Abstract
Huntington’s disease (HD) is a signature polyglutamine disorder. An enduring theory of HD pathogenesis has involved dysregulation of transcription. Indeed, transcriptional regulatory proteins can be modulated to overcome cardinal features of HD-modeled mice, and efforts to move these into human studies are ongoing. Here, we discuss a unifying hypothesis emerging from these studies, which is that HD represents the pathological disruption of evolutionarily conserved adaptive gene programs to counteract oxidative stress, mitochondrial dysfunction and accumulation of misfolded proteins. Transcriptional dyshomeostasis of adaptive genes is further exacerbated by repression of genes involved in normal synaptic activity or growth factor signaling.
Keywords: Huntington’s disease, mutant huntingtin, cellular stress, perturbed transcriptional homeostasis
HD is a neuromotor disorder, characterized by progressive decline in muscle coordination, cognition and psychiatric dysfunction leading inexorably to death. The underlying pathophysiology involves the selective loss of medium spiny projection neurons, sparing interneurons present within the striatum [1]. HD is an autosomal dominant disease attributable to a toxic gain of function of a mutated huntingtin gene (mhtt) with specifically an expanded stretch of CAG repeats within the exon 1 coding region. Elegant protein mapping studies revealed several proteins with microsatellite repeats, including polyglutamine stretches, and that most of these proteins were transcription factors [2]. Accordingly, several groups, including the Cha, Neri, Thompson, Schaffner and Jones labs, provided converging evidence supporting transcriptional dysregulation as a central feature of HD. Here, we review some of these data and provide a unifying theme for how transcriptional dysregulation creates vulnerability to subsets of neurons in HD.
Huntingtin with a toxic gain of function
The mhtt gains a toxic function by unstable expansion of in-frame CAG triplet repeats. The mutant gene encodes a full-length protein that is initially cytosolic. The proteolytic cleavage of mhtt generates N-terminal fragments that are preferentially translocated to the nucleus, where nuclear aggregates form over time [3]. Caspase-6 cleaves mHtt at amino acid 558 into these toxic N-terminal fragments capable of trafficking to the nucleus [4]. Ser-16 phosphorylation can also regulate N-terminal cleavage of mHtt and its nuclear translocation [5]. Recently, Zheng et al. [6] showed that the N-terminal region of Htt itself functions as a nuclear export signal (NES) and mutation of any of the nucleotides in this sequence leads to its enhanced nuclear accumulation. This finding indicates that increases in the polyQ stretch beyond 37–40 repeats in exon-1 of Htt could disrupt the htt NES sequence, resulting in its enhanced nuclear localization. Blocking the nuclear localization of mHtt suppresses neuronal cell death [7], whereas specific targeting of mHtt by including a nuclear localization signal was sufficient for initiation and progression of transcriptional dysregulation and pathogenic behavioral symptoms in a transgenic mouse model of HD [8, 9]. Collectively, these data suggest that nuclear localization of N-terminal fragments of mhtt is necessary and sufficient to induce the dysfunction and death of neurons.
mHtt-induced early molecular changes in HD
Signs of transcriptional dysregulation were found early in the R6/2 model of HD and were coincident with initial behavioral changes but only after visible signs of intranuclear inclusions related to increased caspase-6 activity were observed [10, 11]. Accordingly, mRNA levels of different genes, (growth factor ligands, neurotransmitter receptors and growth factor receptors) with important neuronal functions have been reported to be downregulated both in a HD mouse model as well as patients with HD [12, 13]. Given that progressive changes in protein levels and signs of neuronal cell death were evident only after changes in mRNA levels of affected genes [14], the aggregate of evidence suggests that nuclear events are necessary for at least some features of clinical HD.
