Meiotic HORMA domain proteins prevent untimely centriole disengagement during Caenorhabditis elegans spermatocyte meiosis. Proc Natl Acad Sci U S A. 2013;110:E898–E907. doi: 10.1073/pnas.1213888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lamitina ST, L'Hernault SW. Dominant mutations in the Caenorhabditis elegans Myt1 ortholog wee-1.3 reveal a novel domain that controls M-phase entry during spermatogenesis. Development. 2002;129:5009–5018. doi: 10.1242/dev.129.21.5009. [DOI] [PubMed] [Google Scholar]
- 67.L'Hernault SW. The genetics and cell biology of spermatogenesis in the nematode C. elegans. Mol Cell Endocrinol. 2009;306:59–65. doi: 10.1016/j.mce.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 68.Shakes DC, Ward S. Mutations that disrupt the morphogenesis and localization of a sperm-specific organelle in Caenorhabditis elegans. Dev Biol. 1989;134:307–316. doi: 10.1016/0012-1606(89)90103-6. [DOI] [PubMed] [Google Scholar]
- 69.Machaca K, L'Hernault SW. The Caenorhabditis elegans spe-5 gene is required for morphogenesis of a sperm-specific organelle and is associated with an inherent cold-sensitive phenotype. Genetics. 1997;146:567–581. doi: 10.1093/genetics/146.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arduengo PM, Appleberry OK, Chuang P, L'Hernault SW. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J Cell Sci. 1998;111:3645–3654. doi: 10.1242/jcs.111.24.3645. [DOI] [PubMed] [Google Scholar]
- 71.Zhu GD, L'Hernault SW. The Caenorhabditis elegans spe-39 gene is required for intracellular membrane reorganization during spermatogenesis. Genetics. 2003;165:145–157. doi: 10.1093/genetics/165.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu GD, Salazar G, Zlatic SA, Fiza B, Doucette MM, et al. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol Biol Cell. 2009;20:1223–1240. doi: 10.1091/mbc.E08-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.L'Hernault SW, Benian GM, Emmons RB. Genetic and molecular characterization of the Caenorhabditis elegans spermatogenesis-defective gene spe-17. Genetics. 1993;134:769–780. doi: 10.1093/genetics/134.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelleher JF, Mandell MA, Moulder G, Hill KL, L'Hernault SW, et al. Myosin VI is required for asymmetric segregation of cellular components during C. elegans spermatogenesis. Curr Biol. 2000;10:1489–1496. doi: 10.1016/s0960-9822(00)00828-9. [DOI] [PubMed] [Google Scholar]
- 75.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 76.Lu N, Zhou Z. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int Rev Cell Mol Biol. 2012;293:269–309. doi: 10.1016/B978-0-12-394304-0.00013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J, Wang H, Chen Y, Wang X, Zhang H. Residual body removal during spermatogenesis in C. elegans requires genes that mediate cell corpse clearance. Development. 2012;139:4613–4622. doi: 10.1242/dev.086769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward S, Hogan E, Nelson GA. The initiation of spermiogenesis in the nematode Caenorhabditis elegans. Dev Biol. 1983;98:70–79. doi: 10.1016/0012-1606(83)90336-6. [DOI] [PubMed] [Google Scholar]
- 79.Machaca K, DeFelice LJ, L'Hernault SW. A novel chloride channel localizes to Caenorhabditis elegans spermatids and chloride channel blockers induce spermatid differentiation. Dev Biol. 1996;176:1–16. doi: 10.1006/dbio.1996.9999. [DOI] [PubMed] [Google Scholar]
- 80.Bae YK, Kim E, L'Hernault SW, Barr MM. The CIL-1 PI 5-phosphatase localizes TRP Polycystins to cilia and activates sperm in C. elegans. Curr Biol. 2009;19:1599–1607. doi: 10.1016/j.cub.2009.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Z, Chen L, Shang Y, Huang P, Miao L. The micronutrient element zinc modulates sperm activation through the SPE-8 pathway in Caenorhabditis elegans. Development. 2013;140:2103–2107. doi: 10.1242/dev.091025. [DOI] [PubMed] [Google Scholar]
- 82.Rodriguez MA, LeClaire LL, Roberts TM. Preparing to move: assembly of the MSP amoeboid motility apparatus during spermiogenesis in Ascaris. Cell Motil Cytoskeleton. 2005;60:191–199. doi: 10.1002/cm.20058. [DOI] [PubMed] [Google Scholar]
