Abstract
Sulphur is an essential element for life and exists ubiquitously in living systems1,2. Yet, how the sulphur atom is incorporated in many sulphur-containing secondary metabolites remains poorly understood. For C-S bond formation in primary metabolites, the major ionic sulphur sources are the protein-persulphide and protein-thiocarboxylate3,4. In each case, the persulphide and thiocarboxylate group on these sulphur-carrier (donor) proteins are post-translationally generated through the action of a specific activating enzyme. In all bacterial cases reported thus far, the genes encoding the enzyme that catalyzes the actual C-S bond formation reaction and its cognate sulphur-carrier protein co-exist in the same gene cluster5. To study 2-thiosugar production in BE-7585A, an antibiotic from Amycolatopsis orientalis, we identified a putative 2-thioglucose synthase, BexX, whose protein sequence and mode of action appear similar to those of ThiG, the enzyme catalyzing thiazole formation in thiamin biosynthesis6,7. However, no sulphur-carrier protein gene could be located in the BE-7585A cluster. Subsequent genome sequencing revealed the presence of a few sulphur-carrier proteins likely involved in the biosynthesis of primary metabolites, but surprisingly only a single activating enzyme gene in the entire genome of A. orientalis. Further experiments showed that this activating enzyme is capable of adenylating each of these sulphur-carrier proteins, and likely also catalyzing the subsequent thiolation taking advantage of its rhodanese activity. A proper combination of these sulphur delivery systems is effective for BexX-catalyzed 2-thioglucose production. The ability of BexX to selectively distinguish sulphur-carrier proteins is given a structural basis using X-ray crystallography. These studies represent the first complete characterization of a thiosugar formation in nature and also demonstrate the receptor promiscuity of the sulphur-delivery system in A. orientalis. Our results also provide evidence that exploitation of sulphur-delivery machineries of primary metabolism for the biosynthesis of sulphur-containing natural products is likely a general strategy found in nature.
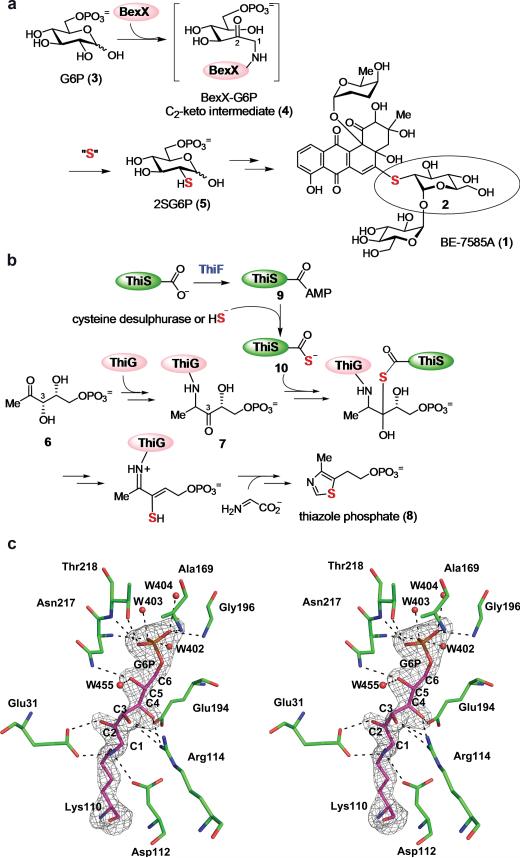
Unusual sugars found in many secondary metabolites play crucial roles in determining the efficacy and specificity of the biological activities of the parent molecules8,9. Despite recent advances made in unusual sugars biosynthesis research, little is known about thiosugar formation due to their rarity in natural products and the limited knowledge of sulphur incorporation in secondary metabolites1,2,10,11. In our studies of the biosynthesis of 2-thiosugar-containing antibiotic BE-7585A (1, Fig. 1a) in A. orientalis subsp. vinearia BA-07585, we identified a putative 2-thioglucose-6-phosphate synthase, BexX6,7, having significant sequence homology to thiazole synthase, ThiG12, which is responsible for the construction of thiazole moiety (8) from DXP (6) in thiamin biosynthesis (Fig. 1b)13,14. Since ThiG catalyzes sulphur insertion into a ThiG-ketosugar adduct (7)12, BexX may play a similar role in the conversion of glucose-6-phosphate (G6P, 3) to 2-thioglucose (2) in A. orientalis (Fig. 1a). The proposed function of BexX is supported by the detection of a covalent adduct (4) between BexX and a 2-ketosugar derived from G6P7. The crystal structure of BexX-substrate complex has now been determined to 2.3 Å resolution (Extended Data Fig. 1) confirming that the covalent attachment of G6P is at Lys110 of BexX (Fig. 1c). However, the absence of genes encoding potential sulphur transfer enzymes, including common sulphur-carrier proteins15, cysteine desulphurases16, and rhodanese-like proteins17, in and around the BE-7585A biosynthetic gene cluster impeded further functional characterization of BexX.
Figure 1. Proposed mechanism for 2-thiosugar formation in BE-7585A biosynthesis.
a, Proposed BexX-catalyzed 2-thiosugar formation. The active site lysine residue (K110)6,7 of BexX initially forms an imine bond with G6P (3) at the C1 position, which is isomerized first to a C1–C2 enamine and then a C2-ketone intermediate (4). Subsequent nucleophilic attack by a sulphur donor occurs at the C2 position of 4 to incorporate a sulphur atom in the 2SG6P (5) product. b, ThiG-catalyzed thiazole phosphate biosynthetic pathway. c, Stereo diagram of BexX active site. Active site side chains and Lys110-G6P intermediate are depicted as sticks with the carbon atoms of residues colored in green and purple, respectively. The Fo-Fc simulated annealing (SA)-omit map of Lys110-G6P intermediate contoured at 4 sigma is shown in gray. Water molecules are shown as red spheres. The carbon atoms of G6P are numbered from C1 to C6.
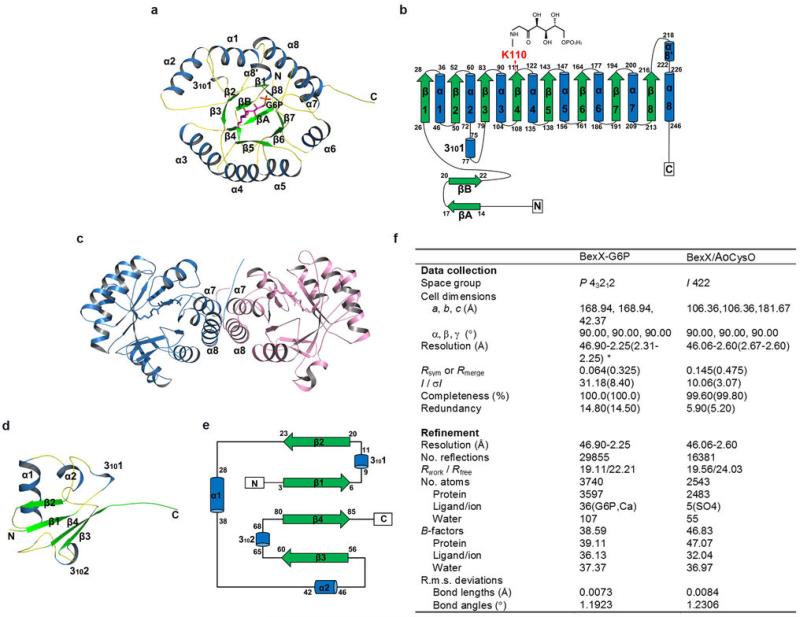
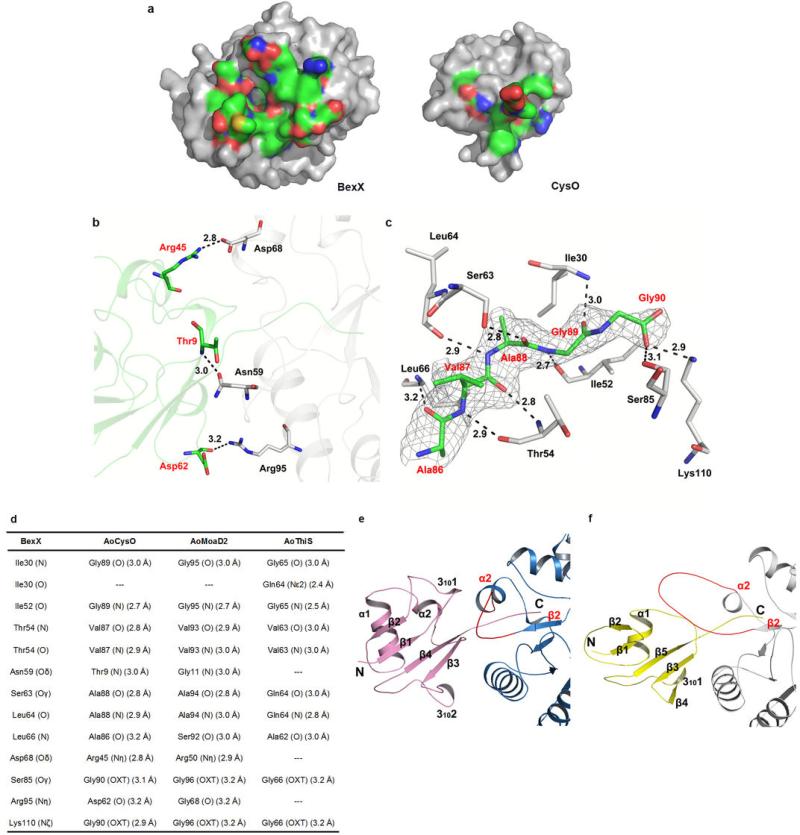
Extended Data Fig. 1. Structures of BexX and CysO from A. orientalis.
a, A stereo ribbon diagram of the (βα)8-barrel fold of BexX is shown with a top view. The α-helices, β-strands and loops are marked in blue, green and yellow, respectively. The ketone-intermediate (4) formed by K110 and G6P is shown as sticks and colored in purple. b, The typical secondary structure composition of the classical (βα)8-barrel is shown as a topology model, the conserved K110 is highlighted in red. c, The quaternary structure of BexX is shown as a ribbon diagram with two monomers colored by chain. d, The ribbon diagram of the AoCysO from BexX/AoCysO structure. Secondary structural elements are colored blue for α-helices, green for β-strands and yellow for loops. e, Topology diagram for AoCysO. f, Data collection and refinement statistics. One crystal was used for each of the two data sets. *Values in parentheses are for highest-resolution shell.
