Summary
Alternative RNA splicing (AS) regulates proteome diversity, including isoform-specific expression of several pluripotency genes. Here, we integrated global gene expression and proteomic analyses and identified a molecular signature suggesting a central role for AS in maintaining human pluripotent stem cell (hPSC) self-renewal. We demonstrate the splicing factor SFRS2 is an OCT4 target gene required for pluripotency. SFRS2 regulates AS of the methyl-CpG-binding protein MBD2, whose isoforms play opposing roles in maintenance of, and reprogramming to, pluripotency. While both MDB2a and MBD2c are enriched at the OCT4 and NANOG promoters, MBD2a preferentially interacts with repressive NuRD chromatin remodeling factors and promotes hPSC differentiation, whereas overexpression of MBD2c enhances reprogramming of fibroblasts to pluripotency. The miR-301 and miR-302 families provide additional regulation by targeting SFRS2 and MDB2a. These data suggest that OCT4, SFRS2, and MBD2 participate in a positive feedback loop, regulating proteome diversity complexity in support of hPSC self-renewal and reprogramming.
Introduction
The transcription factors OCT4, NANOG, and SOX2 are master regulators of pluripotency in embryonic stem cells (ESC) (De Los Angeles et al., 2012), and along with Klf4 and c-Myc, facilitate reprogramming of somatic cells into induced-pluripotent stem cells (iPSC) (Park et al., 2008; Takahashi and Yamanaka, 2006). ESC are indispensable models of early development while iPSC hold great promise as cell-based therapeutics that circumvent the immunologic and ethical hurdles of embryo-derived cells. As a result significant effort has been invested to elucidate the mechanisms that underlie stem cell function, with particular emphasis on these core pluripotent genes. Despite the requirement of OCT4, SOX2, and NANOG in stem cell function (De Los Angeles et al., 2012), discrepancies between ostensibly identical pluripotent cell lines (Gore et al., 2011), in addition to the divergent lineage commitment properties of iPSC derived from different adult tissues (Kim et al., 2010), illustrate that the molecular network balancing self-renewal, pluripotency, and lineage commitment is not yet resolved.
Recently, functional genomics and molecular profiling approaches have been used to explore the broader role of the core pluripotent factors in stem cell biology. These studies expanded the set of genes that support pluripotency (Chia et al., 2010) and defined a biochemical network centered around OCT4, NANOG, and SOX2, which is highly enriched for genes essential for development and stem cell function (Kim et al., 2008). Furthermore, use of ChIP-chip (Boyer et al., 2005) and ChIP-seq (Guenther et al., 2010) has established the landscape of genetic targets for several key pluripotent factors and defined correlations between promoter co-occupancy and transcriptional activation. In parallel, genome-scale molecular measurement technologies have been used to quantify differences in epigenetic modifications (Gifford et al., 2013; Xie et al., 2013), gene expression (Tang et al., 2010), protein translation (Ingolia et al., 2011), in addition to protein expression and phosphorylation (Brill et al., 2009; Phanstiel et al., 2011) between pluripotent stem cells and other cell types. These data provide a rich resource of molecular information, although it remains challenging to generate specific hypotheses from these disparate data types or establish mechanistic links between these molecular profiles and the core pluripotent factors.
Recently alternative splicing (AS) has garnered attention as a possible means by which stem cells regulate the expression of gene and protein isoforms to support pluripotency and self-renewal. Indeed, functional roles for alternatively spliced gene products of NANOG, FOXP1, and Tcf7l1 have been demonstrated (Das et al., 2011; Gabut et al., 2011; Salomonis et al., 2010). In addition, the muscleblind-like family (MBNL) of RNA binding proteins was found to repress pluripotency by mediating expression of several somatic cell-specific protein isoforms, including FOXP1 (Han et al., 2013). These data illustrate a general role for AS in pluripotent cells; however the specific splicing factors and mechanistic links to the core pluripotent genes, which work in concert to reinforce a ground state of self-renewal, remain unresolved.
The splicing factor SFRS2 (also known as SC35) is essential for embryonic development (Xiao et al., 2007) and regulates transcription (Lin et al., 2008). Although several splicing substrates have been identified (Lin et al., 2008), no pluripotency-specific role has been established for SFRS2.