Mechanisms of transcriptional dysregulation
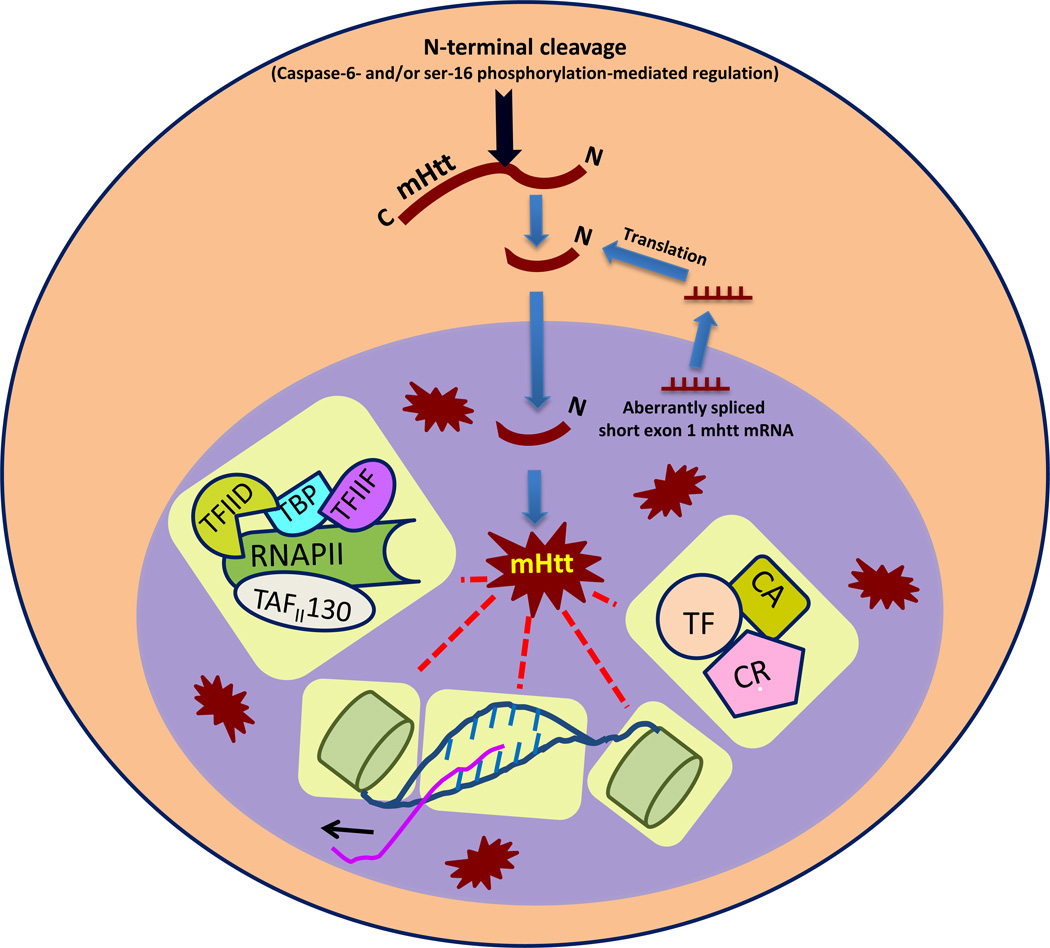
Mechanisms invoked for transcriptional dysregulation in HD are protean. The toxic N-terminal fragments of mHtt can modulate the transcriptional process by having an aberrant protein–protein interaction with the transcriptional machinery, by either modifying chromatin, or through a direct interaction with genomic DNA (Figure 1).
Figure 1.
Known mechanisms of mutant huntingtin (mHtt)-mediated transcriptional dysregulation in Huntington’s disease. After cleavage via either caspase-6 activity or ser-16 phosphorylation, toxic N-terminal fragments of mHtt translocate into the nucleus and form toxic intranuclear aggregates of N-terminal Htt fragments. These, in turn, can influence different components of the transcriptional process such as genomic DNA, histones, basal transcriptional machinery [a complex of RNA polymerase II (RNAP II), transcription initiation factor (TAF)II130, TATA binding protein (TBP), TFIIF, TFIID, etc.], transcription factors (TF), transcriptional corepressors (CR), or transcriptional coactivators (CA) and so on (highlighted in yellow), to cause transcriptional dysregulation.
Aberrant interaction between mHtt and transcription factors, coactivators or corepressors
The polyQ stretch at the N terminus of mHtt provides an appropriate motif for its interaction with glutamine-rich activation domains of different transcription factors, such as cAMP response element-binding protein (CREB), Sp1 and the transcriptional coactivator, CREB binding protein (CBP). Earlier models suggested that polyQ repeats of mHtt form insoluble aggregates [15, 16], which sequester these transcription factors from the cellular pool [17], but Yu et al. [18] showed that the polyglutamine inclusions themselves are unable to deplete the cellular pool of glutamine-rich transcription factors, such as Sp1, significantly. However, the soluble form of mHtt is able to perturb the interaction of these transcription factors (e.g., Sp1) with their transcriptional coactivators [such as transcription initiation factor (TAF)II130] as well as their target DNA [19]. Accordingly, mHtt reduces expression of Sp1 target genes, including the dopamine D2 receptor and nerve growth factor receptor [19, 20]. The co-expression of Sp1 and TAFII130 has been shown to prevent mHtt-mediated toxicity [19]. By contrast, other studies have suggested a pro-death, rather than a pro-survival role for Sp1 in HD [21]. Recent studies reconciled these apparently conflicting observations by demonstrating that agents such as the DNA-binding drug mithramycin that simultaneously inhibits subsets of Sp1-dependent genes (e.g., oncogenes) while activating other subsets (e.g., tumor suppressors) significantly extended lifespan in Drosophila and mice models of HD [22, 23]. The role of Sp1 is highly gene dependent.
