Summary
The Nucleosome Remodeling and Deacetylase (NuRD) complex is essential for embryonic development and pluripotent stem cell differentiation. In this study, we investigated whether NuRD is also involved in the reverse biological process of induction of pluripotency in neural stem cells. By knocking out MBD3, an essential scaffold subunit of the NuRD complex, at different time points in reprogramming, we found that efficient formation of reprogramming intermediates and induced pluripotent stem cells from neural stem cells requires NuRD activity. We also show that reprogramming of epiblast-derived stem cells to naive pluripotency requires NuRD complex function and that increased MBD3/NuRD levels can enhance reprogramming efficiency when coexpressed with the reprogramming factor NANOG. Our results therefore show that the MBD3/NuRD complex plays a key role in reprogramming in certain contexts and that a chromatin complex required for cell differentiation can also promote reversion back to a naive pluripotent cell state.
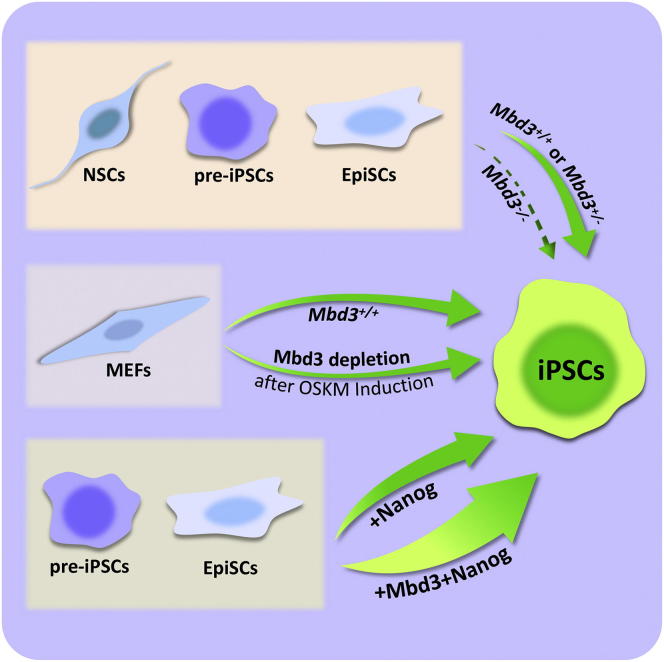
Graphical Abstract
Highlights
-
•
Mbd3 facilitates the initiation of neural stem cell reprogramming
-
•
Mbd3 is also required for efficient iPSC generation from EpiSCs and preiPSCs
-
•
Overexpression of Mbd3/NuRD facilitates reprogramming in a context-dependent manner
dos Santos et al. show that Mbd3/NuRD plays a positive role in reprogramming in certain contexts and that overexpression of Mbd3 facilitates Nanog-mediated reprogramming.
Introduction
Reprogramming of somatic cells to naive pluripotency can be robustly driven by the combined action of transcription factors and culture cues. Among the reprogramming transcription factors, OCT4 plays a central role, as it is sufficient and essential for the induction of pluripotent cells (Kim et al., 2009; Radzisheuskaya et al., 2013; Radzisheuskaya and Silva, 2014). OCT4 interactome studies in embryonic stem cells (ESCs) revealed members of the Nucleosome Remodeling and Deacetylase (NuRD) complex as its highest confidence interactors (Ding et al., 2012; Liang et al., 2008; Pardo et al., 2010; van den Berg et al., 2010). NuRD is composed of six core subunits with at least two enzymatic activities involved in gene regulation: histone deacetylase activity of HDAC1/2 subunits and ATP-dependent chromatin remodeling activity of Mi-2a/β subunits (Lai and Wade, 2011; McDonel et al., 2009). Methyl-CpG binding domain protein 3 (MBD3) is an essential scaffold protein of the NuRD complex, in the absence of which the complex is not assembled (Kaji et al., 2006; Zhang et al., 1999). Embryos lacking MBD3 die shortly after implantation (Hendrich et al., 2001; Kaji et al., 2007) and Mbd3-null ESCs are viable but show severely impaired lineage commitment and exhibit limited differentiation capacity (Kaji et al., 2006; Reynolds et al., 2012a, 2012b). Chromatin remodeling plays an important role in reprogramming to naive pluripotency (Apostolou and Hochedlinger, 2013; Papp and Plath, 2013). Because the NuRD complex is a high confidence interactor of Oct4 and a key regulator of developmental cell state transitions, we have investigated its involvement in the induction of pluripotency.
