Abstract
Mechanisms that regulate regional epithelial cell diversity and pathologic remodeling in airways are poorly understood. We hypothesized that regional differences in cell composition and injury-related tissue remodeling result from the type and composition of local progenitors. We used surface markers and the spatial expression pattern of an SFTPC-GFP transgene to subset epithelial progenitors by airway region. Green fluorescent protein (GFP) expression ranged from undetectable to high in a proximal-to-distal gradient. GFPhi cells were subdivided by CD24 staining into alveolar (CD24neg) and conducting airway (CD24low) populations. This allowed for the segregation of three types of progenitors displaying distinct clonal behavior in vitro. GFPneg and GFPlow progenitors both yielded lumen containing colonies but displayed transcriptomes reflective of pseudostratified and distal conducting airways, respectively. CD24lowGFPhi progenitors were present in an overlapping distribution with GFPlow progenitors in distal airways, yet expressed lower levels of Sox2 and expanded in culture to yield undifferentiated self-renewing progeny. Colony-forming ability was reduced for each progenitor cell type after in vivo bleomycin exposure, but only CD24lowGFPhi progenitors showed robust expansion during tissue remodeling. These data reveal intrinsic differences in the properties of regional progenitors and suggest that their unique responses to tissue damage drive local tissue remodeling. Disclosure of potential conflicts of interest is found at the end of this article.
Keywords: Regional, Progenitor, Epithelium, Lung, Bleomycin
Introduction
The conducting airways and alveoli are lined by epithelia whose cellular composition is tailored to meet functional needs such as mucus clearance, surfactant production, gas exchange, and/or host defense [1]. The differing regional capacities for each of these functions lead to corresponding differences in susceptibility to injury and tissue remodeling in the setting of lung disease [2]. In fibrotic lung diseases such as idiopathic pulmonary fibrosis and bronchiolitis obliterans syndrome, loss or damage to either alveolar epithelial cells and/or distal conducting airway epithelium are common pathological features that contribute to declining lung function [3, 4]. However, despite well described changes that occur to the airway epithelium in conjunction with tissue remodeling, the cellular mechanisms are poorly understood.
Proximal-to-distal patterning of airways to yield regionally distinct epithelial cell types is developmentally specified. An early marker that distinguished prosopective alveolar from conducting airway epithelium is the transcription factor Sox2 [5]. Multipotent lung epithelial progenitor cells are regulated by signals that originate from surrounding mesenchyme [6, 7]. In adult lung, alveolar type 2 cells (AT2) are commonly believed to be progenitor cells responsible for alveolar repair after injury [8, 9]. Recent studies by Chapman et al. suggest that a subset of alveolar cells expressing the laminin receptor α6β4, but little or no surfactant protein C (Sftpc), functions as a progenitor for distal lung epithelium [10]. In the conducting airways, the proportions of nonciliated and ciliated cells that make up the epithelium vary as a function of airway location suggesting that developmental patterning of the lung yields regionally specialized epithelial progenitor cells. The demonstration that chemically resistant epithelial stem/progenitor cells, termed variant Clara cells, restore nascent Clara and ciliated cells of the adult mouse bronchiolar epithelium following naphthalene-induced airway injury provide further evidence of heterogeneity among airway progenitor cells [11, 12]. Luminal progenitor cells of pseudostratified airways share similar characteristics to Clara cells, yet are capable of only limited self-renewal in vivo [13] and are replaced by the progeny of dividing basal cells in both steady-state and repairing conditions [14, 15]. The context with which lung progenitor cells participate in epithelial maintenance has been revealed through the use of somatic chimeras or lineage tracing approaches [13, 16]. However, it has not been established yet whether lung progenitor cells residing at different airway locations have intrinsically distinct differentiation potentials or whether they have equal potential.
In this study, we use fractionation methods to enrich lung epithelial progenitor cells and three-dimensional (3D) culture models to follow their in vitro clonal behavior. We show that there is heterogeneity among progenitor cells that maintain airways and define three distinct progenitor cell types that reside along the proximal-to-distal airway axis. Regionally distinct lung progenitor cell populations show differential responsiveness to bleomycin-induced lung injury and tissue remodeling. These data demonstrate that region-specific airway progenitor cells can be identified and differ in their response to bleomycin exposure. Thus, we provide new tools to evaluate the contribution of epithelial progenitors to distal conducting airway remodeling after parenchymal injury.
Materials And Methods
Reagents
Biotin-conjugated monoclonal antibodies to mouse CD31, CD34, CD45, phycoerythrin-Cy7 (PE-CY7)-conjugated monoclonal antimouse EpCAM, allophycocyanin (APC)-conjugated monoclonal anti-mouse Sca-1, phycoerythrin (PE)-conjugated monoclonal anti-mouse CD24, and APC-eFluor 780 (similar to APC-CY7) - conjugated monoclonal streptavidin were purchased from eBioscience, San Diego, CA, http://www.ebioscience.com). IQ SYBR Green supermix was purchased from Bio-Rad Laboratories (Hercules, CA). Chicken polyclonal anti-green fluorescent protein (GFP) antibody was purchased from Abcam, Cambridge, MA, http://www.abcam.com). SB431542 was purchased from Ascent Scientific, Princeton, NJ, http://www.ascentscientific.com). Growth factor-reduced Matrigel was purchased from BD Pharmingen (Sa Jose, CA). Vectors containing cDNA insert of genes of interest were purchased from Open Biosystems, Huntsville, AL, http://www.openbiosystems.com). Mouse genome 430 2.0 arrays were purchased from Affymetrix, Singapore, http://www.affymetrix.com). Polyclonal anti-mouse Scgb1a1 antibody and polyclonal anti-mouse Sftpc antibody were made in house. Chicken polyclonal to GFP antibody was purchased from Abcam. Polyclonal rabbit antibody against keratin 5 was purchased from Covance, Berkeley, CA, http://www.covance.com). Polyclonal rabbit anti-Sox2 antibody was from Seven Hills Bioreagents, Cincinnati, OH, http://www.sevenhillsbioreagents.com). Mouse IgG1 anti-FoxJ1 clone 2a5 was purchased from eBioscience. Polyclonal anti short Plunc (sPlunc) was kindly provided by Dr. Colin Bingle (The University of Sheffield, U.K.). Molonclonal anti-E-Cadherin (clone 36/E-Cadherin) antibody was purchased from BD Biosciences, Bedford, MA, http://www.bdbiosciences.com).
Animal Husbandry
All mice were maintained in pathogen-free conditions in AAA-LAC approved animal facility at Duke University. Mice were exposed to a 12-hour light/dark cycle and had free access to food and water. Adult mice between the ages of 2–4 months were sacrificed for experiments according to IACUC approved protocols. SFTPC-GFP mice (BALB/c and C57BL/6J mixed background) and FoxJ1-GFP mice (C3H and C57BL/6J mixed background) were kindly provided by Brigid L.M. Hogan (Duke University). Scgb1a1-CreER; Rosa26R-Confetti mice was created by crossing Scgb1a1-CreER mice (From Bridig L.M. Hogan lab, Duke University) with Rosa26R-Confetti mice (The Jackson Laboratory, Bar Harbor, Maine, http://www.jax.org). Scgb1a1-CreER; Rosa26-mT/mG mice were created by crossing Scgb1a1-CreER mice with Rosa26-mT/mG (The Jackson Laboratory). For injury experiments, mice were treated intratracheal instillation with 2.5 U/kg bleomycin (APP Pharmaceuticals LLC, Schaumburg, IL), and three mice were sacrificed at indicated time points.
Flow Cytometry
Lung cell suspensions were prepared for fluorescence-activated cell sorting (FACS) as described previously [17]. Briefly, cells were resuspended in Hanks’ balanced saline solution (HBSS+) buffer (HBSS supplemented with 2% fetal bovine serum (FBS), 10 mM Hepes, 0.1 mM EDTA, 100 IU/ml Penicillin, 100 µg/ml Streptomysin, and 0.25 µg/ml Fungizone). Primary antibodies including CD31-Biotin, CD34-Biotin, CD45-Biotin, CD24-PE, EpCAM-PE-Cy7, and Sca-1-APC were added to incubate cells. Biotin-conjugated antibodies were detected following incubation with streptavidin-APC-Cy7. Dead cells were discriminated by 7-amino-actinomycin D staining.
Microarray Analysis
At least five mice were used per sort and samples were pooled from eight sorts to generate duplicate for GFPneg population and triplicate for GFPlow and GFPhi populations. Total RNA was isolated from pooled epithelial progenitor cells using RNeasy mini kit (Qiagen, Germantown, MD, http://www.qiagen.com), quantified using a nanodrop spectrophotometer (ND-1000, NanoDrop Technologies, Inc., Wilmington, DE, http://www.nanodrop.com), and 0.1 µg of RNA used for Affymetrix Microarray analysis using mouse genome 430 2.0 arrays in the Duke University Microarray Facility. Data were annotated with Affymetrix Expression Console software (Affymetrix). Differentially expressed genes in groups indicated were determined using the GeneSpring GX software (Agilent Technologies, http://www.home.agilent.com/agilent/home.jspx?&cc=US&lc=eng, Santa Clara, CA). Probe sets with a p value <.05 and fold change >1.5 were annotated. Gene expression heat maps were constructed using Treeview (ver.1.60) software (Michael B. Eisen, University of California at Berkeley).