- 83.Washington NL, Ward S. FER-1 regulates Ca2+ -mediated membrane fusion during C. elegans spermatogenesis. J Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- 84.Minniti AN, Sadler C, Ward S. Genetic and molecular analysis of spe-27 a gene required for spermiogenesis in Caenorhabditis elegans hermaphrodites. Genetics. 1996;143:213–223. doi: 10.1093/genetics/143.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geldziler B, Chatterjee I, Singson A. The genetic and molecular analysis of spe-19, a gene required for sperm activation in Caenorhabditis elegans. Dev Biol. 2005;283:424–436. doi: 10.1016/j.ydbio.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 86.Stanfield GM, Villeneuve AM. Regulation of sperm activation by SWM-1 is required for reproductive success of C. elegans males. Curr Biol. 2006;16:252–263. doi: 10.1016/j.cub.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 87.Smith JR, Stanfield GM. TRY-5 Is a Sperm-Activating Protease in Caenorhabditis elegans Seminal Fluid. PLoS Genet. 2011;7:e1002375. doi: 10.1371/journal.pgen.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fitzgerald LA, Foor WE. Ascaris suum: electrophoretic characterization of reproductive tract and perienteric fluid polypeptides, and effects of seminal and uterine fluids on spermiogenesis. Exp Parasitol. 1979;47:313–326. doi: 10.1016/0014-4894(79)90084-5. [DOI] [PubMed] [Google Scholar]
- 89.Zhao Y, Sun W, Zhang P, Chi H, Zhang MJ, et al. Nematode sperm maturation triggered by protease involves sperm-secreted serine protease inhibitor (Serpin) Proc Natl Acad Sci U S A. 2012;109:1542–1547. doi: 10.1073/pnas.1109912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gosney R, Liau WS, LaMunyon CW. A novel function for the presenilin family member spe-4: inhibition of spermatid activation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:44. doi: 10.1186/1471-213X-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liau WS, Nasri U, Elmatari D, Rothman J, LaMunyon CW. Premature sperm activation and defective spermatogenesis caused by loss of spe-46 function in Caenorhabditis elegans. PLoS One. 2013;8:e57266. doi: 10.1371/journal.pone.0057266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST. Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair. Traffic. 2012;13:185–194. doi: 10.1111/j.1600-0854.2011.01267.x. [DOI] [PubMed] [Google Scholar]
- 93.Dou J, Chen L, Hu Y, Miao L. Cholesterol and the biosynthesis of glycosphingolipids are required for sperm activation in Caenorhabditis elegans. Biochim Biophys Acta. 2012;1821:934–942. doi: 10.1016/j.bbalip.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 94.Fraire-Zamora JJ, Tran T, Cardullo RA. Cholesterol-enriched microdomains regulate pseudopod extension in the MSP-based cytoskeleton of amoeboid sperm. Biochem Biophys Res Commun. 2012;427:478–484. doi: 10.1016/j.bbrc.2012.09.071. [DOI] [PubMed] [Google Scholar]
- 95.Liu Z, Wang B, He R, Zhao Y, Miao L. Calcium signaling and the MAPK cascade are required for sperm activation in Caenorhabditis elegans. Biochim Biophys Acta. 2014;1843:299–308. doi: 10.1016/j.bbamcr.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 96.Poiani A. Complexity of seminal fluid: a review. Behav Ecol Sociobiol. 2006;60:289–310. [Google Scholar]
- 97.LaMunyon CW, Ward S. Assessing the viability of mutant and manipulated sperm by artificial insemination of Caenorhabditis elegans. Genetics. 1994;138:689–692. doi: 10.1093/genetics/138.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, et al. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barker DM. Copulatory plugs and paternity assurance in the nematode Caenorhabditis elegans. Anim Behav. 1994;48:147–156. [Google Scholar]
