Abstract
Regression of cervical intraepithelial neoplasia (CIN) 2-3 to CIN 1 or less is associated with immune response as demonstrated by immunohistochemistry in formaldehyde-fixed paraffin-embedded (FFPE) biopsies. Proteomic analysis of water-soluble proteins in supernatants of biopsy samples with LC-MS (LTQ-Orbitrap) was used to identify proteins predictive of CIN2-3 lesions regression. CIN2-3 in the biopsies and persistence (CIN2-3) or regression (≤CIN1) in follow-up cone biopsies was validated histologically by two experienced pathologists. In a learning set of 20 CIN2-3 (10 regressions and 10 persistence cases), supernatants were depleted of seven high abundance proteins prior to unidimensional LC-MS/MS protein analysis. Mean protein concentration was 0.81 mg/mL (range: 0.55–1.14). Multivariate statistical methods were used to identify proteins that were able to discriminate between regressive and persistent CIN2-3. The findings were validated in an independent test set of 20 CIN2-3 (10 regressions and 10 persistence cases). Multistep identification criteria identified 165 proteins. In the learning set, zinc finger protein 441 and phospholipase D6 independently discriminated between regressive and persistent CIN2-3 lesions and correctly classified all 20 patients. Nine regression and all persistence cases were correctly classified in the validation set. Zinc finger protein 441 and phospholipase D6 in supernatant samples detected by LTQ-Orbitrap can predict regression of CIN2-3.
1. Introduction
Among cancers affecting women, cervical cancer has the second highest occurrence worldwide, with an incidence in 2008 of 529,800 cases (14.5% in developed countries and 85.5% in developing countries) and 275,000 estimated deaths [1]. Infection of cervical epithelial cells with high risk human papillomavirus (HPV) is the most important risk factor for development of cervical cancer, as first highlighted by zur Hausen [2]. Noninvasive cervical intraepithelial neoplasia (CIN) precedes the development of invasive cancer and may progress from CIN2-3 to (micro)invasive cancer in 10–25 years on average [3].
Three CIN grades (CIN1, CIN2, and CIN3) are recognized by the World Health Organization to distinguish the degrees of epithelial abnormality and are associated with increasing risks for invasive cancer development. A CIN lesion is, however, not a static event but a dynamic process that can persist and progress but also spontaneously regress [4, 5]. If left untreated, 5–30% of all CIN2-3 lesions (confirmed by a histological punch biopsy) will develop invasive cancer [6]. On the other hand, without cone excision, as many as 32–43% of CIN2-3 lesions will regress spontaneously [7]. Nevertheless, in many countries including Norway, all punch biopsy-confirmed CIN2-3 lesions are usually treated with diathermic cone excision, a fairly aggressive therapy which can have serious adverse side effects [8]. The most serious late-complication is cervical insufficiency which can lead to late abortion and preterm delivery during the second and early third trimester of a future pregnancy [9, 10].
Until recently, regression of CIN2-3 lesions could not be effectively predicted. However, research on functional biomarkers like pRb, p53, and cytokeratin 13/14 has proven to be helpful in predicting regression, especially when combined with local immune response and HPV genotype [11–14]. Furthermore, combined Ki67 and pRb expression can predict which CIN1 lesions will progress to CIN3 [15]. Aggregated information provided by such epithelial biomarkers and local cellular immune response in the microenvironment of CIN2-3 lesions supports prediction of regression/persistence/progression and may result in even more accurate CIN treatment, [16] as well as reducing overtreatment of patients with CIN2-3 lesions.
Unfortunately, the procedures used to obtain formalin-fixed, paraffin-embedded (FFPE) tissue from biopsies irreversibly degrade water-soluble proteins. A protein collection method for small punch biopsy samples that could represent not only the cellular response but also proteins from the cervical neoplasia microenvironment and intracellular compartments may further help define the biology of CIN lesions' dynamic behaviour. We have recently described a method that can preserve and extract water-soluble proteins from punch biopsies [17], how a panel of 3 peaks from SELDI-TOF protein profiles can be used to differentiate normal tissue from CIN tissue samples, and that a discrimination between CIN2 and CIN3 lesions could be obtained using cytokeratin 2 [18].
In the present study we analysed protein samples from CIN2-3 lesions with known regression/persistence status. We have used both SELDI-TOF MS and bottom-up shotgun proteomics [19] approach utilizing nanoflow liquid chromatography coupled to a LTQ-Orbitrap mass spectrometer. The goal was to identify proteins that could be used in prediction of regression or persistence in CIN2-3.
2. Materials and Methods
2.1. Study Population
This study is a subproject from a larger prospective study, approved by the Regional Medical Ethics Committee of Helse Vest, Norway, the Norwegian Data Inspection, and the Health Directorate of Norway, numbers 33.06, 17185, and 07/330. Healthy women aged 25-40 years, with cytological abnormal smears were followed by cervical biopsy and later cone excision. In total, 170 patients with first time onset of CIN2-3 were included from January 2007 to December 2008. The interval between punch biopsy and cone excision was standardized at median 113 days (range: 100–126). This interval was chosen in view of a previous study, which showed that CIN2-3 patients with more than 9-week punch-cone interval have a much higher chance on regression than those with <9-week interval [5]. Regression was defined as CIN1 or less in cone histology and regression rate was 22% (38/170). All patients included in this study were treated according to the national Norwegian population screening quality guidelines [20].
In our cohort of patients we first analysed whether proteins and peptides detected by proteomic LC-MS (LTQ-Orbitrap) could distinguish between CIN2-3 lesions, with and without later regression. Of the 170 patients with cervical punch biopsy samples, a random subset of 20 patients with cervical intraepithelial neoplasia lesions (see below for reviewing details), 10 CIN2 (5 with regression and 5 with persistence) and 10 CIN3 (5 with regression and 5 with persistence), were selected and defined as the learning set. The histological diagnosis was set by two experienced pathologists. The samples were selected so that the whole sampling period was covered and the protein concentration was as close as possible to the average for the whole data set. In a second validation study, another 20 CIN2-3 patients (10 CIN2 cases, 5 with regression and 5 with persistence, and 10 CIN3, 5 with regression and 5 with persistence, defined as the validation set) were selected to test the prognostic value of the proteins found in the learning set.
For the SELDI-TOF study, the sample set from a former investigation was used [21]. These samples were statistically reanalysed with regard to the regression/persistence status. Thus 2 replicates of each of the 5 regression and 40 persistent CIN2-3 samples were included for this part of the study.
2.2. Sample Collection
After colposcopy, punch biopsies and endocervical curettage were taken from the transformation zone and eventually premalignant mucosa. One or two biopsies were immediately placed in polystyrene tubes (Sarsted, Numbrecht, Germany) containing 5 mL RPMI-1640 (Gibco, Carlsbad, USA) tissue culture medium. The biopsies were kept in the tissue culture medium for 24 hours at 4°C before the supernatants were collected, split into aliquots of 500 μL, and stored at −80°C until analysis. Immediately after sample collection, an additional set of biopsies were stored in 4% buffered formaldehyde according to standard procedure.
2.3. Pathology
As described before [17] after 24 hours of incubation in RPMI-1640 medium at 4°C, the biopsies were routinely fixed in buffered 4% formaldehyde, embedded in paraffin, cut at 4 μm, and stained with hematoxylin, eosin, and safran (HES) for routine histological examination. P16 and Ki67 (MIB-1) immunohistochemical (IHC) staining were used to confirm the diagnosis. All HES and IHC sections of the 170 biopsies were reviewed by two independent pathologists, who also used the p16 and Ki67 immunohistochemical information. The participating pathologists were blinded to the original routine clinical findings, histopathological diagnosis, and follow-up. In case of discrepancies the cases were reviewed and diagnosed on a double-head microscope by the two pathologists (Einar Gudlaugsson and Jan Baak) and a consensus diagnosis was obtained.