Matrigel Culture
Lung epithelial cells were cultured in Matrigel as described previously [17]. In brief, sorted lung epithelial cells were mixed with mouse fibroblast MLg cells in Matrigel/basic medium (1:1). Basic medium contains Dulbecco’s modified Eagle’s medium/F12 (Cellgro, Manassas, VA, http://www.cellgro.com) supplemented with insulin/transferrin/selenium (Invitrogen), 10% FBS (Invitrogen), 0.25 µg/ml amphotericin B, 100 IU/ml penicillin, and 100 µg/ml streptomycin. Cells suspended in Matrigel were added to the chamber of 24-well Transwell filter inserts (Becton Dickinson, Franklin Lakes, NJ, http://www.bd.com) and placed in 24-well, flat-bottom culture plates containing basic medium with 10 µM SB431542. Cultures were maintained in a humidified 37°C incubator. To evaluate differentiation of GFPneg, GFPlow, and GFPhi progenitor cells, cultures were maintained with basic medium with SB431542 to allow cell growth for 10 days and the culture medium was switched to basic medium for additional 4 days. To determine self-renewal capacity of GFPneg, GFPlow, and GFPhi progenitor cells, cultures were harvested at 2 weeks by dispase digestion, GFPneg, GFPlow, and GFPhi epithelial cells were resorted by FACS, and seeded for further cultures with the same condition as for passage 0 cells. Colonies were visualized with a Zeiss Axiovert40 inverted fluorescent microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY, http://www.zeiss.com/micro). Colony-forming efficiency was determined by counting the number of colonies with a diameter of ≥50 µm in each culture and representing this number as a percentage of input epithelial cells.
Total RNA Isolation and Real-Time PCR
RNA was extracted from sorted epithelial cells or cells in colonies in Matrigel culture using a Qiagen RNeasy mini kit. Quantitative real-time PCR (qRT-PCR) was performed using the SYBR Green method. PCR runs and fluorescence detection were carried out using an Eppendorf realplex Real-Time PCR System (Eppendorf, Hamburg, Germany, http://www.eppendorf.com). Intronspanning gene-specific primers were designed using Primer-QuestSM software available at http://www.idtdna.com/Scitools/Applications/PrimerQuest/ (Integrated DNA Technologies, Inc, Coralville, IA) and listed in Table 1. Reaction conditions were as follows: an initial cycle of heating at 95°C for 2 minutes, followed by 40 cycles of 95°C for 25 seconds, for denaturation of the PCR products, 60°C for 25 seconds for primer annealing, and 72°C for 20 seconds for extension. Beta-actin was used as a reference gene to normalize all PCRs for the amount of RNA added to the reverse transcription reactions. Melt curves were run to clarify the identity of amplicons.
Table 1.
Sequences of primers for quantitative PCR
| Genes | Forward primer | Reverse primer |
|---|---|---|
| β-Actin | 5′-GGCCAACCGTGAAAAGATGA-3′ | 5′-CAGCCTGGATGGCTACGTACA-3′ |
| Scgb1a1 | 5′-ATCAGAGTCTGGTTATGT-3′ | 5′-ATCCACCAGTCTCTTCAG-3′ |
| Sftpc | 5′-CTCCTGACGGCCTATAAGCC-3′ | 5′-TAGTAGAGTGGTAGCTCTCC-3′ |
| Sox2 | 5′-CACATGAAGGAGCACCCGGATTAT-3′ | 5′-CGGGAAGCGTGTACTTATCCTTCT-3′ |
| Reg3g | 5′-ACACTGGGCTATGAACCCAACAGA-3′ | 5′-ACCACAGTGATTGCCTGAGGAAGA-3′ |
| Sult1d1 | 5′-TGGAACAACTTGGGTCAGTG-3′ | 5′-AAGCTCCATGAATGGTACTCG-3′ |
| Lypd2 | 5′-GCTTGCTATGGCCTTACAG-3′ | 5′- CAGTTGGATGCACTCACA-3′ |
| Kdr | 5′-AGGCCCATTGAGTCCAACTACACA-3′ | 5′-AGACCATGTGGCTCTGTTTCTCCA-3′ |
| Parm1 | 5′-ACCACAGAGAAGAAGCTGTCACCA-3′ | 5′-TCCAAGACTGTGTTCCTCAGGTGT-3′ |
| Slc23a1 | 5′-TCTGGTTCCAGCCAATGCAATCCT-3′ | 5′-TACTGGACACCATGATTGCACCCT-3′ |
| Gjb6 | 5′-TGAGCAGGAGGACTTTGTCTGCAA-3′ | 5′-TGTGAGACACCGGGAAGAAATGGT-3′ |
| Hmox1 | 5′-TAGCCCACTCCCTGTGTTTCCTTT-3′ | 5′-TGCTGGTTTCAAAGTTCAGGCCAC-3′ |
| Iyd | 5′-TGTCATCAAAGCAGCAGGAACAGC-3′ | 5′-TTCAGGTCTGTGACCCATCGCTTT-3′ |
| Jam2 | 5′-AGTATTACTGCGAAGCCCGGAACT-3′ | 5′-TCCTCTGAGCATAGCATGTGCCAA-3′ |
| Aim1l | 5′-TTCCAGAGCCATGGACCCATCTTT-3′ | 5′-AAGGGCACCAATAACTCAGTCCGA-3′ |
| Itih5 | 5′-GGGCAACATTGCCTTTGTCATCCT-3′ | 5′-ACCTCTGCTGTTGGCGATGTAGAA-3′ |
| Lamb3 | 5′-TGCAGTATCTTAGGTGCCCGGAAA-3′ | 5′-TCCTGTAAACTGGTTGCACTGGGA-3′ |
| Ces1g | 5′-TGTGGCAGGCCTACCCAATTCTTA-3′ | 5′-TCTGTCATTGTGGCAGGGTCTTCT-3′ |
| Creb3l1 | 5′-TCCTGCATTCTCTTCCGGCTCAAT-3′ | 5′-AAGGCATCTGACTGGCTGCATAGA-3′ |
| Kif3c | 5′-AGACCGTCCACAACTAAAGTGCGA-3′ | 5′-AGGGTGCAAAGGAACAGAGGTCAT-3′ |
| Pard6g | 5′-TGGGCTACTGGCTGTGAATGATGA-3′ | 5′-GCGGACCACATTGTTCCTTTGGTT-3′ |
| H60c | 5′-ATTGATGGTCTCTGGGCACTGTCA-3′ | 5′-GCCATAACAAGCAGAGCATTGCCA-3′ |
| Slc2a3 | 5′-ACGAAATTGAGGTTGGAGGGAGGT-3′ | 5′-TGACTCACAGGCCACCAGTAACAT-3′ |
Histology and Immunostaining
Colonies were fixed in freshly prepared 4% paraformaldehyde (PFA) and their Matrigel supports were rinsed with phosphate buffered saline (PBS), immobilized in 1.5% agarose, and embedded in paraffin. Five-micrometer sections were collected from either lung tissue or cultures, which were incubated with primary antibodies at 4°C overnight. Sections were washed with PBS and incubated with fluorochrome-conjugated secondary antibody for 2 hours at room temperature. Slides were mounted with Fluoromount G containing 4′-6-diamidino-2-phenylindole (DAPI). Staining was visualized with a Zeiss Axiovert40 inverted fluorescent microscope.
In Situ Hybridization
Antisense or sense digoxygenin (DIG)-labeled probes were transcribed in vitro from the plasmids (Open Biosystems) containing the desired gene using the DIG RNA-labeling kit (Roche, Indianapolis, IN, http://www.roche.com), per the manufacturer’s instructions. The probes were hybridized with lung paraffin sections. DIG-labeled probe was then detected by immunostaining with anti-DIG-horseradish peroxidase (HRP) (Roche) and the signal was amplified with TSA Plus DNP(AP) system (PerkinElmer, Waltham, MA, http://www.perkinelmer.com) according to the manufacturer’s instructions.
Statistical Analysis
Data from three or more independent experiments were collected and analyzed as mean ± SEM. The significance of the results was assessed by a paired t test between two groups. A p value <.05 was considered significant.
