Abstract
Antimicrobial peptides continue to garner attention as potential alternatives to conventional antibiotics. Hipposin is a histone-derived antimicrobial peptide (HDAP) that was previously isolated from Atlantic halibut. Though its potency against several bacterial strains has been documented, its antibacterial mechanism had not been characterized. The mechanism of this peptide is particularly interesting to consider since the full hipposin sequence contains the sequences of parasin and buforin II (BF2), two other known antimicrobial peptides that act via different antibacterial mechanisms. While parasin kills bacteria by inducing membrane permeabilization, buforin II enters cells without causing significant membrane disruption, harming bacteria through interactions with intracellular nucleic acids. In this study, we used a modular approach to characterize hipposin and determine the role of the parasin and buforin II fragments in the overall hipposin mechanism. Our results show that hipposin kills bacteria by inducing membrane permeabilization, and this membrane permeabilization is promoted by the presence of the N-terminal parasin domain. Portions of hipposin lacking the parasin sequence do not cause membrane permeabilization and function more similarly to buforin II. We also determined that the C-terminal portion of hipposin, HipC, is a cell-penetrating peptide that readily enters bacterial cells but has no measurable antimicrobial activity. HipC is the first membrane active histone fragment identified that does not kill bacterial or eukaryotic cells. Together, these results not only characterize hipposin but also provide a useful starting point for considering the activity of chimeric peptides made by combining peptides that operate via differing mechanisms.
Keywords: Histone-derived antimicrobial peptide, hipposin, buforin, parasin, membrane translocation, membrane permeabilization
1.0 Introduction
Over the past decades, there has been increasing interest in the promise of antimicrobial peptides as an alternative to conventional therapeutic agents to treat bacterial infections [1, 2] These peptides have been isolated from many natural sources and are believed to play a role in innate immunity for a diverse array of organisms ranging from bacteria to plants to people. Successful development of these peptides as therapeutic agents will require researchers to have a solid understanding of how these peptides kill bacteria and how mechanisms are influenced by the primary structure of peptides.
Recently, there has been increasing interest in histone-derived antimicrobial peptides (HDAPs) [3, 4]. HDAPs have been isolated from many natural sources and represent fragments of the histone core subunits. Other work has focused on the design of novel HDAPs [5]. Of the HDAPs studied, some of the most active and best characterized HDAPs are derived from the histone H2A subunit [5-9].
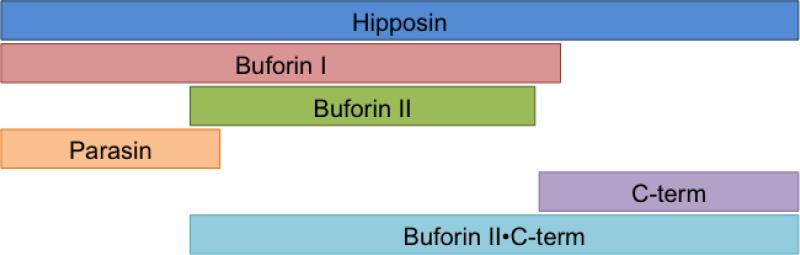
Hipposin is a histone H2A derived antimicrobial peptide isolated from the Atlantic halibut, Hippoglossus hippoglossus L [6, 10]. Although the precise role of hipposin in halibut innate immunity has not been elucidated, its localization in the skin mucus is similar to the location of other antimicrobial peptides isolated from fish [11]. Moreover, its derivation from histone H2A is similar to that of other antimicrobial or antifungal peptides that are produced from the cleavage of larger proteins that serve other cellular roles, such as human thrombin [12] and shrimp hemocyanin [13]. Hipposin is particularly notable as it is a relatively large, 51 residue, antimicrobial peptide whose sequence contains regions essentially identical to two smaller HDAPs with different antibacterial mechanisms of action (Figure 1). The 19 N-terminal residues of hipposin are highly homologous with parasin I, which is known to kill bacteria by inducing membrane permeabilization [8, 14]. A 21 amino acid section located in the middle of the hipposin sequence is identical to buforin II (BF2), a peptide that translocates into cells without causing much membrane permeabilization and is believed to kill bacteria through interactions with intracellular nucleic acids [7, 15-19]. Another naturally occurring peptide, buforin I (BF1) is essentially a fusion of the parasin and BF2 peptides [9]. However, the C-terminal region of hipposin, referred to here as HipC, has not been previously isolated or characterized.
Figure 1.
Schematic showing the primary sequence relationship between hipposin and its naturally occurring fragments, parasin, buforin II (BF2) and buforin I (BF1). Synthetic portions of hipposin considered in this study, including the C-terminal HipC domain, are also shown.
This study investigates the mechanism of hipposin and its related peptides by using a modular approach, characterizing the activity and mechanism of different hipposin fragments. Using this approach, we determined that hipposin kills bacteria by inducing membrane permeabilization, and this membrane permeabilization is promoted by the presence of the N-terminal parasin domain. We also determined that the C-terminal HipC portion is a cell-penetrating peptide that readily enters bacterial cells but has no antimicrobial activity. Thus, HipC is the first membrane active histone fragment identified that does not kill bacterial or eukaryotic cells.