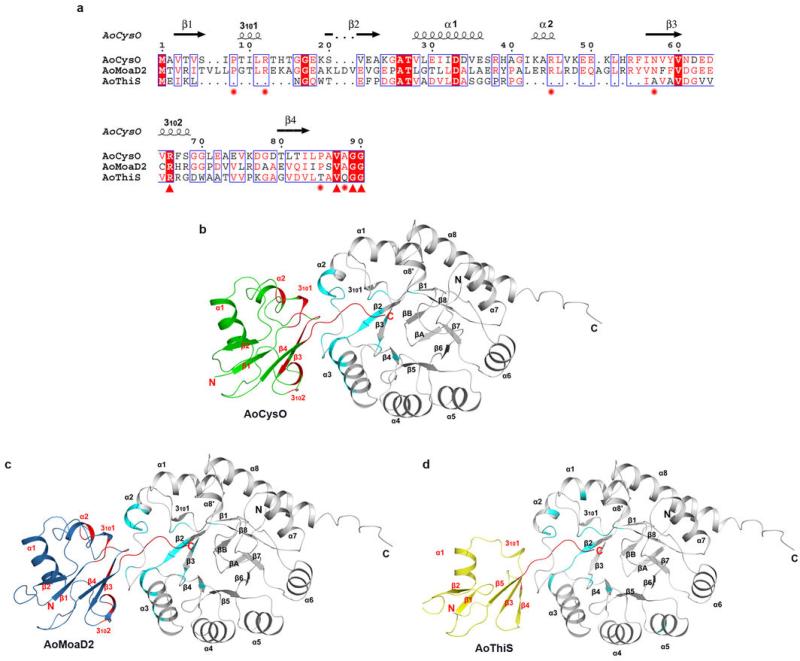
To search for the sulphur-carrier protein required for BexX reaction, the entire genome of A. orientalis was sequenced. A total of 9,210-coding open reading frames in approximately 9.8 Mb genomic DNA was identified, including genes encoding five cysteine desulphurase homologues, five rhodanese homologues, and four sulphur-carrier protein homologues (AoThiS, AoMoaD, AoCysO and AoMoaD2) (Extended Data Table 1). The thiS, moaD and cysO genes are part of the thiamin, molybdopterin and cysteine biosynthetic gene clusters, respectively18,19, whereas moaD2 stands alone with no nearby genes related to any biosynthetic pathway (Extended Data Fig. 2a–d and Supplementary Table 1). While the protein receptor of AoMoaD2 is not immediately apparent, it likely functions as a MoaD homologue due to its high sequence identity to MoaD. In view of the sequence similarity between BexX and ThiG and their mechanistic parallels7, we anticipated that AoThiS, being the cognate sulphur-carrier partner of ThiG13,14, might be recruited for sulphur delivery to the BexX-G6P complex (4) in A. orientalis.
Table 1.
Putative cysteine desulphurases, rhodaneses, and sulphur-carrier proteins found in A. orientalis genome.
| gene (orf#) | Name of the protein | protein with the highest sequence similarity and origin | identity/similarity (%) | protein accession number |
|---|---|---|---|---|
| 10706 | CD1 | cysteine desulphurase/selenocysteine lyase [Amycolalopsis mediterranei U32] | 97/98 | YP_003765029 |
| 11099 | CD2 | cysteine desulphurase [Amycolalopsis mediterranei U32] | 87/91 | YP_003765163 |
| 14916 | CD3 | cysteine desulphurase [Amycolalopsis mediterranei U32] | 88/93 | YP_003766645 |
| 04763 | CD4 | cysteine desulphurase [Amycolalopsis mediterranei U32] | 92/96 | YP_003763873 |
| 09299 | CD5 | cysteine desulphurase [Amycolalopsis mediterranei U32] | 97/99 | YP_003762467 |
| 04658 | RHOI | rhodanese-like protein [Amycolalopsis mediterranei U32] | 89/93 | YP_003763825 |
| 08287 | RH02 | rhodanese-like protein [Amycolalopsis mediterranei U32) | 89/91 | YP_003764615 |
| 09090 | RH03 | rhodanese-like protein [Amycolalopsis mediterranei U32] | 95/96 | YP_003762363 |
| 10524 | RH04 | rhodanese-like protein [Amycolalopsis mediterranei U32] | 81/87 | YP_003763440 |
| 12151 | RH05 | rhodanese-like protein [Amycolalopsis mediterranei U32] | 97/99 | YP_003771104 |
| 02110 | MoeZ | molybdopterin biosynthesis-like protein MoeZ [Amycolalopsis mediterranei U32] | 99/99 | YP_003763336 |
| 13974 | ThiS | thiamin biosynthesis protein ThiS [Amycolalopsis mediterranei U32] | 88/93 | YP_003770674 |
| 13839 | MoaD | ThiS/MoaD family protein [Amycolalopsis mediterranei U32] | 82/90 | YP_003770615 |
| 06461 | CysO | ThiS/MoaD family protein [Amycolalopsis mediterranei U32] | 97/100 | YP_003769822 |
| 10102 | MoaD2 | ThiS/MoaD family protein [Amycolalopsis mediterranei U32) | 91 /94 | YP_003764220 |
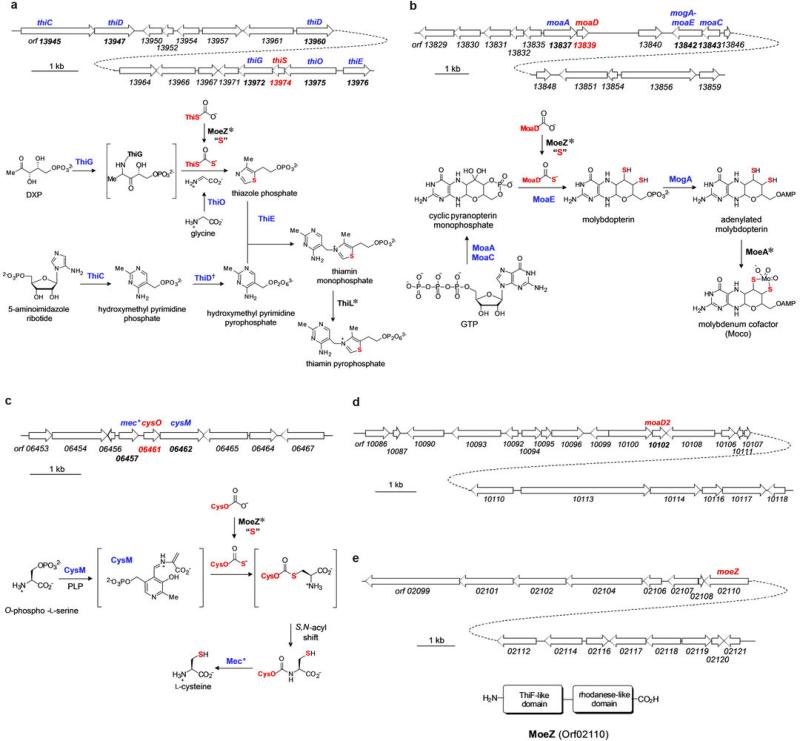
Extended Data Fig. 2. Putative thiamin, molybdenum cofactor, and cysteine biosynthetic genes found in A. orientalis, and their proposed functions.
a, Organization of the putative thiamin biosynthetic gene cluster and proposed thiamin biosynthetic pathway in A. orientalis. *The genes encoding MoeZ and ThiL are not found in the gene cluster. The gene encoding the ThiS-activating enzyme, ThiF, is also absent in the genome. †Two genes encoding proteins homologous to ThiD are found in the gene cluster. b, Organization of the putative molybdopterin biosynthetic gene cluster and proposed molybdenum cofactor biosynthetic pathway in A. orientalis. *The genes encoding MoeZ and MoeA were not found in the gene cluster. The gene encoding the MoaD-activating enzyme, MoeB, is also absent in the genome. c, Organization of the putative cysteine biosynthetic gene cluster and proposed cysteine biosynthetic pathway in A. orientalis. *The gene encoding MoeZ is not found in the gene cluster. d, Organization near the moaD homologue, moaD2, found in the A. orientalis genome. e, Organization near moeZ found in the A. orientalis genome and the conserved domains of AoMoeZ predicted by BLAST analysis.
However, unlike the thiamin biosynthetic gene clusters in Escherichia coli and Bacillus subtilis13,14, the one in A. orientalis does not contain thiF, which encodes the ThiS activating enzyme that is essential for converting ThiS to its thiocarboxylate form (10). The corresponding activating enzymes for AoMoaD and AoCysO are also missing from the respective molybdopterin and cysteine biosynthetic gene clusters. To our surprise, only a single putative activating enzyme is found in the entire genome of A. orientalis: a MoeZ homologue consisting of a ThiF/MoeB-like domain at the N-terminal and a rhodanese homology domain at the C-terminal (Extended Data Fig. 2 and Supplementary Table 1). Due to its unique occurrence in the genome and because it is not part of the molybdopterin gene cluster or associated with any other biosynthetic gene cluster, this protein, AoMoeZ, may be the universal activating catalyst for thiocarboxylate protein production in A. orientalis (Fig. 2a).
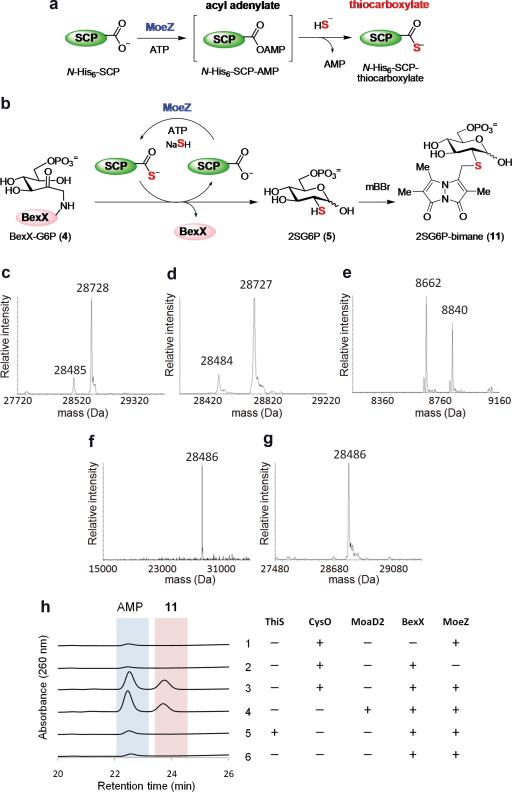
Figure 2. Activation of sulphur-carrier proteins and sulphur transfer to BexX-G6P complex.