- 101.Suh J, Hutter H. A survey of putative secreted and transmembrane proteins encoded in the C. elegans genome. BMC Genomics. 2012;13:333. doi: 10.1186/1471-2164-13-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson GA, Roberts TM, Ward S. Caenorhabditis elegans spermatozoan locomotion: amoeboid movement with almost no actin. J Cell Biol. 1982;92:121–131. doi: 10.1083/jcb.92.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.King KL, Stewart M, Roberts TM, Seavy M. Structure and macromolecular assembly of two isoforms of the major sperm protein (MSP) from the amoeboid sperm of the nematode, Ascaris suum. J Cell Sci. 1992;101:847–857. doi: 10.1242/jcs.101.4.847. [DOI] [PubMed] [Google Scholar]
- 104.Bullock TL, McCoy AJ, Kent HM, Roberts TM, Stewart M. Structural basis for amoeboid motility in nematode sperm. Nat Struct Biol. 1998;5:184–189. doi: 10.1038/nsb0398-184. [DOI] [PubMed] [Google Scholar]
- 105.Roberts TM, Stewart M. Role of major sperm protein (MSP) in the protrusion and retraction of Ascaris sperm. Int Rev Cell Mol Biol. 2012;297:265–293. doi: 10.1016/B978-0-12-394308-8.00007-8. [DOI] [PubMed] [Google Scholar]
- 106.LeClaire LL, Stewart M, Roberts TM. A 48 kDa integral membrane phosphoprotein orchestrates the cytoskeletal dynamics that generate amoeboid cell motility in Ascaris sperm. J Cell Sci. 2003;116:2655–2663. doi: 10.1242/jcs.00469. [DOI] [PubMed] [Google Scholar]
- 107.Batchelder EL, Hollopeter G, Campillo C, Mezanges X, Jorgensen EM, et al. Membrane tension regulates motility by controlling lamellipodium organization. Proc Natl Acad Sci U S A. 2011;108:11429–11434. doi: 10.1073/pnas.1010481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buttery SM, Ekman GC, Seavy M, Stewart M, Roberts TM. Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion. Mol Biol Cell. 2003;14:5082–5088. doi: 10.1091/mbc.E03-04-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miao L, Vanderlinde O, Stewart M, Roberts TM. Retraction in amoeboid cell motility powered by cytoskeletal dynamics. Science. 2003;302:1405–1407. doi: 10.1126/science.1089129. [DOI] [PubMed] [Google Scholar]
- 110.Shimabukuro K, Noda N, Stewart M, Roberts TM. Reconstitution of amoeboid motility in vitro identifies a motor-independent mechanism for cell body retraction. Curr Biol. 2011;21:1727–1731. doi: 10.1016/j.cub.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- 112.McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- 113.Han SM, Cottee PA, Miller MA. Sperm and oocyte communication mechanisms controlling C. elegans fertility. Dev Dyn. 2010;239:1265–1281. doi: 10.1002/dvdy.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim S, Spike C, Greenstein D. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:277–320. doi: 10.1007/978-1-4614-4015-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kubagawa HM, Watts JL, Corrigan C, Edmonds JW, Sztul E, et al. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat Cell Biol. 2006;8:1143–1148. doi: 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- 116.Edmonds JW, Prasain JK, Dorand D, Yang Y, Hoang HD, et al. Insulin/FOXO Signaling Regulates Ovarian Prostaglandins Critical for Reproduction. Dev Cell. 2010;19:858–871. doi: 10.1016/j.devcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoang HD, Prasain JK, Dorand D, Miller MA. A heterogeneous mixture of F-series prostaglandins promotes sperm guidance in the Caenorhabditis elegans reproductive tract. PLoS Genet. 2013;9:e1003271. doi: 10.1371/journal.pgen.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- 119.Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, et al. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- 120.Edmonds JW, McKinney SL, Prasain JK, Miller MA. The gap junctional protein INX-14 functions in oocyte precursors to promote C. elegans sperm guidance. Dev Biol. 2011;359:47–58. doi: 10.1016/j.ydbio.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, et al. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- 122.Kosinski M, McDonald K, Schwartz J, Yamamoto I, Greenstein D. C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development. 2005;132:3357–3369. doi: 10.1242/dev.01916. [DOI] [PubMed] [Google Scholar]