Results
Scgb1a1-Expressing Epithelial Progenitor Cells Maintain and Repair Airways In Vivo and Represent Colony-Forming Cells In Vitro
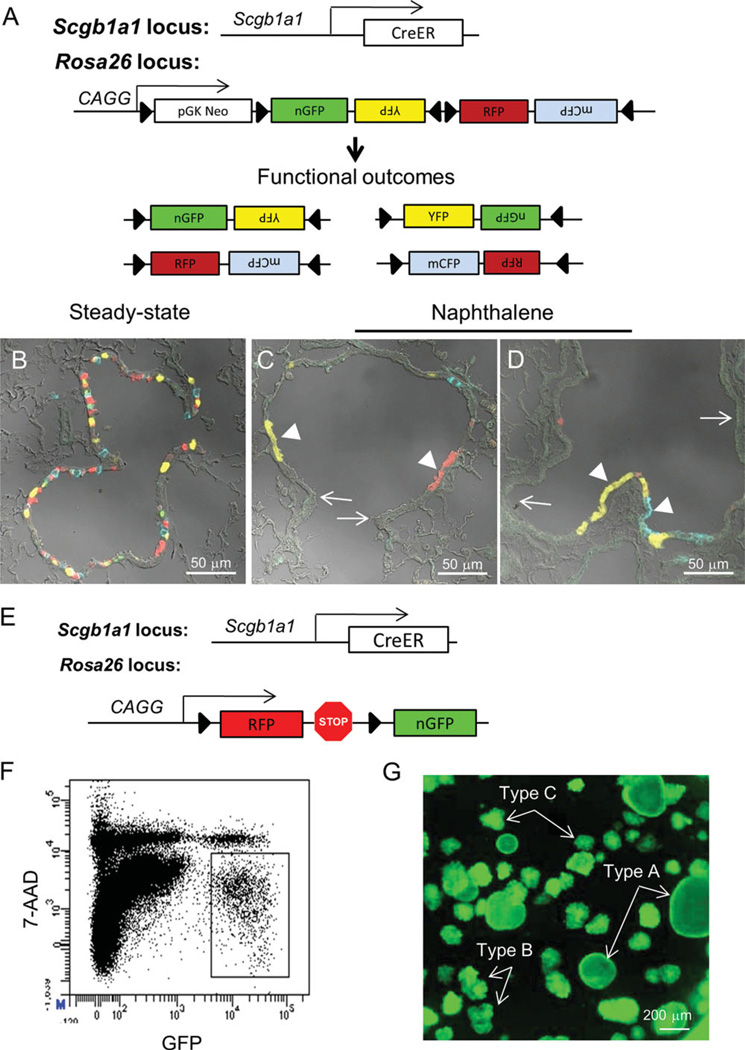
The observation that distinct colony types develop in vitro from EpCAMhigh lung epithelial cells has led to a proposed hierarchy of colony-forming epithelial progenitors [18]. However, the relationship between in vitro clonogenic progenitor cells and epithelial progenitor cells defined in vivo using injury models and lineage tracing approaches has not been well established. To address this issue, we used lineage tracing approaches to identify epithelial progenitor cells in vivo prior to their isolation and characterization in vitro. Previous work by us and others suggests that Scgb1a1-expressing cells fulfill critical roles in epithelial maintenance and repair in vivo [11–13, 19]. To further define the contribution of Scgb1a1-expressing cells to airway repair, we used a tamoxifen-regulated Scgb1a1-CreER driver line, Scgb1a1-CreER; Rosa26-Confetti, to lineage tag these cells in normal and injured airways (Fig. 1A–1D). Lineage tags were efficiently introduced into Scgb1a1-expressing epithelial cells (Fig. 1B). Naphthalene exposure depleted the majority of the Scgb1a1 lineage tagged cells, evident by the presence of large areas of epithelium containing ciliated cells that lacked a lineage tag (Fig. 1C, 1D, arrows). Rare naphthalene-resistant Scgb1a1 lineage tagged cells underwent significant clonal expansion during the 20-day recovery period after naphthalene exposure, leading to the appearance of patches of epithelial cells that shared common fluorescent protein lineage tags (Fig. 1C, 1D, arrowheads). However, nascent epithelium was generally composed of patches that contained more than one color fluorescent protein tag. These data are consistent with the presence of at least two subpopulations of bronchiolar Scgb1a1-expressing cells, Clara cells, and vClara cells and suggest that multiple naphthalene-resistant vClara cells are retained within each microenvironmental niche.
Figure 1.
The airway epithelium is maintained by Scgb1a1-expressing cells. (A): Schematic illustrating Scgb1a1-CreERT2; Rosa26R-Confetti constructs. Upon Cre activation by treatment of mice with tamoxifen, the Confetti construct randomly recombines for active expression of fluorescent protein reporters. Scgb1a1-expressing cells were efficiently lineage labeled after treatment of Scgb1a1-CreERT2; Rosa26R-Confetti mice with tamoxifen (B). Naphthalene-induced lung injury led to clonal expansion of lineage-labeled naphthalene-resistant epithelial progenitor cells within either terminal bronchioles (C) or more proximal bronchioles (D). Scale bars = 50 µm. (E): Schematic illustrating Scgb1a1-CreERT2; Rosa26-mT/mG (Scgb1a1-mT/mG) mice. (F): Lineage labeled cells (GFP+) isolated from Scgb1a1-mT/mG mice were sorted by fluorescence-activated cell sorting and placed in 3D MatriGel culture for 14 days (G). Representative image of the culture shows the formation of type A, B, and C colonies.
To further explore the intrinsic potentials of Scgb1a1-expressing epithelial progenitor cells, we used another Scgb1a1-CreER driver line, Scgb1a1-CreER; Rosa26-mT/mG lineage tracing coupled with in vitro 3D culture of tagged epithelial cells. Cells expressing an activated mEGFP lineage tag (Fig. 1E) were fractionated from dissociated lung tissue by FACS (Fig. 1F) and were placed in 3D cultures to allow for clonal expansion. Clonally derived epithelial colonies were observed after 14 days of culture that exhibited a range of distinct morphologies, type A round luminal, type B, and type C dense colonies, similar to those previously described by McQualter et al. (Fig. 1G and [18]). These data suggest that Scgb1a1 lineage tagged epithelial cells encompass at least three distinct populations of progenitor cells that are regulated in part through cell intrinsic.
Regional Heterogeneity of Lung Epithelial Progenitor Cells
In vivo lineage tracing and injury-repair studies suggest that multiple epithelial progenitor cell populations variably contribute to lung epithelial maintenance [13–15, 19]. Luminal progenitor cells, defined based upon their expression of Scgb1a1, critically maintain the bronchiolar epithelium, yet are dispensable for maintenance of pseudostratified airways [13]. In order to better define the properties of epithelial progenitor cells residing at different airway locations, we took advantage of a transgenic reporter line, SFTPC-GFP, in which the level of GFP expression varies according to airway region (Fig. 2A) [17]. To further define the spatial context of GFP expression in lungs of SFTPC-GFP transgenic mice, we used dual immunofluorescence staining to semiquantitatively assess expression of GFP immunoreactivity within conducting airway and alveolar epithelium (Fig. 2B). Proximal intrapulmonary conducting airway epithelial cells showed no detectable GFP immunoreactivity (GFPneg) (Fig. 2C). In contrast, the majority of Scgb1a1-immunoreactive cells present within terminal bronchioles showed low levels of GFP immunoreactivity (GFPlow) (Fig. 2D). Cells staining most intensely for GFP (GFPhi) were localized to alveoli in a spatial pattern consistent with AT2 cells and were also present in conducting airways in close association with terminal bronchioles (Fig. 2E) and neuroepithelial bodies (Supporting Information Fig. S1). These data are consistent with the distribution of AT2 cells and variant Clara cells and collectively demonstrate that the extent of GFP expression by epithelial cells of SFTPC-GFP transgenic mice differs according to airway location.
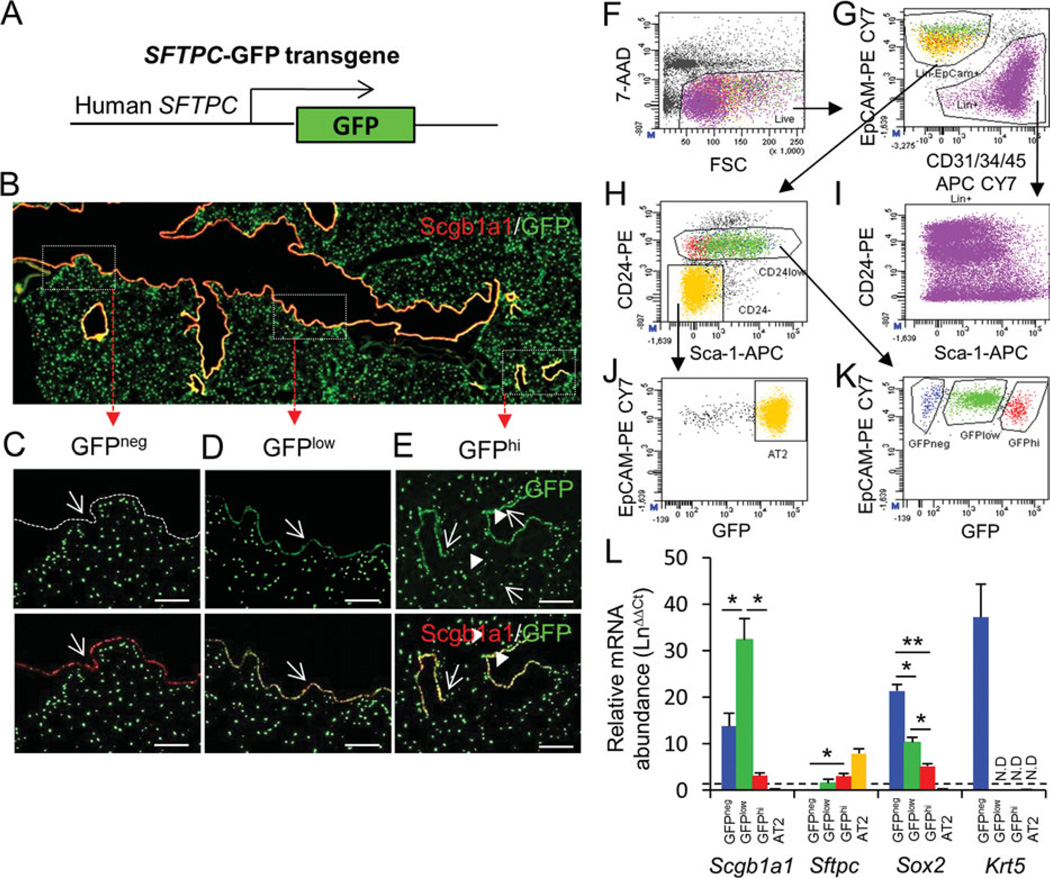
Figure 2.