Results
MBD3 Facilitates the Initiation of Reprogramming from Neural Stem Cells
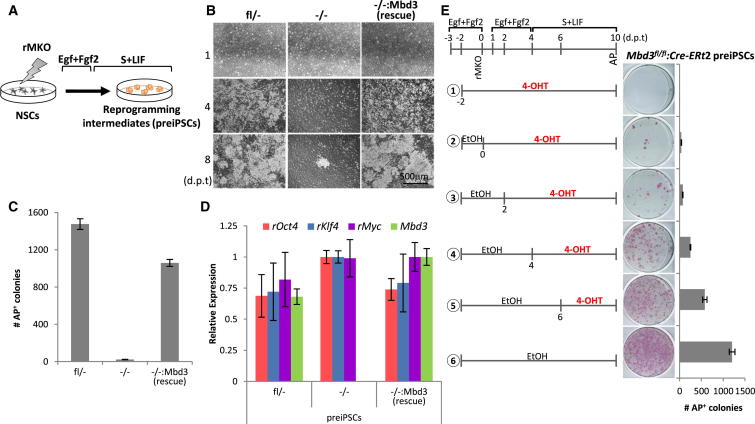
To address the requirement of the NuRD complex in the reprogramming process, we established an Mbd3−/− clonal neural stem cell (NSC) line from Mbd3fl/− NSCs and an Mbd3−/− rescue NSC line (Mbd3−/−:Mbd3) by stable transfection of an Mbd3 transgene (Figures S1A–S1C available online). These NSC lines were transduced with retroviruses encoding cMyc, Klf4, and Oct4 (rMKO) to initiate their reprogramming and were then switched to serum plus LIF (S+LIF) conditions (Figure 1A), which typically results in the formation of highly proliferative reprogramming intermediates, or preiPSCs (Silva et al., 2008). When we used retroviruses encoding GFP (rGFP), equal percentages of GFP+ cells were observed 72 hr after transduction of Mbd3fl/− or Mbd3−/− NSCs, indicating that Mbd3 deletion does not affect transduction efficiency (Figures S1D and S1E). Strikingly, the kinetics of preiPSC emergence was markedly delayed in the Mbd3−/− cells. While Mbd3-expressing preiPSCs dominated the culture by day 4 posttransduction (d.p.t.), Mbd3−/− preiPSCs emerged only by 7–8 d.p.t. (Figure 1B). In addition, the number of emerging alkaline-phosphatase-positive (AP+) Mbd3−/− preiPSC colonies was significantly reduced compared to parental and rescue cell lines (Figure 1C and Figure S1F). Nevertheless, it was possible to establish and expand Mbd3−/− preiPSCs, although less efficiently and with delayed kinetics. Both Mbd3−/− NSCs and Mbd3-null preiPSCs derived from them exhibited slower proliferation, consistent with previous reports of Mbd3−/− ESCs (Kaji et al., 2006; Sims and Wade, 2011) (Figures S1G and S1H). Mbd3−/− preiPSCs expressed slightly higher levels of retroviral transgenes compared to control cells (Figure 1D), suggesting that dosage of reprogramming factors is not the reason for the reduced efficiency of reprogramming initiation that we observed.
Figure 1.
MBD3 Facilitates the Initiation of Reprogramming
(A) Experimental design used to address the kinetics and efficiency of initiation of reprogramming in NSCs with different Mbd3 genotypes. NSCs were transduced with retroviruses encoding cMyc, Klf4, and Oct4 (rMKO), maintained in Egf+Fgf2 medium for 3 days, and then switched to S+LIF medium.
(B) Phase images of the reprogramming intermediates (preiPSCs) emerging from Mbd3fl/−, Mbd3−/−, and Mbd3−/−:Mbd3 (rescue) NSCs at different days posttransduction (d.p.t.).
(C) Efficiency of preiPSC colony formation per 2.5 × 105 NSCs as assessed by alkaline phosphatase (AP) staining at day 9 posttransduction.
(D) qRT-PCR analysis of retroviral transgenes (rOct4, rKlf4, and rMyc) and Mbd3 expression in the obtained preiPSCs maintained in S+LIF. Three independent NSCs transductions were carried out and gene expression was assessed 12 days after transduction. Values are normalized to Gapdh value and shown as relative to the highest value.
(E) Time course of MBD3 requirement during preiPSC formation. Mbd3fl/fl NSCs were stably transfected with pCAG-CreERt2 transgene, transduced with retroviral transgenes, and treated with 4-OHT at indicated time points to induce Cre-mediated deletion of the floxed alleles during reprogramming. Ethanol (EtOH) was used as a control. The encircled numbers correspond to different conditions. PreiPSC colony formation was assessed by AP staining at day 10 posttransduction and is presented as the number of colonies per 7.5 × 104 NSCs. The error bars indicate STDEV.
To further dissect the requirement for MBD3 in the initiation of reprogramming, we analyzed the effect of Mbd3 deletion at different experimental time points. For this experiment, we stably transfected Mbd3fl/fl NSCs with Cre-ERt2, which enabled Cre-mediated excision of the floxed Mbd3 alleles upon addition of 4-hydroxytamoxifen (4-OHT) (Figures S1I–S1L). We found that earlier removal of Mbd3 reduced the number of preiPSC colonies formed (Figure 1E). We also obtained similar results after conditional deletion of Mbd3 exon 1 (ex1fl) which removes all but a small amount of a truncated MBD3 protein isoform (MBD3C) (Aguilera et al., 2011; Kaji et al., 2006) (Figures S1M and S1N).
Taken together, these results demonstrate that lack of a functional NuRD complex strongly impairs the initiation of reprogramming from NSCs.
