Abstract
Background
Survival after cardiac surgery in infancy requires adaptive responses from oxidative stress management and vascular regulation pathways. We tested the hypothesis that genetic variation in these pathways influences post-operative survival in non-syndromic congenital heart disease (CHD) children.
Methods
This is an analysis of a cohort of non-syndromic CHD patients who underwent cardiac surgery with cardiopulmonary bypass before 6 months of age (n=422). Six single nucleotide polymorphisms (SNPs) in 6 genes involved in oxidative stress and vascular response pathways, identified through a priori literature search, were tested for effects on transplant-free survival. Survival curves, adjusting for confounding covariates, were calculated using the Cox Proportional Hazard Models.
Results
Long-term survival was strongly associated with VEGFA SNP rs833069 (p=7.03×10−4) and SOD2 SNP rs2758331 (p=0.019). To test for joint effects of the 2 SNPs on transplant-free survival, the genotypes were grouped to form a risk score reflecting the cumulative number of risk alleles (0–4 alleles/patient). A higher risk score based on the VEGFA and SOD2 SNP genotypes was associated with worse transplant-free survival (p=3.02×10−4) after confounder adjustment. The total burden of risk alleles was additive; individuals with the highest risk score of 4 (n=59 subjects, 14.2% of the cohort) had a total covariate-adjusted HR=15.64 for worse transplant-free survival.
Conclusions
After cardiac surgery, infants who are homozygous for the high-risk alleles for both the VEGFA and SOD2 SNPs have an approximate 16-fold increased risk of death or heart transplant; suggesting that genetic variants are important modifiers of survival after surgery for CHD.
Keywords: Congenital heart disease, CHD; Ischemia/reperfusion injury (myocardial); Genetics, genomics; Genes/polymorphisms/microarrays; Myocardial remodeling (reshaping, constraining, ventriculectomy); Outcomes (including mortality, morbidity, survival, etc.); Statistics, survival analysis
Introduction
Congenital heart defects (CHDs) are the most common human birth defect. Approximately one-third of CHD cases require surgical intervention, with a majority involving cardiopulmonary bypass (CPB). Long-term mortality in the post-operative stages remains considerable, especially for the more severe heart defects(1).
Oxidative stress is considered to be a major factor following cardiac surgery with CPB due to post-operative organ dysfunction(2). The importance of oxidative stress in postoperative outcomes has been demonstrated through the finding that allopurinol, which blocks free radical formation and its resulting oxidative stress, is associated with decreased cardiac event rate after surgery with CPB in in high-risk infants with hypoplastic left heart syndrome (HLHS)(3).
Studies of postsurgical outcomes in pediatric patients have successfully identified several genetic variants involved in vascular response pathways that affect long-term outcomes. First, endothelin-1 missense variant (EDN1 G5665T) has been associated with transplant-free survival in patients with functional single ventricle CHD, with the greatest effects in children with the most severe phenotype, HLHS(4). More recently, a randomized clinical trial reported that missense mutations that up-regulated the Renin-Angiotensin-Aldosterone system (RAAS) were associated with impaired ventricular remodeling, renal function, and somatic growth in infants with functional single ventricle post-cardiac surgery, highlighting the role of vascular response genes on a wide spectrum of postsurgical outcomes(5).
Taken together, these studies suggest that oxidative stress and vascular response play important roles in injury repair and long-term survival in the pediatric CHD population. We sought to examine the effects of specific genetic variants implicated in oxidative stress management and organ recovery on long-term survival in a cohort of children with non-syndromic CHD. Secondarily, we performed an analysis using a genetic risk score, reflecting the number of deleterious alleles each patient has, to determine if the observed genotype effects were independent and additive.
Patients and Methods
Study Design
This is an analysis of a previously described prospective cohort(6–8) of 550 subjects collected to study neurodevelopmental dysfunction following CHD palliation. This specific study sought to identify gene regions related to oxidative stress and vascular response potentially affecting survival in infants after cardiac surgery with non-syndromic CHD. We note that no genome-wide association analyses have been attempted on the phenotype of long-term survival; this is solely a candidate gene study.