The expression of CREB target genes has been reported to be downregulated in both in vitro as well as in vivo models of HD [16, 24]. The expression of one of these CREB target genes [peroxisome proliferator-activated receptor (PPAR) gamma coactivator (PGC)-1α], which has a crucial role in mitochondrial biogenesis, is reduced in HD mice as well as postmortem brains of patients with HD [25, 26]. Other studies have shown that mhtt can disrupt not only nuclear CREB function, but also mitochondrial CREB function [27]. Together, these studies highlight the potential role of mhtt in repressing bioenergetic homeostasis.
CBP functions as a coactivator of many transcription factors, including CREB and Sp1. Aberrant interaction of mHtt with the glutamine-rich domain of CBP leads to perturbation of its coactivator function, which, in turn, disturbs the normal function of associated transcription factors. In addition to its coactivator function, the histone acetyl transferase (HAT) activity of CBP is also disturbed [28]. This causes the cognitive deficit observed in HD [29, 30].
The transcriptional repressor element-1 transcription factor/neuron restrictive silencer factor (REST/NRSF) is a transcriptional corepressor that represses the expression of its target genes, such as brain-derived neurotrophic factor (BDNF), as well as miRNAs [31]. Wild type Htt, a cytosolic protein, interacts with REST and prevents its nuclear translocation and, thus, regulates the expression of its target genes in a positive manner via derepression. By contrast, the affinity of mHtt for REST is weaker, thus leading to its enhanced nuclear entry and, ultimately, to the repression of known REST target genes, such as BDNF and miRNAs [32, 31]. Indeed, expression of BDNF has been found to be compromised in HD models [33–35].
Recently, Wang et al. [36] provided a new perspective on mHtt–transcription factor interactions by showing that Htt itself has putative sequences for different transcription factors, such as Sp1, CREB, signal transducer and activator of transcription (STAT), NRSF, p53, activator protein 1 (AP1), hypoxia-inducible factor (HIF) and nuclear factor (NF)-κB, in its promoter region and, thus, itself provides a regulatory hotspot to the transcription factors so as to play with its own expression. The authors showed that Sp1 can regulate the expression of Htt.
Aberrant interaction between mHtt and basal transcription machinery
mHtt has been shown to interact with many important components of core transcriptional machinery, such as the large subunit of RNA polymerase II, TATA binding protein (TBP), TAFII130, TFIID and TFIIF [19, 37–39], which favors model whereby mHtt disrupts the basic transcriptional machinery in HD subjects. However, it is unclear how this model would account for the regional specificity of transcriptional dysregulation in striatum and cortex despite the presence of mHtt inclusions throughout the brain. It raises a new question that, although mHtt has the potential to interact with many components of transcriptional machinery, we need to understand which of the specific mHtt-mediated interactions account for alterations of gene expression that can explain not only the selective vulnerability, but also the increased expression of some genes.
Histone modification
DNA is present within the nucleus in compact, repeating units called chromatin. The basic unit of chromatin, the nucleosome, comprises DNA packaged by histones and other associated proteins. The specific modification of histones and associated proteins determines the accessibility of the transcriptional machinery to the associated DNA and, thus, has a major role in transcriptional regulation. The post-translational modifications (PTMs) occurring mainly at histone tails are the key modifiers of histone structure and function. Some PTMs, such as acetylation, methylation, ubiquitylation and polyamination, are known to have functional relevance in HD pathobiology [26]. Among these, histone acetylation is the most well studied in HD. Acetylation correlates with increase in transcription, whereas deacetylation correlates with transcriptional repression. Histone acetylation and deacetylation occur through HATs and histone deacetylases (HDACs). HATs increase transcription by loosening the DNA–histone conformation, whereas HDACs function in a reverse way by removing the acetyl group and, thus, repressing the transcriptional process. Histone acetylation remains globally compromised in different HD models [40, 41]. Although mHtt has not been shown to have direct interaction with the nucleosome, it does interact with CBP, which is known to have HAT activity [42]. This might be one of the factors contributing towards the global hypoacetylation evidenced in HD models.