MBD3 Is Required for Efficient iPSC Generation from NSCs, preiPSCs, and EpiSCs
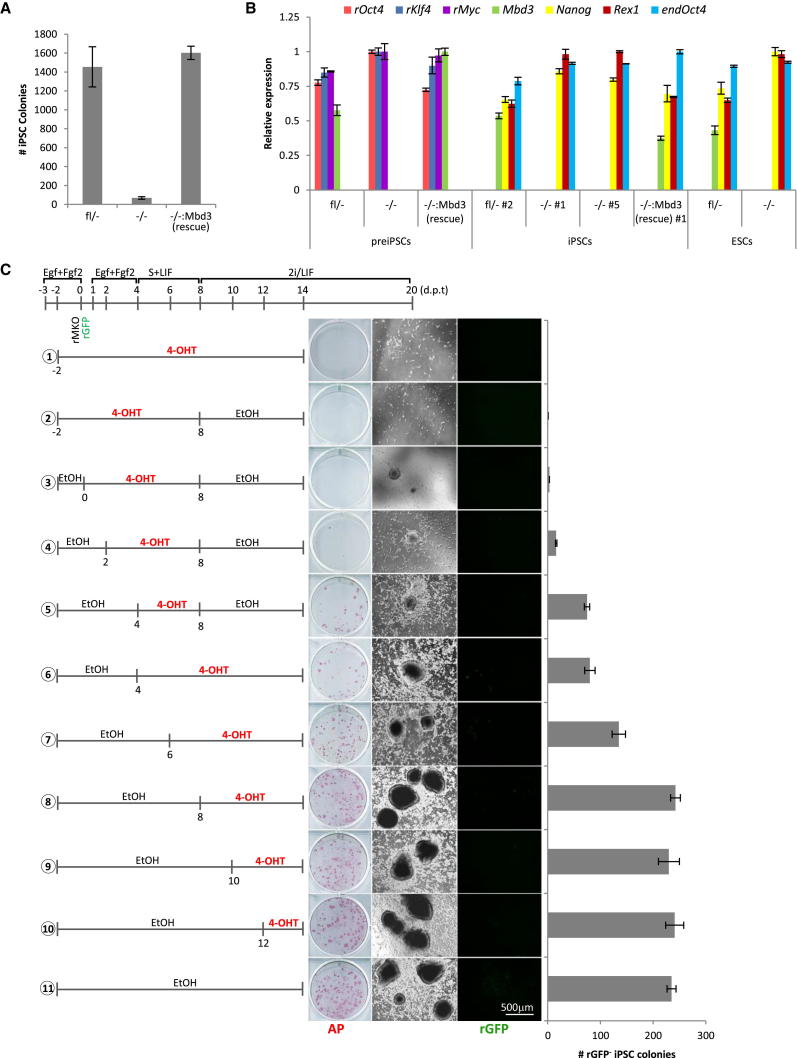
We then evaluated the role of MBD3 in later stages of reprogramming. To induce completion of the reprogramming process, Mbd3fl/−, Mbd3−/−, and rescued Mbd3−/−:Mbd3 preiPSCs were switched to serum-free medium containing LIF and inhibitors of both mitogen-activated protein kinase and glycogen synthase kinase-3 signaling (2i/LIF) (Silva et al., 2008), and the resulting iPSC colonies were scored 12 days later. We observed that the efficiency of conversion to naive pluripotency of Mbd3−/− preiPSCs is strongly reduced compared to Mbd3fl/− and Mbd3−/−:Mbd3 preiPSCs (Figure 2A). The Mbd3fl/−, Mbd3−/−, and Mbd3−/−:Mbd3 iPSCs that were obtained could be expanded clonally in 2i/LIF, and they exhibited reactivation of the pluripotency transcriptional program and silencing of the retroviral reprogramming transgenes as expected (Figure 2B). The Mbd3−/− iPSCs were phenotypically similar to previously reported Mbd3-null ESCs (Kaji et al., 2006), exhibiting impaired embryoid body differentiation and slower proliferation (Figures S2A and S2B). We also observed that Mbd3 deletion in an established preiPSC line before the 2i/LIF medium switch impaired reprogramming to naive pluripotency (Figures S2C and S2D).
Figure 2.
MBD3 Is Required for Efficient iPSC Generation
(A) Quantification of iPSC colonies generated from Mbd3fl/−, Mbd3−/−, and Mbd3−/−:Mbd3 (rescue) preiPSCs after 2i/LIF culture for 12 days. Colony number is per 1.0 × 105 preiPSCs.
(B) qRT-PCR analysis of retroviral transgenes, Mbd3, and pluripotency-associated factors in preiPSCs and corresponding derived iPSCs. qRT-PCR values are normalized to Gapdh value and shown as relative to the highest value.
(C) Mbd3fl/fl:Cre-ERt2 NSCs were transduced with rMKO and rGFP, maintained in Egf+Fgf2 medium for 3 days, switched to S+LIF for 4 more days to allow preiPSC emergence, and then switched to 2i/LIF conditions to induce iPSC formation. 4-OHT was added at different time points (before or after preiPSC emergence) to induce Mbd3-floxed alleles excision. The encircled numbers correspond to different conditions. At day 20 after transfection, GFP− iPSC colonies were counted and subsequently stained for AP. The number of colonies is presented per 7.5 × 104 NSCs. The error bars indicate STDEV.
Next we performed a time course experiment to define the time frame during reprogramming for which MBD3 is required. For this analysis, we transduced Mbd3fl/fl:Cre-ERt2 NSCs with rMKO and rGFP and treated them with 4-OHT at different experimental time points (Figure 2C). The growth medium was changed to S+LIF 4 days after transduction and subsequently, 4 days later, to 2i/LIF. The number of iPSC colonies exhibiting silencing of retroviral GFP expression was assessed 12 days after 2i/LIF medium switch (Figure 2C and Figure S2E). We observed that the number of iPSC colonies formed was proportional to the amount of time cells expressed MBD3 during the initiation phase of reprogramming (prior to 2i/LIF culture). We observed neither a reduction nor a gain of reprogramming efficiency when Mbd3 was deleted at the 2i/LIF stage. Regardless of the stage of Mbd3 deletion, the resulting iPSCs displayed a pluripotency-associated transcriptional signature (Figure S2F). Thus, our data suggest that MBD3 is specifically required for the initiation and intermediate stage of NSC reprogramming rather than establishment of pluripotency.