The methyl-DNA binding protein MBD2 (methyl-CpG binding domain protein 2) comprises two predominant isoforms, MBD2a and MBD2c (Hendrich and Bird, 1998), which share the same methyl-CpG binding (MBD) domain, but differ in the C-terminal region as a result of AS. MBD2 silences gene expression by binding to methylated DNA and recruiting the Nucleosome Remodeling and Deacetylation (NuRD) complex (Zhang et al., 1999). While NuRD has well-established roles in development (Reynolds et al., 2012), the function of MBD2 in stem cells is not well understood. In fact, data from two recent studies are inconsistent with respect to the impact of MBD2 in somatic cell reprogramming (Lee et al., 2012; Onder et al., 2012), although the possibility of isoform-specific function was not considered.
In this study, we establish mechanistic links between OCT4 and SFRS2, and demonstrate that these factors work in concert to regulate AS of MBD2. Expression of specific MBD2 isoforms is further regulated by the microRNA machinery, and we find that the resulting gene products play opposing functional roles with respect to self-renewal of hPSC and reprogramming of fibroblasts. Consistent with these observations, MBD2 isoforms target the promoters of OCT4 and NANOG in human ESC (hESC) but differ dramatically in their ability to biochemically interact with chromatin remodeling proteins. Collectively our results suggest a positive feedback loop comprised of OCT4, SFRS2, and splice products of MBD2, which regulates proteome diversity to support a self-renewing ground state.
Results
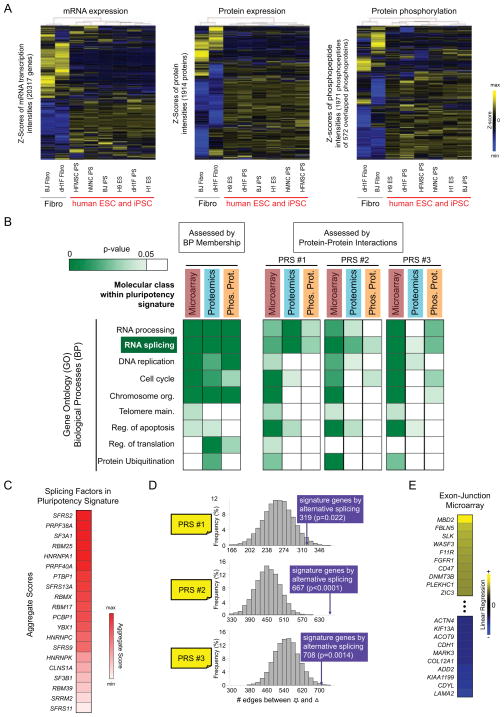
We first sought to identify a molecular signature for pluripotency that integrated gene and protein expression, in addition to protein phosphorylation in cells representing a broad range of genetic backgrounds and cell fates (Fig. S1A, Fig. S2, Table S1). Independent hierarchical clustering of each data type revealed that hPSC from different tissue types exhibit protein phosphorylation, gene transcription, and protein expression profiles that are clearly distinct from differentiated fibroblasts (DF) (Fig. 1A), with each molecular class contributing a subset of unique genes to the signature (Fig. S1B). Notably, the molecular divergence observed between pluripotent cells and DF was considerably higher than hPSC (Fig. S1C); in addition we confirmed that the phosphorylation signature was strongly linked to cell type rather than specific culture conditions (Fig. S1D). As is typical of high-throughput measurements (Brill et al., 2009; Phanstiel et al., 2011; Tang et al., 2010), classification of gene function within the pluripotency signature based on Gene Ontology (GO) biological process revealed enrichment of several disparate pathways (Fig. 1B, left).
Fig. 1. Analysis of the molecular signature associated with hPSC suggests a central regulatory role for RNA splicing.
(A) Independent hierarchical clustering of microarray and proteomic data demonstrated that hPSC are molecularly distinct compared to DF at the level of (left) gene expression, (middle) protein expression, and (right) protein phosphorylation. (B, left) Analysis of pluripotency signature genes according their membership in Gene Ontology (GO) Biological Processes (BP) revealed enrichment of multiple pathways. (B, right) Further analysis of genes within each GO-BP pathway and measurement class (gene, protein, and phosphoprotein) based on physical interactions with three positive reference sets (PRS) of pluripotent factors (see Fig. S1E, Table S2, and Supplemental Methods) suggested that the RNA splicing pathway is strongly associated with pluripotency. Abbreviations: Main., maintenance; Org., organization; Reg., regulation. (C) Splicing factors within the pluripotent molecular signature were individually ranked (see Table S3 and Supplemental Methods), with SFRS2 as the top candidate. (D) Alternatively spliced genes associated with hESC are enriched for physical interactions with positive reference sets (PRS) of pluripotent factors. Null distributions (grey bars) were created by random selection (10,000 iterations) of identical size gene sets from the background of all genes detected by exon-junction microarray. (E) The methyl-DNA binding protein MBD2 displays the strongest alternative splicing pattern between hESC and DF based on the linear regression analysis (see Supplemental Methods).