Acetylation of histones has a reciprocal relation with specific histone methylation marks (i.e., a decrease in acetylation is congruent with increase in methylation). This incidence fits true in HD models. The levels of lysine 9 methylation in histone H3 have been found to be upregulated in HD models [43]. Methylation of histone proteins leads to transcriptional repression. This specific PTM is regulated by histone demethylases and methyl transferases. Moreover, methylation has been reported to be regulated by another PTM (ubiquitylation), particularly, in HD models. Aberrant interaction of mHtt with Bmi, a component of the polycomb complex, increases the level of monoubiquityl histone H2A in HD models. This, in turn, enhances histone H3 lysine 9 methylation and repression [44]. These studies unraveled an intricate network of regulatory mechanisms involved in controlling the chromatin dynamics in HD, which requires us to understand which of these PTMs are being regulated first. Vashishtha et al. [45] have also provided new regulatory insight into the role of histone methylation marks HD. The level of H3K4 trimethylation was found to be downregulated in transcriptionally repressed promoters in HD mouse and human brain and decreasing the levels of demethylases (SMCX/Jarid1c) reversed the downregulation of these genes in primary neurons and was neuroprotective in a Drosophila HD model. Correlating this finding (H3K4 trimethylation) with that of a H3K9 methylation study in a HD model clearly indicates that the regulation of distinct methylation marks is different.
Polyamination, another PTM, also has an important regulatory function with respect to chromatin remodeling by way of adding a strong positive charge on histones and, thus, making the histone–DNA complex more condensed, resulting in transcriptional repression. An enzyme called transglutaminase-2 (TGase-2) can crosslink polyamines to the glutamine tails of histones and, thus, increase the polyamination of histones. The level of polyamines has been reported to be high in the brain of patients with HD [46]. Inhibiting the activity of TGase-2 has been shown to result in correction of transcriptional dysregulation in HD more completely than has inhibition of HDAC [26].
mHtt–DNA interaction
mHtt can also dysregulate transcription by its direct interaction with genomic DNA, altering the conformation of DNA as well as its binding with different transcription factors [46]. Recently, Hogel et al. [47] proposed a model of transcriptional dysregulation based on the interaction of mhtt with specific promoters. Furthermore, these authors showed that these promoters show distinct affinity for mhtt, which explains the fact that some genes are dysregulated at earlier stages, whereas others are dysregulated at late stages. Mithramycin, a DNA-binding drug that interacts with the minor groove of GC-rich regions of DNA, has been shown to significantly prolong lifespan in the R6/2 mice [22]. Although not yet formally tested, mithramycin might serendipitously disrupt interactions of mHtt with specific response elements. New analogs of mithramycin with greater efficacy and less toxicity than the parent compound [23] might be candidates for therapeutics in humans.
HD: a phenomenon of failure of the adaptive transcriptional response to cellular stresses
A host of stresses, including endoplasmic reticulum (ER) stress, mitochondrial (bioenergetic) stress and oxidative stress, has been shown to be persistent in affected neurons in HD. mHtt, because of its expanded polyQ stretch, has a propensity to misfold and form aggregates. These aggregates perturb the proper folding of other proteins inside the lumen of the ER and try to sequester them and form inclusion bodies. This leads to ER stress inside cell. The simultaneous defects in clearance machineries, such as the ubiquitin-proteasome system and autophagic machinery, lead to accumulation of these misfolded proteins and/or inclusion bodies, which, in turn, further aggravates cytoplasmic stress. Concurrent with these cellular events, mHtt also causes oxidative and mitochondrial stress by impairing oxidative phosphorylation [48], Ca2+ handling [49], mitochondrial trafficking to synapses [50] and mitochondrial ultrastructure [51].