- 123.Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Govindan JA, Cheng H, Harris JE, Greenstein D. Galphao/i and Galphas signaling function in parallel with the MSP/Eph receptor to control meiotic diapause in C. elegans. Curr Biol. 2006;16:1257–1268. doi: 10.1016/j.cub.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 125.Brisbin S, Liu J, Boudreau J, Peng J, Evangelista M, et al. A role for C. elegans Eph RTK signaling in PTEN regulation. Dev Cell. 2009;17:459–469. doi: 10.1016/j.devcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 126.Cheng H, Govindan JA, Greenstein D. Regulated trafficking of the MSP/Eph receptor during oocyte meiotic maturation in C. elegans. Curr Biol. 2008;18:705–714. doi: 10.1016/j.cub.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Corrigan C, Subramanian R, Miller MA. Eph and NMDA receptors control Ca2+/calmodulin-dependent protein kinase II activation during C. elegans oocyte meiotic maturation. Development. 2005;132:5225–5237. doi: 10.1242/dev.02083. [DOI] [PubMed] [Google Scholar]
- 128.Yang Y, Han SM, Miller MA. MSP hormonal control of the oocyte MAP kinase cascade and reactive oxygen species signaling. Dev Biol. 2010;342:96–107. doi: 10.1016/j.ydbio.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rose KL, Winfrey VP, Hoffman LH, Hall DH, Furuta T, et al. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1997;192:59–77. doi: 10.1006/dbio.1997.8728. [DOI] [PubMed] [Google Scholar]
- 130.Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136:2211–2221. doi: 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whitten SJ, Miller MA. The role of gap junctions in Caenorhabditis elegans oocyte maturation and fertilization. Dev Biol. 2007;301:432–446. doi: 10.1016/j.ydbio.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 132.Clary LM, Okkema PG. The EGR family gene egrh-1 functions non-autonomously in the control of oocyte meiotic maturation and ovulation in C. elegans. Development. 2010;137:3129–3137. doi: 10.1242/dev.041616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yin X, Gower NJ, Baylis HA, Strange K. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3938–3949. doi: 10.1091/mbc.E04-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McGovern M, Yu L, Kosinski M, Greenstein D, Savage-Dunn C. A role for sperm in regulation of egg-laying in the nematode C. elegans. BMC Dev Biol. 2007;7:41. doi: 10.1186/1471-213X-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Simon JM, Sternberg PW. Evidence of a mate-finding cue in the hermaphrodite nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:1598–1603. doi: 10.1073/pnas.032225799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, et al. The sensory circuitry for sexual attraction in C. elegans males. Curr Biol. 2007;17:1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 137.Barrios A, Nurrish S, Emmons SW. Sensory regulation of C. elegans male mate-searching behavior. Curr Biol. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morsci NS, Haas LA, Barr MM. Sperm status regulates sexual attraction in Caenorhabditis elegans. Genetics. 2011;189:1341–1346. doi: 10.1534/genetics.111.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kleemann GA, Basolo AL. Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self-sperm depletion. Anim Behav. 2007;74:1339–1347. [Google Scholar]
- 140.Marcello MR, Singaravelu G, Singson A. Fertilization. Adv Exp Med Biol. 2013;757:321–350. doi: 10.1007/978-1-4614-4015-4_11. [DOI] [PubMed] [Google Scholar]
- 141.Singson A, Mercer KB, L'Hernault SW. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/s0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- 142.Putiri E, Zannoni S, Kadandale P, Singson A. Functional domains and temperature-sensitive mutations in SPE-9, an EGF repeat-containing protein required for fertility in Caenorhabditis elegans. Dev Biol. 2004;272:448–459. doi: 10.1016/j.ydbio.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 143.Zannoni S, L'Hernault SW, Singson AW. Dynamic localization of SPE-9 in sperm: a protein required for sperm-oocyte interactions in Caenorhabditis elegans. BMC Dev Biol. 2003;3:10. doi: 10.1186/1471-213X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chatterjee I, Richmond A, Putiri E, Shakes DC, Singson A. The Caenorhabditis elegans spe-38 gene encodes a novel four-pass integral membrane protein required for sperm function at fertilization. Development. 2005;132:2795–2808. doi: 10.1242/dev.01868. [DOI] [PubMed] [Google Scholar]
