SUMMARY
CD95 (Fas/APO-1), when bound by its cognate ligand CD95L, induces cells to die by apoptosis. We now show that elimination of CD95 or CD95L results in a form of cell death that is independent of caspase-8, RIPK1/MLKL, and p53, is not inhibited by Bcl-xL expression, and preferentially affects cancer cells. All tumors that formed in mouse models of low-grade serous ovarian cancer or chemically induced liver cancer with tissue specific deletion of CD95 still expressed CD95, suggesting that cancer cannot form in the absence of CD95. Death induced by CD95R/L elimination (DICE) is characterized by an increase in cell size and production of mitochondrial ROS, and DNA damage. It resembles a necrotic form of mitotic catastrophe. No single drug was found to completely block this form of cell death, and it could also not be blocked by the knockdown of a single gene, making it a promising new way to kill cancer cells.
INTRODUCTION
CD95 (Fas/APO-1) is an apoptosis-inducing death receptor (Peter and Krammer, 2003). However, CD95 also plays an apoptosis-independent role in nonimmune cells and it has been implicated in cancer cell growth, migration, and tumor progression (Martin-Villalba et al., 2013; Peter et al., 2007). We previously showed that knockdown of either CD95 or CD95L in multiple cancer cells led to growth reduction (Chen et al., 2010). We also reported reduced tumor load in mouse models of liver cancer and endometrioid ovarian cancer, both with tissue specific deletion of CD95 (Chen et al., 2010).
We now show that the CD95/CD95L system is critical for cancer cell survival with normal cells being less dependent. When either gene was knocked down in a sustained fashion, cancer cells showed substantial death induced by CD95R/L elimination (DICE). An analysis of 12 independently performed genome-scale arrayed shRNA screens identified CD95L as one of 651 essential survival genes. Tumor nodules that grew out of two mouse models of ovarian and liver cancer with tissue specific CD95 deletion still expressed CD95, suggesting a strong selection pressure for cancer cells to maintain CD95 expression.
DICE is characterized by cell swelling and ROS production followed by DNA damage, activation of caspases, and loss of mitochondrial outer membrane potential (MOMP). Cells die by a necrotic form of mitotic catastrophe. We performed a small molecule screen and a genome-wide shRNA screen, but could not find a single drug or a single gene that could either promote or block DICE. Our data suggests that DICE represents multiple death pathways, which indicates that cancer cells may not be able to acquire resistance to DICE by mutations of single genes. This makes DICE an attractive new way to kill cancer cells.
RESULTS
Efficient and Sustained Reduction of CD95 or CD95L Expression Drives Cancer Cells into Cell Death
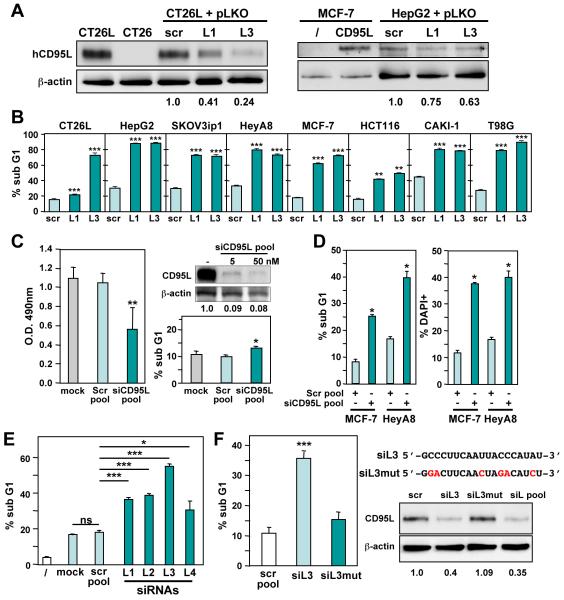
Knockdown of either CD95 or CD95L by introducing either siRNAs or lentiviral shRNAs in various cancer cells caused reduction in growth within 3-5 days (Chen et al., 2010). We now asked whether a profound and sustained knockdown of CD95 or CD95L would cause the cells to die. Two independent CD95L specific shRNAs (L1 and L3) knocked down CD95L as shown for the mouse colon carcinoma cell line CT26 stably expressing human CD95L (CT26L) (Aoki et al., 2001) and the human hepatocellular carcinoma cell line HepG2 (Figure 1A). Paralleling the knockdown efficiency of the different shRNAs, we observed substantial numbers of dead cells in these cell lines and multiple other cancer cell lines representing ovarian, breast, renal, and colon cancer as well as glioblastoma (Figure 1B and Figure S1A). Cell death was quantified nine days after infection with the virus. Knockdown of CD95 using two independent shRNAs also caused induction of cell death in a number of cancer cell lines (Figure S1B, Figure 2, and Table S1). To exclude the possibility of a puromycin effect, we infected T98G and HeyA8 cells either with a nontargeting shRNA lentivirus (pLKO-scr), with L1 or L3, or with the CD95 targeting shRNA R6 in the absence of puromycin (Figure S1C). This resulted in severely reduced growth followed by cell death induction peaking at around 7 days post-infection (Figure S1C). This data suggested that cancer cell lines start dying days after CD95L or CD95 knockdown.