Segregation of region-specific airway epithelial progenitor cells. (A): Schematic of the SFTPC-GFP transgene. (B): Immunolocalization of Scgb1a1 (red) and GFP (green) in lung tissue of SFTPC-GFP lungs defines three distinct airway locations. (C): Proximal conducting airway is lined by epithelial cells that are Scgb1a1pos and GFPneg (arrow). (D): Midlevel bronchioles are lined by epithelium that is Scgb1a1pos and GFPlow (arrow). (E): Terminal bronchioles and alveoli harbor GFPhi epithelial cells that are either Scgb1a1pos (arrows) or Scgb1a1neg (arrowheads), respectively. Scale bars in (C–E) = 50 µm. (F–K): GFPneg, GFPlow, and GFPhi progenitor cells can be fractionated by selection for EpCAMpos, CD24low cells that are CD31neg, CD34neg, and CD45neg. Back-gate of these three populations to the CD24 versus Sca-1 dot plot reveals that GFPneg and GFPlow populations express Sca-1, while GFPhi cells are predominantly Sca-1neg. (I): CD24 antigen is broadly expressed in both epithelial lineage and nonepithelial lineages in the lung. (J): CD24neg Sca-1neg gate in (H) is largely populated by GFPhi type 2 cells. (L): Real-time PCR analysis was performed on total RNA isolated from cell fractions to quantify Scgb1a1, Sftpc, Sox2, and Krt5 mRNA’s. Abbreviations: 7-AAD, 7-amino-actinomycin D; FSC, forward scatter; GFP, green fluorescent protein. *, <0.05; **, <0.01.
We then used a FACS-based strategy to separate lung epithelial cells according to GFP fluorescence level. Lung cells from SFTPC-GFP mice were isolated by elastase digestion and stained with fluorescent antibodies to surface markers and a viability dye (Fig. 2F). We used surface staining for CD31, CD34, and CD45 (collectively referred to as Lin [17, 21]) for negative selection of endothelial, stromal, and hematopoietic cells (Fig. 2G). Epithelial cells were further enriched by surface CD24 and Sca-1 staining (Fig. 2H). CD24 has been used previously to subset mouse mammary and lung epithelial cell populations [18, 22]. Here, we show that CD24 is broadly expressed by both epithelial and nonepithelial cells in the lung (Fig. 2I; Supporting Information Fig. S2). We further show that ciliated cells were CD24hi (Supporting Information Fig. S2), and AT2 cells CD24neg Sca-1neg (Fig. 2J), allowing us to exclude these cell types from the airway epithelial progenitor cell population. The resulting CD24low fraction included both Sca-1pos and Sca-1neg subpopulations of nonciliated airway epithelial cells and was subjected to further fractionation according to levels of GFP fluorescence. Dot plot analysis of this Linneg EpCAMpos CD24low population revealed that SFTPC-GFPneg (GFPneg), SFTPC-GFPlow (GFPlow), and SFTPC-GFPhi (GFPhi) subsets could be resolved based upon GFP fluorescence intensity (Fig. 2K). These cell fractions represented 3.9% ± 2.2%, 24.4% ± 5.1%, and 2.9% ± 0.4% of the LinnegEpCAMpos population, respectively. Back gating of each of these populations to the bivariate CD24 versus Sca-1 dot plot demonstrated that GFPneg and GFPlow populations are Sca-1pos, and that CD24low GFPhi cells are Sca-1neg.
We evaluated the expression of airway epithelial cell markers as a measure of enrichment for progenitor cells within each fraction. Levels of Scgb1a1 mRNA were significantly higher in all three cell fractions relative to total lung RNA, suggesting that they were enriched for nonciliated progenitor cells from conducting airways (Fig. 2L). The conducting airway origin of cells contained within each fraction was further supported by enrichment of Sox2 mRNA. We found an inverse correlation between the abundance of mRNA’s for either Scgb1a1 or Sox2 and Sftpc. This was most apparent for AT2 cells, which showed no detectable Scgb1a1 or Sox2 mRNA’s, but high levels of Sftpc mRNA. Expression of the basal cell marker gene Krt5 was only observed above background levels within total RNA isolated from the GFPneg fraction. Despite the significant enrichment of Krt5-expressing cells within the GFPneg fraction relative to total lung, Krt5-immunoreactive cells represented <1% of the cell types present within this fraction (Supporting Information Fig. S3. Differences in the level of Scgb1a1 mRNA abundance within each cell fraction (Fig. 2L) were associated with marked differences in the fluorescence intensity of Scgb1a1 immunoreactivity (Supporting Information Fig. S3). Most notable were the high levels of Scgb1a1 immunoreactivity observed among cells within the GFPlow cell fraction (Supporting Information Fig. S3C) and the low levels of perinuclear Scgb1a1 immunoreactivity observed among cells within the GFPhi fraction (Supporting Information Fig. S3D). Despite differences in the level of Scgb1a1 expression by cells within each cell fraction, the contribution of Scgb1a1-immunoreactive cells was 88.9% 6 0.1%, 98.9% ± 0.7%, and 96.8% ± 0.5% of total cells within GFPneg, GFPlow, and GFPhi fractions, respectively (Supporting Information Fig. S3). These data demonstrate that epithelial cell fractions generated using a combination of surface marker staining and expression of the SFTPC-GFP transgene represent highly enriched subpopulations of Scgb1a1-immunoreactive cells.
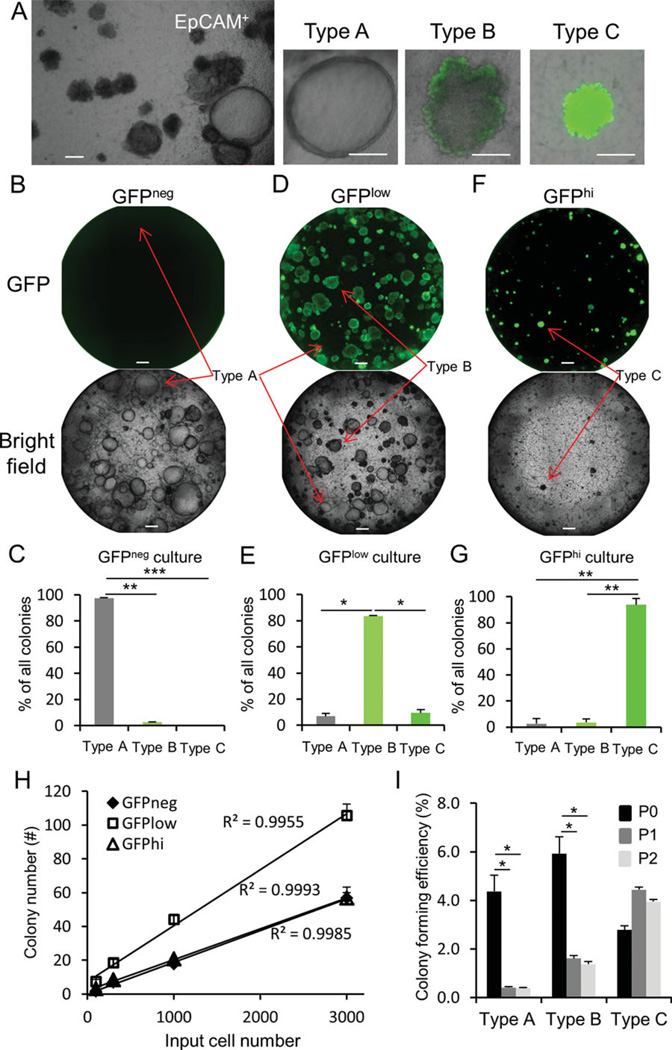
Cultures of total Linneg EpCAMpos cells prepared from SFTPC-GFP mice yielded three clearly discernible colony types that were distinguished based upon both morphology and GFP expression (Fig. 3A). These epithelial colonies exhibited similar morphology to those observed following culture of freshly sorted Scgb1a1 lineage traced cells (cf. Fig. 1G with Fig. 3A). Each subset was then sorted by FACS and cultures established from GFPneg, GFPlow, and CD24low GFPhi fractions (Fig. 3B–3G). GFPneg, GFPlow, and CD24low GFPhi cell fractions resulted in the efficient segregation of colony-forming progenitor cells yielding type A, type B, and type C colonies, respectively (Fig. 3B–3G). A limiting dilution analysis of colony-forming efficiency for GFPneg, GFPlow, and GFPhi cells showed a linear correlation between input cell number and colony number (R2>0.99 for each cell fraction), suggesting that colony-forming ability was an intrinsic property of progenitor cells contained within each fraction (Fig. 3H, Supporting Information Fig. S4). Analysis of the replating efficiency of these cell fractions revealed that only progenitor cells contained within the distal CD24low GFPhi fraction were able to efficiently self-renew in vitro (Fig. 3I). These data suggest that different conducting airway regions harbor functionally distinct epithelial progenitor cells.