2.4. ProteinChip SELDI-TOF MS Analysis
Protein concentrations were assessed using the Bradford methodology. Samples were subjected to SELDI-TOF MS profiling according to the manufacturer's instructions (Ciphergen Biosystems, Fremont, CA, USA). The biopsy supernatants were diluted 1 : 5 with 50 mM sodium acetate (pH 4.3) and then bound to a CM10 ProteinChip array. They were incubated for two hours at room temperature on a platform shaker and then washed twice with 50 mM sodium acetate buffer, followed by two washes of 1 μL energy absorbing molecule (=EAM) solution (consisting of 50% saturated synaptic acid dissolved in 50% acetonitrile and 0.5% trifluoroacetic acid). Two replicates were prepared on different CM10 ProteinChips by two different technicians on two different days. The time-of-flight spectra were generated on the Protein Biological System II mass spectrometer reader (Ciphergen Biosystems, Fremont, CA, USA), using a laser intensity of 170 and a detector sensitivity of seven. Readings were optimized for low molecular weight (2–20 kDa). External mass calibration was performed daily.
2.5. SELDI-TOF MS Data Analysis
The SELDI-TOF MS data analyses were performed in three steps: (1) peak detection, (2) selection of peaks with the highest discriminatory power, and (3) building a multivariate model based on the selection in step (2). The peak detection was done using the Ciphergen Seldi software version 3.2 after internal and external mass calibration followed by normalization (total ion current, TIC, intensity) of all spectra as one group. The mass range from 2000 to 20000 Da contained the majority of the peptides/proteins in the samples and was selected. Masses less than 2000 Da were excluded as these are known to contain adducts and artifacts from the EAM solution and other chemical contaminants. The peak detection includes baseline subtraction, calibration of mass accuracy, and automatic peak detection. Each spectrum was then assigned to one of three groups, normal, regression, or persistence. To select peaks with the highest discriminatory power, the Biomarker Wizard (Ciphergen) was used for peak detection and clustering of all the spectra. This was done using a signal-to-noise (s/n) ratio of 5 and 15% of all spectra for the first pass detection and clustering and an s/n ratio of 2 for the second pass. The cluster results were then imported into SPSS (v17, SPSS Norway AS, Oslo, Norway), CART (Salford, San Diego, CA, USA), and MedCalc (MedCalc Software, Mariakerke, Belgium) for binary logistic regression analysis.
2.6. Immunoaffinity Depletion
The preparation and use of the immunoaffinity column is described in [17]. To deplete samples of the 7 high abundance proteins, 100 μL of RPMI supernatant was diluted with 100 μL Tris-buffered saline (TBS, 0.1 M TRIS-base containing 0.1 M NaCl, pH 8.0), and the solution was injected into a TBS solution with a flow of 0.2 mL/min. The nonretained proteins were trapped on a 4 mm × 2.0 mm (inner diameter, i.d.), C18 security guard cartridge with 300 Å pore size (Phenomenex, Teknolab, Norway) and were eluted by backflushing the security guard cartridge with ethanol at a flow of 0.3 mL/min. The affinity column was washed using 0.1 M glycine at pH 2.5 with a flow of 1.2 mL/min. Both columns were reequilibrated with TBS at a flow of 0.2 mL/min for 5 minutes. The pH adjustments were done using 6 M HCl.
2.7. Protein Digestion and Sample Cleanup
After evaporating the ethanol phase containing the nonretained protein fraction using vacuum centrifugation (Eppendorf Concentrator 5301, VWR, Norway), 100 μL 50 mM ammonium bicarbonate pH 8 was added to the samples. 1 μL of 1 M dithiothreitol (DTT) was added to reduce the proteins. 5 μL of 1 M iodoacetamide (IAA) was then added to alkylate the proteins followed by 5 μL of DTT to stop the alkylation process. For each of these steps, 45-minute incubation time was used. One μg trypsin (Promega) was added, and the samples were kept at 37°C for 18 hours. After digestion with trypsin the samples were purified and concentrated using a C18 ZipTip (Millipore, Norway) procedure. The ZipTips were conditioned by aspirating 30 μL acetonitrile five times and equilibrated with pulling 30 μL 0.1% formic acid (FA) in MilliQ water five times through the stationary phase. Approximately 10 μL of the 0.1% FA solution was left above the stationary phase to avoid drying it. Each sample solution was applied on top of the stationary phase using a pipette and then pushed through the tip using air pressure from the pipette plunger. More sample solution was added when approximately 20 μL of the liquid remained so that the whole volumes of the samples were pushed slowly through the ZipTip. Washing was done by aspiring 30 μL of 0.1% FA five times. Elution of the peptides was done in a total volume of 30 μL of 80 : 20 (v/v) acetonitrile : MilliQ water by aspiring 10 μL of this solution 10 times through the stationary phase. The organic phase was then evaporated using vacuum centrifugation and, to the residual solution, 20 μL 0.1% FA was added prior to the LC-MS/MS analysis.
2.8. LC-MS/MS Analysis
A Dionex Ultimate 3000 nanoflow HPLC equipped with a 300 μm (i.d.) × 0.5 cm length Acclaim PepMap 100 C18 trap column and a 75 μm (i.d.) × 15 cm Acclaim PepMap 100 C18 analytical column (Dionex) was used with a LTQ-Orbitrap hybrid mass spectrometer (Thermo Scientific). 5 μL of the tryptic digests was injected onto the trap column using 0.1% formic acid (VWR) in MilliQ-water at a flow of 2 μL/min. The separation was done using a gradient from 2.5% to 64% acetonitrile in 0.1% FA over 180 minutes at a flow of 300 nL/min. A 10-minute postinjection delay and a 20-minute column reequilibration time were used. The electrospray interface was a PicoTip emitter (SilicaTip, New Objective) with a 10 μm tip without coating. The electrospray voltage was set to 1 kV. No sheath gas was used. The mass spectrometer was used in positive mode. Full scans were performed in the Orbitrap using the m/z range from 200 to 2000. Data dependent MS/MS scans were performed in the LTQ for the five most abundant masses with z≥ 2 and intensity higher than 10,000 counts. Dynamic exclusion for 3 minutes after fragmentation of a given m/z value four times was used. Collision induced dissociation (CID) was used with a collision energy of 35%, activation Q setting of 0.400, and 30 ms activation time for MS. Calibration of the mass spectrometer was done weekly using the calibration solution recommended by Thermo Scientific.
2.9. Bioinformatic Data Analysis
The raw data files were analysed using the Proteome Discoverer 1.0 (Thermo Scientific) with the Sequest algorithm to search against the Homo sapiens (Tax.id: 9606) database at NCBI (531420 sequences) with trypsin as digestion enzyme allowing for 2 missed cleavages. All files were also searched against the human papillomavirus database (Tax.id: 10566) at NCBI (1615 sequences). Precursor ion tolerance was set to 10 ppm, and fragment ion mass tolerance was set to 0.8 Da. Oxidation (M) was set as a dynamic modification and carbamidomethyl (C) was set as a static modification due to the use of DTT and IAA. Phosphorylation (STY) was set as a dynamic modification. A high significance peptide confidence filter was set in Proteome Discoverer (PD) from Thermo, which means that peptide identifications are filtered based on the following combination of charge and Xcorr factor: 1.9 (z = 2), 2.3 (z = 3), and 2.6 (z≥ 4). Additional information for proteins was obtained from the UniProt database entry.