Figure 1. Sustained Knockdown of CD95L Induces Cell Death.
(A) Western blot analysis to monitor the expression of human CD95L and β-actin in cells infected with pLKO scrambled (scr) control, pLKO-L1, or pLKO-L3 after 4 days (CT26L) or 5 days (HepG2, MCF-7-vec, and MCF-7 expressing CD95L). Level of knockdown relative to β-actin is shown.
(B) Viability of 8 cell lines 9 days after infection with shRNA lentiviruses pLKO-L1, L3, or a control virus carrying a scrambled shRNA (scr).
(C) Left: MTS assays of MCF-7 cells 3 days after mock treatment or transfection with 5 nM of either a pool of 4 nontargeting siRNAs (Scr pool) or a pool 4 siRNAs targeting CD95L (siCD95L pool). Right bottom: Nuclear PI staining of MCF-7 cells 4 days after transfection with either Scr or siCD95L pool. Right top: Western blot analysis of CD95L in MCF-7-CD95L cells 48 hrs after transfection with 5 or 50 nM siCD95L.
(D) Quantification of cell death with nuclear PI staining and DAPI staining in MCF-7 and HeyA8 cells transfected three times (3 days, 2 days, 2 days) with either a pool of 4 scrambled oligonucleotides (scr) or a CD95L specific siRNA SmartPool of 4 siRNAs.
(E) MCF-7 cells were transfected with either scr pool or with the 4 individual SmartPool siRNAs siL1-siL4. Transfection was carried out as in (D), and cell death was quantified using nuclear PI staining.
(F) Left: Quantification of cell death in MCF-7 cells transfected with either unmodified siL3 or the same oligonucleotide with 6 single nucleotide replacements (siL3mut). Right: sequences of siL3 and siL3mut and Western blot analysis of MCF-7-CD95L 2 days after transfection with either a scr pool, one of the two oligonucleotides, or the siCD95L SmartPool (siL pool). Level of knockdown relative to actin is shown. P-values were calculated using t-test. * = p<0.01; ** = p<0.001; *** = p<0.0001. ns = not significant.
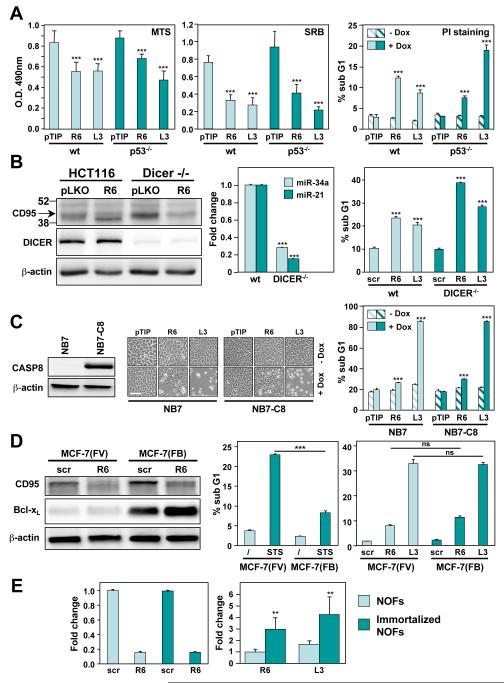
Figure 2. DICE Affects Cancer Cells in a Manner Independent of Common Oncogenic Lesions.