There is growing appreciation that the principles of network theory are applicable to human physiology, whereby extended physical, genetic, or metabolic relationships between biomolecules may have predictive power with respect to biological outcomes (Balázsi et al., 2011; Vidal et al., 2011). Consistent with this notion, we next asked whether interpretation of our molecular signature data within the context of physical interaction networks would highlight specific cellular functions that support self-renewal. Accordingly we assessed the number of physical interactions between constituent genes of the pluripotency signature and three positive reference sets (PRS) of pluripotent factors derived from (i) literature survey, (ii) a recent functional genomics study, and (iii) proteins defined as biochemical interactors of Oct4 or Nanog (Fig. S1E, Supplemental Methods, and Table S2). This analysis revealed that only members of the RNA splicing pathway are consistently enriched across each measurement class (Fig. 1B, right and Table S3). Further analysis (Supplemental Methods) of the splicing factors in our pluripotent molecular signature suggested that the splicing factor SFRS2 might be an important mediator of pluripotency (Fig. 1C, Table S3).
Given the role of SFRS2 in AS, we next compared the levels of spliced isoforms for 16,084 genes in hESC and DF, and found that the spliced products from 2974 genes differed between these cell types (Fig. S3A, Table S4). Strikingly, we observed that 1424 of these were not otherwise represented in the set of pluripotency signature genes (Fig. S3A). As with other molecular classes of the pluripotent signature (Fig. 1B), gene products subject to AS in hESC are enriched for physical interactions with the PRS (Fig. 1D, Table S3). Extension of this analysis to gene GO annotation revealed a consistent enrichment of factors related to transcription regulation and chromatin modification (Fig. S3B, Table S3); in total we observed 236 alternatively spliced genes that spanned these pathways. Within the exon-junction microarray data (Table S4) MBD2 (methyl-CpG binding domain protein 2) had the highest prediction score for AS between hESC and DF (Fig. 1E, Table S4).
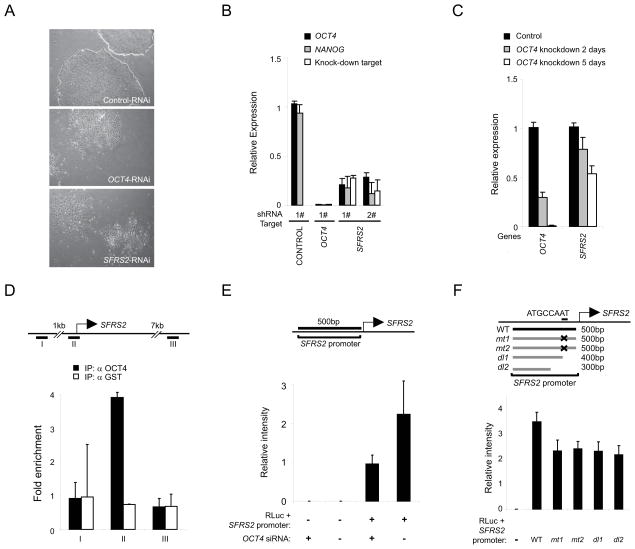
Next we sought to establish specific links between SFRS2, MBD2, and the machinery supporting pluripotency. Depletion of endogenous SFRS2 disrupted self-renewal in hESC as gauged by cell morphology (Fig. 2A), expression of OCT4 and NANOG (Fig. 2B), alkaline phosphatase staining (Fig. S4A), and cell colony integrity (Fig. S4B). We observed a coordinate decrease in expression level of SFRS2 upon OCT4 depletion in hESC (Fig. 2C); importantly, this effect was specific to SFRS2 and not observed for other splicing factors (Fig. S4C). Furthermore, we found that OCT4 bound directly to the promoter of SFRS2 in hESC (Fig. 2D), and drove expression of luciferase downstream of the native SFRS2 promoter in vitro (Fig. 2E). The specificity of this interaction was confirmed by mutation or deletion within the predicted OCT4-binding site of the SFRS2 promoter (Fig. 2F). These data provide evidence for functional and genetic links between OCT4 and SFRS2 in hPSC.