Protein identifications were accepted using one peptide when certain requirements were fulfilled: the Sequest Xcorr factor with regard to charge had to be fulfilled according to the high significance criteria in PD. The peptide had to contain at least 7 amino acids and have at least three consecutive b- and y-ions in the MS2 spectra [22], and it should occur minimum three times in the same sample. In addition, for proteins with only one identified peptide sequence, the peptide sequences were submitted for a BLAST search against the Uniprot Homo sapiens database (htttp://www.uniprot.org/) to confirm that the identification matched the NCBI identification. For proteins listed as unnamed in the NCBI database, the ID mapping tool at UniProt was used to see if the protein was listed with a more descriptive annotation in this database. Only proteins identified in at least 30% of the samples in one of the groups (regression/persistence) were included in the remaining work. Spectral count (SPC) results for the identified peptides were obtained and used for normalization (see (1)) after increasing all numbers with 1 to avoid zeros. Consider
| (1) |
2.10. Multivariate Analysis of the LC-MS/MS Data
The normalized SPC results were imported into Sirius (version 8.1, Pattern Recognition Systems, Bergen, Norway) to perform a partial least-squares discriminant analysis (PLS-DA) using a target projection component, calculations of selectivity ratios (SR), plotting of SR-values, and a discriminating variable (DIVA) test. A binary response variable was added to the SPC dataset to assign all samples to one of the two groups (i.e., regression = 0 and persistence = 1), making it a supervised method. Partial least-squares discriminant analysis (PLS-DA) is a suitable method when the within group variance is comparable or dominant compared to the between group variance [23]. The maximum group discrimination from a PLS-DA model can be represented by a target projection (TP) component that is obtained by combining all PLS components into this single TP component using a latent variable projected onto the response variable [24]. A score value from the target projection model is calculated for each object (sample) with regard to the group variable. A selectivity ratio (SR) plot resembles a spectrum and is a plot of the ratio of explained variance to unexplained variance for each variable [25], where one variable in this case is an identified protein. Variables with high selectivity ratios have high discriminating ability between the two groups. The discriminating variable (DIVA) test is a nonparametric test suitable for small sample sets with group heterogeneity [24]. A correct classification rate (CCR) value is calculated for each variable and will vary between 50% for a variable that provides random classification of the samples and 100% for a variable that gives a complete separation of the two groups. The SR and CCR are closely related in that higher SR should give higher CCR. The DIVA test provides a means of setting boundaries for the selectivity ratio to identify the important discriminating variables (proteins in this case) for a given CCR. More in depth theoretical explanations of all these methods can be found in [23–25]. The model was cross-validated by leaving out a large percentage of the individual samples from both sets in two cross-validation steps. In an outer loop, 20% of the samples were kept out at a time for an external validation. This was repeated 5 times so that all samples were kept out once. In the inner loop, 25% of the samples were kept out at a time, and this was repeated four times to keep out all samples once.
The normalized spectral count data were also imported into SPSS (version 18, SPSS, Oslo, Norway) for a binary logistic regression analysis and CART (Salford, San Diego, CA, USA) for a classification and regression tree analysis, both used as supervised in the sense that a group variable (regression or persistence) was added. The continuous variables were divided into two different subgroups, using a threshold value assessed by receiver-operating curve (ROC) analysis (MedCalc Software, Mariakerke, Belgium).
3. Results
The median age of the patients at inclusion was 29.7 years (range: 25–40), the interval between punch biopsy and cone excision was median, 113 days, and the mean protein concentration of the selected RPMI samples, measured by Bradford, was 0.81 mg/mL (range: 0.55–1.14). The age, punch-cone excision interval, and protein concentration of the RPMI samples of each of the three groups of patients studied (i.e., LC-MC/MS learning set, validation set, and the SELDI-TOF set) were consistent with the overall cohort from which our samples were selected and therefore can be regarded as representative.
3.1. SELDI-TOF MS Results
A total of 40 peaks were detected in the SELDI-TOF spectra using the criteria described in Section 2.5. The development of a binary logistic regression model resulted in one protein peak (m/z 6034) having the best discriminatory power between the regression and persistence samples of the 40 peaks in this dataset. Figure 1 is a scatter plot showing this peak plotted against one of the peaks found as discriminatory between normal and CIN2-3 tissue in the previous study [21]. The figure shows that this SELDI-TOF peak in fact could not discriminate between CIN2-3 lesions with regression and persistence.
Figure 1.
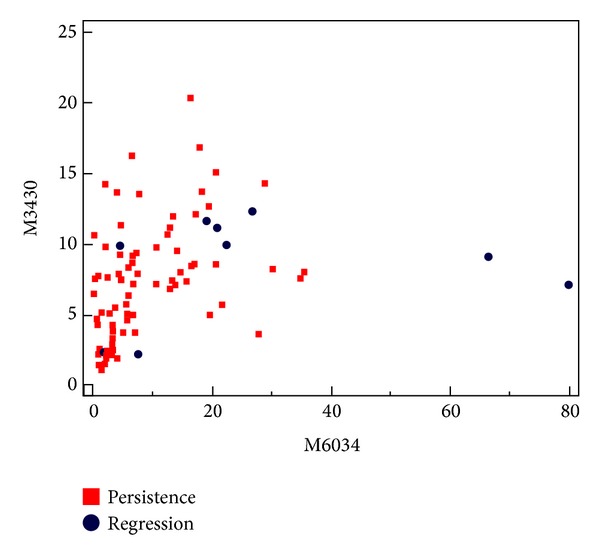
Scatter plot of the SELDI-TOF MS results for the peaks with m/z 6034 and m/z 3430 (samples: n = 5 regression and n = 40 persistence, 2 replicates of each). The figure illustrates that no discrimination is obtained between the regression and persistence groups using these results.
3.2. LC-MS/MS Results
The samples were subjected to depletion of 7 high abundance proteins followed by tryptic digestion and unidimensional LC-MS/MS analysis. Using the high significance peptide confidence filter in Proteome Discoverer and the identification criteria for proteins with only one peptide, a total of 165 protein identifications were included (all listed in Table 1 and more detailed in Supplementary Tables 1 and 2, see Supplementary Material available online at http://dx.doi.org/10.1155/2014/129064): 57 of these were identified with two or more unique peptides and the others with only one unique peptide. Although peptides from human papillomavirus proteins were detected in all samples, none of them gave acceptable protein identification.
Table 1.
List of 165 proteins identified with high significance filter in Proteome Discoverer.
| Accession number | Number of AAs | MW [Da] | Description | ΣCoverage |
|---|---|---|---|---|