In the normal brain, cell stress is counteracted by the engagement of adaptive transcriptional responses that return the brain to a homeostatic state [52]. In HD, these adaptive programs are transcriptionally silenced [e.g., by PGC-1α, Sp1, transcription factor EB (TFEB) and nuclear factor, erythroid 2-like 2 (Nrf-2)], and so cell stress becomes persistent, leading first to neuronal atrophy and then to cell death. This model has several testable predictions. First, strategies to augment adaptive transcriptional responses to mitochondrial, oxidative or proteotoxic stress should extend lifespan and decrease symptoms in HD mice. Indeed, molecular or pharmacological augmentation of PGC-1α can prevent death or enhance lifespan in HD models. Second, forced expression of Sp1 has been shown to counteract oxidative stress; agents that augment Sp1 activity, including mithramycin or HDAC inhibitors, can improve function or enhance lifespan in HD mice; finally, TFEB is a transcriptional activator that can increase expression of genes involved in autophagy; augmentation of TFEB can diminish mhtt toxicity [53]. Together, these studies suggest the search for a strategy by which adaptive programs to many stresses can be activated simultaneously and that, in turn, might lead to the best outcomes. They also predict that vulnerable medium spiny neurons might have higher basal levels of oxidative, mitochondrial or proteotoxic stress than other neurons, or fewer redundant mechanisms for counterminding these stresses.
Targeting different subphenomena of transcriptional dysregulation
Given the many protein players in transcriptional regulation and completion (either genetic or epigenetic) as main players behind transcriptional dysregulation, it represents an exciting point of convergence and replication that manipulations of transcription factors, such as CREB [55, 56], REST [57–59], Sp1 [22] and PPAR-γ [60], or the post-translational modifications of histones (e.g., acetylation [28, 40, 61–67] or methylation [22, 45]) all appear to correct aspects of the transcriptional repression in HD via the use of pharmacological drugs or genetic manipulation. The convergence of these effective therapies continues to suggest that transcriptional dysregulation is a central feature of HD (Table 1).
Table 1.
Scientific attempts targeting specific components of transcription by using different pharmacological drugs or genetic manipulations to correct the transcriptional dysregulation in HD
| Genetic manipulation or pharmacological intervention |
Key findings | Refs |
|---|---|---|
| Rolipram | Treatment with rolipram in rat model of HD for 2 weeks induced CREB activity, which reduced striatal lesion size by 62% | [55] |
| TP10 | Treatment with TP10 in rat model of HD for 2 weeks induced CREB activity, which reduced striatal lesion size by 52% | [56] |
| Quinone-like compound 91 (C91) | C91 induced the expression of REST target genes in Htt-knockdown zebrafish and increased BDNF mRNA in the presence of mHtt | [57] |
| Dominant negative REST (DN:REST) | DN:REST delivery in vivo in motor cortex in two transgenic mice models of HD (BACHD and N171-82Q) restored expression of REST target genes, such as BDNF, Syn1, Penk1 and Chrm4 | [58] |
| REST decoy oligonucleotide | Transfection of REST decoy oligonucleotides in a cellular model of HD restored expression of REST target genes, such as BDNF, Syn1 and Chrm4 | [59] |
| Mithramycin | Pharmacological treatment of the R6/2 mouse model of HD with mithramycin, an established Sp1 inhibitor, extended its survival and prevented the increase in H3 methylation seen in R6/2 mice | [22] |
| Rosiglitazone | Pharmacological treatment of a N171-82Q mouse model of HD with the PPAR-γ agonist, rosiglitazone rescued its motor functions and glucose metabolism dysfunction | [60] |
| SAHA (HDAC inhibitor) | Drosophila expressing the Httex1p Q93 transgene fed with SAHA showed enhanced viability | [28] |
| Sodium butyrate | Sodium butyrate treatment prevented 3-NP-induced oxidative neuronal death in vitro as well as in vivo independent of polyglutamine expansion by enhancing acetylation of Sp1 | [61] |
| Sodium butyrate | Pharmacological treatment of the R6/2 mouse model of HD with sodium butyrate extended survival, and improved body weight and motor function and neuropathological problems | [62] |
| Phenyl butyrate | Treatment of the N171-82Q mouse model of HD with phenyl butyrate extended survival and prevented brain and neuronal atrophy by enhancing histone acetylation and reducing histone methylation | [40] |
| HDAC3 RNAi/deletion allele | Genetic deletion and/or knockdown of HDAC3 in Caenorhabditis elegans expressing HttQ150 suppressed HttQ150-induced neurodegeneration | [63] |
| Rpd3 (Class I HDACs in human)/Sir2 (Class II HDACs in Human) knockdown | Reduction of Rpd3/Sir2 alone or in combination (with additive effect) enhanced neuroprotection in Httex1p Q93 Drosophila | [64] |
| Pimelic diphenylamide, HDACi 4b | Chronic oral administration of pimelic diphenylamide in a R6/2300Q mouse model of HD after the beginning of motor deficit, profoundly improved motor function and body weight, and reduced brain size decline and striatal atrophy | [65] |
| AK-7 (Sirtuin 2 inhibitor) | Pharmacological treatment with AK-7 of two mice models of HD, R6/2 and 140 CAG, improved their motor function, extended their survival, and reduced brain atrophy along with a decrease in aggregated mhtt | [66] |
| Nicotinamide (NAM) | NAM treatment of B6.HDR6/1 mouse model of HD improved motor deficit and enhanced the expression of BDNF and PGC1a but was unable to reduce mhtt aggregation or prevent late-onset weight loss | [67] |
| SMCX/Jarid1c (H3K4 demethylase) knockdown | Either SMCX/Jarid1c knockdown in primary cortical neurons from a BACHD mouse model of HD or single Jarid1c in Drosophila model of HD was protective | [45] |
Abbreviations: 3-NP, 3-nitropropionic acid; Chrm4, cholinergic receptor, muscarinic 4; Penk1, preproenkephalin 1.