Of the 550 original subjects, 56 were removed due to likely genetic syndrome and an additional 72 were removed due to lack of high-quality genotype data, leaving a total of 422 subjects available for analyses. Additional information on data collection (including inclusion/exclusion criteria), operative management, genotyping, and analyses not presented in the main manuscript are found in the Online-Only Appendix Materials.
SNP Selection
To preserve statistical power, we selected 6 candidate SNPs at 6 different genes involved broadly in oxidative and ischemic stress response (see Table 1) a priori based on a systematic literature review of published evidence from other investigators, reporting that variants in these genes have a functional impact potentially relevant to the outcomes (see Appendix Table S1). Four of the 6 genes (EPHX1, SOD2, CYP2E1, and CAT) are involved in oxidative stress. VEGFA and EDN1 are involved in the vascular response to low output and ischemic states associated with CPB; additionally, VEGFA may help promote vascular adaptation to hemodynamic alterations, while the EDN1 missense SNP that has been associated with transplant-free survival in single-ventricle children(4). All studied SNPs were in Hardy-Weinberg equilibrium in controls.
Table 1.
Description of SNPs studied.
| SNP | Genea | Variant Type | Chr:Positionb | Major/Minor Allele | Minor Allele Frequency | Gene Name and Description |
|---|---|---|---|---|---|---|
| rs1051740 | EPHX1 | Tyr113His | 1:224,086,256 | A/G | 0.291 | Epoxide hydrolase 1. Converts epoxides to non-toxic forms |
| rs5370 | EDN1 | Lys198Asn | 6:12,404,241 | T/G | 0.187 | Endothelin 1. Pro-peptide of endothelin 1, a potent vasoconstrictor |
| rs833069 | VEGFA | Intronicc | 6:43,850,557 | A/G | 0.346 | Vascular endothelial growth factor A. Growth factor mediates vascular permeability and endothelium growth/apoptosis inhibition |
| rs2758331 | SOD2 | Intronicd | 6:160,025,060 | T/G | 0.436 | Superoxide dismutase 2. Mitochondrial protein; converts superoxide byproducts to hydrogen peroxide |
| rs10776686 | (CYP2E1) | Intergenic | 10:135,182,921 | A/G | 0.051 | Cytochrome P450 2E1. Endoplasmic reticulum-associated enzyme, involved in varied processes |
| rs1001179 | CAT | 5′UTR | 11:34,416,807 | T/C | 0.159 | Catalase. Key antioxidant heme enzyme present in peroxisome |
3′UTR = 3 prime un-translated region of a gene. 5′UTR = 5 prime un-translated region of a gene. Chr = chromosome. LD = linkage disequilibrium. SNP = single nucleotide polymorphism.
Intergenic SNPs are represented in parentheses naming the nearest gene, e.g. (CYP2E1).
Position information and annotation from reference assembly 36.3.
Intronic SNP, rs833069, is in strong LD (r2 = 0.97) with VEGFA 5′UTR SNP rs2010963, which has been reported to increase expression levels of VEGFA.
Intronic SNP, rs2758331, is in strong LD (r2 = 0.93) with SOD2 Val16Ala missense SNP rs4880.
Analysis
Genotypes were coded using an additive model and all regression models included adjustment for confounders based on genetic ancestry and pertinent clinical covariates. Genetic ancestry was determined using previously described methods(9). Due to the mixed ancestry of the cohort (see Table 2 for demographic information, including genetic ancestry), the first 3 principal component eigenvectors from principal components analysis (PCA) were used as covariates to adjust for potential population stratification(10). We adjusted for multiple contrasts based on the 6 SNPs by applying a Bonferroni correction of α= 0.05/6 = 8.3×10−3.
Table 2.