2.0 Materials and Methods
2.1 Peptide design and synthesis
The peptide sequences used in this study are shown in Table 1. Although the sequence of hipposin is very similar to that of parasin, BF1, and BF2, there are a few single-site differences between these proteins. While most of these changes are rather conservative, the initial N-terminal residue is a more substantial change from Ser 1 in hipposin to Lys 1 in parasin. Since mutations to the Lys 1 residue of parasin alter its activity [8], for consistency in our comparisons we decided to use an S1K mutation in all of our hipposin-derived peptides. We also introduced tryptophan residues at positions 14, 25, and 50 in hipposin and analogous sites in hipposin fragments in order to allow for easier peptide quantification and the lipid vesicle translocation assays of HipC. Initial measurements in our lab showed that the S1K/A14W/F25W/Y50W changes to hipposin did not cause qualitative changes in antimicrobial activity or membrane permeabilization behavior (data not shown). Thus, all data for the full-length hipposin sequence in this paper were collected for the S1K/A14W/F25W/Y50W peptide, which we refer to as HipABC. These substitutions were made in the hipposin fragments used in our modular analysis (Table 1). Table 1 notes how these sequences compare to the physiological sequences of hipposin, buforin I (BF1), buforin II (BF2) and parasin. Notably, the HipB region that was analogous to buforin II (BF2) included the F10W mutation that has been widely used in previous studies and shown to have analogous properties to the wildtype peptide [15, 16, 18, 20, 21].
Table 1.
Amino acid sequences of peptides used in this study. The relationship to physiologically observed peptides is given in parentheses where relevant.
| Peptide | Sequence |
|---|---|
| HipABC (Hipposin S1K/A14W/F25W/Y50W) | KGRGKTGGKARAKWKTRSSRAGLQWPVGRVHRLLRKGNYAHRVGAGAPVWL |
| HipBC | TRSSRAGLQWPVGRVHRLLRKGNYAHRVGAGAPVWL |
| HipC | GNYAHRVGAGAPVWL |
| HipB (BF2 F10W) | TRSSRAGLQWPVGRVHRLLRK |
| HipAB (BF1 A1K/A14W/F25W) | KGRGKQGGKVRAKWKTRSSRAGLQWPVGRVHRLLRKGNY |
Peptides were synthesized both with and without an N-terminally linked biotin at >95% purity by NeoBioSci (Cambridge, MA). All peptide stocks were quantitated from tryptophan absorbance using the average of at least three A280nm measurements.
2.2 Radial diffusion assays
The radial diffusion assay was performed using E. coli (ATCC #25922) and B. subtilis (ATCC #6051) in a manner analogous to previous studies [22]. Cells picked from frozen stocks were incubated overnight at 37°C in 30% w/v tryptic soy broth. The overnight culture is then diluted 1:500 or 1:1000 in fresh tryptic-soy broth (TSB) and grown for 2.5 h. Bacteria were pelleted via centrifugation at 880×g for 10 minutes at 4°C, washed once with 10 mM phosphate buffer (pH 7.4), then pelleted again and resuspended. 4×106 CFU of bacteria are mixed with 10 mL of molten agarose gel (10 mM phosphate, 1% TSB v/v, 1% agarose w/v, pH 7.4) and allowed to solidify on a petri dish. Wells were formed in the solid media using a pipette attached to a bleach trap, 2 μL of 1 × 10−4 M peptide solution was added to each well, and plates were incubated at 37°C for 3 hours. 10 mL of overlay gel (30% w/v TSB, 1% w/v agarose) was poured over the underlay gel and incubated for 12-18 hours at 37 °C. The diameter of bacterial clearing around each well was measured at 7× magnification. The clearance around a well containing only water was 1 mm. Data were collected from at least three independent plates.
2.3 MTT assay
Eukaryotic cell cytotoxicity was determined by the MTT assay as described by Wu et al. [23]. WI-38 human fibroblast cells were seeded in 96-well plates at 3 × 103 cells/well and incubated overnight in RPMI medial lacking phenol red at 37°C and 5% CO2. Cells were then incubated with 1 μM peptide solutions for 48 hours. Each peptide solution was tested in quadruplicate. Cell viability was measured by introducing the 3-(4,5-dimethylthiazolyl-2)-2, 5-diphenyltetrazoliumbromide (MTT) indicator (0.5 mg/mL) (Invitrogen, Carlsbad, CA) into treated cell solutions for 4 hours. MTT is reduced to a purple formazan salt by metabolically active cells; formazan crystals were dissolved after 4 hours by addition of 5% sodium dodecyl sulfate (SDS) (Sigma Aldrich), and mixtures were incubated overnight. Absorbance was read at 570 nm on a Molecular Devices SpectraMax M3 microtiter plate reader (Sunnyvale, CA). Percent cell viability was calculated as the ratio of the A570nm of cells treated with peptide solutions over the A570nm of cells treated with water. Staurosporine, a known cytotoxic agent, was used as a comparison for negative growth. Viability measurements reported are averaged over at least seven samples collected in at least two independent experiments.