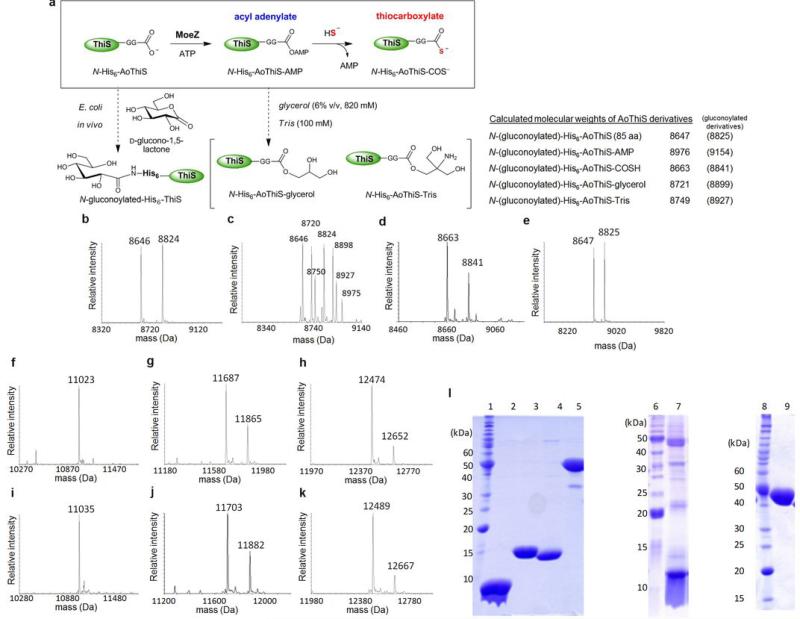
a, AoMoeZ-catalyzed acyl adenylation of a sulphur-carrier protein (SCP, e.g., AoThiS, AoMoaD, AoCysO, AoMoaD2) followed by a nucleophilic attack of bisulphide to yield the corresponding SCP-thiocarboxylate. b, Expected sulphur transfer reaction to BexXG6P (4) using SCP-thiocarboxylate to produce 2SG6P (5), which was further derivatized with mBBr to give 2SG6P-bimane (11). c–g, Deconvoluted ESI-MS analyses of (c) as-isolated C-His6-BexX (28488 calcd), showing the major species is the complex 4 (28730 calcd), and the sulphur transfer reactions using (d) AoThiS [(e) lower mass range of the same reaction showing N-His6-AoThiS-COSH (8663 calcd) and its N-gluconoyl derivatives (8841 calcd), see also Extended Data Fig. 2)], (f) AoCysO, or (g) AoMoaD2. h, HPLC traces of the BexX-catalyzed reaction. In the reaction with AoThiS, the amount of AMP, likely derived from partial decomposition of ATP during incubation, was comparable to that in the control with no added SCP.
To test the proposed function of AoMoeZ, the AoThiS and AoMoeZ of A. orientalis were heterologously expressed in E. coli, each with an N-terminal His6-tag. When AoThiS was incubated with AoMoeZ and ATP, an ESI-MS signal corresponding to the adenylated AoThiS (9) was detected along with few peaks likely derived from reaction of the labile adenylated AoThiS with buffer components (Extended Data Fig. 3c). Upon the addition of excess bisulphide, complete conversion of 9 to its thiocarboxylate form (10) was observed (Extended Data Fig. 3d). Control experiment using bisulphide in the absence of AoMoeZ showed no change of the original AoThiS signals. These results demonstrated that AoMoeZ is capable of charging AoThiS to its ready-to-use thiocarboxylate form (Fig. 2a). The activated AoThiS-COS– was next incubated with the BexX-G6P complex (4). If sulphur transfer occurs and the resulting 2-thiosugar product (5) is released from the enzyme, a shift of the mass signal corresponding to the BexX-G6P complex to that of the free enzyme is anticipated (Fig. 2b). However, no increase of the free BexX in the presence of AoThiSCOS− was observed (Fig. 2c–e), and hence ruled out AoThiS (and bisulphide) as the sulphur donor for BexX in 2-thiosugar formation.
Extended Data Fig. 3. EIS-MS analyses of the AoMoeZ-catalyzed activation of sulphur-carrier proteins and SDS-PAGE of the purified proteins.
a, Reaction scheme of the AoMoeZ-catalyzed activation of AoThiS. b–e, Deconvoluted ESI-MS of as-isolated (b) AoThiS, (c) AoThiS in the presence of AoMoeZ and ATP, (d) AoThiS in the presence of AoMoeZ, ATP, and bisulphide, and (e) AoThiS in the presence of bisulphide (control). The calculated molecular masses are shown as the neutral form in the upper right corner. Analysis of purified N-His6-AoThiS shows two mass signals (obsd, 8646/8824 Da) consistent with the calculated molecular mass of the recombinant enzyme in its native and N-gluconoylated form (calcd, 8647/8825 Da). Gluconoylation of the N-terminal His6-tag is a known post-translational modification when expressing recombinant proteins in E. coli37. Such a modification should not affect AoThiS activity, because the predicted active site for AoThiS is at the C-terminus. Indeed, when N-His6-AoThiS was incubated with N-His6-AoMoeZ and ATP, a MS signal corresponding to the adenylated N-His6-AoThiS (9) was detected along with few peaks likely derived from reaction of the labile adenylated AoThiS with buffer components (see panel c). f–k, Deconvoluted ESI-MS of (f) as-isolated AoMoaD (the calculated molecular mass of N-His6-AoMoaD (105 aa) is 11022 Da), (g) as-isolated AoCysO (the calculated molecular masses of N-His6-AoCysO (109 aa) and its N-gluconoylated derivative are 11688 and 11866, respectively), (h) as-isolated AoMoaD2 (the calculated molecular masses of N-His6-AoMoaD2 (115 aa) and its N-gluconoylated derivative are 12473 and 12651, respectively), (i) AoMoaD incubated with AoMoeZ, ATP, and NaSH (the calculated molecular mass of N-His6-AoMoaD-COSH is 11038 Da), (j) AoCysO incubated with AoMoeZ, ATP, and NaSH (the calculated molecular masses of NHis6-AoCysO-COSH and its N-gluconoylated derivative are 11704 and 11882, respectively), and (k) AoMoaD2 incubated with AoMoeZ, ATP, and NaSH (the calculated molecular masses of N-His6-AoMoaD-COSH and its N-gluconoylated derivative are 12489 and 12667, respectively). l, SDS-PAGE gel of purified sulphur-carrier proteins, AoMoeZ and CD4. NHis6-AoThiS (85 aa, 8.7 kDa, lane 2), N-His6-AoMoaD2 (115 aa, 12.5 kDa, lane 3), N-His6-AoCysO (109 aa, 11.7 kDa, lane 4), N-His6-AoMoeZ (421 aa, 45.0 kDa, lane 5), N-His6-AoMoaD (105 aa, 11.0 kDa, lane 7), and N-His6-CD4 (417 aa, 43.3 kDa, lane 9). The molecular weight marks are 220, 160, 120, 100, 90, 80, 70, 60, 50, 40, 30, 25, 20, 15, and 10 kDa (top to bottom, lane 1, 6 and 8). The protein AoMoaD was not expressed well, and the partially purified protein solution contained significant amounts of endogenous proteins from the E. coli host.
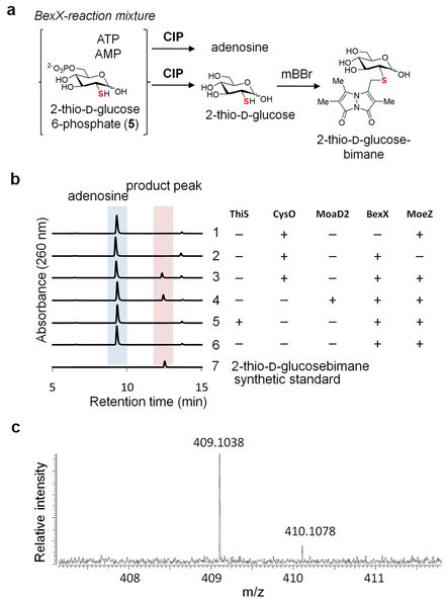
To assess the competence of AoMoaD, AoCysO and AoMoaD2 as sulphur-carrier proteins in BexX-catalyzed reaction, the N-His6-tagged AoMoaD, AoCysO and AoMoaD2 were prepared. Similar to the case of AoThiS, thiocarboxylation of each protein in the presence of AoMoeZ, ATP and NaSH was confirmed by MS analysis (Extended Data Fig. 3f–k). Since the activated AoMoaD was generated in low quantity and purity, only AoCysO and AoMoaD2 were used in the incubation with the BexX-G6P complex. Relative intensities of mass signals corresponding to BexX-G6P (4) and the free enzyme were monitored before and after the addition of the activated sulphur-carrier proteins. To our delight, only signal ascribed to free BexX was discernible after treatment with AoCysO and AoMoaD2 (Fig. 2f, g). To gain further evidence, the thiosugar product (5) was derivatized with monobromobimane (mBBr) prior to HPLC analysis to give 11 to facilitate detection (Fig. 2b). Indeed, when AoCysO or AoMoaD2 was used, a clear appearance of a new peak (product peak) along with the increase in AMP production was observed (Fig. 2h, trace 3, 4). The product peak was isolated and characterized as 2-thioglucose-6-phosphate-bimane (11) by ESI-MS and NMR (Supplementary Information). Each assay sample was also treated with alkaline phosphatase, and the dephosphorylated product matches well with the synthetic standard (Extended Data Fig. 4). As expected, no thiosugar product was detected in the sample of AoThiS. These results firmly established that BexX-catalyzed 2-thiosugar formation could proceed in the presence of either AoCysO-COS− or AoMoaD2-COS−, but not AoThiS-COS−. These two examples clearly reveal the moonlighting capability of some of the sulphur-delivery enzymes in natural product biosynthesis and nicely bridge the biosynthetic pathways of primary and secondary metabolites.