Baseline and operative characteristics of the cohort
| Baseline Characteristics | Cohort Subset (n=422) |
|---|---|
| Gender, n (%) | |
| Female | 176 (41.7%) |
| Male | 246 (58.3%) |
| Ethnicity, n (%) | |
| Asian/Pacific Islander, Hispanic, or other ancestry | 51 (12.1%) |
| African ancestry, not Hispanic | 98 (23.2%) |
| European ancestry, not Hispanic | 273 (64.7%) |
| Gestational Age, weeks, mean ± SD | 38.5 ± 2.05 |
| Gestational Weight, kg, mean ± SD | 3.15 ± 0.62 |
| Diagnostic Class, n (%) | |
| I: 2 ventricles, no arch obstruction | 204 (48.3%) |
| II: 2 ventricles, arch obstruction | 41 (9.7%) |
| III: 1 ventricle, no arch obstruction | 46 (10.9%) |
| IV: 1 ventricle, arch obstruction | 131 (31.1%) |
| Specific CHD Diagnoses, n (%) | |
| Hypoplastic Left Heart Syndrome | 130 (30.8%) |
| Tetralogy of Fallot | 64 (15.2%) |
| Transposition of the Great Arteries | 34 (8.1%) |
| Ventricle Septal Defect | 40 (9.5%) |
| Ventricle Septal Defect, Coarctation of Aorta | 19 (4.5%) |
| Single Ventricle | 30 (7.1%) |
| Other Distinct CHD Diagnoses | 105 (24.9%) |
| Preoperative intubation, n (%) | 119 (28.2%) |
| Preoperative length of stay, days, mean ± SD | 2.14 ± 2.61 |
| Age at first operation, days, mean ± SD | 42.1 ± 54.7 |
| Weight at first operation, kg, mean ± SD | 3.82 ± 1.25 |
| Total cardiopulmonary bypass time in first operation, min, mean ± SD | 67.1 ± 40.3 |
| Use of DHCA, n (%) | 259 (61.4%) |
| Total DHCA time in first operation, min, mean ± SD | 41.4 ± 18.3 |
| Hematocrit level after hemodilution in first operation, %, mean ± SD | 27.8 ± 4.1 |
| Genetic risk score categorya | |
| 0 risk alleles | 11 (2.6%) |
| 1 risk allele | 64 (15.2%) |
| 2 risk alleles | 138 (32.7%) |
| 3 risk alleles | 150 (35.5%) |
| 4 risk alleles | 59 (14.0%) |
| Long-term mortality, n (%) | 47 (11.1%) |
| Time to mortality, years, mean ± SD | 9.32 ± 3.48 |
| Heart Transplants, n (%) | 4 (1.0%) |
CHD = congenital heart defect. DHCA = deep hypothermic circulatory arrest.
Number of VEGFA SNP rs833069 or SOD2 SNP rs2758331 major alleles, which are both associated with worse long-term survival.
Survival analyses and graphics were performed in R (http://www.r-project.org/). Time to long-term mortality was calculated from date of initial surgery to date of death; these data include all deaths, including operative deaths. A Cox Proportional Hazards (CPH) model was used to evaluate the joint effect of the studied SNP and potential covariates. Output from the CPH model was used for plotting of survival curves. An additional analysis included cardiac transplant as an endpoint in addition to any death using the aforementioned methods. Survival analyses were adjusted for the previously reported confounding variables: the first 3 principal component eigenvectors for race, gender, gestational age, gestational weight, diagnostic class(11), preoperative intubation, preoperative LOS, age at first operation, weight at first operation, total CPB time, use of deep hypothermic circulatory arrest (DHCA), total DHCA time, and hematocrit at first operation(12). A European ancestry (EA) only sensitivity analysis for all outcomes was performed to ensure the direction of SNP effects was consistent within the largest genetic ancestry subgroup. A separate, diagnostic-class stratified analysis was performed for transplant free survival to ensure consistency of SNP effects across the physiologic range of heart defects.