Fig. 2. OCT4 and SFRS2 display interdependent functional links in hPSC.
(A–B) SFRS2 is required to support self-renewal. Lentiviral shRNA-mediated depletion of SFRS2 disrupted pluripotency in H1 ESC as monitored by (A) colony morphology and (B) expression of OCT4 and NANOG. (C) Depletion of OCT4 in H1 ESC for 2 and 5 days led to a coordinate decrease in the expression of SFRS2. (D) OCT4 selectively binds the proximal promoter region of SFRS2 in H1 ESC. (E) OCT4 depletion in human clone 9 iPSC disrupts luciferase expression downstream of the native SFRS2 promoter. (F) Mutation or deletion of the predicted OCT4 binding motif (ATGCCAAT) in the proximal SFRS2 promoter region decreased downstream luciferase expression in clone 9 iPSC. See also Fig. S4. Error bars represent the mean ± SEM.
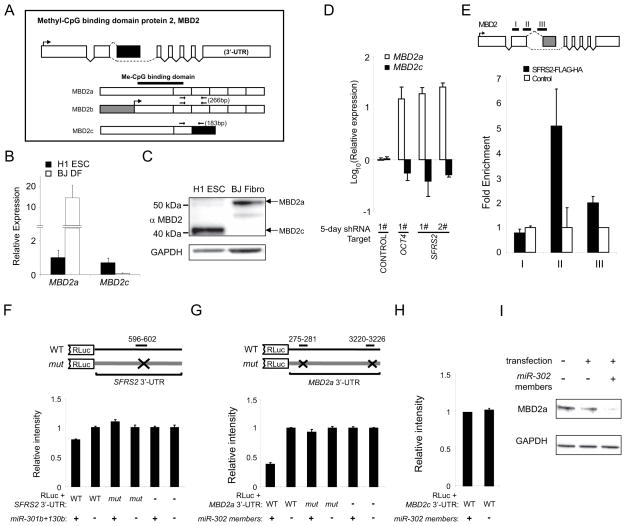
The methyl-DNA binding protein MBD2 comprises multiple isoforms (Hendrich and Bird, 1998) (Fig. 3A). We detected preferential gene- and protein-level expression of the MBD2c and MBD2a isoforms in H1 ESC and BJ fibroblasts, respectively (Fig. 3B–C). Interestingly, depletion of endogenous SFRS2 or OCT4 in hESC led to a dramatic increase in expression of MBD2a and a reduction in MBD2c (Fig. 3D and Fig. S4D). Next we probed for a direct biochemical interaction between SFRS2 and MBD2 pre-mRNA by assaying RNA that co-precipitated with exogenously expressed SFRS2-FLAG-HA. We observed that SFRS2 bound to MBD2 pre-mRNA specifically at intron 2, preceding exon 3 that is unique to the ESC-predominant MBD2c isoform (Fig. 3E), suggesting that SFRS2 may mediate alternative splicing of this methyl-DNA binding protein in hPSC.
Fig. 3. MBD2 isoform expression is independently regulated by the splicing factor SFRS2 and the miR-302 family of microRNAs in hPSC.
(A) Exon and protein graph for the methyl-CpG binding protein MBD2. Dashed lines indicate splice sites. Protein segments corresponding to each exon are annotated with predicted functional domains and primer locations. (B–C) Verification of the distinct MBD2 isoforms in H1 ESC and BJ DF by (B) qRT-PCR and (C) western blot. (D) Lentiviral shRNA-mediated depletion of OCT4 and SFRS2 independently led to a significant increase in MBD2a expression along with reduced levels of MBD2c after 5 days in H1 ESC. (E) Exogenously expressed SFRS2-FLAG-HA preferentially binds to MBD2 pre-mRNA at intron 2 (primer pairs II and III, each spanning into exon 2 and exon 3, respectively) but not inside exon 2 (primer pair I) in H1 ESC. (F) miR-301b and miR-130b suppress luciferase expression in the context of wild-type but not mutated sequences corresponding to the 3′-UTR of SFRS2 in HeLa cells. (G–I) miR-302 cluster members target the 3′-UTR of MBD2 in an isoform-specific manner. (G) miR-302 cluster members specifically suppressed luciferase expression upstream of the wild-type but not mutated MBD2a 3′-UTR sequence in HeLa cells. (H) miR-302 cluster members did not affect luciferase expression upstream of the MBD2c 3′-UTR sequence in HeLa cells. (I) Overexpression of miR-302 cluster members in 293T cells reduced expression of endogenous MBD2a. Error bars represent the mean ± SEM.