| 25777600 | 953 | 105769 | 26S proteasome non-ATPase regulatory subunit 1 | 1,1 |
| 92090990 | 757 | 86306 | Activating signal cointegrator 1 complex subunit 2 | 1,7 |
| 119631843 | 438 | 48626 | Activin A receptor, type I | 7,3 |
| 90819233 | 1651 | 187556 | Afadin isoform 2 | 1,5 |
| 112877 | 201 | 23497 | Alpha-1-acid glycoprotein 1 | 15,9 |
| 119598593 | 366 | 39172 | Alpha-2-HS-glycoprotein | 5,7 |
| 224809474 | 951 | 106839 | Ankycorbin isoform B | 3,5 |
| 119573007 | 92 | 10573 | Apolipoprotein A-II | 52,2 |
| 119621207 | 3000 | 338317 | Apolipoprotein B (including Ag(x) antigen) | 0,8 |
| 73622085 | 665 | 72088 | ATPase WRNIP1; AltName: Werner helicase-interacting protein 1 | 2,4 |
| 85700402 | 1704 | 191239 | ATP-binding cassette subfamily A member 3 | 2,1 |
| 14916956 | 633 | 69181 | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial | 3,2 |
| 260763963 | 652 | 73680 | B-cell scaffold protein with ankyrin repeats isoform 3 | 2,8 |
| 410564 | 27 | 2891 | Beta-trace {N-terminal} | 74,1 |
| 119630319 | 616 | 69142 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | 3,4 |
| 110618250 | 651 | 69567 | Cadherin-related family member 5 isoform 3 | 2 |
| 93204551 | 754 | 85485 | Calpain-7-like protein | 2,3 |
| 119621019 | 2225 | 242829 | Carbamoyl phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | 0,6 |
| 215274265 | 626 | 70812 | Carnitine O-acetyltransferase | 2,1 |
| 119588550 | 361 | 39391 | CD44 antigen (Indian blood group) | 3,3 |
| 151101301 | 550 | 60746 | Centrosomal protein POC5 isoform 2 | 6,7 |
| 171184451 | 3117 | 350716 | Centrosome-associated protein 350 | 0,9 |
| 283135365 | 21 | 2382 | Chain A, human insulin | 100 |
| 289526593 | 120 | 13375 | Chain B, crystal structure of monomeric human cystatin C | 9,2 |
| 296278495 | 120 | 13562 | Chain B, crystal structure of the second bromodomain of human polybromo (protein polybromo-1) | 14,2 |
| 295789308 | 153 | 15779 | Chain B, human sod1 D124v variant | 64,7 |
| 293651901 | 243 | 28061 | Chain B, solution structure of double super helix model (apolipoprotein A-I) | 33,3 |
| 289526844 | 220 | 24615 | Chain B, structural basis of membrane-targeting by dock180 (dedicator of cytokinesis protein 1) | 8,6 |
| 291463533 | 127 | 13753 | Chain B, wild type human transthyretin (Ttr) | 38,6 |
| 289526762 | 30 | 3428 | Chain D, enhancing the therapeutic properties of a protein by a desig binding site | 100 |
| 294979722 | 141 | 0 | Chain E, deoxy human normal adult hemoglobin | 56,7 |
| 294979723 | 146 | 0 | Chain G, deoxy human normal adult hemoglobin | 89 |
| 295321918 | 374 | 41579 | Chain J, model of alpha-actinin Ch1 bound to F-actin | 9,1 |
| 119630802 | 740 | 78794 | Chromosome 20 open reading frame 75 | 2,7 |
| 119568512 | 153 | 17486 | Chromosome 6 open reading frame 75 | 10,5 |
| 10518503 | 444 | 49288 | Coagulation factor VII isoform B precursor | 3,4 |
| 119581366 | 1261 | 135504 | Cordon-bleu homolog (mouse) | 2,3 |
| 83582815 | 89 | 9881 | Cornifin-B | 19,1 |
| 119602496 | 311 | 35475 | Cysteine/histidine-rich 1 | 4,2 |
| 10719963 | 520 | 59956 | Cytochrome P450 4F8 | 3,1 |
| 119581796 | 712 | 78265 | Cytoplasmic polyadenylation element binding protein 4 | 2,4 |
| 119610301 | 2073 | 237519 | Dedicator of cytokinesis 11 | 0,9 |
| 20141302 | 110 | 11277 | Dermcidin | 25,5 |
| 55749932 | 470 | 53503 | Desmin | 10,9 |
| 63054852 | 508 | 58371 | DNA nucleotidylexotransferase isoform 2 | 5,3 |
| 119614620 | 977 | 109666 | Endoplasmic reticulum to nucleus signalling 1 | 1,6 |
| 24418674 | 974 | 110429 | Exocyst complex component 4 | 1,5 |
| 17369686 | 1087 | 123828 | Exportin-7 | 1,5 |
| 22749363 | 434 | 49418 | F-box only protein 15 isoform 1 | 3,5 |
| 119570461 | 1769 | 200655 | Fer-1-like 3, myoferlin (C. elegans) | 0,9 |
| 119568019 | 1322 | 142839 | Fibronectin type III domain containing 1 | 1,5 |
| 13129018 | 188 | 20994 | Gamma-glutamylcyclotransferase | 10,1 |
| 119596338 | 147 | 16448 | Ganglioside-induced differentiation-associated protein 1-like 1 | 10,2 |
| 46409304 | 443 | 48955 | Glutamate-rich protein 1 | 3,8 |
| 119625129 | 215 | 24427 | Glycoprotein M6A | 7,4 |
| 119621332 | 947 | 107158 | GREB1 protein | 1,5 |
| 152031617 | 669 | 72408 | GTP-binding protein 1 | 2,1 |
| 119625298 | 455 | 51153 | Guanylate cyclase 1, soluble, alpha 3 | 2,9 |
| 119583777 | 947 | 106793 | hCG1641824 | 2 |
| 119584015 | 69 | 7604 | hCG1813122 | 17,4 |
| 119597394 | 4919 | 535890 | hCG19253 | 0,4 |
| 119582778 | 87 | 10173 | hCG1981126 | 12,6 |
| 119610862 | 1925 | 215078 | hCG1986053 | 1,6 |
| 119568453 | 491 | 54322 | hCG2030297 | 5,1 |
| 119572490 | 576 | 62385 | hCG2040584 | 4,3 |
| 119598528 | 57 | 6808 | hCG2045397 | 29,8 |
| 4504517 | 205 | 22768 | Heat shock protein beta-1 | 21 |
| 119589211 | 147 | 16045 | Hemoglobin, delta | 62,6 |
| 119589125 | 462 | 51643 | Hemopexin | 7,6 |
| 121925 | 221 | 22336 | Histone H1.3 | 5,9 |
| 5901922 | 378 | 44440 | Hsp90 cochaperone Cdc37 | 3,7 |
| 164684901 | 94 | 11002 | Immunoglobulin heavy chain variable region | 18,1 |
| 17366467 | 2758 | 313745 | Inositol 1,4,5-trisphosphate receptor type 1 | 0,6 |
| 119623087 | 1257 | 133685 | Insulin receptor substrate 4 | 1 |
| 226694184 | 1179 | 130077 | Integrin alpha-E | 1,7 |
| 261878618 | 623 | 69452 | Inter-alpha-trypsin inhibitor heavy chain H1 isoform C | 2,3 |
| 27477074 | 502 | 55848 | Interleukin-17 receptor B precursor | 2,6 |
| 46397807 | 449 | 49715 | Keratin, type I cuticular Ha7 | 3,3 |
| 195972866 | 584 | 58766 | Keratin, type I cytoskeletal 10 | 22,6 |
| 239938886 | 623 | 62027 | Keratin, type I cytoskeletal 9 | 28,3 |
| 238054406 | 644 | 65999 | Keratin, type II cytoskeletal 1 | 30,1 |
| 239938650 | 639 | 65393 | Keratin, type II cytoskeletal 2 epidermal | 7,5 |
| 143811411 | 590 | 62340 | Keratin, type II cytoskeletal 5 | 11,9 |
| 5031839 | 564 | 60008 | Keratin, type II cytoskeletal 6A | 14,2 |
| 238054404 | 564 | 60030 | Keratin, type II cytoskeletal 6B | 9 |
| 59803089 | 564 | 59988 | Keratin, type II cytoskeletal 6C | 16,3 |
| 90110027 | 483 | 53671 | Keratin, type II cytoskeletal 8 | 7,9 |
| 119619543 | 916 | 102928 | KIAA1024 protein | 2,1 |
| 119600758 | 1047 | 116840 | KIAA1128 | 4,3 |
| 119583893 | 1420 | 158295 | Kinesin family member 13B | 2,9 |
| 231569458 | 629 | 70939 | Lactoperoxidase isoform 3 preproprotein | 2,5 |
| 27436948 | 634 | 70618 | Lamin-A/C isoform 3 | 8,2 |
| 119599090 | 222 | 25734 | Latexin | 5,4 |
| 119628276 | 394 | 43749 | Mannosidase, alpha, class 1C, member 1 | 6,4 |
| 290457624 | 397 | 41734 | Mesoderm posterior protein 2 | 4,8 |
| 115502451 | 496 | 53464 | Mothers against decapentaplegic homolog 6 | 4 |
| 119573924 | 271 | 28644 | Myeloid cell leukemia sequence 1 (BCL2-related) | 7,4 |