Extended Data Fig. 4. BexX-catalyzed 2-thio-D-glucose 6-phosphate formation followed by alkaline phosphatase treatment.
a, Reaction scheme to make the expected bimane derivative. b, HPLC traces of the C-His6-BexX-catalyzed reactions utilizing N-His6-AoThiS, N-His6-AoCysO or N-His6-AoMoaD2, and the control reactions. The thiosugar product was treated with alkaline phosphatase (CIP) and then derivatized with mBBr. HPLC analysis of the synthetic standard of 2-thio-D-glucose-bimane is shown on the bottom trace (trace 7). c, High-resolution ESI-MS (positive) of the isolated product peak, calculated for 2-thio-D-glucose-bimane C16H22N2O7S+ [M + Na]+ 409.1040, observed 409.1038.
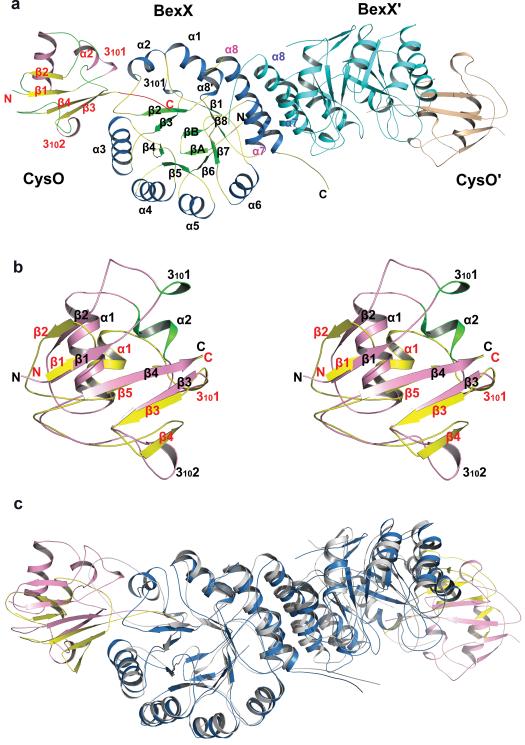
Thiocarboxylated sulphur-carrier proteins recognize their partners through specific protein/protein interactions.20-22 Because BexX shares 37% sequence identity with ThiG22 and the two enzymes are structurally homologous, it was surprising to find that AoThiS does not serve as a sulphur-carrier protein for BexX. To understand the sulphur-carrier protein specificity of BexX, the BexX/AoCysO structure was determined to 2.6 Å resolution (Fig. 3a, Extended Data Fig. 1f). We were unable to crystallize BexX/AoMoaD2; however, because of the compact ubiquitin-like fold and similar sizes of AoCysO (90 residues) and AoMoaD2 (96 residues), we were able to construct a reliable homology model for BexX/AoMoaD2 using the BexX/AoCysO structure as a template. We also constructed a hypothetical model of BexX/AoThiS using ThiS from Thermus thermophilus (PDB code: 2HTM) as a guide. AoCysO and AoMoaD2 superimpose very well with a root mean square deviation (rmsd) of 0.1 Å for 80 Cα carbon atoms. In contrast, AoCysO and AoThiS show significant differences, especially in the loop regions, with an rmsd of 2.7 Å for 43 Cα carbon atoms (Fig. 3b, Extended Data Fig. 1d, e and 5). The most significant difference between either AoCysO or AoMoaD2, and AoThiS (66 residues) is the insertion of two additional α-helices, which are located in the BexX/sulphur-carrier protein interface (Extended Data Fig. 5). As a result the amount of accessible surface area buried upon complex formation is ~1000 Å2 for BexX/AoCysO and BexX/AoMoaD2 compared to only ~600 Å2 in BexX/AoThiS (Extended Data Fig. 5, 6a). AoCysO contributes 19 residues and BexX contributes 26 interface residues to the interface, which is similar to 16 AoMoaD2 residues and 23 BexX residues in BexX/AoMoaD2. In contrast, only eight AoThiS residues contribute to the interface in the BexX/AoThiS model. Ten interface residues are conserved between AoCysO and AoMoaD2, but only four of these are conserved in AoThiS (Extended Data Fig. 5a). The hydrogen bonding scheme for BexX/AoCysO is also conserved in BexX/AoMoaD2 (Extended Data Fig. 6b–d). A comparison of the BexX/AoCysO complex and the Bacillus subtilis ThiG/ThiS (PDB ID: 1TYG)22 complex provides further insight. Superposition of BexX/AoCysO and ThiG/ThiS results in an rmsd of 1.7 Å for the BexX-ThiG core (Fig. 3c); however, AoCysO and ThiS do not overlay well (rmsd more than 40 Å). Thus, even though the overall sulphur-carrier protein folds are similar, and each sulphur-carrier protein is positioned to insert its C-terminal tail into the active site of its partner (Extended Data Fig. 6e, f), the selection of AoCysO or AoMoaD2 by BexX is clearly determined by the interface interactions.
Figure 3. Structure of BexX/AoCysO from A. orientalis.
a, The ribbon diagram of the BexX/AoCysO heterotetramer generated using twofold crystallographic symmetry. BexX′ and AoCysO′ are colored in cyan and wheat, respectively. Secondary structural elements of BexX and AoCysO are colored in blue and pink for α-helices, green and yellow for β-strands, yellow and green for loops, respectively. The C-terminal tail (AVAGG) of AoCysO is highlighted in red. The helices α7 and α8 from BexX and BexX′ are labeled in magenta and blue, respectively. b, Stereo diagram of the superposition of AoCysO (pink) and AoThiS (yellow). Secondary structural elements are labeled in black for AoCysO and red for AoThiS. The two major insertions of AoCysO, 3101 and α2, are highlighted in green. c, Comparison of BexX/AoCysO dimer and B. subtilis ThiG/ThiS dimer. Monomers are colored in blue for BexX, pink for AoCysO, gray for ThiG, and yellow for ThiS.
Extended Data Fig. 5. Sequence alignment of AoCysO, AoMoaD2 and AoThiS and hydrophobic interactions for BexX complexes.
a, Sequence alignment was performed based on structural supersession using the programs 3D-Coffee38, MultAlin39 and ESPript40. Main differences between AoCysO (or AoMoaD2) and AoThiS result from an insertion of ten residues between β1 and β2 of AoCysO (or AoMoaD2) and an insertion of 14 (or 15) residues between α1 and β3 of AoCysO (or AoMoaD2). The first insertion includes the short helix 3101 and the second includes helix α2. Both of these insertions are involved in the BexX-AoCysO (or BexX-AoMoaD2) interface. Ten interface residues (marked with red star and red triangle) are conserved between AoCysO and AoMoaD2, however, only four residues are conserved in AoThiS (marked with red triangle). Two differences between AoCysO and AoMoaD2 represent conservative substitutions; while Thr9 and Ala86 in AoCysO is replaced by Gly11 and Ser92 in AoMoaD2, the interface interaction is contributed by hydrogen bonds that formed by the backbone atom. b–d, Hydrophobic interactions of (b) BexX/AoCysO, (c) BexX/AoMoaD2, and (d) BexX/AoThiS. BexX monomers are shown as gray ribbon diagrams with hydrophobic interaction regions colored in cyan. AoCysO, AoMoaD2 and AoThiS are shown as cartoon and colored in green, skyblue and yellow, respectively. Hydrophobic interaction regions in sulphur carrier protein are colored in red. The α-helices and β-strands in BexX and sulphur carrier proteins are labeled in black and red, respectively.
Extended Data Fig. 6. The BexX/AoCysO interface, predicted hydrogen bonds in BexX with sulphur-carrier proteins, and comparison of the BexX/AoCysO interface to the Bacillus subtilis ThiG/ThiS interface.
a, Interacting surfaces of BexX (left) and AoCysO (right). The surface is color coded by atom type (oxygen, red; nitrogen, blue; carbon, green). Non-interacting surfaces are color coded gray. b, Hydrogen bonds on the surface of BexX with AoCysO are shown as black dashes. c, Hydrogen bonds formed by the C-terminal tail of AoCysO and the surrounding residues from BexX are shown as black dashes. The Fo-Fc simulated annealing (SA)-omit map of the C-terminal residues (AVAGG) is rendered in gray and contoured at 3.0 sigma. Residues are shown as sticks with the carbon atoms colored gray for BexX and green for AoCysO. AoCysO residues are labeled in red; BexX residues are labeled in black. d, Predicted hydrogen bonds in BexX with other sulphur-carrier proteins. The hydrogen bonding scheme for BexX/AoCysO complex (9 of 12 involving the C-terminal tail) is conserved in the model of BexX/AoMoaD2 complex. e, The interface between BexX (skyblue) and AoCysO (pink). The secondary structural elements of AoCysO are labeled in black, the β2 and α2 elements in BexX are labeled in red. f, The interface between ThiG (gray) and ThiS (yellow) from Bacillus subtilis. The secondary structural elements of ThiS are labeled in black, the β2 and α2 elements in ThiG are labeled in red. The β2-α2 loop region in BexX and ThiG is highlighted in red. For CysO, 31011 and α2 form hydrophobic contacts with the β2-α2 loop and α2 of BexX. ThiG also uses its β2-α2 loop to interact with ThiS; however, ThiS uses two different loop regions to form the interface. In addition the β2-α2 loop of BexX is closer to the (βα)8 barrel compared to ThiG for which the β2-α2 loop extends toward out and covers the top of the ThiS.