In addition, close inspection of the 3′-UTR of SFRS2 and MBD2a (but not MBD2c) revealed potential binding motifs for miR-301 and miR-302, microRNA families that are functionally associated with lineage commitment and self-renewal (Fig. S4E) (Bar et al., 2008). We confirmed that overexpression of miR-301b and miR-130b reduced luciferase driven by the wild-type SFRS2 3′-UTR, while mutation of the miR-301 motif restored luciferase expression (Fig. 3F). Similarly, miR-302 specifically targeted the 3′-UTR of MBD2a (Fig. 3G), but not that of MBD2c (Fig. 3H). Indeed, we confirmed that exogenous expression of miR-302 reduced levels of MBD2a in vivo (Fig. 3I). These data suggest that the miR-301/302 families may independently regulate SFRS2 and MBD2 to “fine-tune” the expression of MBD2 isoforms.
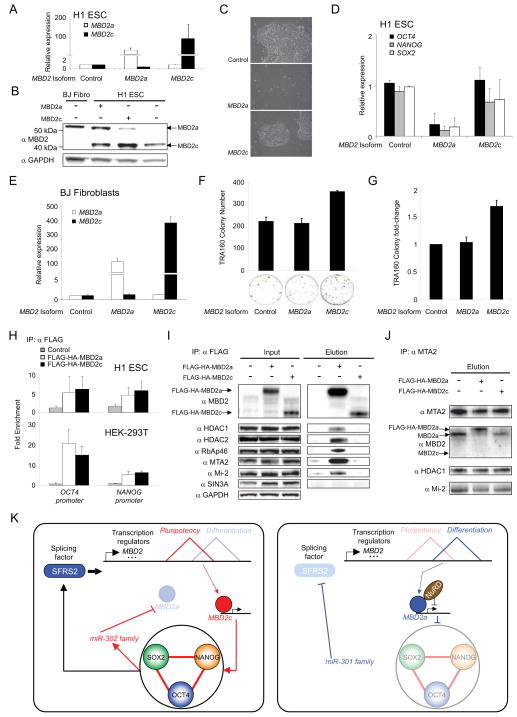
We next investigated the functional roles of MBD2 isoforms in hPSC. Overexpression of MBD2a (Fig. 4A–B) disrupted pluripotency as evidenced by cell morphology (Fig. 4C) in addition to reduced expression of OCT4, NANOG, and SOX2 (Fig. 4D). In contrast, increased MBD2c levels had no effect in hESC based on these measures. However, addition of the ESC-specific MBD2c isoform (Fig. 4E) to a cocktail of reprogramming factors enhanced reprogramming efficiency in BJ fibroblasts, while exogenous expression of MBD2a had no effect (Fig. 4F–G). These data suggested that MBD2a and MBD2c play opposing roles in pluripotency. Chromatin-immunoprecipitation indicated that MBD2a and MBD2c were enriched at OCT4 and NANOG promoter regions in 293T in addition to H1 ESC cells (Fig. 4H). Interestingly, co- and reverse-immunoprecipitation followed by western blot (Fig. 4I–J) revealed that the somatic cell-specific MBD2a isoform exhibits much higher affinity for interaction with members of the transcriptionally repressive NuRD complex, including: HDAC1, HDAC2, RbAp46, MTA2, and Mi-2 (Zhang et al., 1999). The specificity of the MBD2a-NURD interaction was further confirmed by probing for SIN3A, a co-repressor (Zhang et al., 2005) independent of NuRD that did not biochemically interact with either MBD2 isoform (Fig. 4I).
Fig. 4. A general model for regulation of proteome diversity that supports self-renewal in hPSC.