| 119610415 | 1737 | 199228 | Myosin, heavy polypeptide 8, skeletal muscle, perinatal | 1,2 |
| 33667040 | 1048 | 119352 | NACHT, LRR, and PYD domains-containing protein 8 | 2,7 |
| 33624861 | 429 | 48129 | Nesprin-2 isoform 2 | 4 |
| 93204871 | 628 | 70024 | Netrin-4 precursor | 2,4 |
| 160332335 | 5890 | 628699 | Neuroblast differentiation-associated protein AHNAK | 4,5 |
| 119625194 | 527 | 60213 | NIMA- (never in mitosis gene A-) related kinase 1 | 2,9 |
| 156632525 | 714 | 80686 | Nuclear protein MDM1 | 2,4 |
| 119570426 | 800 | 92490 | Nucleolar complex associated 3 homolog (S. cerevisiae) | 1,6 |
| 74749412 | 510 | 57244 | Olfactomedin-4 | 2,8 |
| 24430183 | 638 | 73293 | Outer dense fiber protein 2 isoform 2 | 2,7 |
| 21735584 | 820 | 93729 | Oxysterol-binding protein-related protein 3 isoform D | 1,7 |
| 296439282 | 595 | 68541 | P2X purinoceptor 7 | 2,2 |
| 237757297 | 643 | 70596 | Pannexin-2 isoform 2 | 2 |
| 119626787 | 301 | 34291 | Phosphatidylinositol glycan, class K | 6,3 |
| 74730959 | 315 | 35934 | PIH1 domain-containing protein 2 | 4,1 |
| 74730663 | 189 | 20681 | Plasma cell-induced resident endoplasmic reticulum protein | 18,5 |
| 296439496 | 1271 | 139580 | Pleckstrin homology domain-containing family G member 4B | 1,1 |
| 119602579 | 2105 | 233975 | Plectin 1, intermediate filament binding protein 500 kDa | 1 |
| 150421625 | 764 | 83232 | Polymeric immunoglobulin receptor | 2,8 |
| 123402 | 479 | 51177 | POU domain, class 2, transcription factor 2 | 2,9 |
| 22261792 | 1499 | 167582 | Probable phospholipid-transporting ATPase VA | 3,7 |
| 32171249 | 190 | 21015 | Prostaglandin-H2 D-isomerase | 17,4 |
| 119578886 | 247 | 26680 | Protease, serine, 3 (mesotrypsin) | 5,3 |
| 122801 | 352 | 38974 | Protein AMBP; AltName: Full = Alpha-1 microglycoprotein | 6,5 |
| 162416266 | 758 | 87316 | Protein dpy-19 homolog 2 | 1,7 |
| 257743264 | 606 | 69420 | Protein THEMIS isoform 3 | 3,1 |
| 296452931 | 1208 | 138572 | Protein timeless homolog | 1,3 |
| 74733527 | 701 | 80649 | Pseudouridylate synthase 7 homolog-like protein | 2 |
| 167016536 | 107 | 12037 | Putative nucleosome assembly protein 1-like 6 | 10,3 |
| 74760358 | 247 | 26522 | Putative trypsin-6 | 12,2 |
| 74712786 | 109 | 11855 | Putative uncharacterized protein FP588 | 18,4 |
| 119631914 | 1896 | 211398 | RAP1 interacting factor homolog (yeast) | 0,8 |
| 125987856 | 611 | 69368 | Rho-related BTB domain-containing protein 3 | 2,5 |
| 119620511 | 39 | 4319 | Ribosomal protein S27a | 41 |
| 126215690 | 3280 | 373742 | RING finger protein 213 | 0,7 |
| 296452978 | 1393 | 152659 | RNA polymerase II-associated protein 1 | 0,9 |
| 119573716 | 93 | 10828 | S100 calcium binding protein A8 (calgranulin A) | 11,8 |
| 119573719 | 114 | 13234 | S100 calcium binding protein A9 (calgranulin B) | 28,1 |
| 4506027 | 307 | 35057 | Serine/threonine-protein phosphatase 4 catalytic subunit | 4,9 |
| 113576 | 609 | 69321 | Serum albumin precursor | 34,3 |
| 119569088 | 107 | 12318 | SH3 domain binding glutamic acid-rich protein like 2 | 14 |
| 119576122 | 262 | 29185 | Similar to CG12314 gene product | 7,3 |
| 119620924 | 753 | 82996 | Similar to RIKEN cDNA 4632412N22 gene | 2,5 |
| 296452999 | 2785 | 318182 | Small subunit processome component 20 homolog | 0,9 |
| 119589494 | 452 | 50195 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 | 4,4 |
| 119596056 | 314 | 33833 | Solute carrier family 9 (sodium/hydrogen exchanger), member 8 | 4,1 |
| 119599799 | 450 | 51876 | Sorting nexin 4 | 4,2 |
| 119628509 | 259 | 29439 | Steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 9,7 |
| 119599133 | 506 | 58721 | TCDD-inducible poly(ADP-ribose) polymerase | 2,2 |
| 109895218 | 526 | 57476 | Thymocyte selection-associated high mobility group box protein TOX | 6,1 |
| 291045225 | 33423 | 3711285 | Titin isoform N2-A | 0,3 |
| 119605952 | 641 | 71538 | TNF receptor-associated factor 7 | 2,7 |
| 585328 | 80 | 8635 | Trefoil factor 3 | 22,5 |
| 119573617 | 242 | 28050 | Tropomyosin 3 | 14,5 |
| 14389309 | 449 | 49863 | Tubulin alpha-1C chain | 7,4 |
| 47157315 | 1124 | 125019 | Tyrosine-protein kinase JAK3 | 1,3 |
| 55977767 | 466 | 53619 | Vimentin | 19,7 |
| 32483410 | 474 | 52883 | Vitamin D-binding protein precursor | 17,5 |
| 119568661 | 331 | 36884 | WNT1 inducible signaling pathway protein 3 | 4,5 |
| 20532312 | 653 | 74145 | Zinc finger protein 274 | 11,6 |
| 149588643 | 679 | 77891 | Zinc finger protein 283 | 1,9 |
| 140560957 | 590 | 68217 | Zinc finger protein 285A | 2,4 |
| 30580627 | 626 | 72145 | Zinc finger protein 441 | 2,2 |
| 74759403 | 364 | 41163 | Zinc finger protein 589 | 4,7 |
| 119603081 | 647 | 74123 | Zinc finger protein 595 | 2,5 |
| 119598822 | 485 | 56018 | Zinc finger protein 639 | 2,5 |
| 187671927 | 394 | 46069 | Zinc finger protein 763 | 3,8 |
| 74758703 | 808 | 93088 | Zinc finger protein 841 | 2 |
Figure 2 shows a plot of the target projection score results from the complete dataset. The discrimination between the regression and persistence group is 95% since all persistence samples have score values with a positive sign, and 19 out of the 20 regression samples have a negative score value.
Figure 2.
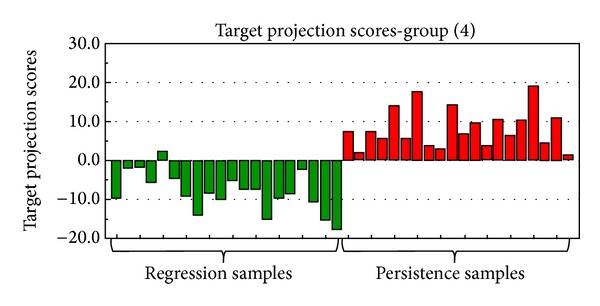
Score plot of discrimination between the regression group and persistence group after the target projection analysis.
Figure 3 shows a selectivity ratio plot for all the identified proteins resulting from doing a DIVA test with 90% correct classification rate set as an objective goal. This resulted in a selectivity ratio of ±1.26 as the limit for a variable to be significant in discriminating the groups. These limits are shown as solid horizontal lines in the figure.
Figure 3.
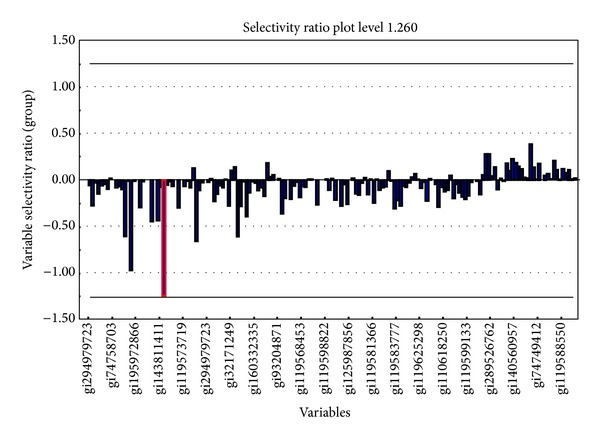
Selectivity ratio plot for all identified proteins. The selectivity ratio limit for a 90% mean corrected classification rate is shown as horizontal lines. The zinc finger protein 441 is marked with red square box.
Only one protein, the zinc finger protein 441 (ZNF441) (gi308153532) (red box), had a selectivity ratio of 1.26.
The CART analysis of the learning set resulted in a two-node model in which the ZNF441 was used as the primary group discriminator and, “similar to CG12314 gene product”, as the second most contributing discriminator. This protein was identified with one peptide (RVLITGSLNWTTQAIQNNR, precursor m/z: 2265.1714 Da, charge: +2). A Blast search against the UniProt human database with this sequence gave only one hit, phospholipase D6 (PLD6) (UniProt identifier: Q8N2A8). A search using the ID-mapping tool at the UniProt website gave no results. However, a UniProt Blast search for the complete sequence from the NCBI entry resulted in a unique hit with 100% identity score, PLD6.