(A) Quantification of growth inhibition (MTS and SRB assay) and cell death (nuclear PI staining) of HCT116 parental and HCT116 p53−/− cells infected with inducible shRNA constructs against CD95 (R6) or CD95L (L3) after incubation with or without Dox for 5 days. Values represent mean ± s.d. from three independent experiments. P-values were calculated using t-test.
(B) Left: Western blot analysis for CD95, DICER, and actin in HCT116 parental and HCT116 DICER−/− cells 5 days after infection with the pLKO-R6 or pLKO-L3 constructs. Center: real time PCR quantification of miR-34a and miR-21 in HCT116 parental and HCT116 DICER−/− cells. Right: Quantification of DNA degradation in HCT116 parental and HCT116 DICER−/− cells after infection with pLKO-R6 or pLKO-L3 for 10 days. P-values were calculated by ANOVA.
(C) NB7 parental and NB7-caspase-8 cells were infected with an inducible shRNA construct against CD95 (R6), CD95L (L3), or control virus (pTIP). Western blot analysis of caspase-8 (left), phase contrast image (middle), and MTS assay (right) were performed 3 days after Dox treatment. P-values were calculated using t-test. Scale bar = 100 μm.
(D) Left: Western blot analysis for CD95 and Bcl-xL in MCF-7 Fas-vector (FV) and MCF-7 Fas-Bcl-xL (FB) cells 5 days after infection with pLKO-scr, pLKO-R6, or pLKO-L3. Center: Quantification of DNA degradation in MCF-7(FV) and MCF-7(FB) cells after treatment with 1 μM staurosporine (STS) for 18 hours, or right: after infection with pLKO-scr, pLKO-R6, or pLKO-L3 for 10 days. P-values were calculated by ANOVA.
(E) Normal ovarian fibroblasts (NOFs) or immortalized NOFs were infected with either pLKO-scr, pLKO-R6 or pLKO-L3. Left: Real-time PCR analysis of CD95 mRNA in the cells 7 days after infection. Right: Cell death quantified by Trypan blue staining 7 days after infection (including 2 days of puromycin selection). Shown is the fold change relative to the cells infected with pLKO-scr. P-value was calculated using t-test.
** = p<0.001; *** = p<0.0001; ns = not significant.
To exclude a contribution of lentiviral gene products to the observed cell death, we tested whether targeting CD95L with siRNAs would also induce cell death in cancer cells. Transfecting MCF-7 cells once with a low amount of CD95L targeting siRNA SmartPool (5 nM) caused a significant reduction of CD95L protein resulting in about 40% growth inhibition (as assessed by MTS assay) after 3 days with little cell death detected even after 4 days of transfection (as assessed by nuclear PI staining) (Figure 1C), suggesting different thresholds for growth inhibition and cell death induction. To achieve a sustained knockdown, we serially transfected MCF-7 or HeyA8 cells three times with either a CD95L specific siRNA SmartPool, or a control pool of four nontargeting (scr) siRNAs. Both MCF-7 and HeyA8 cells responded to the CD95L knockdown with substantial cell death (Figure 1D).
A widely used control to establish specificity of knockdown is to test more than one siRNA/shRNA. A summary of all si/shRNAs tested in this study and their target sites in either the CD95L or CD95 mRNA is shown in Figure S2A. In case of CD95L a total of 12 si/shRNAs targeting nonoverlapping sequences across the entire CD95L mRNA induced cell death in multiple cell lines at levels up to 100%. We tested the 4 siRNAs that were part of the CD95L specific SmartPool individually in MCF-7 cells (Figure 1E). Each of the 4 CD95L specific siRNAs significantly induced cell death. The most active siRNA was siL3. In order to further establish specificity of the CD95L targeting, we mutated this oligonucleotide in 6 positions (Figure 1F). This reduced the ability to induce cell death by >80%. While wt siL3 significantly knocked down CD95L in MCF-7-CD95L cells, the mutated version did not, supporting the activity of siL3 to induce cell death (Figure 1F). These data confirm that in order to kill cancer cells by eliminating CD95L, knockdown with either shRNAs or siRNAs needs to be both efficient and sustained.