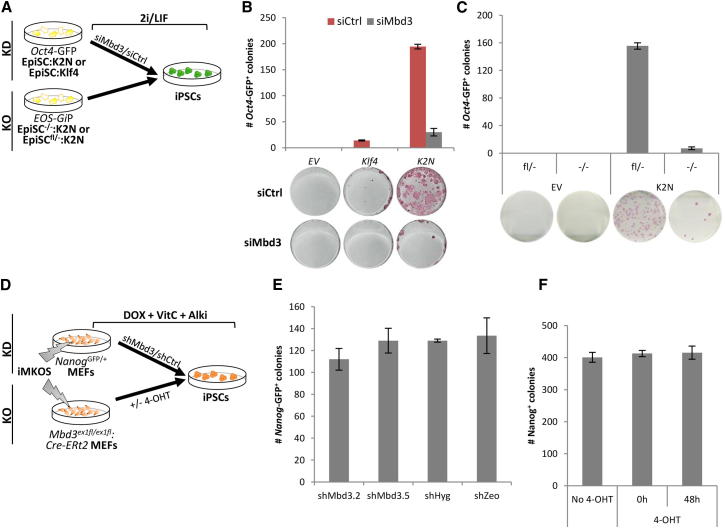
Epiblast stem cells (EpiSCs) can be reprogrammed to naive pluripotency by a combination of overexpression of at least one transcription factor, such as KLF4, KLF2, or NANOG, and the use of serum-free 2i/LIF medium, which not only promotes reprogramming of EpiSCs but also blocks their self-renewal (Guo et al., 2009; Silva et al., 2009). To examine the role of MBD3 in reprogramming in this context, we stably transfected wild-type EpiSCs carrying an Oct4-GFP reporter with piggyBac (PB) vectors constitutively expressing Klf2 and Nanog (K2N) or Klf4, and we then transfected these with either small interfering RNA (siRNA) against Mbd3 or control siRNA (Figure 3A). Strikingly, Mbd3 knockdown led to a complete impairment of KLF4-mediated reprogramming and to a 6-fold reduction in the reprogramming ability of K2N (Figure 3B and Figures S3A and S3B). Similar results were obtained when EpiSCs with Mbd3 genetic knockout were used (Figure 3C and Figures S3C and S3D).
Figure 3.
Requirement of MBD3 in Other Reprogramming Systems
(A) Experimental designs used to analyze the effect of Mbd3 KD and KO on EpiSC reprogramming efficiency. For the KD experiments, wild-type EpiSCs (carrying an Oct4-GFP cassette), stably transfected with pPB-CAG-Klf2.2A.Nanog (K2N) or pPB-CAG-Klf4, were transfected with either siMbd3 or siControl (siCtrl) and, after 24 hr, were plated in 2i/LIF for 12 days. For the KO experiments, Mbd3−/− or Mbd3fl− EpiSCs carrying an Oct4-GFP reporter (EOS-GiP), stably transfected with K2N (or empty vector control, EV), were plated in 2i/LIF for 12 days.
(B and C) The efficiency of EpiSC reprogramming after Mbd3 removal, either by KD (B) or KO (C), was assessed by counting Oct4-GFP+ colonies. Representative AP stained plates are also indicated. 1.0 × 104 EpiSCs were plated in (B). 1.5 × 104 EpiSCs were plated in (C).
(D) Experimental designs used to analyze the effect of Mbd3 KD and Mbd3 exon 1 KO. For the KD experiments, Nanog-GFP MEFs transfected with doxycycline-inducible MKOS piggyBac transposon (iMKOS) were cultured in S+LIF + DOX + vitamin C (vitC) + Alki 24 hr before lentiviral infections of shMbd3 or shControls. For the KO experiments, Mbd3ex1fl/ex1fl MEFs transfected with iMKOS were infected with pMX-Cre-ERt2. Reprogramming was carried out in S+LIF + DOX + vitC + Alki, and 4-OHT was added either at the time of DOX administration (0h) or 48 hr later (48h).
(E) Number of Nanog-GFP+ iPSC colonies at day 13 of reprogramming upon infection of indicated shRNAs.
(F) Number of Nanog+ colony numbers determined by immunofluorescence after 13 days of reprogramming of Mbd3ex1fl/ex1fl:Cre-ERt2 MEFs. The error bars indicate STDEV. Typical iMKOS positive cell number at day 2 of reprogramming is 1.0–3.0 × 104 cells per well, providing 1%–2% reprogramming efficiency.