Figure 4 shows a scatter plot using the spectral count results for these two proteins of the two sets and illustrates the discrimination obtained.
Figure 4.
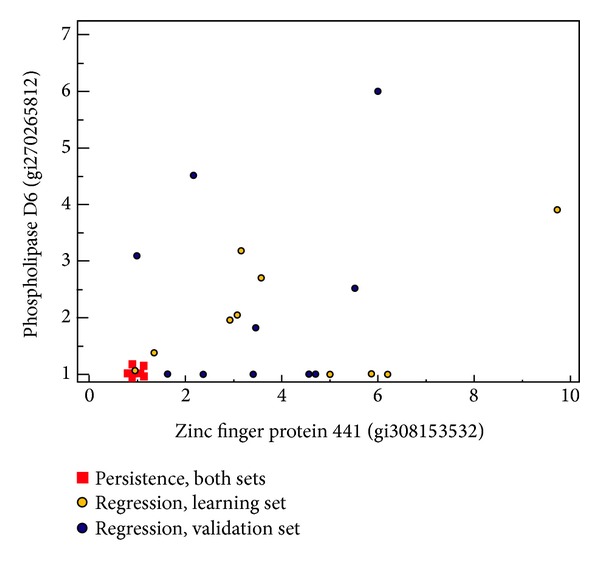
Scatter plot of the spectral count data for the zinc finger protein 441 and phospholipase D6 protein showing the discrimination between regression and persistence samples. The regression samples are shown further subdivided in learning (yellow circles) and validation (blue circles) sets.
The binary logistic regression model also resulted in ZNF441 having highest discriminatory power (results not shown).
ZNF441, identified using one highly significant peptide (QCGKALSHLKSFQR), was found in 10 and 9 of the 10 regression samples in the learning and validation set, respectively, and in none of the persistence samples. The PLD6 protein was also identified using only one high significance peptide and occurred in 7 and 5 of the 10 regression samples in the two sets.
Figure 5 shows the peptide sequence, the MS2 spectrum, and the y- and b-series for the ZNF441 peptide.
Figure 5.
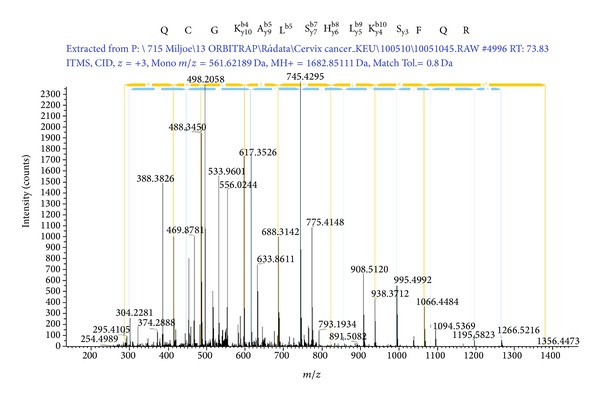
MS2 spectrum and amino acid sequence for the identified peptide from zinc finger protein 441. The y- and b-series are also shown.
A Blast search using the peptide sequence against the human UniProt database gave the two ZNF441 isomers as the only proteins with a 100% sequence similarity and the best scores from the search. ZNF441 (gi308153532, UniProt: Q8N8Z8) is a nuclear protein with 693 amino acids which belongs to the Krueppel C2H2-type zinc finger protein family, and it contains 19 C2H2-type zinc fingers and 1 KRAB-domain (UniProt entry).
ROC curve analysis showed that the optimal threshold for both ZNF441 and PLD6 was ≤1 versus >1. Using these thresholds, all regression and persistence cases of the learning set were correctly classified. In the validation set, 9 of the 10 regression and all 10 persistence cases were correctly classified. Figure 4 illustrates the power of the two proteins to distinguish between regressive and persistent CIN2-3 lesions for all cases in both the learning and the test set.
4. Discussion
This study describes the results from analysis of three different datasets regarding regression or persistence of CIN2-3 lesions: one dataset from SELDI-TOF MS and two datasets from LC-MS/MS analysis.
For the SELDI-TOF MS study, supernatants from a total of 45 patient samples (5 CIN2-3 with regression and 40 CIN2-3 with persistence) were analysed. One discriminatory peak was found by developing a binary logistic regression model using the SELDI-TOF MS dataset, but no discrimination between CIN2-3 lesions with regression or persistence could be obtained. Other binding conditions for the CM10 chip could have been used, as well as other chip types, but this was not pursued further as obtaining protein identification from a SELDI-TOF MS peak proved challenging.
LC-MS/MS analysis was much more promising than SELDI-TOF. All three multivariate statistical methods applied on the normalized spectral count results gave the same result, indicating that ZNF441 can discriminate between regressive and persistent CIN2-3 lesions. To our knowledge the exact function of ZNF441 has not yet been revealed, but the large family of transcriptional regulators of KRAB-containing zinc finger proteins are known to act as tumour suppressors [26]. In general, zinc finger proteins are a highly abundant group of proteins that varies in both structure and function [27]. They are involved in several cellular activities, including development, differentiation, and tumour suppression [28]. A zinc finger is a peptide domain whose secondary structure is stabilized by a bound zinc ion and a zinc finger protein can contain between 1 and 40 such domains [27]. The C2H2-domain is considered the “classical” zinc finger and is among the most abundent ones of the zink finger domains [28]. Zinc fingers were originally considered only as DNA-binding domains, but their role in protein-protein interactions has eventually been recognized [29]. Proteins with multiple zinc fingers can have two to three different types of binding activity through different fingers [28].
The Krueppel-associated box (KRAB-domain) is located near the N-terminal end of the protein, spans across 50–75 amino acids, and is divided into two boxes (A and B). KRAB-containing proteins are transcriptional repressors and use the zinc fingers to bind DNA [29]. KRAB-containing proteins are critical to cell differentiation, proliferation, apoptosis, and neoplastic transformation. Increased expression of the ZNF23 has been found to induce apoptosis in ovarian cancer cell lines [30]. ZNF431 functions as a transcriptional repressor for Patched1 (PTCH1) through binding to the target promoter sequence [31]. PTCH1 is a member of the Hedgehog (HH) family and acts as a negative regulator of the HH pathway. This pathway is important during embryonic development but has also been shown to be active during cancer development in adults. [32]. Repression of PTCH1 in a gastric cell line [33] was shown to correlate with high level of methylation of CpG islands at regulatory sequences and this could be associated with the development of gastric cancer. Another zinc finger protein, ZNF411, was found to suppress the MAP kinase signalling pathway [34], which is important for cell cycle checkpoints [35]. Overexpression of this pathway has been reported in different squamous cell carcinomas [36, 37]. The relationship between CIN grade and the MAP kinase pathway has also been investigated and was found to be an early marker for cervical carcinogenesis but not related to virus clearance [38]. Furthermore, the oncogenic E6 and E7, expressed in high risk HPV and known to play an important role in CIN tumour progression, also contain zinc finger domains, as recently reviewed by Ruttkay-Nedecky et al. [39]. In fact, new cell-permeable artificial zinc finger proteins (AZPs) have been launched as potential antiviral drug candidates that are able to reduce HPV replication [40, 41].
Phospholipase D6 (PLD6) was only reported by the CART analysis as contributing to the discrimination. In general, phospholipase D (PLD) proteins have been implicated in membrane trafficking [42, 43], cytoskeletal reorganization [44], endocytosis, exocytosis, cell migration, and cell proliferation [45]. The mouse homologue Zucchini (mZuc), also known as PLD6, has been shown to possess single strand-specific nuclease activity. This endoribonuclease has been shown to be essential for primary piRNA biogenesis [46, 47]. piRNAs are a distinct class of small RNAs, called Piwi-interacting RNAs, and have been discovered in both mammalian [48] and Drosophila germline [49]. They cluster at transposon loci in male germline stem cells and it has been suggested that piRNAs and their associated Piwi proteins are involved in epigenetic mechanisms like methylation and chromatin modifications [49]. A piRNA population has also been identified in the He-La cervical cancer cell line [50]. In germline stem cells these components are critical for silencing mobile genetic elements via DNA methylation. [51]. Furthermore, piRNAs have been detected in human cancer and somatic cells, and epigenetic disruption of the PIWI/piRNA pathway is indeed a hallmark for cancer development in testis [52]. Diminished piRNA expression has been found in testicular tumours as compared to normal testis.