Results
Demographic and clinical variables are presented in Table 2. Of the 422 non-syndromic children studied, 16 died operatively and 31 additional children died during the follow-up period, with an average time to mortality of 9.32 years, while an additional 4 patients underwent cardiac transplant. Number of transplants or death by diagnostic class was 8 (3.9%) for class 1, 2 (4.9%) for class 2, 7 (15.2%) for class 3, and 34 (25.9%) for class 4. Median follow-up time for all subjects was 10.18 years.
Survival analyses demonstrated that SNPs at two different loci were associated with long-term survival: VEGFA intronic SNP rs833069 (HR=0.37, p=7.03×10−4) and SOD2 intronic SNP rs2758331 (HR=0.52, p=0.019; Table 3). For both SNPs, each copy of the minor allele was associated with an increase in long-term survival. The VEGFA effect was significant after Bonferroni correction, while the SOD2 effect was not. In the case of VEGFA SNP rs833069, each “G” allele led to a dose-dependent increase in survival probability (Figure 1A). A similar effect was noted for SOD2 SNP rs2758331 (Figure 1B). We note that when testing for transplant-free survival, both VEGFA and SOD2 remained significant (see Appendix Table S2).
Table 3.
SNP results (p ≤ 0.05) for long-term mortality.
| SNP | Gene | EA-only HR (95% CI)a | Full Cohort HR (95% CI)a | EA-only Pb | Full Cohort P |
|---|---|---|---|---|---|
| rs833069 | VEGFA | 0.25 (0.105–0.609) | 0.37 (0.211–0.659) | 0.0021 | 7.03×10−4 |
| rs2758331 | SOD2 | 0.29 (0.133–0.636) | 0.52 (0.304–0.900) | 0.0019 | 0.019 |
CI = Confidence Interval. EA = European Ancestry, non-Hispanic. HR = Hazard Ratio from survival analyses.
HR and 95% CI were calculated using Cox proportional hazards methods for the outcome of long-term survival, adjusting for the covariates listed in the methods.
Analyses performed on the majority genetic ancestry group (EA) of the cohort (n=273 subjects), using analysis methods outlined in a.
Figure 1. Vascular response related VEGFA SNP rs833069 (a) and oxidative stress related SOD2 SNP rs2758331 (b) genotypes predict long-term survival.
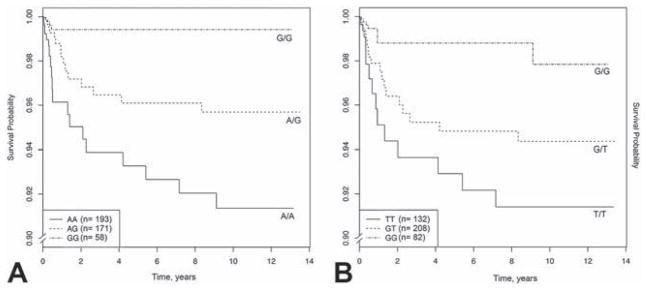
The survival curves show a dose dependent effect of each SNP. Note the range of the y-axis, “Survival Probability”. VEGFA = vascular endothelial growth factor A; SOD2 = superoxide dismutase 2.