Figure 3.
Segregation of regional progenitor cells based upon SFTPC-GFP expression. (A): Culture of total EpCAM+ cells from SFTPC-GFP mice reveals three distinct types of epithelial colonies. (B, C): Two-week cultures of GFPneg, GFPlow, and GFPhi cells indicate that type A colonies are enriched within the GFPneg epithelial fraction. (D, E): Type B colonies are enriched in the GFPlow epithelial fraction. (F, G): Type C colonies are enriched within the GFPhi epithelial fraction. (H): Quantitative analysis of the colony-forming ability of GFPneg, GFPlow, and GFPhi cells. (I): Self-renewal capacity of progenitors yielding type A, B, and C colonies was examined. p0, passage 0; p1, passage 1; p2, passage 2. Data represent combined results from three independent experiments. *, <.05; **, <.01; ***, <.001. Scale bar = 200 µm. Abbreviation: GFP, green fluorescent protein.
Progenitor Cells Are Programmed to Generate Region-Specific Specialized Progeny
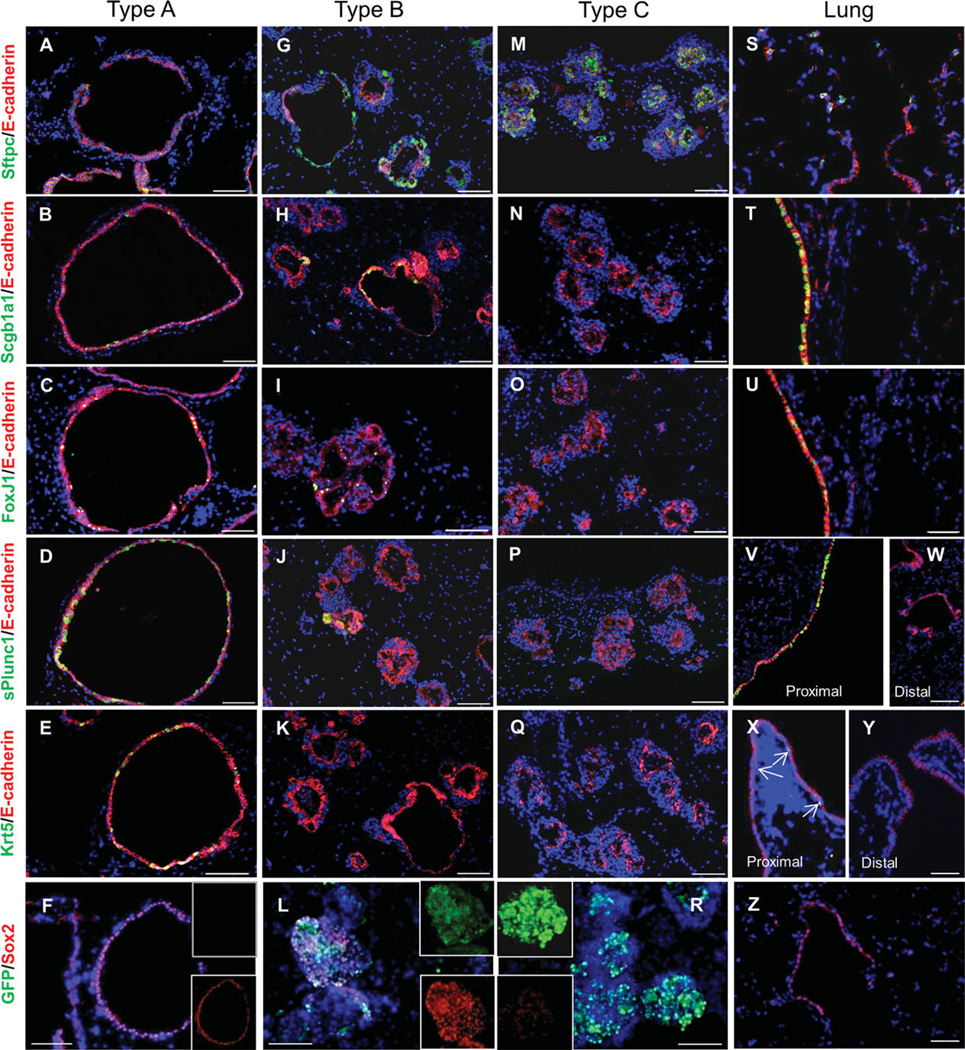
To better understand the intrinsic potentials of distinct colony- forming progenitor cells, cultures were established in the presence of the transforming growth factor-β (TGFβ)-inhibitor SB431542 and after 7 days changed to SB431542-free medium to promote epithelial cell differentiation. Cultures were then harvested to evaluate expression of genes that distinguish specialized cell types that normally reside in different airway locations in vivo. The differentiation potential of epithelial progenitor cells was determined by immunophenotypic analysis of epithelial organoids harvested from 3D cultures. We evaluated expression of Sftpc, a marker for AT2 cells and subpopulations of distal conducting airway cells, Scgb1a1 and Foxj1, markers of luminal epithelium of conducting airways, sPlunc, a marker for secretory cells of proximal airways, Krt5, a marker for basal cells in pseudostratified airways, and Sox2, a marker that distinguishes conducting airway epithelium from alveolar epithelium (Fig. 4). In all the cultures, we distinguished epithelial cells from surrounding stromal support cells based upon positive immunofluorescent staining for Ecadherin. We found that differences in colony morphology were associated with the presence of unique populations of specialized epithelial cells within each colony type (Fig. 4). Type A colonies, which were preferentially derived from the GFPneg fraction of SFTPC-GFP lung cells, showed the broadest differentiation potential (Fig. 4A–4F). Epithelial cells that expressed Scgb1a1, sPlunc, Foxj1, and Krt5, indicative of the presence of Clara, serous, ciliated, and basal cells, respectively, were present. Type A colonies also displayed the lowest level of Sftpc immunoreactivity (Fig. 4A). The epithelium of type B colonies (Fig. 4G–4L) included abundant populations of Scgb1a1 and Sftpc-immunoreactive cells, contained Foxj1-immunoreactive cells, but lacked significant numbers of cells showing sPlunc and Krt5 immunoreactivity. Epithelial cells of type C colonies showed positive immunoreactivity for Sftpc (Fig. 4M) but none of the other markers that define specialized cell types of conducting airways (Fig. 4N–4Q). These data suggest that epithelial cells contained within type A colonies more closely resemble those of the pseudostratified epithelium of large conducting airways (Fig. 4U, 4X). In contrast, epithelial cells of type B and type C colonies share similarities with distal conducting airways, with the epithelium of type B colonies resembling bronchioles. Expression patterns of Sftpc, FoxJ1, sPLUNC, and Krt5 among type A, type B, and type C colonies by immunofluorescent staining were further agreed by the analysis of mRNA abundance of these genes in each type of colonies by quantitative RT-PCR (qRT-PCR) (Supporting Information Fig. S5). Epithelial cells of each colony type were positive for Sox2 immunoreactivity (Fig. 4F, 4L, 4R). This finding was expected for the epithelium generated by type A and B colonies due to their expression of markers of specialized conducting airway epithelial cell types. However, expression of low levels of Sox2 by epithelial cells of type C colonies was unexpected. Epithelium of type C colonies expressed Sftpc but failed to express markers of specialized conducting airway epithelium. These data provide further support for the notion that airways harbor region-specific progenitor cells and that these progenitors retain elements of their positional identity in culture.
Figure 4.
In vitro differentiation of airway regional progenitor cells. Green fluorescent protein (GFPneg), GFPlow, and GFPhi progenitor cells were cultured in presence of SB431542 for 10 days and switched to SB431542-deficient medium for 4 days. Paraffin sections of each culture type (GFPneg, A–E; GFPlow, G–K; GFPhi, M–Q) and normal adult mouse lung tissue (S–Y) were stained with Sftpc, Scgb1a1, FoxJ1, sPlunc1, and Krt5 by dual immunofluorescence for E-caderin, respectively. Rare Krt5-positive cells are indicated in proximal airway by arrows in (X). Dual immunofluorescence stain of GFP and Sox2 on each culture type (F, L, and R) and normal lung section (Z) indicated a variable level of Sox2 among these colonies. Scale bar = 50 µm.