In the current study, PLD6 was found to be expressed in most regression cases (12/20) but not in the persistent cases. The exact mechanisms for the epigenetic silencing exerted by the piRNA-PIWI pathway components remain unsolved and identification of additional protein components is crucial for a better understanding of the role of piRNAs in cancer [53].
This study and a previous study [17] show that CIN biopsies shed a complex mixture of proteins into a cell culture medium when placed at 4°C for 24 hours. For the LC-MS/MS study, supernatants from two series of 20 patient samples each (10 CIN2-3 with regression and 10 with persistence in each series) were analysed using a bottom-up shotgun proteomics approach [54] in which the proteins were digested into smaller peptides using trypsin. The peptide mixture was then analysed using unidimensional LC-MS/MS. Samples were pretreated by an immunoaffinity adsorbent which was previously validated by SDS-PAGE and LC-MS/MS (Supplementary Figure 1). Despite the depletion of seven high abundance proteins including immunoglobulins and albumin, peptides from these proteins were detected, while transferrin was not found at all after depletion. In addition, not unexpectedly, hemoglobins constitute a relatively large part of the identified proteins (cervical tissues with CIN2-3 are usually richly vascularised) and should be included in future depletion work. The complexity of the depleted fraction is still a challenge. Further fractionation of the depleted samples prior to the LC-MS/MS analysis would be an advantage to increase the supernatant proteome coverage and also possibly the sequence coverage of the identified proteins. This fractionation could be obtained using 2D-gel separation of the protein mixture or, for example, a cation exchange fractionation of the peptides after digestion. Another option is enrichment of subproteomes like phosphorylated proteins or glycosylated proteins. The results in this study should of course be validated by analysing a larger number of samples and also by analyses using other methodology like immunohistochemistry.
5. Conclusions
Using three different statistical methods to analyse normalized spectral count data, this study has identified zinc finger protein 441 as a highly discriminating factor between CIN2-3 regressive and persistent lesions. Phospholipase D6 contributes to the discrimination.
Interestingly the two proposed proteins are important factors for repression of tumour growth. Zinc finger proteins constitute the largest family of transcriptional regulators in mammals with important DNA binding domains and are also involved in protein-protein interactions. Their ability to induce apoptosis has been shown, as well as their function as nuclear transcriptional repressors of genes involved in signal transduction important for development of carcinogenesis. PLD6 is involved in biogenesis of piRNAs, small noncoding RNAs involved in hypermethylation events and important for transcriptional, epigenetic, and signalling pathways alterations. In line with these findings, both PLD6 and ZNF441 were almost absent in the persistent CIN2-3 cases contrary to the regression cases (Figure 4).
The depletion of seven high abundance proteins followed by a unidimensional separation of tryptic digests of nondepleted protein mixtures shows the potential of the described method for collection of proteins from CIN biopsies. From a prognostic aspect, the findings are promising tools for further investigation and understanding of the biology behind regression of precancerous cervical lesions.
Supplementary Material
Excel spreadsheets containing peptide identification details for all proteins from the complete dataset with regression, table 1, and without regression, table 2
Acknowledgments
Thanks are due to Pattern Recognition Systems (Bergen, Norway) and Professor Olav Kvalheim at the University of Bergen (Bergen, Norway) for making Sirius version 8.1 available for this work. Thanks are also due to Akvamiljø AS for providing access to both the SELDI-TOF MS and the LTQ-Orbitrap. Funding for this research was provided by the Folke Hermansen Foundation in 2007 and Helse Vest in 2011.
Conflict of Interests
The authors declare that they have no conflict of interests regarding the publication of this paper.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians . 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses in the causation of human cancers—a brief historical account. Virology. 2009;384(2):260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Insinga RP, Dasbach EJ, Elbasha EH. Epidemiologic natural history and clinical management of human papillomavirus (HPV) Disease: a critical and systematic review of the literature in the development of an HPV dynamic transmission model. BMC Infectious Diseases. 2009;9, article 119 doi: 10.1186/1471-2334-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baak JP, Kruse AJ, Robboy SJ, Janssen EA, van Diermen B, Skaland I. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. Journal of Clinical Pathology. 2006;59(10):1017–1028. doi: 10.1136/jcp.2005.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munk AC, Kruse AJ, van Diermen B, et al. Cervical intraepithelial neoplasia grade 3 lesions can regress. APMIS. 2007;115(12):1409–1414. doi: 10.1111/j.1600-0463.2007.00769.x. [DOI] [PubMed] [Google Scholar]
- 6.McCredie MR, Paul C, Sharples KJ, et al. Consequences in women of participating in a study of the natural history of cervical intraepithelial neoplasia 3. Australian and New Zealand Journal of Obstetrics and Gynaecology. 2010;50(4):363–370. doi: 10.1111/j.1479-828X.2010.01170.x. [DOI] [PubMed] [Google Scholar]
- 7.Ostor AG. Natural history of cervical intraepithelial neoplasia—a critical review. International Journal of Gynecological Pathology. 1993;12(2):186–192. [PubMed] [Google Scholar]
- 8.Martin-Hirsch PP, Paraskevaidis E, Bryant A, Dickinson HO, Keep SL. Surgery for cervical intraepithelial neoplasia. Cochrane Database of Systematic Reviews. 2010;16(6) doi: 10.1002/14651858.CD001318.pub2.CD001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyrgiou M, Koliopoulos G, Martin-Hirsch P, Arbyn M, Prendiville W, Paraskevaidis E. Obstetric outcomes after conservative treatment for intraepithelial or early invasive cervical lesions: systematic review and meta-analysis. The Lancet. 2006;367(9509):489–498. doi: 10.1016/S0140-6736(06)68181-6. [DOI] [PubMed] [Google Scholar]
- 10.Sadler L, Saftlas A, Wang W, Exeter M, Whittaker J, McCowan L. Treatment for cervical intraepithelial neoplasia and risk of preterm delivery. The Journal of the American Medical Association. 2004;291(17):2100–2106. doi: 10.1001/jama.291.17.2100. [DOI] [PubMed] [Google Scholar]
- 11.Kadish AS, Timmins P, Wang Y, et al. Regression of cervical intraepithelial neoplasia and loss of human papillomavirus (HPV) infection is associated with cell-mediated immune responses to an HPV type 16 E7 peptide. Cancer Epidemiology Biomarkers and Prevention. 2002;11(5):483–488. [PubMed] [Google Scholar]
- 12.Munk AC, Gudlaugsson E, Ovestad IT, et al. Interaction of epithelial biomarkers, local immune response and condom use in cervical intraepithelial neoplasia 2-3 regression. Gynecologic Oncology. 2012;127(3):489–494. doi: 10.1016/j.ygyno.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Ovestad IT, Gudlaugsson E, Skaland I, et al. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. Journal of Clinical Pathology. 2011;64(4):303–307. doi: 10.1136/jcp.2010.083626. [DOI] [PubMed] [Google Scholar]
- 14.Ovestad IT, Gudlaugsson E, Skaland I, et al. Local immune response in the microenvironment of CIN2-3 with and without spontaneous regression. Modern Pathology. 2010;23(9):1231–1240. doi: 10.1038/modpathol.2010.109. [DOI] [PubMed] [Google Scholar]
- 15.Kruse AJ, Skaland I, Janssen EA, et al. Quantitative molecular parameters to identify low-risk and high-risk early CIN lesions: role of markers of proliferative activity and differentiation and Rb availability. International Journal of Gynecological Pathology. 2004;23(2):100–109. doi: 10.1097/00004347-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Baak JP, Kruse AJ. Use of biomarkers in the evaluation of CIN grade and progression of early CIN. Methods in Molecular Medicine. 2005;119:85–99. doi: 10.1385/1-59259-982-6:085. [DOI] [PubMed] [Google Scholar]
- 17.Uleberg KE, Munk AC, Brede C, et al. Identification of water soluble proteins recovered from biopsies with high-grade intraepithelial neoplasia. Journal of Proteomics. 2011;9(36) doi: 10.1186/1477-5956-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uleberg KE, Munk AC, Brede C, et al. Discrimination of grade 2 and 3 cervical intraepithelial neoplasia by means of analysis of water soluble proteins recovered from cervical biopsies. Proteome Science. 2011;9, article 36 doi: 10.1186/1477-5956-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen EI, Yates JR., III Cancer proteomics by quantitative shotgun proteomics. Molecular Oncology. 2007;1(2):144–159. doi: 10.1016/j.molonc.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quality assurance manual. Norwegian Cancer Registry, National advisory board for the Norwegian cervical cancer screening program, 2012, http://www.kreftregisteret.no/no/Forebyggende/Masseundersokelsen-mot-livmorhalskreft.