2.4 Propidium iodide uptake assay
E. coli (ATCC #25922) bacteria picked from frozen stock were grown overnight in TSB media (30% w/v TSB) at 37°C. The overnight culture was diluted 1:1000 in fresh TSB and allowed to grow 2.5 hours to mid-log phase. Bacteria were pelleted via centrifuge at 880xg for 10 minutes at 4°C, washed once with 10 mM phosphate buffer (pH 7.4), pelleted again, and resuspended to an optical density of 0.5. Propidium iodide was added to a concentration of 20 μg/mL and the system was allowed to equilibrate. PI/DNA complexation was measured at an excitation of 535 nm and emission of 617 nm on a Varian Cary Eclipse fluorescence spectrophotometer (Agilent Technologies, Santa Clara, CA). Following equilibration, peptide solution was added to a concentration of 2 μM and fluorescence was monitored until new equilibrium was reached. Addition of an equal volume of water without peptide was used as a negative control. Increase in fluorescence due to the peptide's presence was measured by comparing the fluorescence 5 minutes after peptide addition to the averaged fluorescence during the one minute before peptide addition. Averages of at least three independent experiments are reported.
2.5 Confocal microscopy imaging of bacteria
Peptide translocation into E. coli (Top10 containing a pET45b plasmid for ampicillin resistance) was visualized as described by Park et al. [19]. Overnight cultures from frozen bacterial stocks were diluted 1:100 in TSB liquid media and allowed to grow to mid-logarithmic phase. Bacteria were then pelleted by centrifugation at approximately 880xg and resuspended in sterile 10 mM sodium phosphate buffer (pH 7.4). Bacterial cells at a concentration of 107 CFU/mL were exposed to 4 μg/mL biotinylated peptide solutions for 30 minutes at 37 °C. Cell/peptide solutions were then placed on a poly-L-lysine coated glass slide and treated with 0.066% Triton-X for 1 – 2 minutes. Biotinylated peptides were rendered visible by the addition of streptavidin conjugated with AlexaFluor 488 (Invitrogen) at a final concentration of 5 μg/mL. Cells were visualized with a Leica TCS SP5 laser scanning confocal microscope with excitation at 488 nm by an argon laser at 20% output. All 8-bit 512×512 images represent the average of six scans at 63X magnification (Leica Plan-Apochromat objective; numerical aperture 1.40). Composite images were produced by Leica LAS AF software (Buffalo Grove, IL). Z-stacks of 0.07 – 0.09 μm thickness were analyzed using ImageJ image analysis software (NIH) in conjunction with the LOCI BioFormats plug-in (LOCI, Madison, WI). Unlabeled images were evaluated by outside individuals for location of peptide fluorescence within the cell to prevent bias in the reading of the data.
2.6 Lipid vesicle membrane translocation assay
Measurement of translocation of HipC into lipid vesicles was performed using the technique proposed by Matsuzaki and co-workers [16, 18] as implemented in Spinella et al [24]. Phospholipids dissolved in chloroform were obtained from Avanti Polar Lipids (Alabaster, AL). Mixtures of phosphtidylcholine (POPC), phosphatidylglycerol (POPG), and 5-dimethylaminonapthlene-1-sulfonyl phosphatidylethanolamine (DNS-POPE) were made in a 75:20:5 ratio. Chloroform was evaporated using a nitrogen gas stream. Lipid cakes were desiccated overnight. The vesicles were rehydrated in either HEPES buffer (10mM HEPES, 45mM NaCl, 1 mM EDTA, pH 7.4) containing 0.2mM porcine trypsin (Sigma Aldrich, St. Louis, MO) for the experimental condition or in HEPES buffer containing both 0.2 mM porcine trypsin as well as 2.0 mM Bowman-Birk trypsin inhibitor (BBI) (Sigma Aldrich) for the control. Vesicles were then subjected to 5 freeze-thaw cycles before extrusion through a nucleopore track etch membrane with 0.1 μm pores (Whatman) to generate uniform vesicle size. To remove phosphorus salts from control vesicles, an estimated 0.3 μM of control vesicles were spun at 13,000 rpm for ten minutes three times in Pall Corporation Nanosep 10K OmegaTM microcentrifuge tubes and rinsed with HEPES buffer after each spin. Vesicle concentration was determined by measuring phosphorus content in triplicate using a standard curve of phosphate solutions following the previously described procedure (http://www.avantilipids.com/index.php?%20option=com_content&view=article&id=168 6&Itemid=405).
Quantitated vesicles were then diluted to 0.25 mM in a solution with trypsin inhibitor at ten times the trypsin concentration. 2 μl of peptides prepared at 2.5×104 M were added to each well of an opaque 96 well plate (VWR, Radnor, PA). 198 μL of vesicle solution was added to each well, and fluorescence was monitored for 25 minutes with an excitation wavelength of 280 nm and an emission wavelength of 525 nm, using a SpectraMax M3 multi-mode microplate reader (Molecular Devices, Sunnyvale, CA). The final average fluorescence value (Favg) for both control and experimental conditions was calculated by dividing the average fluorescence values during the last minute of the experiment by the fluorescence 10 s after combining peptide and vesicle samples (F0). The F0 was taken at 10 s to reduce artifacts in the fluorescence signal that can occur immediately after mixing in the sample. A quantitative translocation ratio (TR) was calculated for the peptide by dividing the Favg for the control vesicles by the Favg for the experimental vesicles. Six replicates of this experiment were performed for HipC using three independent preparations of experimental and control vesicles.