All the results described above indicate that MBD3 is critical for efficient reprogramming in the contexts that we examined, contrasting with previous reports (Luo et al., 2013; Rais et al., 2013). To examine whether this difference is a reflection of the specific reprogramming systems that we used, we performed PB-mediated reprogramming of mouse embryonic fibroblasts (MEFs) combined with Mbd3 depletion using two different approaches (Figure 3D). First, we used an Mbd3 knockdown system in which Nanog-GFP MEFs were treated with doxycycline (DOX) for the induction of the MKOS or STEMCCA reprogramming cassettes (Kaji et al., 2009; Sommer et al., 2009) and cultured in S+LIF medium supplemented with vitamin C and Alki (Tgfβ signaling inhibitor). Twenty-four hours after induction they were transduced with lentiviruses expressing shRNA against Mbd3 (Figure 3E and Figures S3E–S3G). Second, we depleted Mbd3 by treating Cre-ERt2-transduced Mbd3ex1fl/ex1fl MEFs with 4-OHT at 0 hr or 48 hrs after induction of reprogramming factor expression (Figure 3F and Figures S3H and S3I). While both systems demonstrated about 80% downregulation of MBD3 protein, neither impacted on the efficiency of MEF reprogramming. However, depletion of MBD3 protein would take a few days from the time of 4-OHT administration, so it is possible that in this system cells go through the most critical stage of reprogramming with MBD3 protein still present at a sufficient level.
From our experiments we therefore found that, depending on the reprogramming context, MBD3/NuRD depletion can either have no apparent effect on reprogramming or significantly impair the transition to naive pluripotency.
Overexpression of MBD3/NuRD Can Facilitate Reprogramming
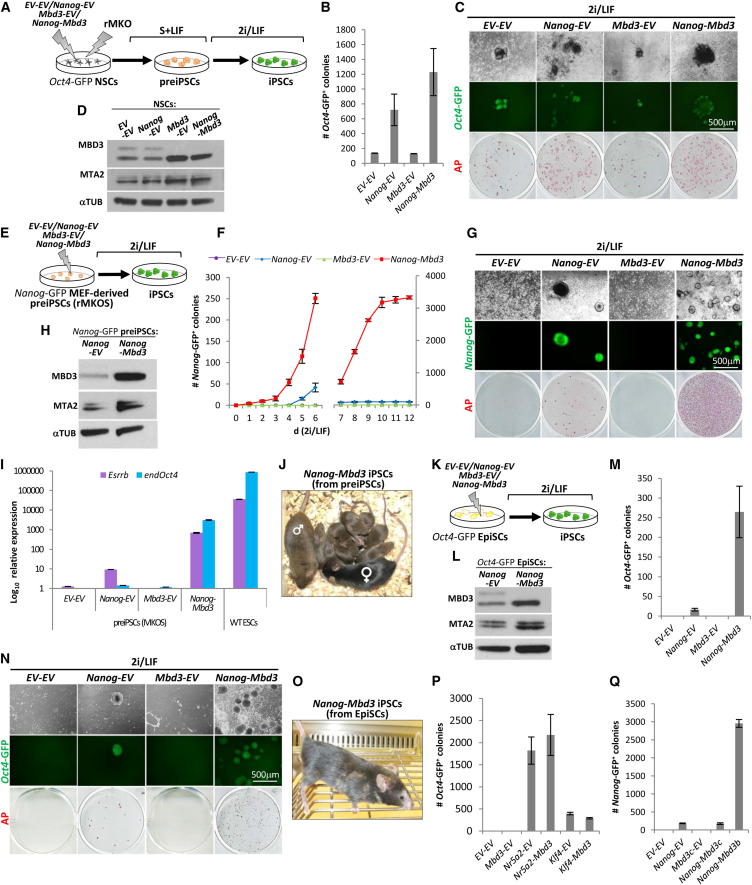
Because the complete removal of Mbd3 or a decrease in its expression can significantly impair the generation of both preiPSCs and iPSCs from NSCs, and iPSCs from preiPSCs and EpiSCs, we tested whether MBD3 levels are limiting for reprogramming. For that, Oct4-GFP reporter NSCs and Nanog-GFP MEF-derived preiPSCs were stably transfected with Mbd3 (Figures 4A and 4E). MBD3 overexpression had neither a positive nor detrimental effect on the efficiency of iPSC formation in both systems (Figures 4B, 4C, 4F and 4G). However, combined overexpression of MBD3 and NANOG in MEF-derived preiPSCs led to accelerated reprogramming kinetics and an up to 30-fold increase in reprogramming efficiency compared to Nanog-Empty vector (EV) control (Figures 4F and 4G and Figures S4A and S4B). This synergistic effect correlated with the upregulation of both Esrrβ and endogenous Oct4 expression, to 5% and 3% of the expression levels of wild-type ESCs, respectively, prior to induction of pluripotency by 2i/LIF medium switch (Figure 4I). Interestingly, MBD3 overexpression in these cell lines caused an increase in protein levels of MTA2, a core subunit of the NuRD complex degraded in the absence of MBD3 (Figures 4D and 4H) (Kaji et al., 2006). This suggests that the effects of MBD3 overexpression are potentially attributable to the total amount, and subsequently total activity, of the NuRD complex. We also found that EpiSCs overexpressing both Nanog and MBD3 showed a 30-fold increase in the ability to generate iPSCs relative to a Nanog only control (Figures 4K–4N). Importantly, all iPSCs generated by the overexpression of NANOG and MBD3 exhibited the molecular properties expected for naive pluripotent cells (Figures S4C–S4F) as well as chimera and germline competence after the excision of reprogramming transgenes (Figures 4J and 4O). We did not see reprogramming synergy with the NuRD complex for two other known reprogramming factors, KLF4 and NR5A2 (Figure 4P and Figure S4G).
Figure 4.