Concluding remarks
Transcriptional dysregulation has been well established as a pathological process in HD. Some groups emphasize the role of protein–protein interactions involving mHtt and different transcription factors, coactivators or other transcriptional regulators, some groups focus mainly on histone modification, whereas others concentrate on direct mHtt–DNA interactions as a causal phenomenon involved in transcriptional dysregulation in HD pathobiology. A narrow focus on these separate subphenomena seems to be a possible reason for the failure to devise an appropriate therapeutic approach. Perhaps a search for convergent regulators of all of these phenomena will provide a ripe therapeutic target (e.g., mhtt itself). A compelling notion is that, whatever the specific domain of transcription that is interrupted, the fact that mhtt disables the compensatory adaptive programs to cell stress in medium spiny neurons, offers a therapeutically relevant and testable model (Figure 2). This model continues to gain currency as we recognize the ability of neurotransmitters and growth factors, also silenced in HD, to drive adaptive gene expression to bioenergetic, oxidative and proteotoxic stress [68].
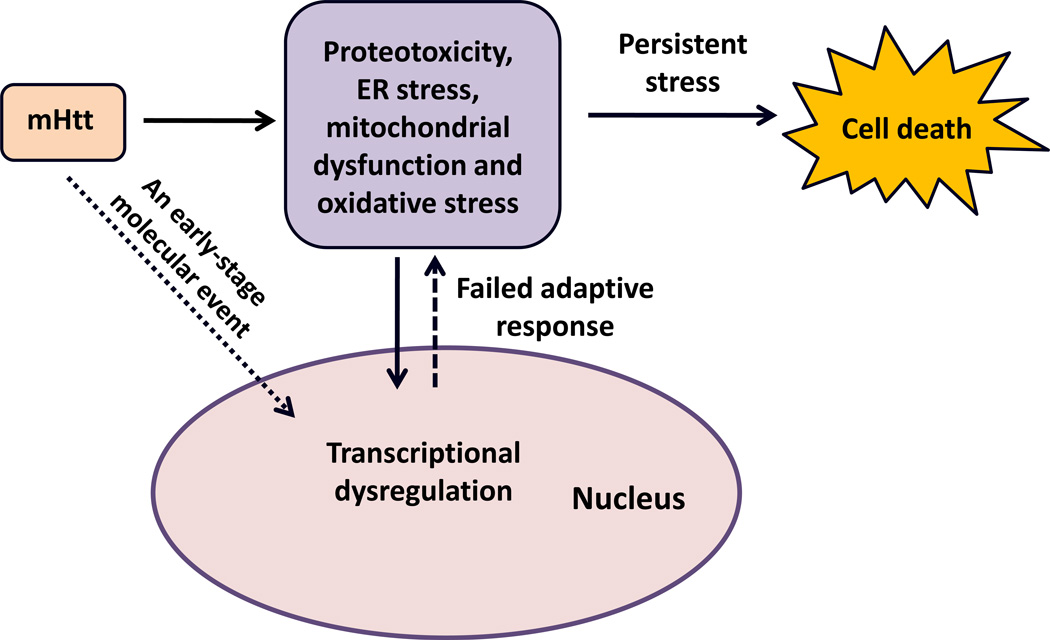
Figure 2.
Model depicting Huntington’s disease as a failure of adaptive transcriptional homeostasis leading to persistent bioenergetic, oxidative and proteotoxic stress. Abbreviations: ER, endoplasmic reticulum; mHtt, mutant Huntingtin.