- 21.Uleberg KE, Munk AC, Skaland I, et al. A protein profile study to discriminate CIN lesions from normal cervical epithelium. Cellular Oncology. 2011;34(5):443–450. doi: 10.1007/s13402-011-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zegels G, van Raemdonck GA, Coen EP, Tjalma WA, van Ostade XWM. Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Science. 2009;7, article 17 doi: 10.1186/1477-5956-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajalahti T, Kroksveen AC, Arneberg R, et al. A multivariate approach to reveal biomarker signatures for disease classification: application to mass spectral profiles of cerebrospinal fluid from patients with multiple sclerosis. Journal of Proteome Research. 2010;9(7):3608–3620. doi: 10.1021/pr100142m. [DOI] [PubMed] [Google Scholar]
- 24.Rajalahti T, Arneberg R, Kroksveen AC, Berle M, Myhr K, Kvalheim OM. Discriminating variable test and selectivity ratio plot: quantitative tools for interpretation and variable (biomarker) selection in complex spectral or chromatographic profiles. Analytical Chemistry. 2009;81(7):2581–2590. doi: 10.1021/ac802514y. [DOI] [PubMed] [Google Scholar]
- 25.Rajalahti T, Arneberg R, Berven FS, Myhr KM, Ulvik RJ, Kvalheim OM. Biomarker discovery in mass spectral profiles by means of selectivity ratio plot. Chemometrics and Intelligent Laboratory Systems. 2009;95(1):35–48. [Google Scholar]
- 26.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biology. 2003;4(10, article 231) doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Current Opinion in Structural Biology. 2001;11(1):39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 28.Iuchi S. Three classes of C2H2 zinc finger proteins. Cellular and Molecular Life Sciences. 2001;58(4):625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochemistry and Biophysics. 2008;50(3):111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, Yang S, Ge R, Sun H, Shen F, Wang Y. ZNF23 induces apoptosis in human ovarian cancer cells. Cancer Letters. 2008;266(2):135–143. doi: 10.1016/j.canlet.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 31.He Z, Cai J, Lim JW, Kroll K, Ma L. A novel KRAB domain-containing zinc finger transcription factor ZNF431 directly represses Patched1 transcription. The Journal of Biological Chemistry. 2011;286(9):7279–7289. doi: 10.1074/jbc.M110.178780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merchant JL, Saqui-Salces M, El-Zaatari M. Hedgehog signaling in gastric physiology and cancer. Progress in Molecular Biology and Translational Science. 2010;96:133–156. doi: 10.1016/B978-0-12-381280-3.00006-3. [DOI] [PubMed] [Google Scholar]
- 33.Zuo Y, Song Y. Detection and analysis of the methylation status of PTCH1 gene involved in the hedgehog signaling pathway in a human gastric cancer cell line. Experimental and Therapeutic Medicine. 2013;6(6):1365–1368. doi: 10.3892/etm.2013.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, Zhu C, Luo J, et al. ZNF411, a novel KRAB-containing zinc-finger protein, suppresses MAP kinase signaling pathway. Biochemical and Biophysical Research Communications. 2004;320(1):45–53. doi: 10.1016/j.bbrc.2004.05.130. [DOI] [PubMed] [Google Scholar]
- 35.Poehlmann A, Roessner A. Importance of DNA damage checkpoints in the pathogenesis of human cancers. Pathology Research and Practice. 2010;206(9):591–601. doi: 10.1016/j.prp.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Lessard JL, Robinson RA, Hoffman HT. Differential expression of ras signal transduction mediators in verrucous and squamous cell carcinomas of the upper aerodigestive tract. Archives of Pathology and Laboratory Medicine. 2001;125(9):1200–1203. doi: 10.5858/2001-125-1200-DEORST. [DOI] [PubMed] [Google Scholar]
- 37.Mishima K, Yamada E, Masui K, et al. Overexpression of the ERK/MAP kinases in oral squamous cell carcinoma. Modern Pathology. 1998;11(9):886–891. [PubMed] [Google Scholar]
- 38.Branca M, Ciotti M, Santini D, et al. Activation of the ERK/MAP kinase pathway in cervical intraepithelial neoplasia is related to grade of the lesion but not to high-risk human papillomavirus, virus clearance, or prognosis in cervical cancer. American Journal of Clinical Pathology. 2004;122(6):902–911. doi: 10.1309/VQXF-T880-JXC7-QD2W. [DOI] [PubMed] [Google Scholar]
- 39.Ruttkay-Nedecky B, Jimenez Jimenez AM, Nejdl L, et al. Relevance of infection with human papillomavirus: the role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins (Review) International Journal of Oncology. 2013;43(6):1754–1762. doi: 10.3892/ijo.2013.2105. [DOI] [PubMed] [Google Scholar]
- 40.Mino T, Mori T, Aoyama Y, Sera T. Gene- and protein-delivered zinc finger-staphylococcal nuclease hybrid for inhibition of DNA replication of human papillomavirus. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0056633.e56633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mino T, Mori T, Aoyama Y, Sera T. Inhibition of human papillomavirus replication by using artificial zinc-finger nucleases. Nucleic Acids Symposium Series. 2008;(52):185–186. doi: 10.1093/nass/nrn094. [DOI] [PubMed] [Google Scholar]
- 42.Donaldson JG. Phospholipase D in endocytosis and endosomal recycling pathways. Biochimica et Biophysica Acta—Molecular and Cell Biology of Lipids. 2009;1791(9):845–849. doi: 10.1016/j.bbalip.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth MG. Molecular mechanisms of PLD function in membrane traffic. Traffic. 2008;9(8):1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- 44.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochemistry and Cell Biology. 2004;82(1):225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 45.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Molecular Cancer Research. 2003;1(11):789–800. [PubMed] [Google Scholar]
- 46.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491(7423):279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimasu H, Ishizu H, Saito K, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491(7423):284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 48.Esposito T, Magliocca S, Formicola D, Gianfrancesco F. PiR_015520 belongs to Piwi-associated RNAs regulates expression of the human melatonin receptor 1A gene. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0022727.e22727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H, Yin H. A novel epigenetic mechanism in Drosophila somatic cells mediated by PIWI and piRNAs. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:273–281. doi: 10.1101/sqb.2008.73.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu Y, Li C, Zhang K, et al. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Reports. 2010;43(9):635–641. doi: 10.5483/BMBRep.2010.43.9.635. [DOI] [PubMed] [Google Scholar]
- 51.Kim VN. Small RNAs just got bigger: piwi-interacting RNAs (piRNAs) in mammalian testes. Genes and Development. 2006;20(15):1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 52.Ferreira HJ, Heyn H, Garcia del Muro X, et al. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics. 2013;9(1):113–118. doi: 10.4161/epi.27237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siddiqi S, Terry M, Matushansky I. Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PloS ONE. 2012;7(3) doi: 10.1371/journal.pone.0033711.e33711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson SK, Washburn MP. The continuing evolution of shotgun proteomics. Drug Discovery Today. 2005;10(10):719–725. doi: 10.1016/S1359-6446(05)03450-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel spreadsheets containing peptide identification details for all proteins from the complete dataset with regression, table 1, and without regression, table 2