New Molecular Markers that Distinguish Region-Specific Epithelial Progenitor Cells
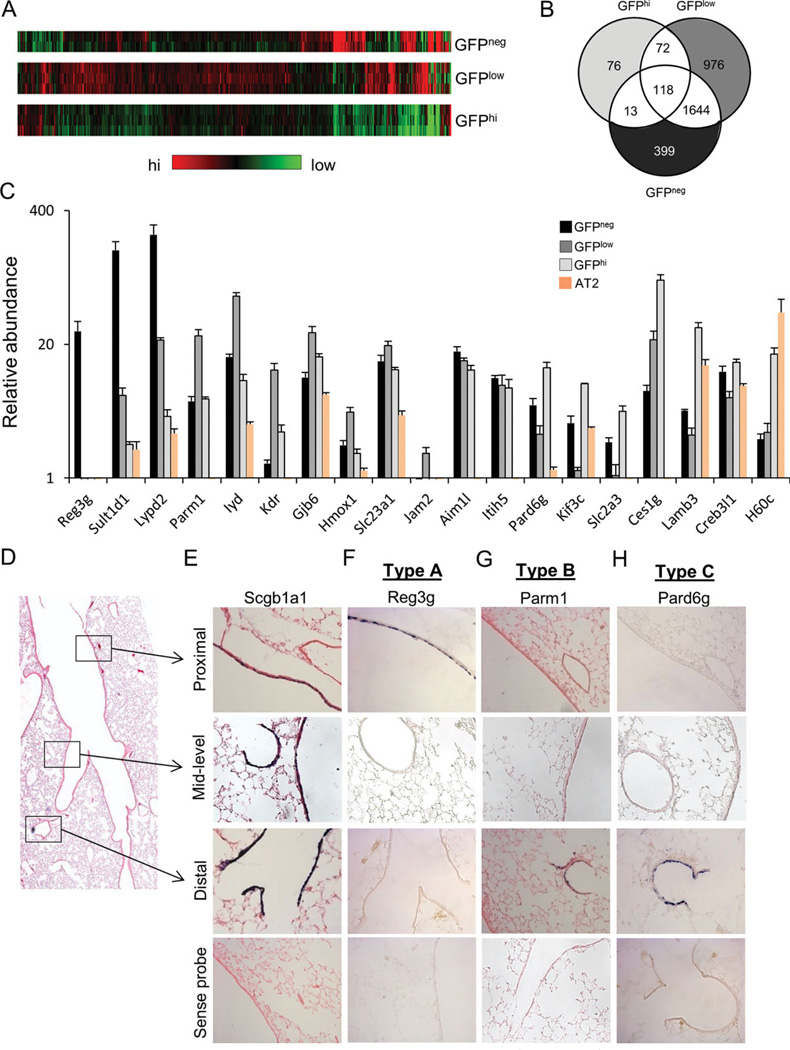
Our demonstration that progenitor cells of proximal and distal conducting airways show distinct functional characteristics in vitro is consistent with previous reports of differences in the renewal potential of Scgb1a1-expressing progenitors from these airway locations [13]. However, there are no unique marker genes that distinguish distal progenitor cell subsets or their specialized progeny from their proximal conducting airway counterparts. To address this deficiency, we next sought to define new molecular markers that had potential to distinguish these progenitor cell subsets. To accomplish this, we used microarray analysis to compare transcriptomes of freshly isolated GFPneg, GFPlow, and CD24lowGFPhi cell populations from SFTPC-GFP mice. Included in this analysis were enriched populations of AT2 cells that were selected based upon their Sca1neg CD24neg GFPhi phenotype (Fig. 2J). Microarray analysis was performed for total RNA isolated from each cell fraction using Affymetrix mouse genome 430 2.0 chips. Using this approach, heatmaps were built to show unique gene expression patterns obtained for each cell fraction (Fig. 5A). Results of bioinformatics analysis are summarized as a Venn diagram (Fig. 5B). A total of 399 transcripts were enriched within total RNA isolated from GFPneg cells, 976 transcripts were enriched within total RNA of GFPlow cells, and 76 transcripts were enriched within total RNA of CD24low GFPhi cells (Fig. 5B). Transcripts enriched in GFPneg cells included sPlunc, Muc5Ac, Muc5B, lactotransferrin, and trefoil factor 2 that have previously been associated with secretory cells of proximal conducting airways [23–25]. Sca-1 mRNA was also enriched in this population, which is consistent with the relatively high level of surface Sca1 immunoreactivity observed by flow cytometry (Fig. 2H). Other transcripts that were more abundant within the GFPneg cell fraction included Reg3g, Sult1d1, Lypd2, Slc15a2, and an unknown gene sequence U46068. Validation of three of these genes, Reg3g, Sult1d1, and Lypd2, by qRT-PCR confirmed that their mRNA’s are highly enriched within RNA isolated from GFPneg cells compared to GFPlow cells (38-, 25-, and 11-fold, respectively), CD24low GFPhi cells (134-, 80-, and 58-fold, respectively), or AT2 cells (sorted lung cells with the phenotype of EpCAMpos CD24neg Sca-1neg GFPhi from SFTPC-GFP mice) (268-, 86-, and 86-fold, respectively) (Fig. 5C). qRT-PCR analysis confirmed enrichment of Parm1, Iyd, and Kdr in total RNA isolated from the GFPlow fraction compared to GFPneg cells (four, six, and fivefold, respectively), CD24low GFPhi cells (four, four, and ninefold, respectively), or AT2 cells (80-, 18-, and 112-fold, respectively) (Fig. 5C). Other transcripts that were enriched in GFPlow cells compared to all other cell fractions included Gjb6, Hmox1, Slc23a1, and Jam2 in GFPlow cells. No differences were observed in the abundance of transcripts for Aim1l and Itih5 between each of the cell fractions (Fig. 5C). Validation of Pard6g, Kif3c, Slc2a3, and Ces1g confirmed their enrichment in total RNA isolated from the CD24low GFPhi cell fraction compared to GFPneg cells (2-, 2-, 2-, and 12-fold, respectively), GFPlow cells (five, seven, four, and fourfold, respectively), or AT2 cells (10-, 3-, 45-, and 140-fold, respectively) (Fig. 5C). Transcripts for Slc2a3 and Ces1g were enriched within total RNA from CD24low GFPhi cells; however, transcripts for Lamb3 and H60c in CD24low GFPhi cells could not be distinguished from AT2 cells (Fig. 5C).
Figure 5.
Novel genes that define region-specific progenitor cells. (A): Sorted GFPneg, GFPlow, and GFPhi progenitor cells are lyzed to extract RNA, which was then processed for Affymetrix Microarray analysis. The heatmap was made to show distinct gene signatures of these progenitor cells. (B): Venn diagram showing differentially expressed genes in GFPhi, GFPlow, and GFPneg progenitor cells with a 1.2-fold as a cutoff using the GeneSpring GX software. (C): Quantitative PCR confirmed that Reg3g, Parm1, and Pard6g are highly enriched in GFPneg, GFPlow, and GFPhi airway progenitor cells. Data are presented as mRNA abundance relative to total lung RNA. Data show the average of three samples, representing two independent experiments. The spatial distribution of selected mRNA’s was determined by in situ hybridization of cRNA probes to normal adult mouse lung tissue. (D): An adjacent lung tissue section was stained with H&E to reveal the branching pattern of conducting airways. (E–H): The spatial expression pattern was determined for mRNA’s corresponding to Scgb1a1 (positive control), Reg3g, Parm1, and Pard6g. Abbreviation: GFP, green fluorescent protein.
We used in situ hybridization to determine the spatial distribution of genes that define each cell subset within the intact lung. Antisense DIG-labeled probes to Scgb1a1 (pan-airway progenitors), Reg3g (putative proximal airway progenitors), Parm1, and Pard6g (putative distal conducting airway progenitors) were hybridized to lung tissue sections and their localization determined by histochemistry (Fig. 5D–5H). Sense DIG-labeled probes were synthesized and used in parallel hybridizations as negative controls. Expression patterns for each transcript were compared to that obtained for localization of Scgb1a1 mRNA. In situ localization of Scgb1a1 mRNA revealed abundant populations of epithelial cells that localize to both proximal and distal conducting airways (Fig. 5E). Reg3g mRNA was restricted to abundant population of epithelial cells of proximal airways (Fig. 5F). This distribution was broader than that of rare intralobar Krt5 basal cells (Fig. 4X) or pulmonary neuroendocrine cells (not shown), and was not observed within isolated ciliated cells (Supporting Information Fig. S5), suggesting that its expression defined the nonciliated progenitor cell population of proximal airways. In contrast, Parm1 and Pard6g were each undetectable within the epithelium of proximal airways but showed overlapping patterns of mRNA localization within distal conducting airways (Fig. 5G, 5H). Neither Parm1 nor Pard6g were present at detectable levels within alveolar epithelial cells. These data demonstrate that both GFPlow and GFPhi cell fractions harbor cell types that reside in the distal conducting airway. We used qRT-PCR to demonstrate that enriched CD24hi ciliated cells lack expression of either Parm1 or Pard6g (Supporting Information Fig. S6). These data demonstrate that nonciliated epithelial progenitor cells of terminal airways can be defined based upon their expression of Parm1 or Pard6g.