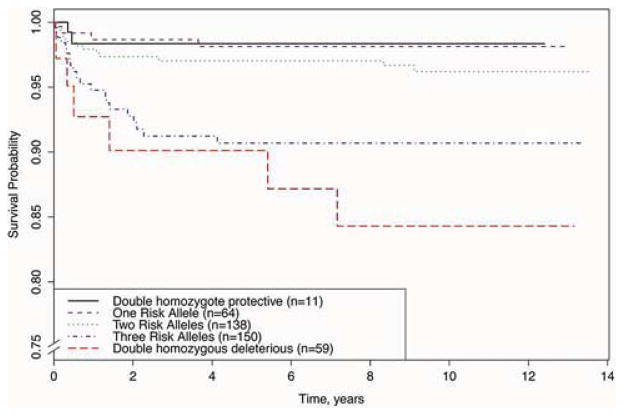
To test for joint effects of the 2 SNPs in VEGFA and SOD2 on long-term transplant-free survival, the genotypes were grouped to form a risk score reflecting the cumulative number of risk alleles (0–4 alleles/patient). For both VEGFA SNP rs833069 and SOD2 SNP rs2758331, the minor allele was low-risk, while the more common variant was the high-risk allele for survival (see Table 3 and Figure 1). A higher risk score based on the VEGFA and SOD2 SNP genotypes was associated with worse transplant-free survival (p=3.02×10−4), adjusting for confounders. The total burden of risk alleles was additive; individuals with the highest risk score of 4 (n=59 subjects, 14.2% of the cohort) had a total covariate-adjusted HR=15.64 for worse transplant-free survival (see Figure 2).
Figure 2. Genetic risk score reflecting number of VEGFA and SOD2 risk alleles carried by each patient is predictive of long-term survival.
Note the range of the y-axis, “Survival Probability”.
To rule out effects of residual population stratification, we performed a separate sensitivity analysis on the majority European Ancestry (EA) subset of the cohort (n=273 subjects) for the significant SNPs (Table 3). These EA only analyses show consistency of the effect size and directions with the complete cohort. Thus, our results are unlikely to be secondary to population stratification.
To examine the potential for genotype confounding, whereby our identified VEGFA and SOD2 SNPs are associated with less severe CHD, therefore leading to increased survival, we performed separate Cochran-Armitrage tests for both genotypes and diagnostic class. From these analyses, we found that the VEGFA SNP rs833069 minor allele was associated with higher-risk diagnostic class subgroups (p=9.12×10−8, see Appendix Table S3). Further analysis of specific diagnoses found that the VEGFA minor allele was in excess in HLHS (all diagnostic class 4), while the VEGFA major allele was positively associated with both Tetralogy of Fallot (TOF) and Transposition of the Great Arteries (TGA)(see Appendix Tables S4–S6). The vast majority of both the TOF (67/69) and TGA (49/52) subgroups were categorized into diagnostic class 1. SOD2 genotype was not associated with diagnostic class.
We conducted a sensitivity analysis, stratifying the primary analysis by diagnostic class, to evaluate potential confounding (see Appendix Table S7) by severity of CHD. From these generally underpowered analyses, we noted that VEGFA and SOD2 genotypes remained marginally significant (VEGFA p=0.037 and SOD2 p=0.066) while the genetic risk score remained highly significant (p=0.0067) for class 4 subjects. Genetic risk score was also significant in class 3 subjects (p=0.017). The other effects were not significant, though coefficients trended in the correct direction for the remainder of diagnostic classes. Given this and that class was a covariate in our original regression model, the association of VEGFA genotype with CHD class did not account for the association of risk score with survival.
Comment
We have performed a candidate SNP analysis for genes involved in oxidative stress and injury repair pathways in the prediction of long-term survival in children with non-syndromic CHD. SNPs at two genes, vascular endothelial growth factor A, VEGFA, and superoxide dismutase 2, SOD2, were associated with long-term survival. Moreover, we have shown through a genetic risk score that the effects of the VEGFA and SOD2 SNPs are cumulative – the more copies of the deleterious alleles of either SNP a patient has, the lower their probability of survival is. Through exclusion of subjects with chromosomal or genetic anomalies, we have obtained results that are more accurate representations of the pediatric population with non-syndromic CHD, as genetic anomalies are frequently associated with poorer outcomes in children with CHD(8).