Highlights.
This is a review commissioned for Drug Discovery Today: Strategies
The article forms part of an issue on Huntington’s Disease
This is a review commissioned for Drug Discovery Today: Strategies
Acknowledgments
Research reported in this review was supported by the National Institute on Aging of the National Institutes of Health under award number P01 AG14930 and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrante RJ, et al. Selective sparing of a class of striatal neuron in Huntington’s disease. Science. 1985;230:561–564. doi: 10.1126/science.2931802. [DOI] [PubMed] [Google Scholar]
- 2.Gerber HP, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 3.DiFiglia M, et al. Aggregation of Huntingtin in neuronal intranuclear inclusions and dystrophic neuritis in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 4.Warby SC, et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum. Mol. Genet. 2008;17:2390–2404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 5.Havel LS, et al. Preferential accumulation of N-terminal mutant huntingtin in the nuclei of striatal neurons is regulated by phosphorylation. Hum. Mol. Genet. 2011;20:1424–1437. doi: 10.1093/hmg/ddr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Z, et al. An N-terminal nuclear export signal regulates trafficking and aggregation of Huntingtin (Htt) protein exon 1. J. Biol. Chem. 2013;288:6063–6071. doi: 10.1074/jbc.M112.413575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saudou F, et al. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 8.Schilling G, et al. Nuclear-targeting of mutant huntingtin fragments produces Huntington’s disease-like phenotypes in transgenic mice. Hum. Mol. Genet. 2004;13:1599–1610. doi: 10.1093/hmg/ddh175. [DOI] [PubMed] [Google Scholar]
- 9.Benn CL, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum. Mol. Genet. 2005;14:3065–3078. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 10.Luthi-Carter R, et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 2002a;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 11.Graham RK, et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J. Neurosci. 2010;30:15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum. Mol. Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 13.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 14.Luthi-Carter R, Cha J-HJ. Mechanisms of transcriptional dysregulation in Huntington’s disease. Clin. Neurosci. Res. 2003;3:165–177. [Google Scholar]
- 15.Kazantsev A, et al. Insoluble detergent-resistant aggregates form between pathological and non-pathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nucifora FC, Jr, et al. Interference by huntingtin and atrophin-1 with CBP mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 17.Sadri-Vakili G, et al. Huntingtin inclusions do not downregulate specific genes in the R6/2 Huntington’s disease mouse. Eur. J. Neurosci. 2006;23:3171–3175. doi: 10.1111/j.1460-9568.2006.04871.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu ZX, et al. Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum. Mol. Genet. 2002;11:905–914. doi: 10.1093/hmg/11.8.905. [DOI] [PubMed] [Google Scholar]
- 19.Dunah AW, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 20.Chen-Plotkin AS, et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington’s disease. Neurobiol. Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Qiu Z, et al. Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J. Biol. Chem. 2006;281:16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- 22.Ferrante RJ, et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J. Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleiman SF, et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugars KL, et al. Decreased cAMP response element mediated transcription: an early event in exon 1 and full-length cell models of Huntington’s disease that contributes to polyglutamine pathogenesis. J. Biol. Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 25.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 26.McConoughey SJ, et al. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med. 2010;2:349–370. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, et al. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J. Biol. Chem. 2005;280:40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in. Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 29.Giralt A, et al. Long-term memory deficits in Huntington’s disease are associated with reduced CBP histone acetylase activity. Hum. Mol. Genet. 2012;21:1203–1216. doi: 10.1093/hmg/ddr552. [DOI] [PubMed] [Google Scholar]
- 30.Sadri-Vakili G, Cha JH. Mechanisms of disease: histone modifications in Huntington’s disease. Nat. Clin. Pract. Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 31.Bithell A, et al. Transcriptional dysregulation of coding and non-coding genes in cellular models of Huntington’s disease. Biochem. Soc. Trans. 2009;37:1270–1275. doi: 10.1042/BST0371270. [DOI] [PubMed] [Google Scholar]
- 32.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 33.Zuccato C, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 34.Johnson R, et al. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol. Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Packer A, et al. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang R, et al. Sp1 regulates human huntingtin gene expression. J. Mol. Neurosci. 2012;47:311–321. doi: 10.1007/s12031-012-9739-z. [DOI] [PubMed] [Google Scholar]