Response of Region-Specific Progenitor Cells to Bleomycin-Induced Injury
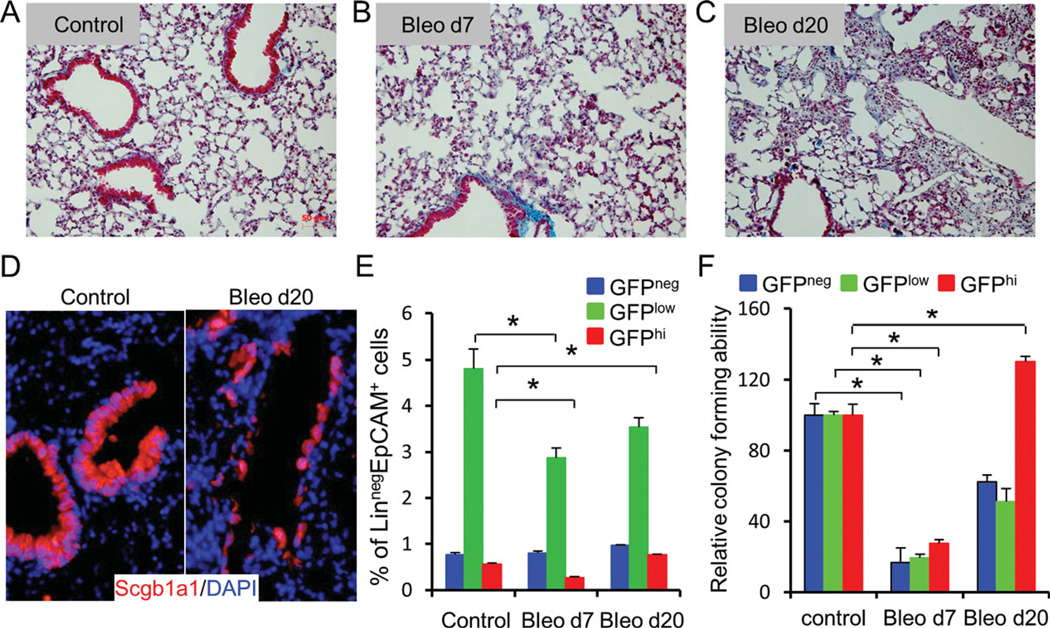
Progenitor cell function may be compromised in the setting of lung injury through either direct effects or by changes to their surrounding microenvironment. The impact of local tissue remodeling on gene expression and function among the newly identified populations of epithelial progenitor cells was evaluated in a bleomycin model of lung fibrosis. Bleomycin exposure resulted in remodeling of distal conducting airway and alveolar compartments (Fig. 6A–6C). This was associated with changes in the morphology and in the level of Scgb1a1 immunoreactivity within epithelial cells of terminal bronchioles at the 20-day recovery time point (Fig. 6D). GFPneg, GFPlow, and CD24low GFPhi cells were sorted from SFTPC-GFP mice at 7 days and 20 days after intratracheal instillation of bleomycin. No significant changes were observed in the abundance of GFPneg cells within the total Linneg EpCAMpos population (Fig. 6E). However, both distal airway epithelial cell fractions showed early decreases in cell number relative to the total Linneg EpCAMpos population (Fig. 6E). In contrast to the GFPlow fraction, the abundance of cells in the CD24low GFPhi fraction recovered at a greater rate from day 7 to day 20 following bleomycin exposure (Fig. 6E). CD24neg GFPhi AT2 cells showed a similar decline in abundance to CD24low GFPhi cells 7 days after bleomycin exposure but showed no evidence of recovery by the 20-day recovery time point (not shown).
Figure 6.
Response of region-specific progenitor cells to bleomycin injury. (A–C): Trichrome stain of lung sections from control SFTPC-GFP mice and those treated with bleomycin. (D): Immunostain of lung sections from control or bleomycin-treated group (day 20) with Scgb1a1. (E): Percentage of GFPneg, GFPlow, and GFPhi fraction relative to LinnegEpCAM+ population. (F): After bleomycin injury, colony-forming efficiency (CFE) of GFPneg, GFPlow, and GFPhi progenitor cells was determined. CFEs are shown relative to the control group. Data represent average 6 SEM from three mice in control mice or bleomycin-treated mice. Abbreviations: DAPI, 4′-6-diamidino-2-phenylindole; GFP, green fluorescent protein.
To evaluate progenitor cell function, GFPneg, GFPlow, and CD24low GFPhi cells of control and bleomycin-exposed SFTPC-GFP transgenic mice were placed in 3D culture (Fig. 6F). Colony-forming efficiency of progenitor cells within each cell fraction was reduced by 70%–80% compared to untreated controls 7 days after bleomycin exposure (Fig. 6F). Twenty days after bleomycin exposure, the colony-forming efficiency of progenitor cells contained within the GFPneg and GFPlow fractions increased compared to 7 days but remained significantly lower than that of uninjured controls (Fig. 6F). In contrast, despite an initial decline in the colony-forming efficiency of progenitor cells within the CD24low GFPhi population, colony-forming efficiency was 30% higher than the uninjured control group by the 20-day recovery time point (Fig. 6F).
Conclusion
Our results demonstrate that epithelial progenitor cells residing at different airway locations show intrinsic differences in their clonal behaviors and differentiation potentials. These findings support a model in which local hierarchies of epithelial progenitor cells yield regionally appropriate specialized cells for maintenance of epithelial diversity along the proximodistal airway axis. We define novel marker genes that distinguish these progenitor cell subtypes allowing further analysis of their individual contributions to tissue maintenance and remodeling.
Discussion
Our findings suggest that the airway epithelium is maintained during homeostasis and repair by distinct populations of progenitor cells that can be distinguished based on their regionof- origin and unique gene expression signatures. Proximal airway progenitor cells have the broadest differentiation potential, yielding three specialized cell types in culture that included luminal secretory and ciliated cells and subluminal basal cells. Two types of distal airway progenitor cells were identified; one with capacity for bipotential differentiation to yield luminal ciliated and secretory cells, and a second progenitor cell type with a high capacity for self-renewal but limited in vitro differentiation potential. Unlike proximal airway progenitor cells, both distal airway progenitor cell types yielded Sftpc-expressing progeny. Our findings demonstrate that airway progenitor cells retain elements of their positional identity following isolation and during in vitro culture, suggesting that they are programmed to yield specialized functional cell types reflective of their region-of-origin.
Analyses of either somatic chimeras or mice with inducible genetic lineage tags suggest that endogenous epithelial progenitor cells play critical roles in normal tissue maintenance [13, 15, 16, 20]. These and related studies [11, 12] also show that Scgb1a1-expressing cells include chemically resistant progenitor cells that participate in epithelial renewal after injury (Fig. 1). We now show that the Scgb1a1-expressing pool of epithelial progenitor cells is heterogeneous in its cellular composition and that this heterogeneity is fundamental to the preservation of functionally distinct airway zones that reside along the proximal-to-distal axis of the airway. Our observation that adult epithelial progenitor cells are intrinsically programmed to assume defined cellular fates is consistent with results investigating fate determination in the developing embryonic lung. Shannon et al. used epithelial-mesenchymal recombinants to demonstrate that the developmental fate of lung endoderm is dynamically regulated through both paracrine and cell intrinsic mechanisms [6, 26, 27]. Their study demonstrated that the fate of early lung endoderm is regulated by mesenchymally derived factors, yet endoderm isolated from late embryonic lung acquired the intrinsic capacity to assume region-specific fates independently of the influence of surrounding mesenchyme [27].
Our finding that progenitor cells yielding morphologically distinct colony types reside within different anatomic regions of the airway supports a model of airway maintenance by regional progenitor cells rather than a common progenitor cell hierarchy. However, McQualter et al. did provide evidence for local hierarchies of progenitor cells involved in maintenance of distal airways [18]. In their study, they showed that lung epithelial progenitor cells could yield multiple colony types in vitro and presented evidence that one of these colony types, described as mixed bronchiolar/alveolar, may represent a multipotent progenitor for distal bronchioles and alveoli; similar to the previously described bronchioalveolar stem cell [28]. This model can be applied to the two distal bronchiolar progenitor cell types identified in our study, those progenitors that yielded type B and type C colonies, due to their overlapping patterns of spatial localization. Colonies generated by the GFPlow and CD24low GFPhi cell fractions closely resemble the two colonies, respectively, described by McQualter et al. [18]. However, both of these progenitors express Scgb1a1 and also generate Sox2-expressing epithelial colonies in vitro suggestive of a conducting airway rather than alveolar phenotype. Our identification of Parm1 and Pard6g as new molecular markers that distinguish the GFPlow and CD24low GFPhi progenitor cell subsets will allow the creation of new tools, such as fluorescent protein reporter lines or CreER lineage tracing systems, to better define their contribution toward normal epithelial maintenance and remodeling of the postnatal airway. Proximal conducting airways are also maintained by more than one type of epithelial progenitor; nonciliated luminal and basal cells [14, 15, 29]. Our data suggest that both populations of proximal airway progenitor are present within the GFPneg cell fraction and most likely contribute to the development of type A colonies in 3D culture. In light of the rarity of basal cells within intralobar airways of the mouse lung, our finding that Reg3g expression defines an abundant population of nonciliated cells within proximal intralobar airways suggests that this marker defines the nonciliated luminal progenitor. Further studies using expression of Reg3g as a tool to identify this progenitor cell population are needed to fully characterize their contribution to maintenance and repair of proximal airway epithelium.