In our analyses, we have identified minor alleles at VEGFA and SOD2 as both being significantly protective against long-term mortality (VEGFA p=7.03×10−4 and SOD2 p=0.019). We also have demonstrated that the effects of both SNPs are independent and additive when considered together as a genetic risk score [p=3.02×10−4, HR=15.64 in the highest risk group (n=59) compared to the group with no risk alleles (n=11)]. The VEGFA SNP studied, rs833069, is in strong linkage disequilibrium (LD; r2=0.97) with one 5′UTR SNP, rs2010963, whose minor allele has been associated with higher VEGFA expression(13); thus the SNP studied is associated with higher VEGFA expression. Prior experiments injecting exogenous vascular endothelial growth factor (VEGF) into rats found an increase in cardiac vascular permeability and cellular damage, which is hypothesized to be a mechanism through which VEGF causes injury post myocardial infarction (MI)(14). However, other studies have found that lower endogenous VEGF levels are associated with an increased risk of adverse cardiovascular events following MI(15) and that exogenous VEGF can rescue cardiac function after MI(16). Recent animal evidence suggests that while long-term VEGFA expression (e.g., from a plasmid) is detrimental to heart remodeling after MI due to the side-effects of hypotension and edema, short-term, pulse-like VEGFA expression that more closely matches in vivo VEGF dynamics improves survival at 1-year follow-up while avoiding the previous VEGF-associated side-effects(17). Taken together, we hypothesize that our findings reflect a spatiotemporally restricted increased level of VEGFA gene expression in patients with the protective allele, which, when endogenously expressed and regulated, improves vascular response to ischemic conditions and increases myocardial function after surgery, leading to improved long-term outcomes. Within this context, our findings could represent a potential therapeutic target using the aforementioned short-term, pulse-like VEGF administration for clinical follow-up after surgical palliation of non-syndromic CHD. The SOD2 intronic variant analyzed, rs2758331 was studied due to strong LD (r2=0.93) with a well-studied SOD2 Val16Ala missense SNP, rs4880, which has been reported to increase enzyme activity approximately 33%(18), consistent with a protective improvement in the oxidative stress response.
Post-hoc literature review found a prior report of the VEGFA major allele (associated with decreased VEGFA expression(13), and decreased survival probability in our data) being associated with TOF(19). Our data replicates this finding, as the VEGFA major allele was associated with both TOF and TGA, which represented relatively minor (low morbidity/low mortality) heart defects in our cohort (vast majority diagnostic class 1). Additionally, the VEGFA minor allele, which is associated with increased VEGFA expression(13) and increased survival in our data, was associated with the most severe heart defect, HLHS (all 130 subjects with HLHS were in the highest risk diagnostic class 4). Had the VEGFA minor allele been associated with both lower class, less severe CHD and improved survival, the class association might have led to a false positive survival association. However, we find that the VEGFA minor allele is associated with the most severe CHD, and independently (as demonstrated by both the adjustment for diagnostic class in the original analyses and the significant and protective effects of VEGFA in the stratified sensitivity analysis of class 4 subjects) is protective against death or heart transplant in these highest-risk children. Thus, we conclude that there are not confounding effects of VEGFA (or SOD2, which was not associated with diagnostic class) on survival, as the protective effects of VEGFA were strongest in children with the most severe CHD. Further research into the mechanisms through which VEGFA expression influences the type and severity of CHD is now warranted.
Some limitations of this study must be considered. First, power was limited due to the size of the cohort and the lack of comparable cohorts with which to consider pooling data. We addressed this by limiting the number of hypotheses, including not attempting genome-wide analyses. As noted, all SNPs tested had or tagged known functional effects, improving the probability of true associations. However, true positives with smaller effect sizes may have been missed. Second, although the cohort is largely of European ancestry, we used data from subsets of all genetic ancestries and adjusted for this through usage of a standard statistical method for adjusting for population stratification(10) and performed sensitivity analyses in the majority EA subset of the cohort. These analyses suggest that the observed associations are not due to population stratification. Finally, our literature-identified and tested SNPs do not represent all biologically plausible candidates. We could only test variants of interest that were represented or in high LD with SNPs on our genotype chip, limiting hypotheses tested.