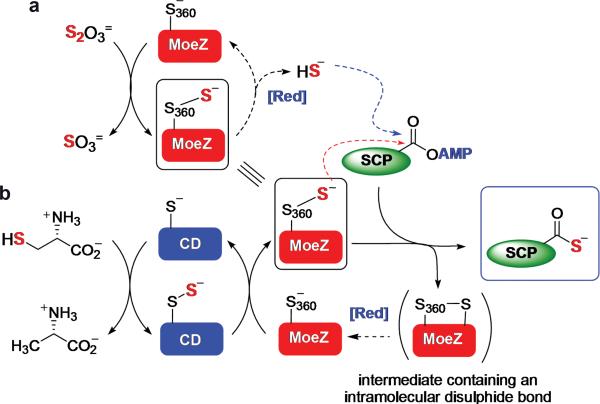
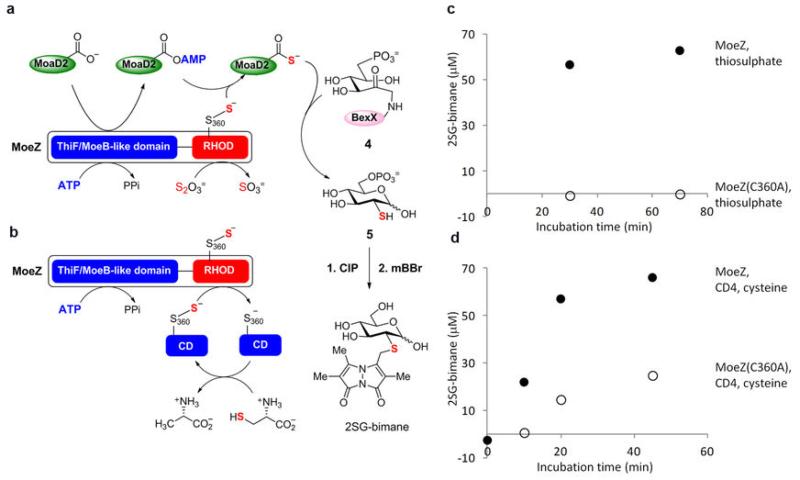
Finally we also examined whether the C-terminal rhodanese domain of AoMoeZ plays a role in sulphur transfer. In a typical rhodanese reaction, the conserved cysteine residue in rhodanese (e.g., C360 in AoMoeZ) is converted to a persulphide group in the presence of thiosulphate or through the action of a cysteine desulphurase using L-cysteine as the sulphur source17. Since the resulting persulphide is a known sulphur donor3, it can be used to charge the adenylated sulphur-carrier proteins to the thiocarboxylate forms (Fig. 4). To test the potential second role of AoMoeZ as a sulphur donor, incubation of AoMoeZ and AoCysO or AoMoaD2 was first carried out in the presence of ATP and thiosulphate (with no addition of reducing agent to prevent bisulphide formation). In both cases, thiolation of AoCysO and AoMoaD2 catalyzed by AoMoeZ was observed but not with the AoMoeZ(C360A) mutant (Extended Data Fig. 7) which retained a similar level of adenylation activity (Extended Data Fig. 8). These observations are consistent with the involvement of the C-terminal rhodanese domain of AoMoeZ in sulphur transfer. Next, BexX-catalyzed 2-thiosugar formation was performed with AoMoeZ and AoMoaD2 in the presence of ATP using either thiosulphate (Fig. 4a) or L-cysteine and a cysteine desulphurase (CD4, Supplementary Table S2) from A. orientalis (Fig. 4b) as the primary sulphur sources. As expected, the 2-thioglucose product was detected in both cases in the absence of reducing agents (Extended Data Fig. 9). Taken together, these results support the likely dual role of AoMoeZ in catalyzing both adenylation and thiolation of the sulphur-carrier proteins.
Figure 4. Possible involvement of the rhodanese domain of AoMoeZ in thiolation of sulphur-carrier proteins.
a, b, The C-terminal rhodanese domain (RHOD) of AoMoeZ catalyzes thiolation of the adenylated sulphur-carrier protein (SCP). The sulphur source for charging the rhodanese domain can be from (a) thiosulphate, or derived from (b) L-cysteine mediated by a cysteine desulphurase (CD). Nucleophilic attack (red dashed line) to the adenylated sulphur-carrier protein followed by intramolecular disulphide bond formation (with another cysteine residue in MoeZ) allows sulphur transfer from the persulphide group to SCP. The protein persulphide intermediate can be reduced to release bisulphide, which can also attack adenylated sulphur-carrier proteins (blue dashed line). To prevent such complications, the experiments were carried out in the absence of reducing agents.
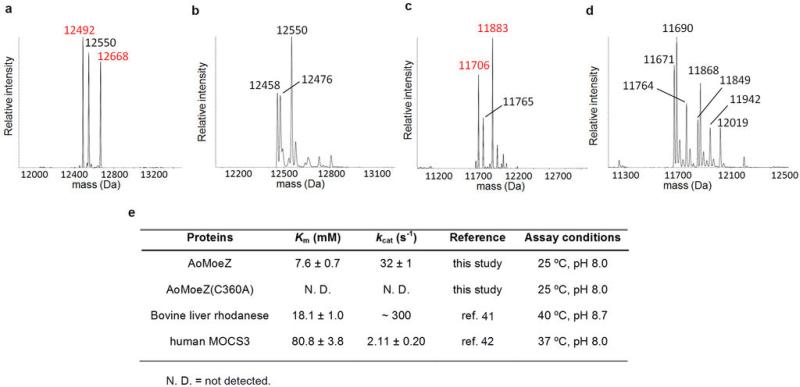
Extended Data Fig. 7. AoMoeZ-dependent protein thiocarboxylate formation of sulphur carrier proteins using thiosulphate as the sulphur source.
a–d, Deconvoluted ESI-MS of (a) AoMoaD2 incubated with AoMoeZ (the observed peaks are consistent with the calculated molecular masses of N-His6-AoMoaD2-COSH (12489), N-His6-AoMoaD2-glycerol (12547), and N-gluconoylated-His6-AoMoaD2-COSH (12667)), (b) AoMoaD2 incubated with AoMoeZ(C360A) mutant (the observed peaks are consistent with the calculated molecular masses of N-His6-AoMoaD2 (12473) and N-His6-AoMoaD2-glycerol (12547)), (c) AoCysO incubated with AoMoeZ (the observed peaks are consistent with the calculated molecular masses of N-His6-AoCysO-COSH (11704), N-His6-AoCysO-glycerol (11762), and N-gluconoylated-His6-AoMoaD2-COSH (11882)), and (d) AoCysO incubated with AoMoeZ(C360A) mutant (the observed peaks are consistent with the calculated molecular masses of N-His6-AoCysO (11688), N-His6-AoCysO-glycerol (11762), their N-gluconoylated derivatives (11866, and 11940), N-His6-AoCysO-AMP (12017)). Observed masses corresponding to protein thiocarboxylate are shown in red. Two peaks corresponding to the dehydration of N-His6-AoMoaD2 and N-His6-AoCysO were likely caused by in-source CID during the ESI-MS analysis. e, Kinetic parameters for the thiosulphate: cyanide sulphurtransferase activity of MoeZ from A. orientalis. Bovine liver rhodanese is a typical rhodanese enzyme. Compared to the bovine rhodanese, human molybdopterin synthase sulphurase (human MOCS3) displayed much lower thiosulphate:cyanide sulphurtransferase activity. In the case of human MOCS3, L-cysteine and cysteine desulphurase are proposed as the physiological sulphur source over thiosulphate because of its lower rhodanese activity43,44. However, this may not be the case for MoeZ from A. orientalis (AoMoeZ) since its rhodanese activity is comparable to bovine liver rhodanese as shown above.
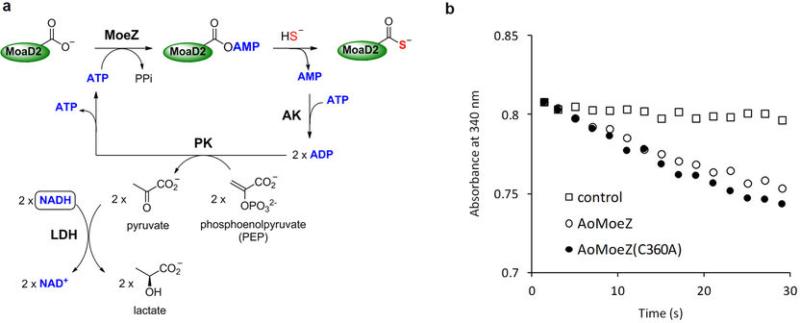
Extended Data Fig. 8. Relative adenylation activitiy of AoMoeZ and AoMoeZ(C360A) mutant.
a, Reaction scheme of the AoMoeZ-catalyzed adenylation activity assay. The adenylation activities of AoMoeZ and its C360A mutant were inferred using a colorimetric assay to monitor the production of AMP (indicated by the decrease of NADH at 340 nm) when AoMoeZ or its C360A mutant was co-incubated with a sulphur carrier protein (AoMoaD2) in the presence of ATP, sodium hydrosulphide, adenylate kinase (AK), pyruvate kinase (PK), and lactate dehydratase (LDH). b, Relative adenylation activity of AoMoeZ (open circle) and its C360A mutant (closed circle) as well as no AoMoeZ/AoMoeZ(C360A) control (open square) were measured by the coupled enzyme assay as described above. Little difference of the decrease of absorpotion at 340 nm was observed for AoMoeZ and its C360A mutant (compare to the control with no AoMoeZ), suggesting that mutation at Cys360 had little effect on the adenylation activity of AoMoeZ.
Extended Data Fig. 9. BexX-catalyzed 2-thiosugar formation using various sulphur sources.
a, b, Reaction scheme of C-His6-BexX-catalyzed 2-thiosugar formation utilizing NHis6-AoMoeZ, N-His6-AoMoaD2 and (a) thiosulphate or (b) L-cysteine and cysteine desulphurase (CD4) from A. orientalis. The reactions were carried out in the absence of reducing agent to avoid complications from generation of bisulphide from protein persulphide (see also below*). Under this condition, AoMoeZ cannot be regenerated after single-turnover. The thiosugar product was derivatized with mBBr and then treated with alkaline phosphatase (CIP) to yield 2-thio-D-glucose-bimane (2SG-bimane). c, d, The 2SG-bimane product concentrations in different incubation time points with (c) thiosulphate or (d) L-cysteine and CD4 as the sulphur source were estimated based on the product peak area of each HPLC trace. The 2SG-bimane synthetic standard (10, 25, 50, 77, 100, 200 μM) was used for the calibration. The closed and open circles denote product formation from the incubation with N-His6-AoMoeZ and N-His6-AoMoeZ(C360A) mutant, respectively. *The observed minor product formation with MoeZ(C360A) mutant, L-cysteine and CD4 (see d, open circles) is likely caused by the formation of bisulphide, which could be generated upon reduction of CD4-persulphide in the presence of free cysteine molecules. In fact, small amount of bisulphide could be detected under similar conditions with L-cysteine and CD4 (in the absence of other proteins and reducing agents) within 15-min incubation by the methylene blue assay45.