- 145.Xu XZ, Sternberg PW. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003;114:285–297. doi: 10.1016/s0092-8674(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 146.Singaravelu G, Chatterjee I, Rahimi S, Druzhinina MK, Kang L, et al. The sperm surface localization of the TRP-3/SPE-41 Ca2+ -permeable channel depends on SPE-38 function in Caenorhabditis elegans. Dev Biol. 2012;365:376–383. doi: 10.1016/j.ydbio.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kroft TL, Gleason EJ, L'Hernault SW. The spe-42 gene is required for sperm-egg interactions during C. elegans fertilization and encodes a sperm-specific transmembrane protein. Dev Biol. 2005;286:169–181. doi: 10.1016/j.ydbio.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 148.Wilson LD, Sackett JM, Mieczkowski BD, Richie AL, Thoemke K, et al. Fertilization in C. elegans requires an intact C-terminal RING finger in sperm protein SPE-42. BMC Dev Biol. 2011;11:10. doi: 10.1186/1471-213X-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Singson A, Hill KL, L'Hernault SW. Sperm competition in the absence of fertilization in Caenorhabditis elegans. Genetics. 1999;152:201–208. doi: 10.1093/genetics/152.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Argon Y, Ward S. Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics. 1980;96:413–433. doi: 10.1093/genetics/96.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wilson KL, Fitch KR, Bafus BT, Wakimoto BT. Sperm plasma membrane breakdown during Drosophila fertilization requires sneaky, an acrosomal membrane protein. Development. 2006;133:4871–4879. doi: 10.1242/dev.02671. [DOI] [PubMed] [Google Scholar]
- 152.Parry JM, Singson A. EGG molecules couple the oocyte-to-embryo transition with cell cycle progression. Results Probl Cell Differ. 2011;53:135–151. doi: 10.1007/978-3-642-19065-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- 154.O'Connell KF, Maxwell KN, White JG. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev Biol. 2000;222:55–70. doi: 10.1006/dbio.2000.9714. [DOI] [PubMed] [Google Scholar]
- 155.Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 156.Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- 157.Hill DP, Shakes DC, Ward S, Strome S. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Dev Biol. 1989;136:154–166. doi: 10.1016/0012-1606(89)90138-3. [DOI] [PubMed] [Google Scholar]
- 158.McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Dev Biol. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 159.Johnston WL, Krizus A, Dennis JW. Eggshell chitin and chitin-interacting proteins prevent polyspermy in C. elegans. Curr Biol. 2010;20:1932–1937. doi: 10.1016/j.cub.2010.09.059. [DOI] [PubMed] [Google Scholar]
- 160.Browning H, Strome S. A sperm-supplied factor required for embryogenesis in C. elegans. Development. 1996;122:391–404. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- 161.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 162.Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- 163.Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, et al. A novel sperm-delivered toxin causes late-stage embryo lethality and transmission ratio distortion in C. elegans. PLoS Biol. 2011;9: e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Darwin C. The Descent of Man and Selection in Relation to Sex. London: John Murray; 1871. [Google Scholar]
- 166.Birkhead TR, Immler S. Making sperm: design, quality control and sperm competition. Soc Reprod Fertil Suppl. 2007;65:175–181. [PubMed] [Google Scholar]
- 167.LaMunyon CW, Ward S. Sperm precedence in a hermaphroditic nematode (Caenorhabditis elegans) is due to competitive superiority of male sperm. Experientia. 1995;51:817–823. doi: 10.1007/BF01922436. [DOI] [PubMed] [Google Scholar]