More work must be done to establish these findings before they can be implemented into the clinical setting. First, these results should be replicated in an independent cohort; unfortunately, no comparable genetic study of non-syndromic CHD patients is available at this time. Moreover, it must be noted that any replication of our work would require an inception cohort with DNA obtained prior to the first surgery. Failure to obtain DNA prior to the first surgery will likely lead to a survivor bias, as those with the deleterious alleles identified in our work will die at a rate disproportionate to the rest of the cohort.
In conclusion, the results presented offer evidence that long-term survival in children with non-syndromic CHD is likely affected by variation in genes involved in oxidative stress and vascular response mechanisms. Given the high incidence of CHD and the frequent need for surgical palliation, further molecular follow-up of these genes is imperative. Identification of these candidate genes and their differential susceptibility to oxidative and ischemic stress provides a potential window into novel pathways that can aid in the development of therapies and preventative strategies to aid in decreasing the morbidity and mortality of necessary cardiac surgery in infants with non-syndromic CHD.
Supplementary Material
Acknowledgments
We would like to thank all subjects and families for their participation. Genotyping was performed by the Center for Applied Genomics at the Children’s Hospital of Philadelphia.
This work was supported by a grant from the Fannie E. Rippel Foundation, an American Heart Association National Grant-in-Aid (9950480N), HL071834 from the National Institutes of Health, and a Washington State Life Sciences Discovery Award to the Northwest Institute for Genetic Medicine. DSK was supported by 1F31MH101905-01.
Footnotes
Meeting Presentation: J. Maxwell Chamberlain Memorial Paper for Congenital Heart Surgery: Vascular Endothelial Growth Factor and Superoxide Dismutase Gene Variants Have an Additive Adverse Effect on Covariate-Adjusted Transplant Free Survival after Surgery for Isolated Congenital Heart Disease. Society of Thoracic Surgeons 50th Annual Meeting, Orlando, FL, January 27th, 2014
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, et al. Hypoplastic left heart syndrome: current considerations and expectations. Journal of the American College of Cardiology. 2012 Jan 3;59(1 Suppl):S1–42. doi: 10.1016/j.jacc.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laffey JG, Boylan JF, Cheng DCH. The systemic inflammatory response to cardiac surgery: implications for the anesthesiologist. Anesthesiology. 2002 Jul;97(1):215–52. doi: 10.1097/00000542-200207000-00030. [DOI] [PubMed] [Google Scholar]
- 3.Clancy RR, McGaurn SA, Goin JE, Hirtz DG, Norwood WI, Gaynor JW, et al. Allopurinol neurocardiac protection trial in infants undergoing heart surgery using deep hypothermic circulatory arrest. PEDIATRICS. 2001 Jul;108(1):61–70. doi: 10.1542/peds.108.1.61. [DOI] [PubMed] [Google Scholar]
- 4.Kirshbom PM, Mahle WT, Joyner RW, Leong T, Wilson M, Kogon BE, et al. The endothelin-1 G5665T polymorphism impacts transplant-free survival for single ventricle patients. The Journal of Thoracic and Cardiovascular Surgery. 2008 Jul;136(1):117–22. doi: 10.1016/j.jtcvs.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 5.Mital S, Chung WK, Colan SD, Sleeper LA, Manlhiot C, Arrington CB, et al. Renin-angiotensin-aldosterone genotype influences ventricular remodeling in infants with single ventricle. Circulation. 2011 May 31;123(21):2353–62. doi: 10.1161/CIRCULATIONAHA.110.004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynor JW, Gerdes M, Zackai EH, Bernbaum J, Wernovsky G, Clancy RR, et al. Apolipoprotein E genotype and neurodevelopmental sequelae of infant cardiac surgery. The Journal of Thoracic and Cardiovascular Surgery. 2003 Dec;126(6):1736–45. doi: 10.1016/s0022-5223(03)01188-7. [DOI] [PubMed] [Google Scholar]