Overexpression of MBD3/NuRD Facilitates NANOG-Mediated Reprogramming
(A) Experimental design used to address the effect of MBD3 overexpression on NSC reprogramming. NSCs carrying an Oct4-GFP cassette were stably transfected with pPB-CAG-Nanog and pPB-CAG-Mbd3b or pPB-CAG-empty controls, transduced with rOKM, cultured in Egf+Fgf2 medium for 3 days, switched to S+LIF medium for 6 days, and then switched to 2i/LIF conditions.
(B) Quantification of Oct4-GFP+ colonies after 12 days in 2i/LIF conditions. Colony number is per 1.0 × 105 NSCs.
(C) Phase and GFP images and AP staining of the iPSCs obtained from NSCs overexpressing respective transgenes.
(D) Western blot analysis of MBD3, MTA2, and TUBULIN (TUB) protein expression in NSCs overexpressing the indicated transgene combinations.
(E) Experimental design used to address the effect of MBD3/NuRD overexpression on the conversion of preiPSCs to iPSCs. PreiPSCs (carrying a Nanog-GFP) were stably transfected with the same transgene combinations as in (A) and plated in 2i/LIF conditions for 12 days.
(F) The kinetics of the emergence of Nanog-GFP+ colonies from the transgenic preiPSCs during a 12 day culture in 2i/LIF conditions (y axis scale changes at day 7). Colony number is per 1.0 × 105 preiPSCs.
(G) Phase and GFP images and AP staining of the iPSCs formed from preiPSCs overexpressing respective transgenes.
(H) Western blot analysis of MBD3, MTA2, and TUBULIN (TUB) protein expression in preiPSCs overexpressing NANOG or NANOG and MBD3.
(I) qRT-PCR analysis of Esrrβ and endogenous (end) Oct4 expression in preiPSCs 12 days after stable transgene transfection and culture in S+LIF (y axes in log10 scale). The expression levels of Esrrβ and endogenous Oct4 in these Nanog-Mbd3 preiPSCs are 5% and 3%, respectively, of the expression levels of WT ESCs in 2i/LIF. The Esrrβ expression level is also approximately 80 times greater than that of Nanog-EV preiPSCs and 700 times greater than that of Mbd3-EV and EV-EV preiPSCs. The endogenous Oct4 expression level is approximately 2,000 times greater than that of Nanog-EV preiPSCs and 3,000 times greater than that of Mbd3-EV and EV-EV preiPSCs. qRT-PCR values are normalized to Gapdh value and shown as relative to the highest value.
(J) Germ line contribution of Nanog-Mbd3 iPSCs generated from preiPSCs (brown color). Cells were treated with TAT-Cre for reprogramming transgene excision prior to blastocyst injection. Chimeric father, C57BL/6 mother, and pups resulting from cross can be viewed.
(K) Experimental design used to address the effect of MBD3/NuRD overexpression on EpiSC reprogramming. EpiSCs (carrying an Oct4-GFP) were stably transfected with the same transgene combinations as in (A) and (E) and plated in 2i/LIF conditions for 12 days.
(L) Western blot analysis of MBD3, MTA2, and TUBULIN (TUB) protein expression in EpiSCs overexpressing NANOG or NANOG and MBD3.
(M) Quantification of Oct4-GFP+ colonies after 12 days of 2i/LIF culture. Colony numbers are per 2.0 × 104 EpiSCs.
(N) Phase and GFP images and AP staining of the iPSCs formed from EpiSCs overexpressing respective transgenes.
(O) Chimera of Nanog-Mbd3 iPSCs generated from EpiSCs (brown color). Cells were treated with TAT-Cre for reprogramming transgene excision prior to blastocyst injection.
(P) Quantification of Oct4-GFP+ colonies after 12 days of 2i/LIF culture, generated from EpiSCs transfected with Klf4 or Nr5a2 (together or not with Mbd3). Colony numbers are per 2.0 × 104 EpiSCs.
(Q) Quantification of Nanog-GFP+ colonies after 12 days of 2i/LIF culture generated from MEF-derived preiPSCs stably transfected with Nanog alone, or Nanog together with Mbd3b or Mbd3c. Colony numbers are per 1.0 × 105 preiPSCs. The error bars indicate STDEV.
The MBD3 isoform that we used for the rescue and overexpression experiments was MBD3B, the most abundant isoform by protein levels in ESCs (Figure S4K). In contrast to MBD3B, we found that MBD3C did not synergize with NANOG (Figure 4Q and Figures S4H–S4J). The two isoforms differ in only the first 60 N-terminal amino acids (Figure S4H), indicating that this region is of importance for NANOG-dependent MBD3 ability to facilitate reprogramming.
These results demonstrate that MBD3 overexpression does not impair induction of naive pluripotency and that it can in fact facilitate reprogramming in conjunction with enhanced NANOG expression.
Discussion
In this study we have identified a positive facilitator role for MBD3/NuRD in transcription-factor-mediated reprogramming of NSCs and EpiSCs. In our analyses, we found that genetic or siRNA-mediated depletion of Mbd3 led to a reduction in the efficiency of reprogramming in these contexts, but not in reprogramming of MEFs. More specifically, we found through time course experiments that MBD3/NuRD function is particularly important during the initiation phase of reprogramming of NSCs and is more dispensable in the later stages when the pluripotency network is becoming more stably established.