In summary, we have performed whole genome sequencing of A. orientalis and demonstrated that sulphur delivery for 2-thiosugar production in the biosynthesis of BE-7585A is achieved by hijacking sulphur transfer systems from primary metabolism. Although the overall reaction mechanism of 2-thiosugar formation resembles that of thiamin biosynthesis, BexX cannot utilize the corresponding sulphur-carrier protein, AoThiS, from the thiamin pathway. Instead, sulphur-carrier proteins likely involved in cysteine (AoCysO) and molybdopterin (AoMoaD2) biosynthesis are recruited to transfer their C-terminal thiocarboxylate sulphur to the BexX-G6P complex (4). Two structural snap shots, which represent the BexX-G6P ketone intermediate (4) and the BexX/AoCysO heterotetramer, provide significant insights into the proposed sulphur incorporation mechanism as well as the structural basis by which sulphur-carrier proteins are selected. These results indicate that a functional alliance between a sulphur-carrier protein and its acceptor protein is not specific, but is not entirely random. The assembly of operational sulphur transfer machinery by using components from sulphur–carrier systems of primary metabolism to deliver sulphur atom to make 2-thiosugar represents an efficient strategy for the biosynthesis of a relatively rare metabolite. Such an ad hoc approach of sulphur transfer may represent a paradigm for as yet undiscovered pathways of sulphur-containing natural product biosynthesis. The revelation that AoMoeZ is the universal activating enzyme for all known sulphur-carrier proteins in A. orientalis is another significant finding. The unique presence of only a single ThiF-type enzyme in the entire genome has also been noted in several other microorganisms (Extended Data Table 2). It appears that the charging of multiple sulphur-carrier proteins in different biosynthetic pathways by one activating-enzyme may be a common phenomenon in nature (at least in Actinomycetales)5,23,24. In addition, the fact that functional pairs of sulphur-carrier proteins and their acceptor proteins are not necessarily located in the same gene cluster raises the possibility that some cryptic gene clusters in various genomes may actually encode pathways for the biosynthesis of sulphur-containing natural products. Such a possibility has generally been overlooked in current efforts to deconvolute genomic information.
Table 2.
BLAST-P analysis of E1-like proteins in genomes of selected strains of Actinomycetales.
| Family | # of E-1 like protein | Name of bacterial strain | protein accession number |
|---|---|---|---|
| Streptomycetaceae | 1 | Streptomyces coelicolor A3(2) | MoeZ (NP_629326) |
| Streptomycetaceae | 1 | Streptomyces avermitilis MA-4680 | MoeZ (NP_824258) |
| Streptomycetaceae | 1 | Streptomyces griseus subsp. griseus NBRC 13350 | MoeZ (YP_001823859) |
| Streptomycetaceae | 1 | Streptomyces caltleya NRRL 8057 | MoeZ (YP_004913535) |
| Streptomycetaceae | 1 | Streptomyces violaceusniger Tu 4113 | MoeZ (YP_004811516) |
| Mycobacterium tuberculosis H37Rv | MoeZ (YP_177942) | ||
| Mycobacteriaceae | 4 | MoeB (YP_177929) | |
| Rv2338c(NP_216854) | |||
| Rv1355c(NP_215871) | |||
| Mycobacterium bovis AF2122/97 | MoeZ (NP_856876) | ||
| Mycobacteriaceae | 4 | MoeB (NP_856788) | |
| MB2366c(NP_856015) | |||
| Mb1390c (NP_855044) | |||
| Mycobacteriaceae | 2 | Mycobacterium avium subsp. paratuberculosis K-10 | MoeZ (YP_962240) |
| MoeY ? (YP_960282) | |||
| Mycobacteria ceae | 2 | Mycobacterium abscessus ATCC 19977 | MoeZ(YP_001704255) |
| E1 family (YP_001702828) | |||
| Corynebacteriaceae | 2 | Corynebacterium glutamicum ATCC 13032 | MoeZ? (NP_599461) |
| MoeZ? (NP_601246) | |||
| Corynebacleriaceae | 1 | Corynebacterium jeikeium ATCC 43734 | MoeZ (ZP_05845904) |
| Nocardiaceae | 2 | Nocardia farcinica IFM 10152 | MoeZ (YP_120782) |
| nfa49170 (YP_121133) | |||
| Nocardiaceae | 2 | Rhodococcus jostil RHA1 | MoeZ (YP_706296) |
| RHA1_ro05752 (YP_705688) | |||
| Pseudonocardiaceae | 1 | Saccharopolyspora erythraea NRRL 2338 | MoeZ (YP_001103327) |
| MoeZ(YP_003763336) | |||
| MoeB?(YP_003764340) | |||
| Pseudonocardiaceae | 4 | Amycolalopsis mediterranei U32 | AMED 124T (YP_003763458) |
| E1-family (YP_003766676) | |||
| MoeZfYP_716188) | |||
| Frankiaceae | 5 | Frankia alni ACN14a | HesA ? (YP_716927) |
| HesA2 ? (YP_716575) | |||
| E1-family (YP_711122) | |||
| E1-family (YP_715173) | |||
| Micrococcaceae | 1 | Anthrobacter chlorophenolicus A6 | MoeZ (YP_002488550) |
| Microbactehaceae | 1 | Clavibacler michiganensis subsp. michiganensis NCPPB 382 | MoeZ (YP_001223108) |
| Micromonosporaceae | 2 | Micromonospora auranliaca ATCC 27029 | MoeZ(YP_003838663) |
| E1-family fYP_003839126) | |||
| Micromonosporaceae | 3 | Salinispora arenicola CNS-205 | MoeZ(YP_001535374) |
| E1-family(YP_001536912) | |||
| E1-family (YP_001538897) | |||
| Nocardioidaceae | 1 | Nocardioides sp. JS614 | MoeZ (YP_922675) |
| Propionibacteriaceae | 1 | Microlunatus phosphovorus NM-1 | MoeZ (YP_004573609) |
Methods
Whole-genome sequencing and analysis
Spores of A. orientalis were inoculated into 10 mL of soybean-casein digest (TSB) medium and grown in a rotary incubator at 30 °C and 250 rpm for 2 days. The resultant seed culture (4 mL) was transferred to 100 mL of TSB medium and grown under the same conditions for 2 days. The growth culture (25 mL) was centrifuged at 5,000 × g for 20 min at 4 °C, and the cells were washed with 25 mL of 10 mM EDTA. After another centrifugation, the cells were stored at −80 °C until use. The cells were resuspended in 5 mL of 1 mM EDTA, and the suspension was divided into 1.5 mL-tubes (0.4 mL each). The genomic DNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen) according to the manufacturer's instructions. The resultant DNA solution (0.5 μg/μL, 50 μL from each tube) was subjected to massive parallel sequencing using Roche 454 GS FLX Titanium and Illumina Genome Analyzer IIx at the High Throughput Sequencing Core Facility of the Academia Sinica in Taiwan. Primary assembly was carried out using Roche 454 Newbler software. Contig extension and closing of short gaps was achieved by scripts built in-house at the core. Genome annotation was performed using Glimmer (Gene Locator and Interpolated Markov ModelER) version 3.025, tRNAscan-SE26, and RNAmmer27. Homologous protein sequences were identified in the NCBI database using the basic local alignment search tool (BLAST).
Preparation of proteins
C-His6-BexX was prepared as described before.7 The thiS (orf13974), moaD (orf13839), cysO (orf06461), moaD2 (orf10102), moeZ (orf02110), and cd4 (orf04763) genes were PCR-amplified from A. oreintalis genomic DNA using primers with engineered NdeI and HindIII restriction sites. The sequences of the primers are described in the Supplementary information. The PCR-amplified gene fragments were purified, digested with NdeI and HindIII and ligated into pET28b(+) vector (Novagen) that was also digested with the same enzymes. For crystallization studies, the bexX gene was also subcloned into pET28b(+) and produced as an N-terminal His6-tagged protein. In addition, the cysO gene was subcloned into IMPACT™pTYB1 vector (New England Biolab) digested with NdeI and SapI for production of AoCysO thiocarboxylate.28 The resultant plasmids were used to transform E. coli BL21 star (DE3) strain (Invitrogen) for protein overexpression. An overnight culture of E. coli transformants grown in the LB medium (10 mL) containing 50 μg/mL of kanamycin at 37 °C were used to inoculate 1 L of the same growth medium. The culture was incubated at 37 °C with shaking (230 rpm) until the OD600 reached ~0.5. Protein expression was then induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM, and the cells were allowed to grow at 18 °C and 125 rpm for an additional 24 h. The cells were harvested by centrifugation at 4,500 × g for 15 min and stored at –80 °C until lysis. All purification steps were carried out at 4 °C using Ni-NTA resin according to the manufacturer's protocol. The proteins were eluted using 250 mM imidazole buffer containing 10% glycerol except those for crystallization studies. The collected protein solution was dialyzed against 3 × 1-L of 50 mM Tris·HCl buffer (pH 8) containing 300 mM NaCl and 15% glycerol. The protein solution was then flash-frozen in liquid nitrogen and stored at −80 °C until use. For crystallization studies, N-His6-BexX eluted with 250 mM imidazole buffer was incubated with 2 mM glucose 6-phophate (G6P) and 2 mM dithiothreitol for 1 h at 4 °C and then further purified in 10 mM Tris·HCl buffer (pH 8.0) containing 50 mM NaCl by using a Superdex G200 column (GE Healthcare). In case of cysO/pTYB1, the cell lysate were loaded onto a column of chitin beads (New England Biolabs, 10 mL) at a flow rate of 0.8 mL/min. The column was then washed with 15 column volumes of column buffer at a flow rate of 2 mL/min. Intein mediated cleavage of the protein was carried out at 18 °C for 12 h with 30 mL of 50 mM dithiothreitol to yield AoCysO or with Na2S to yield AoCysO-thiocarboxylate.28 AoCysO was further purified in 50 mM NaCl, 10 mM Tris·HCl buffer (pH 8.0) containing 50 mM NaCl by using a Superdex G75 column (GE Healthcare). Protein concentration was determined by the Bradford assay using bovine serum albumin as the standard.29 The molecular mass and purity (> 90% except N-His6-AoMoaD) of the proteins were estimated by SDS-PAGE analyses (Extended Data Fig. 3l).