The molecular basis for functional differences between progenitor cells in the lung is not known. We show that levels of Sox2 expression vary between epithelial progenitor cell types of adult conducting airways and between the daughter cells generated through in vitro clonal expansion of different subsets of these epithelial progenitor cells. Sox2 is a transcription factor that maintains the pluripotency of embryonic stem cells and is an important regulator of organ development [4, 30]. Its expression has been localized to adult tissue stem cells found in a number of adult tissues including the stomach, cervix, anus, testes, lens, and multiple glands, where it fulfills important roles in adult tissue regeneration and stem cell maintenance [31]. Conditional loss of Sox2 function in either early or late embryonic lung endoderm significantly impacts renewal and differentiation of developmental progenitor cells [5, 32]. However, functional roles for Sox2 in the regulation of adult lung progenitor cells have yet to be elucidated. Interestingly, dose-dependent effects of Sox2 have been observed on the differentiation and renewal potentials of retinal neural progenitor cells [33] and amplification of the Sox2 locus is a common finding in aggressive colon and lung cancers [34– 36]. The potential regulatory influence of different levels of Sox2 expression is further suggested by the observation of ectopic airway epithelium, including K14-expressing basal cells, in alveoli following forced expression of either Sox2 or the related family member, Sox17 [5, 37]. We found that bleomycin exposure results in the selective depletion of epithelial cell types present within the distal lung cell fractions and acute loss of colony-forming ability by remaining progenitor cells within each of the three cell fractions. The combined loss of epithelial cells and colony-forming ability in distal airways may account for the preferential effect of bleomycin on distal lung tissue remodeling. Furthermore, recovery postbleomycin exposure was associated with the preferential expansion of progenitors that yield distal airway Sox2low type C colonies. These progenitor cells were unique in their inability to form a well-defined lumen and the relatively undifferentiated phenotype of epithelial cells generated by their clonal expansion. These findings highlight differences between regional airway progenitor cells in their response to bleomycin-induced lung injury and tissue remodeling. Additional work is needed to understand how Sox2 regulates adult progenitor cells and how CD24low Sox2low epithelial progenitors contribute to tissue remodeling after bleomycin exposure.
Pathological changes in the cellular composition and physiological functions of airway epithelium represent general features of a number of lung diseases including asthma, chronic obstructive pulmonary disease, cancer, and cystic fibrosis as well as pulmonary fibrosis [38]. Novel markers identified for epithelial progenitor cells in this study will enhance our efforts to understand the biology of region-specific epithelial stem/progenitor cells in such pulmonary diseases and ultimately help in developing more specific and effective cellular, molecular, and genetic therapeutic interventions.
Acknowledgments
We are grateful to Lixia Luo, Karen Terry, Jeffrey Drake, and Ningshan Liu for technical assistance. This work was supported by NHLBI Grants 1U01HL099997-01 (BRS and CFK), RO1 HL090136, U01 HL100402 (CFK), and HL064888, HL090146, and HL089141 (BRS).
Footnotes
Author contributions: C.H., M.K., and S.B.R.: conception and design; S.B.R.: financial support; S.B.R.: provision of study material or patients; C.H., M.K., B.B.L., L.J., and S.B.R.: collection and/or assembly of data; C.H., M.K., R.C.R., B.B.L., K.C.F., L.J.H., and S.B.R.: data analysis and interpretation; C.H., M.K., R.C.R., J.D., N.P.W., R.S.H., K.C.F., and S.B.R.: manuscript writing; S.B.R.: final approval of manuscript.
H.C. and K.M. contributed equally to this article.
Disclosure Of Potential Conflicts Of Interest
The authors indicate no potential conflicts of interest.
See www.StemCells.com for supporting information available online.
References
- 1.Snyder JC, Teisanu RM, Stripp BR. Endogenous lung stem cells and contribution to disease. J Pathol. 2009;217:254–264. doi: 10.1002/path.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proud D, Leigh R. Epithelial cells and airway diseases. Immunol Rev. 242:186–204. doi: 10.1111/j.1600-065X.2011.01033.x. [DOI] [PubMed] [Google Scholar]
- 3.Noble PW, Jiang D. Matrix regulation of lung injury, inflammation, and repair: The role of innate immunity. Proc Am Thorac Soc. 2006;3:401–404. doi: 10.1513/pats.200604-097AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato M, Keshavjee S. Bronchiolitis obliterans syndrome: Alloimmune-dependent and -independent injury with aberrant tissue remodeling. Semin Thorac Cardiovasc Surg. 2008;20:173–182. doi: 10.1053/j.semtcvs.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Que J, Okubo T, Goldenring JR, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon JM. Induction of alveolar type II cell differentiation in fetal tracheal epithelium by grafted distal lung mesenchyme. Dev Biol. 1994;166:600–614. doi: 10.1006/dbio.1994.1340. [DOI] [PubMed] [Google Scholar]
- 7.Warburton D, Schwarz M, Tefft D, et al. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 8.Evans MJ, Cabral LJ, Stephens RJ, et al. Renewal of alveolar epithelium in the rat following exposure to NO2. Am J Pathol. 1973;70:175–198. [PMC free article] [PubMed] [Google Scholar]
- 9.Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab Invest. 1974;30:35–42. [PubMed] [Google Scholar]
- 10.Chapman HA, Li X, Alexander JP, et al. Integrin a6b4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol. 2002;161:173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong KU, Reynolds SD, Giangreco A, et al. Clara cell secretory protein- expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Respir Cell Mol Biol. 2001;24:671–681. doi: 10.1165/ajrcmb.24.6.4498. [DOI] [PubMed] [Google Scholar]
- 13.Rawlins EL, Okubo T, Xue Y, et al. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong KU, Reynolds SD, Watkins S, et al. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giangreco A, Arwert EN, Rosewell IR, et al. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA. 2009;106:9286–9291. doi: 10.1073/pnas.0900668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teisanu RM, Chen H, Matsumoto K, et al. Functional analysis of two distinct bronchiolar progenitors during lung injury and repair. Am J Respir Cell Mol Biol. 2011;44:794–803. doi: 10.1165/rcmb.2010-0098OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McQualter JL, Yuen K, Williams B, et al. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds SD, Giangreco A, Power JH, et al. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol. 2000;156:269–278. doi: 10.1016/S0002-9440(10)64727-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong KU, Reynolds SD, Watkins S, et al. In vivo differentiation potential of tracheal basal cells: Evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2004;286:L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 21.Teisanu RM, Lagasse E, Whitesides JF, et al. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells. 2009;27:612–622. doi: 10.1634/stemcells.2008-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 23.Whitsett JA, Budden A, Hull WM, et al. Transforming growth factor-beta inhibits surfactant protein A expression in vitro. Biochim Biophys Acta. 1992;1123:257–262. doi: 10.1016/0005-2760(92)90004-f. [DOI] [PubMed] [Google Scholar]
- 24.Evans CM, Williams OW, Tuvim MJ, et al. Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol. 2004;31:382–394. doi: 10.1165/rcmb.2004-0060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmai C, Sutherland RE, Kim KK, et al. Alveolar epithelial cells express mesenchymal proteins in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2011;301:L71–L78. doi: 10.1152/ajplung.00212.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon JM, Gebb SA, Nielsen LD. Induction of alveolar type II cell differentiation in embryonic tracheal epithelium in mesenchyme-free culture. Development. 1999;126:1675–1688. doi: 10.1242/dev.126.8.1675. [DOI] [PubMed] [Google Scholar]
- 27.Shannon JM, Nielsen LD, Gebb SA, et al. Mesenchyme specifies epithelial differentiation in reciprocal recombinants of embryonic lung and trachea. Dev Dyn. 1998;212:482–494. doi: 10.1002/(SICI)1097-0177(199808)212:4<482::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 28.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 29.Evans MJ, Shami SG, Cabral-Anderson LJ, et al. Role of nonciliated cells in renewal of the bronchial epithelium of rats exposed to NO2. Am J Pathol. 1986;123:126–133. [PMC free article] [PubMed] [Google Scholar]
- 30.Dejima K, Randell SH, Stutts MJ, et al. Potential role of abnormal ion transport in the pathogenesis of chronic sinusitis. Arch Otolaryngol Head Neck Surg. 2006;132:1352–1362. doi: 10.1001/archotol.132.12.1352. [DOI] [PubMed] [Google Scholar]
- 31.Randell SH, Boucher RC. Effective mucus clearance is essential for respiratory health. Am J Respir Cell Mol Biol. 2006;35:20–28. doi: 10.1165/rcmb.2006-0082SF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tompkins DH, Besnard V, Lange AW, et al. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS One. 2009;4:e8248. doi: 10.1371/journal.pone.0008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taranova OV, Magness ST, Fagan BM, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donohue JF, Ohar JA. Effects of corticosteroids on lung function in asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2004;1:152–160. doi: 10.1513/pats.200402-003MS. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y, Futtner C, Rock JR, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warheit DB, Salley SO, Barnhart MI. Thrombin-stimulated effects on megakaryocytopoiesis and pulmonary-platelet interactions. Gen Physiol Biophys. 1989;8:611–631. [PubMed] [Google Scholar]
- 37.Warheit DB, Kelly DP, Carakostas MC, et al. A 90-day inhalation toxicity study with benomyl in rats. Fundam Appl Toxicol. 1989;12:333–345. doi: 10.1016/0272-0590(89)90050-x. [DOI] [PubMed] [Google Scholar]
- 38.Rock JR, Hogan BLM. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu Rev Cell Dev Biol. 2011;27:493–512. doi: 10.1146/annurev-cellbio-100109-104040. [DOI] [PubMed] [Google Scholar]