- 37.Luthi-Carter R, et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington’s disease mouse models reveal context independent effects. Hum. Mol. Genet. 2002b;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- 38.Suhr ST, et al. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J. Cell. Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhai W, et al. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 40.Gardian G, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J. Biol. Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 41.McFarland KN, et al. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington’s disease. PLoS ONE. 2012;7:e41423. doi: 10.1371/journal.pone.0041423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steffan JS, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu H, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc. Natl. Acad. Sci. U.SA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim MO, et al. Altered histone monoubiquitylation mediated by mutant huntingtin induces transcriptional dysregulation. J. Neurosci. 2008;28:3947–3957. doi: 10.1523/JNEUROSCI.5667-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vashishtha M, et al. Targeting H3K4 trimethylation in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3027–E3036. doi: 10.1073/pnas.1311323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeitner TM, et al. Increased levels of gamma-glutamylamines in Huntington disease CSF. J. Neurochem. 2008;106:37–44. doi: 10.1111/j.1471-4159.2008.05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benn CL, et al. Huntingtin modulates transcription, occupies gene promoters in vivo and binds directly to DNA in a polyglutamine-dependent manner. J. Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hogel M, et al. Promoters are differentially sensitive to N-Terminal mutant Huntingtin-mediated transcriptional repression. PLoS ONE. 2012;7:e41152. doi: 10.1371/journal.pone.0041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milakovic T, Johnson GV. Mitochondrial respiration and ATP production are significantly impaired in striatal cells expressing mutant huntingtin. J. Biol. Chem. 2005;280:30773–30782. doi: 10.1074/jbc.M504749200. [DOI] [PubMed] [Google Scholar]
- 50.Milakovic T, et al. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J. Biol. Chem. 2006;281:34785–34795. doi: 10.1074/jbc.M603845200. [DOI] [PubMed] [Google Scholar]
- 51.Li XJ, et al. Impaired mitochondrial trafficking in Huntington’s disease. Biochim. Biophys. Acta. 2010;1802:62–65. doi: 10.1016/j.bbadis.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Squitieri F, et al. Severe ultrastructural mitochondrial changes in lymphoblasts homozygous for Huntington disease mutation. Mech. Ageing Dev. 2006;127:217–220. doi: 10.1016/j.mad.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Ratan RR, et al. Harnessing hypoxic adaptation to prevent, treat, and repair stroke. J. Mol. Med. 2007;85:1331–1338. doi: 10.1007/s00109-007-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsunemi T, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 2012;4:142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeMarch Z, et al. Beneficial effects of rolipram in a quinolinic acid model of striatal excitotoxicity. Neurobiol. Dis. 2007;25(2):266–273. doi: 10.1016/j.nbd.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Giampa C, et al. Phosphodiesterase 10 inhibition reduces striatal excitotoxicity in the quinolinic acid model of Huntington’s disease. Neurobiol. Dis. 2009;34:450–456. doi: 10.1016/j.nbd.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 57.Conforti P, et al. Binding of the repressor complex REST-mSIN3b by small molecules restores neuronal gene transcription in Huntington’s disease models. J. Neurochem. 2013 doi: 10.1111/jnc.12348. http://dx.doi.org/10.1111/jnc.12348. [DOI] [PubMed] [Google Scholar]
- 58.Conforti P, et al. In vivo delivery of DN:REST improves transcriptional changes of REST-regulated genes in HD mice. Gene Ther. 2013b;20:678–685. doi: 10.1038/gt.2012.84. [DOI] [PubMed] [Google Scholar]
- 59.Soldati C, et al. Rescue of gene expression by modified REST decoy oligonucleotides in a cellular model of Huntington’s disease. J. Neurochem. 2011;116:415–425. doi: 10.1111/j.1471-4159.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 60.Jin J, et al. Neuroprotective effects of PPAR-γ agonist rosiglitazone in N171-82Q mouse model of Huntington’s disease. J. Neurochem. 2013;125:410–419. doi: 10.1111/jnc.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryu H, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J. Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bates EA, et al. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J. Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum. Mol. Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas EA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chopra V, et al. The Sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington disease mouse models. Cell Rep. 2013;2:1492–1497. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hathorn T, et al. Nicotinamide improves motor deficits and upregulates PGC-1alpha and BDNF gene expression in a mouse model of Huntington’s disease. Neurobiol. Dis. 2011;41:43–50. doi: 10.1016/j.nbd.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bell KFS, Hardingham GE. The influence of synaptic activity on neuronal health. Curr. Opin. Neurobiol. 2011;21:299–305. doi: 10.1016/j.conb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]