3.0 Results and Discussion
3.1 Hipposin appears to kill bacteria through membrane permeabilization
As observed for hipposin [10], HipABC is active against both gram-negative (E. coli) and gram-positive (B. subtilis) bacteria (Table 2). This activity appears selective for bacteria over eukaryotic cells, as fibroblast cells exposed to a comparatively high (1 μM) concentration of HipABC remain viable (Table 2).
Table 2.
Activity of peptides against bacterial and eukaryotic cells. Antibacterial activity is shown as diameters of clearance (in mm) from radial diffusion assays. Cytotoxicity against eukaryotic cells is shown as the percent viability of WI-38 fibroblasts cells exposed to 1 μM of peptide measured using an MTT assay. Error is expressed as standard deviation.
| Peptide | Radial diffusion assay data | MTT % viability | |
|---|---|---|---|
| E. coli | B. subtilis | WI-38 | |
| HipABC | 7.5±0.4 | 8.5±1.2 | 94±7 |
| HipBC | 7.3±0.2 | 6.5±0.3 | 91±5 |
| HipC | 1.1±0.1 | |1.3±0.1 | 88±7 |
| HipB | 9.1±0.7 | 7.2±0.6 | 91±6 |
| HipAB | 8.7±0.4 | 7.7±1.2 | 94±5 |
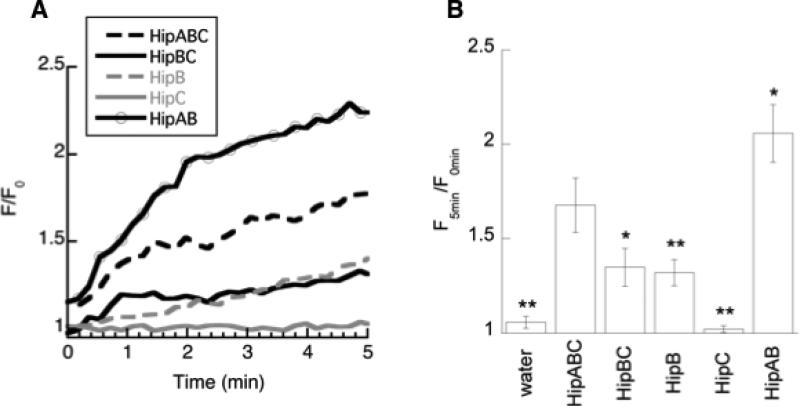
HipABC is observed to kill bacteria through membrane permeabilization. Bacterial cells exposed to HipABC in the presence of propidium iodide (PI) show dramatically increased fluorescence due to entry of PI into cells (Fig. 2). Notably, HipABC induced even more permeabilization than magainin under analogous conditions [25], a peptide often considered a prototypical membrane permeabilizing peptide [26]. Localization of biotinylated HipABC using streptavidin-AlexaFluor conjugates is observed around the cellular membrane with a relative lack of fluorescence inside the cell (Fig. 3, Supplemental Figure 1), echoing patterns of fluorescence localization in other histone-derived peptides such as parasin that are known to cause permeabilization [8]. We also noted a significant amount of cellular debris present in cellular samples treated with HipABC, which likely resulted from cells that had already been fragmented by the peptide at the time of imaging.
Figure 2.
Membrane permeabilization in E. coli caused by the addition of peptides measured using a propidium iodide uptake assay. A) Representative data for peptides considered in this study. Data is plotted as fluorescence at a given time after peptide addition relative to the average fluorescence in the minute before peptide was added (F/F0). B) Permeabilization results averaged over multiple trials, reported as a ratio of the fluorescence five minutes after peptide addition divided by the fluorescence averaged for the minute before peptide was added. Average of at least three trials is shown with variation provided as standard error. Measurements that were significantly different from HipABC at the p<0.10 (*) and p<0.05 (**) level are denoted with asterisk.
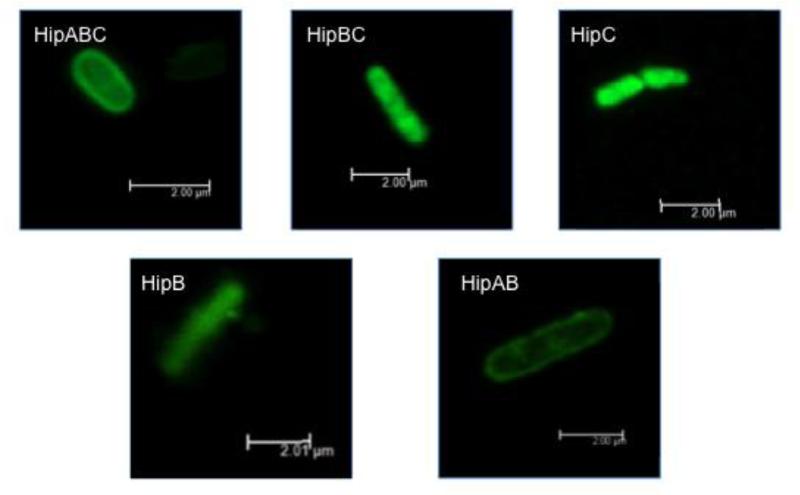
Figure 3.