(A–D) Lentiviral-mediated expression of MBD2a but not MBD2c disrupts pluripotency in H1 ESC. Expression of MBD2 isoforms as monitored by (A) qRT-PCR and (B) western blot. Pluripotency in H1 ESC was assessed by (C) colony morphology and (D) expression of OCT4, NANOG, and SOX2. (E–G) Exogenous expression of MBD2c but not MBD2a enhances reprogramming efficiency in BJ DF. (E) Expression of MBD2 isoforms in infected BJ DF as monitored by qRT-PCR. Reprogramming efficiency was assessed by the (F) number and (G) fold change of TRA-1-60 colonies measured across biological replicates. (H) Exogenous MBD2a and MBD2c independently bind to OCT4 and NANOG promoter regions in (top) H1 ESC and (bottom) 293T cells. (I–J) MBD2 interacts with the NuRD complex in an isoform-specific manner. (I) Members of the NuRD transcription repressor complex (HDAC1, HDAC2, MTA2, Mi-2, and RbAp46) co-immunoprecipitate with exogenous FLAG-HA-MBD2a but not FLAG-HA-MBD2c in 293T cells. Neither MBD2 isoform interacts with the SIN3A-Histone deacetylase complex. (J) Co-immunoprecipitation of MTA2 in 293T cells overexpressing FLAG-HA-tagged MBD2 isoforms confirmed the preferential interaction between MBD2a and MTA2, a core NuRD complex member. (K) Proposed model illustrating a putative positive feedback loop, in which the splicing factor SFRS2 along with microRNAs, controlled by the core pluripotency genes, regulate the expression of MBD2 isoforms that either support (MBD2c) or oppose (MBD2a) expression of OCT4, NANOG, and SOX2 through recruitment of the NuRD complex. Error bars represent the mean ± SEM.
Discussion
Pluripotent stem cells are phenotypically well-defined but exhibit significant molecular heterogeneity (Kim et al., 2010). These observations suggest that the core pluripotent factors, OCT4, SOX2, and NANOG, must balance a stochastic transcriptional ground state and yet respond rapidly to exogenous cues in order to properly orchestrate the cell lineages required for life, all from a relatively modest number of protein-coding genes (Wu et al., 2010). Alternative splicing represents a likely pathway whereby the core pluripotency factors can dynamically regulate proteome diversity to support high-fidelity lineage commitment (Wang et al., 2008). Although several examples of alternatively spliced gene products have been functionally validated in pluripotent cells (Das et al., 2011; Gabut et al., 2011; Han et al., 2013; Salomonis et al., 2010), a general framework that mechanistically links OCT4, NANOG, or SOX2 with specific splicing factors, pre-mRNA substrates, and canonical regulators of gene transcription, has yet to be described.
We found that the splicing factor SFRS2 was strongly represented within the pluripotent molecular signature, and moreover that OCT4 bound to SFRS2 promoters in vivo and drove luciferase expression in vitro. These data establish interdependent genetic and functional links between OCT4 and SFRS2 in hPSC. We confirmed a cell type-specific expression pattern for MBD2 isoforms, and found that SFRS2 biochemically targets the pre-mRNA of this methyl-DNA binding protein. We also observed a reciprocal link between OCT4 and MBD2a, manifested at the level of gene expression and pluripotent phenotype. Interestingly, hESC displayed distinct morphologies in response to depletion of SRFR2 or overexpression of MBD2a, suggesting that the splicing factor likely targets additional gene products; indeed it is intriguing to speculate that the pool of pluripotent-specific, alternatively spliced transcripts in our exon-junction microarray data may be rich in previously unrecognized gene isoforms that support self-renewal. Similarly, use of next-generation DNA sequencing technologies may provide an exhaustive set of pluripotent-specific gene isoforms and splicing factor gene targets. Notwithstanding a comprehensive analysis of SFRS2 gene targets, our current results provide compelling mechanistic evidence that the functional role of OCT4 in pluripotent cells extends to the pathways that regulate gene splicing.