ESI-MS Analyses of proteins
The purified sulphur carrier protein (i.e., N-His6-AoThiS (90 μM), N-His6-AoMoaD (50 μM), N-His6-AoCysO (60 μM), or N-His6-AoMoaD2 (90 μM)) in 5 or 100 mM Tris·HCl buffer (pH 8.0) was subjected to ESI-MS analysis, which was performed at the Mass spectroscopy was performed at the Mass Spectrometry core facility in the College of Pharmacy at the University of Texas, Austin (Extended Data Fig. 3b, f–h). In the case of N-His6-AoThiS, the protein was also incubated with N-His6-AoMoeZ (80 μM) and ATP (5 mM) in 100 mM Tris·HCl buffer (pH 8.0) containing MgCl2 (5 mM) at 30 °C for 0.5 h (Extended Data Fig. 3c). Additionally, to aliquots of the above solution was added sodium sulphide (NaSH, 10 mM), and the resulting solution was subjected to ESI-MS analysis after incubation at 30 °C for 0.5 h (Extended Data Fig. 3d). As a control, a reaction containing only N-His6-AoThiS (90 μM) and NaSH (5 mM) in 100 mM Tris·HCl buffer (pH 8.0) was similarly analyzed (Extended Data Fig. 3e). The other sulphur carrier proteins (i.e., N-His6-AoMoaD (50 μM), N-His6-AoCysO (60 μM), and N-His6-AoMoaD2 (90 μM)) were separately incubated with N-His6-AoMoeZ (7 μM), ATP (5 mM), and NaSH (10 mM) in 50 mM Tris·HCl buffer (pH 8.0) containing MgCl2 (5 mM) at 30 °C for 0.5 h and subjected to ESI-MS analysis (Extended Data Fig. 3i–k). Finally, each sulphur carrier protein (i.e., NHis6-AoThiS (90 μM), N-His6-AoCysO (60 μM) or, N-His6-AoMoaD2 (90 μM)), was also incubated with C-His6-BexX (100 μM) and N-His6-AoMoeZ (7 μM) in 50 mM Tris·HCl buffer (pH 8.0) containing ATP (5 mM), NaSH (10 mM), and MgCl2 (5 mM) at 30 °C for 0.5 h. The resultant solution was subjected to ESI-MS analysis (Fig. 2d–g).
2-Thiosugar Formation and its Detection
The typical BexX reaction mixture (50 μL) contained C-His6-BexX (100 μM), sulphur carrier proteins (N-His6-AoCysO (30 μM), NHis6-AoMoaD2 (45 μM), or N-His6-AoThiS (45 μM)), N-His6-AoMoeZ (15 μM), ATP (2 mM), glucose-6-phosphate (G6P, 2 mM), NaSH (5 mM) in 50 mM NH4·HCO3 buffer (pH 8.0) containing MgCl2 (5 mM). The resulting reaction mixture was incubated at room temperature for 8 h and stored at −20 °C until analysis. To the reaction mixture (10 μL) prepared above was then added 10 μL of 10 mM monobromobimane (mBBr) methanol solution (final concentration of mBBr was 5 mM) and incubated at room temperature for 5 min. The mixture was centrifuged at 16,000 × g for 5 min to remove the precipitant, and 10 μL of the supernatant was transferred to a new tube. The solution was evaporated in vacuo using a Speedvac SC100 (Savant). The resulting residue was redissolved in 100 μL of 50 mM NH4·HCO3 buffer (pH 8.0) and subjected to HPLC analysis using a Dionex CarboPac PA1 analytical column (4 × 250 mm). The sample was eluted with a gradient of water (solvent A) and 1 M NH4OAc (solvent B). The gradient was run from 5 to 15% B over 5 min, 15−30% B over 15 min, 30−100% B over 7 min with a 5 min wash at 100% B, and 100−5% B over 3 min, followed by re-equilibration at 5% B for 5 min. The flow rate was 1 mL/min, and the detector was set at 260 nm (Fig. 2h). The peak corresponding to the enzymatic reaction product was isolated and subjected to ESI-MS and NMR analyses (see Supplementary information for the structural characterization). Alternatively, the reaction mixture stored at −20 °C was thawed and treated with calf intestinal alkaline phosphatase (CIP) (0.2 μL, 2 units) and incubated at 37 °C for 1 h. The precipitant that appeared during the incubation was removed by centrifugation at 16,000 × g for 2 min, and 2 μL of a 100 mM monobromobimane (mBBr) methanol solution was added to the reaction solution (final concentration of mBBr was 5 mM). The resulting mixture was incubated at room temperature for 5 min, and the supernatant (5 μL) was diluted with deionized water (95 μL) prior to HPLC analysis using an analytical C18 column (4 × 250 mm). The sample (20 μL) was eluted with a gradient of water (solvent A) and 80% acetonitrile (solvent B). The gradient was run from 5 to 30% B over 15 min, 30−80% B over 5 min, 80−5% B over 5 min, followed by reequilibration at 5% B for 10 min. The flow rate was 1 mL/min, and the detector was set at 260 nm. The 2-thio-D-glucose-bimane standard (0.1 mM) was prepared from the chemically synthesized 2-thio-D-glucose6 incubated with mBBr at room temperature for 5 min. The peak corresponding to the enzymatic reaction product was also isolated and subjected to ESI-MS analysis (Extended Data Fig. 4).
Determination of Rhodanese Activity of AoMoeZ
The site-specific C360A mutant of AoMoeZ were constructed according to the mutagenesis protocol using moeZ/pET28b(+) as DNA template. The forward primer 5’-GATCGTCCTGCACGCCAAGTCGGGC-3’ and the reverse primer 5’-GCGGGCGCCCGACTTGGCGTGCAGG-3’ were used in the PCR amplification. The resulting plasmids moeZ(C360A)/pET28b(+) were used to transform the E. coli BL21 star (DE3) strain for protein overexpression. The rhodanese activity of AoMoeZ was determined using the assay developed by Sörbo30. A typical assay mixture contained 50 mM Tris·HCl (pH 8.0), 50 mM KCN, approximately 2 μM of AoMoeZ or AoMoeZ(C360A), and a variable amount of sodium thiosulfate (from 0 to 35 mM) in a volume of 100 μL. The reaction was initiated by the addition of AoMoeZ and was quenched after 10-s incubation at 25 °C by the addition of 50 μL of reagent A (15% formaldehyde). Then 150 μL of reagent B (1 g Fe(NO3)3·9H2O and 2 mL 65% HNO3 in 13 mL H2O) was added for color development. Formation of SCN− in the reaction was quantified using the extinction coefficient for Fe(SCN)3 (4200 M−1 cm−1 at 460 nm). The steady state kinetic parameters were determined in triplicates by fitting the experimental data using Michaelis–Menten equation (Extended Data Fig. 7e). Assay for protein thiocarboxylate formation was performed in the anaerobic chamber to minimize the oxidation of AoMoeZ or AoMoeZ(C360A). A typical reaction mixture contained 80 μM AoMoeZ or AoMoeZ(C360A), 4 mM ATP, 5 mM MgCl2, 5 mM Na2S2O3 in 50 mM HEPES (pH 8.0) with 500 mM glycerol (from enzyme stock solution), and 100 μM of one of the sulphur carrier proteins, N-His6-AoMoaD2 or N-His6-AoCysO. The reaction was incubated in the glove-box at ~30 °C for 40 min and then quenched by flash freezing in liquid nitrogen. Samples were then submitted to ESI-MS for analysis (Extended Data Fig. 7a–d).
Spectrophotometric analysis of the adenylation reaction catalyzed by AoMoeZ and its C360A mutant
The adenylation of sulphur-carrier proteins catalyzed by MoeZ and its C360A mutant was monitored using a coupled enzyme assay in the presence of an excess of NaSH (Extended Data Fig. 8a). The coupled enzyme reaction was monitored by detecting the consumption of NADH (ε340 = 6220 M−1cm−1) at 340 nm. A typical reaction mixture (120 μL) contained AoMoeZ or its C360A mutant (3 μM), MoaD2 (10 μM), ATP (80 μM), NaSH (3 mM), phosphoenolpyruvate (PEP, 2 mM), NADH (0.16 mM), and ~1.5 unit each of adenylate kinase (AK), pyruvate kinase (PK), and lactate dehydratase (LDH) in 50 mM NH4·HCO3 buffer (pH 8.0) containing MgCl2 (2.5 mM). The reaction was initiated upon addition of ATP at time 0, and absorbance at 340 nm was monitored for 30 sec (Extended Data Fig. 8b).
2-Thiosugar Formation using other sulphur sources
A typical reaction mixture (40 μL) contained C-His6-BexX (15 μM), N-His6-AoMoaD2 (18 μM), N-His6-AoMoeZ or its C360A mutant (100 or 90 μM), ATP (2.5 mM), glucose-6-phosphate (G6P, 2.5 mM), and different sulphur sources [(i) 0.2 mM Na2S2O3, or (ii) 0.15 mM L-cysteine and 25 µM cysteine desulphurase CD4 with 0.25 mM pyridoxal 5-phosphate (PLP)] in 50 mM NH4·HCO3 buffer (pH 8.0) containing MgCl2 (5 mM). The resulting reaction mixture was incubated at 30 °C for 30 or 70 min for (i) and 0, 10, 20, or 50 min for (ii). The reaction was quenched by adding an equal volume of acetonitrile, and to the collected supernatant was added mBBr (~2 mM final concentration). After incubation at room temperature for 30 min, the reaction mixture was dried by speed vacuum. The residue was re-dissolved in 50 mM NH4·HCO3 (pH 8.0) and treated with 3 unit of CIP at 37 °C for 1.5 h. The CIP enzyme was removed by centrifugation after precipitation with acetonitrile (50% v/v in final concentration), and the collected supernatant was dried by speed vacuum. The residue was then dissolved in 40 µL of deionized water prior to HPLC analysis with a C18 column (4 × 250 mm). Each sample (20 μL) was eluted with a gradient of water (solvent A) and acetonitrile (solvent B). The gradient was run from 4 to 24% B over 15 min, 24−64% B over 5 min, 64−4% B over 5 min, followed by re-equilibration at 5% B for 8 min. The flow rate and the detector setting were the same as described above (1 mL/min, 260 nm). The 2-thio- D-glucose-bimane standard (10, 25, 50, 77, 100, 200 μM) described above was also injected into HPLC for calibration of the peak area (Extended Data Fig. 9).
Crystallization of BexX-G6P
Crystals of BexX-G6P were grown using the vapor diffusion hanging drop method. A solution containing 10 mg/mL of BexX in 10 mM Tris, 50 mM NaCl, pH 8.0 was preincubated on ice with G6P at final concentration of 2 mM for about 1 h. Hanging drops were formed by mixing 1.5 μL of protein solution and 1.5 μL of well solution containing 40% (v/v) PEG300, 0.1 M sodium cacodylate/HCl, pH 6.5 and 0.2 M calcium acetate. Rod shape crystals grew in about 6 days to their maximum size of 0.2-0.3 mm × 0.1-0.2 mm. Preliminary X-ray analysis showed that the crystals belonged to the space group P41212 or P43212 with unit cell dimensions of a = 168.9 Å and c = 42.4 Å. The Matthew's coefficient assuming two monomers of BexX per asymmetric is 2.8 Å3/Da and corresponds to a solvent content of 56.1%.