Composite confocal microscopy images of E. coli after exposure to HipABC, HipBC, HipC, HipB, and HipAB. Biotinylated peptides were visualized with streptavidin-AlexaFluor 488 excited by an argon laser at 488 nm. Composite images created by overlaying individual z-stack slices taken at 0.07 – 0.1 μm with a six frame-average. Scale bars indicate 2 μm.
3.2 The N-terminal portion of hipposin analogous to parasin causes membrane permeabilization
The full hipposin peptide consists of three regions: the N-terminal parasin domain, the middle buforin II (BF2) domain and the C-terminal region of unknown function (Fig. 1). In our modular analysis, we have termed these HipA (N-terminal), HipB (middle) and HipC (C-terminal), respectively. Since parasin has previously been observed to induce membrane permeabilization in bacteria [8], we hypothesized that the HipA region might promote the membrane permeabilization activity of hipposin. To test this hypothesis, we measured the membrane permeabilization of a hipposin fragment omitting the parasin region, which we refer to as HipBC (Table 1).
As expected, HipBC induced very little membrane permeabilization (Fig. 2). However, despite its inability to disrupt membranes, HipBC did maintain similar antimicrobial activity to that observed for HipABC (Table 2). We expect that this behavior resulted from the increased membrane translocation of HipBC, which was observed to readily enter cells visualized using confocal microscopy (Fig. 3). Notably, HipBC was seen inside cells and not only localized at the membranes as determined when considering z-stack images taken through the z-plane of cells of interest (Supplemental Figure 2). The tendency of HipBC to enter cells is similar to that observed in BF2 (Fig. 3, Supplemental Figure 3).
3.3 The hipposin C-terminal region, HipC, readily enters bacterial cells without causing cell death
Although the HipA (parasin) and HipB (BF2) sections of hipposin have been characterized, previous work had not attributed any function to the C-terminal region of hipposin, HipC. Thus, we were interested in whether this fragment had any antimicrobial activity or membrane interactive properties of its own. Radial diffusion assays indicate that HipC had no activity greater than water against either E. coli or B. subtilis (Table 2). HipC also shows no measurable cytotoxicity against mammalian cells (Table 2).
However, despite its lack of activity, confocal microscopy suggests that HipC readily enters bacterial cells (Fig. 3). Analysis of the z-plane in HipC-treated bacterial cells demonstrates localization throughout the cell (Supplemental Figure 4), indicating an ability to translocate into cells without causing significant membrane permeabilization, as confirmed by propidium iodide uptake assays (Fig. 2). In fact, HipC caused even less membrane permeabilization than HipB (BF2), and was essentially indistinguishable from the water control in our PI assay.
In order to determine whether HipC requires any cellular receptors for membrane translocation we also considered its ability to translocate into lipid vesicles. These experiments utilized a translocation assay used previously for BF2 and other HDAPs [16, 18, 25]. Lipid vesicles containing trypsin are produced with membranes doped with dansylated-PE lipids. The dansylated-PE lipids decorating the membrane have a FRET response when intact peptides associate to the membrane. However, as peptides translocate into the vesicles they are proteolyzed by the encapsulated trypsin, decreasing the FRET response. Degradation of peptides interacting with vesicles containing inhibited trypsin was examined to control for incomplete inhibition of trypsin outside experimental vesicles. A decrease in FRET signal in experimental conditions relative to this control indicates peptide translocation into the vesicle.