We also found that MBD3 overexpression, with resulting higher levels of the NuRD complex, facilitates reprogramming of MEF-derived preiPSCs and EpiSCs when combined with expression of NANOG, but not with other tested reprogramming factors, which is consistent with previous observations that NuRD complex subunits are high confidence protein interactors of both OCT4 and NANOG (Costa et al., 2013; Ding et al., 2012; Gagliardi et al., 2013; Liang et al., 2008; Pardo et al., 2010; van den Berg et al., 2010). In our experiments the N teminus of the MBD3B isoform, which has previously been suggested to be required for protein-protein interactions (Aguilera et al., 2011), appeared to be required for the observed synergistic effect with NANOG in reprogramming (Figure 4Q). In the future we will aim to understand how NANOG and MBD3 work together to drive cells (preiPSCs) that are arrested in the reprogramming process toward pluripotency.
Our data suggest that the NuRD complex might be facilitating gene activation during reprogramming. Interestingly, MBD3 was recently shown to localize to the regulatory sequences of active genes (Günther et al., 2013; Reynolds et al., 2013; Shimbo et al., 2013), including ESC super-enhancers (Hnisz et al., 2013). Moreover, genome-wide expression analysis revealed that 61% of differentially expressed genes are downregulated after Mbd3 deletion in ESCs (Reynolds et al., 2012b). Although some of this decrease in transcription might be due to indirect effects, it seems likely that the NuRD complex acts at enhancers as a mediator of transcription-factor-induced gene activation and thus could also interact with pluripotency factors such as NANOG to support genome-wide reprogramming. In addition, NuRD has been proposed to mediate transient mTOR downregulation and subsequent activation of autophagy, a key step during early stages of reprogramming (Wang et al., 2013).
Our results are in apparent disagreement with two recent reports that suggested an inhibitory role for MBD3 in reprogramming (Luo et al., 2013; Rais et al., 2013), including one (Rais et al., 2013) that argued that reduction or deletion of Mbd3 leads to rapid deterministic reprogramming with 100% efficiency. There are a number of differences between our study and these two previous reports, including the choice of reprogramming cassettes and the reprogramming culture conditions. In contrast to our study, Rais et al. (2013) used a secondary system for somatic cell reprogramming and lentiviral cassette delivery, and it has been reported that both of these factors influence iPSC generation efficiency (Stadtfeld and Hochedlinger, 2010). Moreover, distinct reprogramming factor stoichiometry can provide varying intracellular environments, which may show different dependencies on MBD3 activity for reprogramming. In addition, in our hands heterozygous Mbd3fl/− ESCs express MBD3 at nearly wild-type levels (Figures S4K–S4M; Reynolds et al., 2012b), but Rais et al. (2013) reported that their Mbd3fl/− ESCs expressed MBD3 at 20% of wild-type levels. Further examination of these and other practical and procedural differences between our study and the previous work should help clarify the basis of the apparent differences seen.
Overall, taking into account the results that we report here and previous studies, our conclusion is that at least in some contexts MBD3/NuRD plays a positive role in reprogramming, and that loss of MBD3 expression leads to a reduction in the efficiency of the reprogramming process.
NuRD plays well-documented roles in controlling gene expression and developmental transitions in a wide variety of different metazoan systems (Ahringer, 2000; McDonel et al., 2009; Reynolds et al., 2013). MBD3 is known to be required for embryonic development and pluripotent cell differentiation (Kaji et al., 2006, 2007), and the composition of the complex or specific interactions of its individual subunits may regulate different aspects of its function (Allen et al., 2013; Reynolds et al., 2013). Further insights into the function of the NuRD complex during different cell state transitions will help us understand the process of induced pluripotency as well as embryonic development.
Experimental Procedures
Cell Culture
Platinum-E, preiPSCs, and MEFs were cultured in GMEM (Sigma-Aldrich) supplemented with 10% FCS, 1× NEAA, 1× Pen/Strep, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, and 20 ng/ml of LIF (homemade), indicated as S+LIF medium throughout. ESCs and iPSCs were maintained in N2B27-based medium (DMEM/F12 and Neurobasal [both Life Technologies] in 1:1 ratio, 1× Pen/Strep, 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, 1:200 N2 [PAA], and 1:100 B27 [Life Technologies]) supplemented 20 ng/ml of LIF and 2i inhibitors: CHIR99021 (3 μM) and PD0325901 (1 μM), indicated as 2i/LIF throughout (Ying et al., 2008). NSCs were cultured in DMEM/F12 (GIBCO) supplemented with 1× NEAA, 0.1 mM 2-mercaptoethanol, 1× Pen/Strep, 1:100 B27, 1:200 N2 supplement, 4.5 μM HEPES, 0.03 M glucose, 120 μg/ml BSA, 10 ng/ml of Egf (Peprotech), and 20 ng/ml of Fgf2 (homemade), indicated as Egf+Fgf2 medium throughout. EpiSCs were maintained in N2B27-based medium containing 12 ng/ml of Fgf2 and 20 ng/ml of Activin A (homemade), indicated as Fgf2/Act.A medium throughout. EpiSCs and NSCs were cultured on plastic coated with fibronectin (10 μg/ml, Millipore) or laminin (10 μg/ml, Sigma-Aldrich), respectively. All other cell types were grown on gelatine. All cell types were maintained at 7% CO2. For Cre-mediated transgene excision, cells were treated with 500 nM of 4-OHT.