Although the editing of pre-mRNA transcripts can be reconstituted in vitro, it has become clear that gene splicing in vivo is intimately linked to transcription, chromatin structure, and histone modifications (Braunschweig et al., 2013; Lin et al., 2008). NuRD is a chromatin remodeling complex that is thought to promote lineage commitment of ESCs via silencing of pluripotency genes (Reynolds et al., 2012). While previous work suggested that NuRD was recruited to methylated DNA by MBD2 (Zhang et al., 1999), we found that although both MBD2a and MBD2c bound to the promoter regions of OCT4 and NANOG in hPSC, only the somatic cell-specific MBD2a isoform biochemically interacts with NuRD. Such isoform-specific recruitment of NuRD may enable pluripotent cells to rapidly regulate their transcriptional profiles in response to specific differentiation cues. Our finding was recently corroborated in murine ESC, along with data suggesting that the region of Mbd2a immediately N-terminal to the MBD domain, but absent in Mbd2c, mediates interaction with NuRD members (Baubec et al., 2013). Interestingly, two recent computational studies suggested that tissue-specific alternative splicing may mediate protein-protein interactions en masse to support distinct phenotypes (Buljan et al., 2012; Ellis et al., 2012). Our data provide a specific example that fits this model, whereby the activity of a chromatin remodeling factor (NuRD) is mediated through interactions with protein isoforms (MBD2) expressed in a cell type-specific manner. Our analysis further revealed that the pluripotent-specific MBD2c isoform augmented reprogramming efficiency of somatic cells, while MBD2a had no effect. This observation is consistent with a strongly repressive role for endogenous MBD2a-NuRD complexes in somatic cells and potentially reconciles discrepancies reported for the role of MBD2 in pluripotent cells (Lee et al., 2012; Onder et al., 2012). Systematic titration of MBD2a levels in the context of enforced MBD2c expression in somatic cells may fully delineate the interplay of these isoforms and reveal whether MBD2a represents a key hurdle to reprogramming. The function of MBD family proteins and isoforms in NuRD and in reprogramming are likely complex, as exemplified by a recent report demonstrating that depletion of the methyl-CpG binding domain protein 3 (MBD3), also a component of NuRD, renders reprogramming deterministic and highly efficient (Rais et al., 2013). Defining the dynamics of these mutually exclusive MBD family-NuRD complexes (Le Guezennec et al., 2006) along with their regulatory target genes in hPSC should shed further light on the mechanisms of somatic cell reprogramming.
Recent evidence suggests that the repressive activity of NuRD is opposed by signaling pathways that support expression of pluripotent factors, hence maintaining a stochastic ground state in which ESC self-renew but are transcriptionally poised for lineage-specific differentiation (Hu and Wade, 2012). We found that several serine residues on SFRS2 were preferentially phosphorylated in pluripotent cells (Table S1). The question of which signaling axis (e.g., AKT/SRPK (Zhou et al., 2012)) mediates phosphorylation on SFRS2, and whether this activity represents a general mechanism to reinforce expression of pluripotent-specific gene isoforms in hPSC, is worthy of future study.
Non-coding RNA has emerged as an important post-transcriptional regulatory pathway in pluripotent cells, with functional links established between specific micro- or long non-coding (lnc)- RNAs and master regulatory transcription factors (Loewer et al., 2010; Marson et al., 2008). The alternatively spliced MBD2 isoforms harbor differences in both their 3′-UTR and protein-coding sequences. As a result, the somatic cell-specific MBD2a isoform is targeted by miR-302 family members. To our knowledge the 3′-UTR of MBD2c is not subject to miR-mediated suppression, although we did observe modest regulation of SFRS2 by miR-301 family members in vitro. These results are consistent with the notion that the microRNA machinery may act synergistically with splicing factors and gene isoforms to either enforce a self-renewing ground state or rapidly translate lineage commitment signals into appropriate transcriptional programs. Further analysis will be required to determine the full extent of microRNA-mediated regulation of proteome diversity and whether lncRNA (Wang et al., 2013) or other non-coding sequences are also involved. Collectively these data may allow a quantitative assessment of the network topology including the relative contribution of each node comprising a putative feedback loop linking the core pluripotent genes with the alternative splicing apparatus and specific gene isoforms.
In summary, we delineate genetic, biochemical, and functional links consistent with a general model (Fig. 4K), in which the master regulators of pluripotency (e.g., OCT4) act in concert with splicing factors (e.g., SFRS2) and the microRNA machinery to mediate protein diversity via alternative splicing (e.g, MBD2), ultimately enforcing a pluripotent ground state.
Experimental Procedures
Cell culture
hPSCs for proteomics and phosphoproteomics were maintained in mTeSR media (Stemcell Technologies) on 6-well plates pre-coated with matrigel (BD Bioscience) as previously described (Park et al., 2008).
Chromatin Immunoprecipitation (ChIP)
ChIP was performed in H1 ESC using anti-OCT4 (Santa Cruz).
Coimmunoprecipitation (CoIP)
Protein CoIP was performed in 293T cells using anti-FLAG gel (Sigma).
Genome-wide expression and alternative splicing data
Gene expression (Table S1) and alternative splicing profiling (Table S4) was performed using Affymetrix arrays. All array data have been deposited in the GEO database under the accession# GSE55673.
Proteomic data
Samples were processed (Ficarro et al., 2009) for protein expression and phosphorylation analyses by 3D RP-SAX-RP LC-MS/MS (Ficarro et al., 2011). Native mass spectrometry data are available for download at: http://blaispathways.dfci.harvard.edu/mz/.