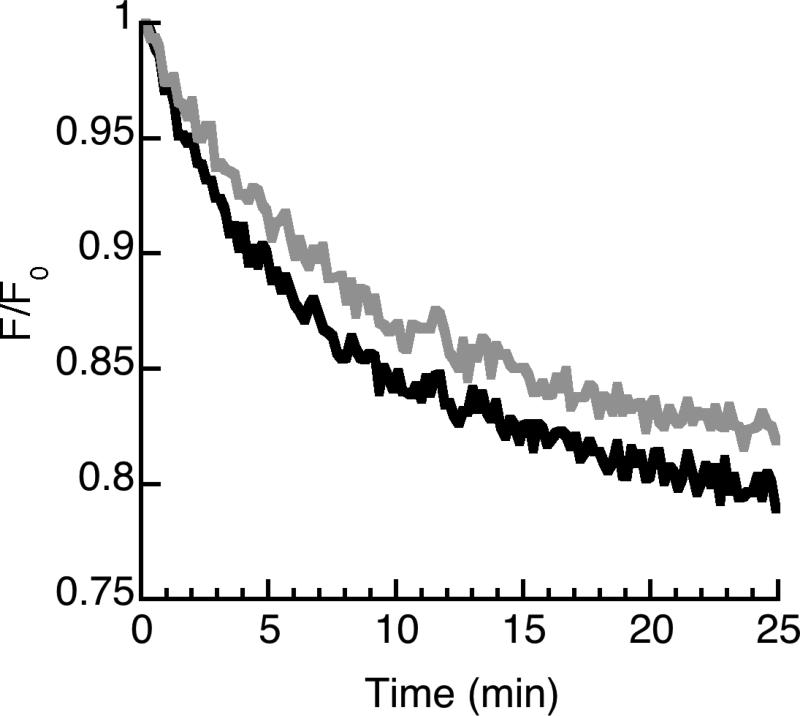
Our data are consistent with the entry of HipC into lipid vesicles, as shown by the decreased FRET response in vesicles containing active trypsin compared to vesicles with inhibited trypsin (Fig. 4, Table 3). In particular, the average translocation ratio for HipC (1.020±0.007) is greater than unity and significantly greater than that of DesHDAP3, a non-translocating peptide considered in previous studies [25] (Table 3). However, the translocation ratio of HipC is rather low and significantly less than that of the previously studied translocating peptides, buforin II and DesHDAP1 (Table 3) [25]. Thus, the translocation of HipC into vesicles appears relatively modest, particularly compared to its entry into bacteria (Fig. 3). Nonetheless, the lack of antibacterial activity and low levels of membrane permeabilization suggest that HipC may represent a promising peptide for cellular transfection applications.
Figure 4.
Representative measurement of HipC translocation into lipid vesicles using an encapsulated trypsin assay. Peptide was exposed to lipid vesicles containing trypsin. Data are shown as the FRET signal at 525 nm (black) relative to the initial fluorescent signal (F/Fo). In the control trace (gray), peptides were exposed to vesicles containing both trypsin and trypsin inhibitor to control for incomplete inhibition of trypsin outside the vesicle under experimental conditions. The lower fluorescence for the experimental condition denotes translocation.
Table 3.
Translocation of HipC into lipid vesicles compared to histone-derived antimicrobial peptides (BF2, DesHDAP1 and DesHDAP3) characterized in previous work [25]. The average translocation ratios over a minimum of three experiments are expressed as [(F0/F)control/(F0/F)experimental], with errors expressed as standard error. Ratios that were significantly different than HipC at p<0.10 (*) or p<0.05 (**) are denoted with asterisk.
| Peptide | Translocation ratio |
|---|---|
| HipC | 1.02±0.01 |
| BF2 | 1.07±0.03 (*) |
| DesHDAP1 | 1.13±0.03 (**) |
| DesHDAP3 | 0.97±0.02 (*) |
To evaluate the role of the HipC region on the overall activity of hipposin, we tested the activity and mechanism of HipAB, which lacks the HipC region (Table 1). This HipAB peptide is homologous to the buforin I peptide considered in previous work [9]. HipAB demonstrated similar activity to HipABC in radial diffusion assays (Table 2), implying that HipC has relatively little impact on the activity of the larger hipposin peptide. Consistent with this observation, the presence of the HipC region in HipABC compared to HipAB generally did not seem to alter the mechanisms of peptide action. Both peptides caused relatively large amounts of membrane permeabilization in PI assays, although the presence of the HipC domain in HipABC appears to inhibit permeabilization (Fig. 2). Peptide localization studies suggest similar patterns of localization to the bacterial membrane for both peptides (Fig. 3, Supplemental Figure 5), although the presence of HipC may somewhat enhance the amount of cellular entry observed for HipABC compared to HipAB.
4.0 Conclusions
Despite the initial identification of hipposin as a broadly effective antimicrobial peptide almost a decade ago [6, 10], its mechanism of action had not been previously characterized. This study shows that hipposin kills bacteria through a mechanism involving significant membrane permeabilization. Interestingly, this overall mechanism arises in a peptide that is made from the combination of two peptides, parasin (HipA) and BF2 (HipB), that independently operate via different mechanisms. In the context of hipposin, it appears that the presence of the permeabilizing HipA domain effectively dictates the mechanism of the larger peptide. The removal of this domain results in a peptide (HipBC) that causes less membrane permeabilization and improved translocation into cells. In fact, the presence of the N-terminal HipA domain generally appears to increase membrane permeabilization, as observed for HipABC and HipAB versus the other peptides considered (Fig. 2). HipABC and HipAB also showed increased membrane localization and less translocation compared to the other peptides (Fig. 3). These observations are consistent with the overall membrane permeabilizing mechanism observed in previous studies for parasin, which is analogous to HipA [8].
This study also identified a novel peptide derived from the C-terminal region of hipposin, HipC, that translocates across cell membranes without causing significant membrane permeabilization. This region is not required for translocation in hipposin fragments lacking HipA, as the HipB fragment can translocate on its own. However, the presence of HipC does seem to modulate the membrane permeabilization properties of the HipA fragment (Fig. 2) and may increase translocation slightly in HipABC compared to HipAB (Fig. 3). Unlike BF2, HipC shows no antibacterial activity, and thus bears similarity to other cell-penetrating peptides, such as Tat or penetratin [27, 28]. Future work should further explore the translocation mechanism of HipC, particularly in light of its potential applications in systems where antimicrobial activity would be problematic, such as the introduction of genetic material into bacterial cells.