Derivation of Cell Lines
NSCs
Brains from Mbd3fl/fl and Mbd3ex1fl/ex1fl E13.5 embryos were dissected, dissociated in Egf+Fgf2 medium, and plated onto the laminin-coated cell culture flasks. Mbd3fl/− NSCs were derived from ESCs as described (Pollard et al., 2006). Briefly, ESCs were seeded on gelatinized 10 cm dishes in N2B27 medium for 7 days. After this period, cells were trypsinized and plated on nongelatinized dishes for 3 days in Egf+Fgf2 medium. The emergent neurospheres were then seeded on gelatinized plates and maintained in monolayer in Egf+Fgf2 medium. For Cre-excision of the Mbd3 floxed allele, Mbd3fl/− NSCs were nucleofected with a pCAG-Cre-ires-Puro plasmid and clonal lines of Mbd3−/− NSCs were expanded.
MEFs
Organ-deprived carcasses from E12.5 or E13.5 embryos were dissociated into small pieces, trypsinized, and plated in S+LIF medium.
EpiSCs
Mbd3fl/− and Mbd3−/− EpiSCs were derived from ESCs as previously described (Guo et al., 2009). Briefly, ESCs transfected with pPB-EOS-GFP-ires-Puro (EOS-GiP; GFPiresPuro under the control of early transposon promoter and Oct4 and Sox2 enhancers) were cultured in Fgf2/Act.A medium for at least 10 passages before analysis. To obtain a pure EpiSC culture, GFP+ cells were removed by FACS.
Acknowledgments
We acknowledge N. Reynolds and J. Ramalho-Santos for advice and helpful discussions. We are also grateful to Y. Costa for critical reading of the manuscript. This study was supported by a Wellcome Trust Fellowship (WT101861), an ERC starting grant, and the Anne Rowling Clinic. J.C.R.S. and B.H. are Wellcome Trust Senior Research Fellows in the Basic Biomedical Sciences. R.L.S. is a recipient of a Ph.D. fellowship from the Portuguese Foundation for Sciences and Technology, FCT (SFRH/BD/51198/2010). L.T. is a recipient of a Ph.D. fellowship from The College of Medicine and Veterinary Medicine, University of Edinburgh.
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Supplemental Information
References
- Aguilera C., Nakagawa K., Sancho R., Chakraborty A., Hendrich B., Behrens A. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 2011;469:231–235. doi: 10.1038/nature09607. [DOI] [PubMed] [Google Scholar]
- Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–356. doi: 10.1016/s0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- Allen H.F., Wade P.A., Kutateladze T.G. The NuRD architecture. Cell. Mol. Life Sci. 2013;70:3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Xu H., Faiola F., Ma’ayan A., Wang J. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012;22:155–167. doi: 10.1038/cr.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi A., Mullin N.P., Ying Tan Z., Colby D., Kousa A.I., Halbritter F., Weiss J.T., Felker A., Bezstarosti K., Favaro R. A direct physical interaction between Nanog and Sox2 regulates embryonic stem cell self-renewal. EMBO J. 2013;32:2231–2247. doi: 10.1038/emboj.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther K., Rust M., Leers J., Boettger T., Scharfe M., Jarek M., Bartkuhn M., Renkawitz R. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013;41:3010–3021. doi: 10.1093/nar/gkt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Guy J., Ramsahoye B., Wilson V.A., Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Caballero I.M., MacLeod R., Nichols J., Wilson V.A., Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K., Nichols J., Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Sebastiano V., Wu G., Araúzo-Bravo M.J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Lai A.Y., Wade P.A. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P.L., Xin H.W., Jung S.Y., Qin J., Wong J.M., Cooney A.J., Liu D., Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Luo M., Ling T., Xie W., Sun H., Zhou Y., Zhu Q., Shen M., Zong L., Lyu G., Zhao Y. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31:1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- McDonel P., Costello I., Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. Int. J. Biochem. Cell Biol. 2009;41:108–116. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M.M., Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard S.M., Benchoua A., Lowell S. Neural stem cells, neurons, and glia. In: Klimanskaya I.L.R., editor. Embryonic Stem Cells. Elsevier Academic Press; Boston: 2006. pp. 151–169. [Google Scholar]
- Radzisheuskaya A., Silva J.C. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A., Chia Gle.B., dos Santos R.L., Theunissen T.W., Castro L.F., Nichols J., Silva J.C. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat. Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y., Zviran A., Geula S., Gafni O., Chomsky E., Viukov S., Mansour A.A., Caspi I., Krupalnik V., Zerbib M. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O’Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., Salmon-Divon M., Dvinge H., Hynes-Allen A., Balasooriya G., Leaford D., Behrens A., Bertone P., Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N., O’Shaughnessy A., Hendrich B. Transcriptional repressors: multifaceted regulators of gene expression. Development. 2013;140:505–512. doi: 10.1242/dev.083105. [DOI] [PubMed] [Google Scholar]
- Shimbo T., Du Y., Grimm S.A., Dhasarathy A., Mav D., Shah R.R., Shi H., Wade P.A. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 2013;9:e1004028. doi: 10.1371/journal.pgen.1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J.K., Wade P.A. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol. Biol. Cell. 2011;22:3094–3102. doi: 10.1091/mbc.E11-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D.L.C., Snoek T., Mullin N.P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R.A. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xia P., Ye B., Huang G., Liu J., Fan Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13:617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.