Quantitative Reverse-Transcription-Polymerase Chain Reaction
For quantitative RT-PCR assays, relative gene expression levels in BJ DF or infected H1 ESC were calculated based on the internal standard gene TBP and were normalized to those in either wild-type H1 ESC or in H1 ESC with virus infection control, respectively.
RNA Immunoprecipitation (RIP)
Relative occupancy values (fold enrichments) were calculated by determining the IP efficiency (ratios of the amount of immunoprecipitated RNA to that of the input sample) and normalized to the level observed by immunoprecipitation using non-specific IgG, which was defined as 1.0.
Statistical Methods
The Student’s t test was used to estimate statistical significance (p < 0.05).
Supplementary Material
Highlights.
The splicing factor SFRS2 supports human PSC self-renewal and is a target of OCT4.
SFRS2 mediates splicing of MBD2 isoforms that play opposing roles in pluripotency.
MBD2a but not MBD2c can interact with the NuRD chromatin remodeling complex.
OCT4, SFRS2, and MBD2 comprise a positive feedback loop in human PSC.
Acknowledgments
We thank J. Sahalie for technical assistance, M.W. Lensch for teratoma interpretation, and G. Adelmant for critical reading of the manuscript. Y.-H.L. is supported by the A*Star Investigator research award. H.L. is supported by Mayo Clinic Center for Individualized Medicine and Mayo Clinic Center for Regenerative Medicine. Generous support for this work was provided (to C.J.L) by the Pathology Department of the Brigham and Women’s Hospital and the National Blood Foundation. J.J.C. is supported by Howard Hughes Medical Institute, SysCODE (Systems-based Consortium for Organ Design & Engineering) and NIH grant # RL1DE019021. Generous support for this work was provided (to J.A.M.) by the Dana-Farber Cancer Institute, the Harvard Stem Cell Institute (#P01GM099117), and the NIH/NINDS #P01NS047572. Y.L. acknowledges support from T32HL66987-11. G.Q.D. is supported by the Howard Hughes Medical Institute. G.Q.D., J.J.C., and J.A.M. acknowledge support from NIH/NHLBI #5RC2HL102815-02. G.Q.D. serves on the scientific advisory boards of iPierian, Inc., Verastem, Inc., and MPM Capital.
Footnotes
Y.L., Y.-H.L., H.L., M.C., C.J.L., J.J.C., G.Q.D, and J.A.M. designed research; Y.L., Y.-H.L., M.C., S.B.F., and C.-X.T. performed experiments; H.L., J.R.P., and N.S. performed bioinformatics analysis; S.T.A. derived the HFMSC-iPS cell line; Y.L. and J.A.M. coordinated the project and wrote the manuscript. All authors provided critical editing of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Ivánek R, Lienert F, Schübeler D. Methylation-Dependent and -Independent Genomic Targeting Principles of the MBD Protein Family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Das S, Jena S, Levasseur DN. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J Biol Chem. 2011;286:42690–42703. doi: 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O’Hanlon D, Kim PM, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- Ficarro SB, Adelmant G, Tomar MN, Zhang Y, Cheng VJ, Marto JA. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, Garg B, Adelmant G, Webber JT, Luckey CJ, Marto JA. Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Mol Cell Proteomics. 2011;10:O111.011064. doi: 10.1074/mcp.O111.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. An Alternative Splicing Switch Regulates Embryonic Stem Cell Pluripotency and Reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WAM, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung H-K, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013 doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wade PA. NuRD and Pluripotency: A Complex Balancing Act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LIR, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Prasain N, Chae HD, Kim YJ, Mantel C, Yoder MC, Broxmeyer HE. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells. 2012;31:666–681. doi: 10.1002/stem.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Loh YH, Tesar PJ, Daley GQ. Accessing naïve human pluripotency. Curr Opin Genet Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Brumbaugh J, Wenger CD, Tian S, Probasco MD, Bailey DJ, Swaney DL, Tervo MA, Bolin JM, Ruotti V, et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O’Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010;107:10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, Wu M, Xiong J, Guo X, Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. PNAS. 2010 doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fatima N, Dufau ML. Coordinated changes in DNA methylation and histone modifications regulate silencing/derepression of luteinizing hormone receptor gene transcription. Mol Cell Biol. 2005;25:7929–7939. doi: 10.1128/MCB.25.18.7929-7939.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.