More generally, this study of hipposin gives potential insight into the possibility of designing custom antimicrobial peptides in a modular fashion by combining peptides of known antimicrobial function. The combination of different peptides is particularly attractive as the number of characterized antimicrobial peptides has increased dramatically over the past several years [29]. Previous work has suggested that some antimicrobial peptides might act synergistically if used in combinations against cells. Some examples of this have utilized “cocktails” consisting of different peptides, such as tachyplesin and magainin [30]. Other studies have proposed using hybrid chimeric peptides as a way of enhancing activity. For example, researchers recently found that chimeric peptides combining lactoferricin and lactoferrin [31] or the human β-defensins 2 and 3 (HBD2 and HBD3) had increased activity relative to an equimolar mixture of its two components [32]. These previous studies have focused on combining peptides that operate via a permeabilizing mechanism, although an increasing number of antimicrobial peptides have been identified that operate via mechanisms other than membrane permeabilization. Other research has focused on combining cell-penetrating peptides, such as TAT, with other bioactive moieties that would promote release from endosomes upon cell entry [33, 34].
Our results provide a useful starting point for systematically considering the relationship between modules in a larger peptide. A simplistic reading of the results in this study could lead to the inference that combining a permeabilizing peptide “module” and translocating peptide “module” will lead to a chimeric peptide that operates via a permeabilizing mechanism. However, the relative ordering and spacing of different modules in a chimeric protein may impact the mechanism of designed peptides in more subtle ways. Thus, more systematic considerations of different peptide combinations will be necessary to determine how much this relationship can be generalized to other peptides.
Supplementary Material
Highlights.
Hipposin kills bacteria through a membrane permeabilizing mechanism
The parasin domain promotes the membrane permeabilization of hipposin
Portions of hipposin lacking the parasin domain translocate membranes
HipC, the C-terminal domain of hipposin, is a cell-penetrating peptide
HipC is the first membrane active histone fragment that does not kill bacteria
Acknowledgements
Research funded by National Institute of Allergy and Infectious Diseases (NIH-NIAID) award R15AI079685, the Camille and Henry Dreyfus Foundation, and the Wellesley College Staley Fund. D. E. E. is a Henry Dreyfus Teacher-Scholar.
Abbreviations
- AMP
Antimicrobial peptide
- HDAP
Histone-derived antimicrobial peptide
- BF1
Buforin I
- BF2
Buforin II
- DesHDAP
Designed histone-derived antimicrobial peptide
- MTT
3-(4,5-dimethylthiazolyl-2)-2, 5-diphenyltetrazoliumbromid
- PI
propidium iodide
- POPC
phosphatidylcholine
- POPG
Phosphatidylglycerol
- DNS-POPE
5-dimethylaminonaphthalene-1-sufonyl phosphatidylethanolamine
- SDS
sodium dodecyl sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hancock RE, Patrzykat A. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr Drug Targets Infect Disord. 2002;2:79–83. doi: 10.2174/1568005024605855. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Iwamuro S. Potential roles of histones in host defense as antimicrobial agents. Infect Disord Drug Targets. 2008;8:195–205. doi: 10.2174/1871526510808030195. [DOI] [PubMed] [Google Scholar]
- 4.Smith VJ, Desbois AP, Dyrynda EA. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar Drugs. 2010;8:1213–1262. doi: 10.3390/md8041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao HS, Spinella SA, Lee AT, Elmore DE. Design of novel histone-derived antimicrobial peptides. Peptides. 2009;30:2168–2173. doi: 10.1016/j.peptides.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Birkemo GA, Luders T, Andersen O, Nes IF, Nissen-Meyer J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.) Biochim Biophys Acta. 2003;1646:207–215. doi: 10.1016/s1570-9639(03)00018-9. [DOI] [PubMed] [Google Scholar]
- 7.Cho JH, Sung BH, Kim SC. Buforins: Histone H2A-derived antimicrobial peptides from toad stomach. Biochimica Et Biophysica Acta-Biomembranes. 2009;1788:1564–1569. doi: 10.1016/j.bbamem.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 8.Koo YS, Kim JM, Park IY, Yu BJ, Jang SA, Kim KS, Park CB, Cho JH, Kim SC. Structure-activity relations of parasin I, a histone H2A-derived antimicrobial peptide. Peptides. 2008;29:1102–1108. doi: 10.1016/j.peptides.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Park CB, Kim MS, Kim SC. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochemical and Biophysical Research Communications. 1996;218:408–413. doi: 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- 10.Birkemo GA, Mantzilas D, Luders T, Nes IF, Nissen-Meyer J. Identification and structural analysis of the antimicrobial domain in hipposin, a 51-mer antimicrobial peptide isolated from Atlantic halibut. Biochim Biophys Acta. 2004;1699:221–227. doi: 10.1016/j.bbapap.