Crystallization of BexX/AoCysO Complex
Crystals of BexX/AoCysO were grown using the vapor diffusion hanging drop method. Both AoCysO and AoCysO thiocarboxylate were used for crystallization trials; however, AoCysO consistently gave better crystals and was used for the structures reported here. The BexX/AoCysO complex was formed by preincubating 0.55 mL of BexX at 15 mg/mL and 1.0 mL of AoCysO at 10 mg/mL in 10 mM Tris, 50 mM NaCl, pH 8.0 for 1 h. Hanging drops were formed by mixing 1.5 μL of protein solution and 1.5 μL of well solution containing 28% PEG4000, 0.1 M Tris, pH 8.0 and 0.2 M LiSO4. Plate shape crystals appeared within 5 days and grew to their maximum size of 0.5 mm × 0.4 mm × 0.02 mm in about two weeks. The crystals belonged to space group I422 with unit cell dimensions of a = 106.4 Å and c = 181.7 Å. The Mathew's coefficient is 3.5 Å3/Da assuming one monomer of BexX and one monomer of AoCysO per asymmetric unit and corresponds to a solvent content of 64.5%.
X-ray Data Collection and Processing
X-ray diffraction data for BexX-G6P were collected at beamline A1 at the Cornell High Energy Synchrotron Source (CHESS) using an Area Detector Systems Corporation (ADSC) Quantum 210 CCD detector with a crystal to detector distance of 200 mm and a wavelength of 0.9767 Å. The data collection temperature was 100 K. A total of 180° of data was collected with an oscillation range of 0.5° per frame and an exposure time of 3 s per frame. Data for BexX-G6P/AoCysO were collected at NE-CAT beamline 24-ID-C at the Advanced Photon Source (APS) using an ADSC Q315 CCD detector. The wavelength was 0.9791 Å, the data collection temperature was 100 K, and the detector distance was 400 mm. Individual frames were collected over a range of 180° using 1 s for each 1.0°. X-ray diffraction data were indexed, integrated, scaled, and merged using the program HKL200031.
Structure Determination and Refinement
The structure of BexX was determined by molecular replacement using Phaser32 as implemented in the PHENIX33 program package. A monomer of thiamin thiazole synthase (ThiG) from Bacillus subtilis (PDB ID: 1TYG)22, which shares 37% sequence identity with BexX, was modified using Chainsaw in CCP434 to generate a search model. The initial molecular replacement solution was refined to an R-factor of 40.0% and R-free of 44.0%. All side chains were added and the model was manually adjusted using COOT35. After several cycles of refinement using PHENIX33 and Refmac536, G6P and water molecules were added. The final model was refined to an R-factor of 19.1% and R-free of 22.2%. The Ramachandran plot shows 94.7% of residues in the most favorable regions and 5.3% in the allowed regions. No residues were in generously allowed regions or disallowed regions. The structure of BexX/AoCysO was determined by molecular replacement using a monomer of BexX from the BexX-G6P complex and a monomer of CysO from Mycobacterium tuberculosis (PDB ID: 3DWM)20 as the search models. An initial model, corresponding to one monomer of BexX and one monomer of AoCysO, was generated by Phaser32 as implemented in PHENIX33. Packing analysis showed that a BexX/AoCysO dimer is formed by crystallographic two-fold symmetry. The initial refinement resulted in an R-factor of 33.8% and R-free of 39.5%. Subsequent cycles of model building in COOT35 and refinement in PHENIX33 and Refmac536 resulted in a final R-factor of 19.6% and R-free of 24.0%. The Ramachandran plot shows that 90.0% of residues are located in the most favorable regions, 9.7% in the allowed regions, and 0.3% in the generously allowed regions. No residues were in the disallowed regions.
Supplementary Material
Acknowledgements
We thank Professor Chi-Huey Wong, Dr. Rachel Chen, and funds provided by Academia Sinica for the whole-genome draft sequencing, the staff at Analytical Instrumentation Facility of the College of Pharmacy (University of Texas), APS NE-CAT and the CHESS beamlines (Cornell University) for their assistance in data collection. We also thank David Kim for assistance with the early biochemical experiments, Dr. Cynthia Kinsland of the Cornell Protein Production Facility for providing the cysO/pTYB1 clone, Dr. Yang Zhang for the great help on structure determination of BexX/AoCysO, and Leslie Kinsland for help in preparing the manuscript. This work was supported in part by grants from the National Institutes of Health (GM035906 to H.-w.L. and DK67081 to S.E.E.) and the Welch Foundation (F-1511 to H.-w.L.). The X-ray crystallography work was conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award GM103403 from the National Institute of General Medical Sciences at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The use of the Cornell High Energy Synchrotron Source is supported by the National Science Foundation (DMR-0936384), and National Institutes of Health/National Institute of General Medical Sciences (GM103485).
Footnotes
Author Contributions H.-w.L. provided the scientific direction and the overall experimental design for the studies. E.S. and H.S. designed and performed the biochemical experiments. X.Z. and S.E.E. were responsible for the crystal structure studies. M.-Y.J.L., T.-l.L., A.O., J.-y.L., and Y.-h.C. carried out the whole genome sequencing and gene annotation. E.S., X.Z., H.S., S.E.E. and H.-w.L. wrote the manuscript.
The nucleotide sequences reported herein have been deposited into the GenBank database under the accession numbers JN602207, JN602208, JN602209, JN602210, and JN602211. The Brookhaven Protein Data Bank codes for BexX-G6P and BexX/AoCysO are 4N6F and 4N6E, respectively. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Parry RJ. Comprehensive Natural Products Chemistry. Vol. 1. Elsevier Science Ltd; Oxford: 1999. Biosynthesis of sulfur-containing natural products. pp. 825–863. [Google Scholar]
- 2.Fontecave M, Ollagnier-de-Choudens S, Mulliez E. Biological radical sulfur insertion reactions. Chem. Rev. 2003;103:2149–2166. doi: 10.1021/cr020427j. [DOI] [PubMed] [Google Scholar]
- 3.Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 4.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs AM, Iyer LM, Aravind L. Natural history of the E1-like superfamily: Implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki E, Ogasawara Y, Liu H.-w. A biosynthetic pathway for BE-7585A, a 2-thiosugar-containing angucycline-type natural product. J. Am. Chem. Soc. 2010;132:7405–7417. doi: 10.1021/ja1014037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki E, Liu H.-w. Mechanistic studies of the biosynthesis of 2-thiosugar: evidence for the formation of an enzyme-bound 2-ketohexose intermediate in BexX-catalyzed reaction. J. Am. Chem. Soc. 2010;132:15544–15546. doi: 10.1021/ja108061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thibodeaux CJ, Melançon CE, Liu H.-w. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 9.Thibodeaux CJ, Melançon CE, Liu H.-w. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C-I, McCarty RM, Liu H.-w. The biosynthesis of nitrogen-, sulfur- and high-carbon chain-containing sugars. Chem Soc. Rev. 2013;42:4377–4407. doi: 10.1039/c2cs35438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braunshausen A, Seebeck FP. Identification and characterization of the first ovothiol biosynthetic enzyme. J. Am. Chem. Soc. 2011;133:1757–1759. doi: 10.1021/ja109378e. [DOI] [PubMed] [Google Scholar]
- 12.Park JH, et al. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (Vitamin B1). Biochemistry. 2003;42:12430–12438. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]
- 13.Begley TP. Cofactor biosynthesis: an organic chemist's treasure trove. Nat. Prod. Rep. 2006;23:15–25. doi: 10.1039/b207131m. [DOI] [PubMed] [Google Scholar]
- 14.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Ann. Rev. Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 17.Cipollone R, Ascenzi P, Visca P. Common themes and variations in the rhodanese superfamily. IUBMB Life. 2007;59:51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 19.Burns KE, et al. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 2005;127:11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurgenson CT, Burns KE, Begley TP, Ealick SE. Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis. Biochemistry. 2008;47:10354–10364. doi: 10.1021/bi800915j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat. Struct. Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 22.Settembre EC, et al. Thiamin biosynthesis in Bacillus subtilis: structure of the thiazole synthase/sulfur carrier protein complex. Biochemistry. 2004;43:11647–11657. doi: 10.1021/bi0488911. [DOI] [PubMed] [Google Scholar]
- 23.Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 2008;27:3267–3278. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voss M, Nimtz M, Leimkühler S. Elucidation of the dual role of mycobacterial MoeZR in molybdenum cofactor biosynthesis and cysteine biosynthesis. PLoS One. 2011;6:e28170. doi: 10.1371/journal.pone.0028170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinsland C, Taylor SV, Kelleher NL, McLafferty FW, Begley TP. Overexpression of recombinant proteins with a C-terminal thiocarboxylate: implications for protein semisynthesis and thiamin biosynthesis. Protein science : a publication of the Protein Society. 1998;7:1839–1842. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 30.Sörbo B. Enzymic transfer of sulfur from mercatopyruvate to sulfate or sulfinates. Biochim. Biophys. Acta. 1957;24:324–329. doi: 10.1016/0006-3002(57)90201-9. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski ZM, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J. Applied Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geoghegan KF, et al. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: The cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 1999;267:169–184. doi: 10.1006/abio.1998.2990. [DOI] [PubMed] [Google Scholar]
- 38.Tommaso P, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic acids research. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger P, Westley J. An expanded mechanism for rhodanese catalysis. J. Biol. Chem. 1974;249:780–788. [PubMed] [Google Scholar]
- 42.Chowdhury MM, Dosche C, Löhmannsröben H, Leimkühler S. Dual role of the molybdenum cofactor biosynthesis protein MOCS3 in tRNA thiolation and molybdenum cofactor biosynthesis in humans. J. Biol. Chem. 2012;287:17297–17307. doi: 10.1074/jbc.M112.351429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthies A, Rajagopalan KV, Mendel RR. Leimkühler, S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5946–5951. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marelja Z, Stöcklein W, Nimtz M, Leimkühler S. A novel role for human Nfs1 in the Cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 2008;283:25178–25185. doi: 10.1074/jbc.M804064200. [DOI] [PubMed] [Google Scholar]
- 45.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.