- 168.LaMunyon CW, Ward S. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proc Biol Sci. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.LaMunyon CW, Ward S. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proc Biol Sci. 1999;266:263–267. doi: 10.1098/rspb.1999.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.LaMunyon CW, Ward S. Evolution of larger sperm in response to experimentally increased sperm competition in Caenorhabditis elegans. Proc Biol Sci. 2002;269:1125–1128. doi: 10.1098/rspb.2002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Barriere A, Felix MA. Natural variation and population genetics of Caenorhabditis elegans. In: WormBook, editor. The C. elegans Research Community. 2005. Wormbook.org. pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sivasundar A, Hey J. Sampling from natural populations with RNAi reveals high outcrossing and population structure in Caenorhabditis elegans. Curr Biol. 2005;15:1598–1602. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 173.Baldi C, Viviano J, Ellis RE. A bias caused by ectopic development produces sexually dimorphic sperm in nematodes. Curr Biol. 2011;21:1416–1420. doi: 10.1016/j.cub.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 175.Taylor MJ, Hoerauf A. Wolbachia bacteria of filarial nematodes. Parasitol Today. 1999;15:437–442. doi: 10.1016/s0169-4758(99)01533-1. [DOI] [PubMed] [Google Scholar]
- 176.Sinkins SP. Invertebrate post-segregation distorters: a new embryo-killing gene. PLoS Biol. 2011;9:e1001114. doi: 10.1371/journal.pbio.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gems D, Riddle DL. Longevity in Caenorhabditis elegans reduced by mating but not gamete production. Nature. 1996;379:723–725. doi: 10.1038/379723a0. [DOI] [PubMed] [Google Scholar]
- 178.Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, et al. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science. 2014;343:536–540. doi: 10.1126/science.1242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Eberhard WG. Postcopulatory sexual selection: Darwin's omission and its consequences. Proc Natl Acad Sci U S A. 2009;106(Suppl 1):10025–10032. doi: 10.1073/pnas.0901217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hosken DJ, Stockley P, Tregenza T, Wedell N. Monogamy and the battle of the sexes. Annu Rev Entomol. 2009;54:361–378. doi: 10.1146/annurev.ento.54.110807.090608. [DOI] [PubMed] [Google Scholar]
- 182.Hill KL, L'Hernault SW. Analyses of reproductive interactions that occur after heterospecific matings within the genus Caenorhabditis. Dev Biol. 2001;232:105–114. doi: 10.1006/dbio.2000.0136. [DOI] [PubMed] [Google Scholar]
- 183.Baldi C, Cho S, Ellis RE. Mutations in two independent pathways are sufficient to create hermaphroditic nematodes. Science. 2009;326:1002–1005. doi: 10.1126/science.1176013. [DOI] [PubMed] [Google Scholar]
- 184.Baird SE, Sutherlin ME, Emmons SW. Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution. 1992;46:585–594. doi: 10.1111/j.1558-5646.1992.tb02067.x. [DOI] [PubMed] [Google Scholar]
- 185.Baird SE. Haldane's rule by sexual transformation in Caenorhabditis. Genetics. 2002;161:1349–1353. doi: 10.1093/genetics/161.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Woodruff GC, Eke O, Baird SE, Felix MA, Haag ES. Insights Into Species Divergence and the Evolution of Hermaphroditism from Fertile Interspecies Hybrids of Caenorhabditis Nematodes. Genetics. 2010;186:997–1012. doi: 10.1534/genetics.110.120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lo TW, Pickle CS, Lin S, Ralston EJ, Gurling M, et al. PhodUsing TALENs and CRISPR/Cas9 to Engineer Insertions and Deletions. Genetics. 2013;195:331–348. doi: 10.1534/genetics.113.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE. Rapid creation of forward-genetics tools for C. briggsae using TALENs: lessons for nonmodel organisms. Mol Biol Evol. 2014;31:468–473. doi: 10.1093/molbev/mst213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 192.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]