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Smith VJ, Desbois AP, Dyrynda EA. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar Drugs. 2010;8:1213–1262. doi: 10.3390/md8041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papareddy P, Rydengard V, Pasupuleti M, Walse B, Morgelin M, Chalupka A, Malmsten M, Schmidtchen A. Proteolysis of human thrombin generates novel host defense peptides. PLoS Pathog. 2010;6:e1000857. doi: 10.1371/journal.ppat.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Destoumieux-Garzon D, Saulnier D, Garnier J, Jouffrey C, Bulet P, Bachere E. Crustacean immunity. Antifungal peptides are generated from the C terminus of shrimp hemocyanin in response to microbial challenge. J Biol Chem. 2001;276:47070–47077. doi: 10.1074/jbc.M103817200. [DOI] [PubMed] [Google Scholar]
- 14.Park IY, Park CB, Kim MS, Kim SC. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998;437:258–262. doi: 10.1016/s0014-5793(98)01238-1. [DOI] [PubMed] [Google Scholar]
- 15.Uyterhoeven ET, Butler CH, Ko D, Elmore DE. Investigating the nucleic acid interactions and antimicrobial mechanism of buforin II. FEBS Lett. 2008;582:1715–1718. doi: 10.1016/j.febslet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, Chikushi A, Tougu S, Imura Y, Nishida M, Yano Y, Matsuzaki K. Membrane translocation mechanism of the antimicrobial peptide buforin 2. Biochemistry. 2004;43:15610–15616. doi: 10.1021/bi048206q. [DOI] [PubMed] [Google Scholar]
- 17.Park CB, Kim HS, Kim SC. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochemical and Biophysical Research Communications. 1998;244:253–257. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi S, Takeshima K, Park CB, Kim SC, Matsuzaki K. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: Proline as a translocation promoting factor. Biochemistry. 2000;39:8648–8654. doi: 10.1021/bi0004549. [DOI] [PubMed] [Google Scholar]
- 19.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is reponsible for the cell-penetrating ability of buforin II. Proc Natl Acad Sci USA. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming E, Maharaj NP, Chen JL, Nelson RB, Elmore DE. Effect of lipid composition on buforin II structure and membrane entry. Proteins-Structure Function and Bioinformatics. 2008;73:480–491. doi: 10.1002/prot.22074. [DOI] [PubMed] [Google Scholar]
- 21.Xie Y, Fleming E, Chen JL, Elmore DE. Effect of proline position on the antimicrobial mechanism of buforin II. Peptides. 2011;32:677–682. doi: 10.1016/j.peptides.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 23.Wu JM. Structure and function of a custom anticacner peptide. CB1a, Peptides. 2009;30:839–848. doi: 10.1016/j.peptides.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Spinella SA, Nelson RB, Elmore DE. Measuring peptide translocation into large unilamellar vesicles. J Vis Exp. 2012:e3571. doi: 10.3791/3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavia KE, Spinella SA, Elmore DE. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim Biophys Acta. 2012;1818:869–876. doi: 10.1016/j.bbamem.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 27.Dupont E, Prochiantz A, Joliot A. Penetratin story: an overview. Methods Mol Biol. 2011;683:21–29. doi: 10.1007/978-1-60761-919-2_2. [DOI] [PubMed] [Google Scholar]
- 28.Jones AT, Sayers EJ. Cell entry of cell penetrating peptides: tales of tails wagging dogs. J Control Release. 2012;161:582–591. doi: 10.1016/j.jconrel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi S, Hirakura Y, Matsuzaki K. Bacteria-selective synergism between the antimicrobial peptides alpha-helical magainin 2 and cyclic beta-sheet tachyplesin I: toward cocktail therapy. Biochemistry. 2001;40:14330–14335. doi: 10.1021/bi015626w. [DOI] [PubMed] [Google Scholar]
- 31.Haney EF, Nazmi K, Bolscher JG, Vogel HJ. Structural and biophysical characterization of an antimicrobial peptide chimera comprised of lactoferricin and lactoferrampin. Biochim Biophys Acta. 2012;1818:762–775. doi: 10.1016/j.bbamem.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Jung S, Mysliwy J, Spudy B, Lorenzen I, Reiss K, Gelhaus C, Podschun R, Leippe M, Grotzinger J. Human beta-defensin 2 and beta-defensin 3 chimeric peptides reveal the structural basis of the pathogen specificity of their parent molecules. Antimicrob Agents Chemother. 2011;55:954–960. doi: 10.1128/AAC.00872-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YJ, Johnson G, Pellois JP. Modeling of the endosomolytic activity of HA2-TAT peptides with red blood cells and ghosts. Biochemistry. 2010;49:7854–7866. doi: 10.1021/bi1008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomone F, Cardarelli F, Di Luca M, Boccardi C, Nifosi R, Bardi G, Di Bari L, Serresi M, Beltram F. A novel chimeric cell-penetrating peptide with membrane-disruptive properties for efficient endosomal escape. J Control Release. 2012;163:293–303. doi: 10.1016/j.jconrel.2012.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.