- 7.Gaynor JW, Nord AS, Wernovsky G, Bernbaum J, Solot CB, Burnham N, et al. Apolipoprotein E genotype modifies the risk of behavior problems after infant cardiac surgery. PEDIATRICS. 2009 Jul;124(1):241–50. doi: 10.1542/peds.2008-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DS, Stanaway IB, Rajagopalan R, Bernbaum JC, Solot CB, Burnham N, et al. Results of genome-wide analyses on neurodevelopmental phenotypes at four-year follow-up following cardiac surgery in infancy. PLoS ONE Public Library of Science. 2012;7(9):e45936. doi: 10.1371/journal.pone.0045936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009 Jan;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006 Jul 23;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 11.Clancy RR, McGaurn SA, Wernovsky G, Spray TL, Norwood WI, Jacobs ML, et al. Preoperative risk-of-death prediction model in heart surgery with deep hypothermic circulatory arrest in the neonate. The Journal of Thoracic and Cardiovascular Surgery. 2000 Feb;119(2):347–57. doi: 10.1016/S0022-5223(00)70191-7. [DOI] [PubMed] [Google Scholar]
- 12.Gaynor JW, Mahle WT, Cohen MI, Ittenbach RF, DeCampli WM, Steven JM, et al. Risk factors for mortality after the Norwood procedure. Eur J Cardiothorac Surg. 2002 Jul;22(1):82–9. doi: 10.1016/s1010-7940(02)00198-7. [DOI] [PubMed] [Google Scholar]
- 13.Vailati FB, Crispim D, Sortica DA, Souza BM, Brondani LA, Canani LH. The C Allele of -634G/C Polymorphism in the VEGFA Gene Is Associated with Increased VEGFA Gene Expression in Human Retinal Tissue. Investigative Ophthalmology & Visual Science. 2012 Sep 4;53(10):6411–5. doi: 10.1167/iovs.12-9727. [DOI] [PubMed] [Google Scholar]
- 14.Weis S, Shintani S, Weber A, Kirchmair R, Wood M, Cravens A, et al. Src blockade stabilizes a Flk/cadherin complex, reducing edema and tissue injury following myocardial infarction. J Clin Invest. 2004 Mar 15;113(6):885–94. doi: 10.1172/JCI20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsudaira K, Maeda K, Okumura N, Yoshikawa D, Morita Y, Mitsuhashi H, et al. Impact of Low Levels of Vascular Endothelial Growth Factor After Myocardial Infarction on 6-Month Clinical Outcome. Circ J. 2012;76(6):1509–16. doi: 10.1253/circj.cj-11-1127. [DOI] [PubMed] [Google Scholar]
- 16.Guo H-D, Cui G-H, Yang J-J, Wang C, Zhu J, Zhang L-S, et al. Sustained delivery of VEGF from designer self-assembling peptides improves cardiac function after myocardial infarction. Biochemical and Biophysical Research Communications. 2012 Jul 20;424(1):105–11. doi: 10.1016/j.bbrc.2012.06.080. [DOI] [PubMed] [Google Scholar]
- 17.Zangi L, Lui KO, Gise von A, Ma Q, Ebina W, Ptaszek LM, et al. Modified mrNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nature Biotechnology Nature Publishing Group. 2013 Sep 8;31(10):899–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastaki M, Huen K, Manzanillo P, Chande N, Chen C, Balmes JR, et al. Genotype-activity relationship for Mn-superoxide dismutase, glutathione peroxidase 1 and catalase in humans. Pharmacogenet Genomics. 2006 Apr;16(4):279–86. doi: 10.1097/01.fpc.0000199498.08725.9c. [DOI] [PubMed] [Google Scholar]
- 19.Lambrechts D. Low expression VEGF haplotype increases the risk for tetralogy of Fallot: a family based association study. J Med Genet. 2005 Jun 1;42(6):519–22. doi: 10.1136/jmg.2004.026443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.