Arrayed shRNA Lethality Screens Identify CD95L and CD95 as Genes Critical for the Survival of Cancer Cells
We collaborated with the HTS Core Facility at Memorial Sloan-Kettering Cancer Center (MSKCC) to investigate whether CD95 and CD95L were ever nominated as lethality gene candidates in their screens, as they are among the very few groups in the world performing arrayed shRNA hairpin screening against the TRC1 library of 80,598 shRNAs targeting 16,039 human genes (Bhinder et al., 2013). Gene nomination was very stringent and required at least 3 active hairpins out of the total of 5 hairpins (i.e. an H score of at least 60) (Bhinder and Djaballah, 2012). At this level of stringency, CD95L was detected as a survival gene in multiple cancer cell line screens (Figure S2B). 4 out of 5 shRNAs targeting different regions of the CD95L mRNA killed at least 9 of the 12 cancer cell lines tested. Knockdown of CD95 was also effective, killing 7 of the 12 cancer cell lines. The CD95L shRNA was found to have an average H-score of 86. CD95L was identified as one of 651 genes (4.1% of all tested genes) to be critical for the survival of 75% of the cancer cell lines (data not shown). Many of the survival genes that were identified were predicted due to their critical role in cell cycle/cell division (Table S2). The combined data indicated that cancer cell dependency on CD95L/CD95 for survival results in substantial cell death when either CD95 or CD95L are eliminated. For simplicity, we refer to cell death resulting from loss of CD95 or CD95L as DICE (Death Induced by CD95R/L Elimination).
DICE Induction Is not Affected by Common Oncogenic Lesions
To induce DICE in a more controlled fashion, we used a Tet-inducible lentiviral vector expressing GFP (pTIG) and also modified it to express a puromycin resistance gene instead of GFP (pTIP). Both vectors include cassettes for expression of the CD95 specific shRNA R6 (i.e. pTIP-R6) or the CD95L specific shRNAs L1 or L3 (i.e. pTIP-L1 or pTIP-L3). Addition of the tetracycline analog doxycycline (Dox) induced efficient knockdown of CD95 or CD95L in different cell lines (Figure S3A, S3B, and S3C). In most cell lines, partial knockdown of either CD95 or CD95L was detectable 1 day after addition of Dox, reaching a maximum after 4 days (Figure S3A, S3B, S3C, and data not shown). Similar to the cells infected with CD95 and CD95L specific lentiviral shRNAs, Dox-induced knockdown of CD95 or CD95L in multiple cell lines caused substantial cell death as evidenced by the appearance of rounded up, detached cells and DNA fragmentation as well as reduced viability as measured by MTS or SRB assays (Figures S3D-G, and data not shown) with no cell death in Dox treated cells expressing a pTIP control vector (Figure S3E, S3F and Figure 2A and 2C) or pTIP-scrambled (data not shown).
To test whether DICE can be effective in cancer cells carrying common oncogenic lesions, we induced DICE in cells lacking expression of p53, DICER, or caspase-8 or overexpressing Bcl-xL. First, we infected parental HCT116 and HCT116 p53−/− cells with the pTIP-R6 or the pTIP-L3 virus. Addition of Dox induced cell death in both cell lines within 3 days, monitored in three different ways (Figure 2A), consistent with the fact that DICE could be induced in 15 tested cell lines irrespective of their p53 status (Table S1). Cancer cells often delete or downregulate the miRNA processing enzyme DICER resulting in globally reduced expression of miRNAs (Zhang et al., 2006). DICE was effectively induced by both knockdown of CD95 or CD95L in DICER deficient HCT116 cells (Figure 2B). Interestingly, DICER deficiency did not prevent processing of the expressed shRNAs as evidenced by the efficient knockdown of endogenous CD95 in DICER deficient HCT116 cells yet two expressed miRNAs (miR-21 and miR-34a) were downregulated. To test the effect of loss of caspase-8 on DICE induction, we generated NB7 cells (a neuroblastoma cell line that carries a deletion of both CASP8 alleles (Teitz et al., 2000)) inducibly expressing shR6 or shL3. Knockdown of CD95L resulted in the same level of dead cells in parental NB7 cells and NB7 cells reconstituted with full length caspase-8 (Figure 2C).
After having tested the role of three tumor suppressors in the execution of DICE, we tested the effect of expression of Bcl-xL, which is frequently overexpressed in cancer. We used MCF-7(FB) cells expressing both exogenous CD95 and Bcl-xL (Figure 2D). While apoptosis induction by treatment with staurosporine (STS) was significantly inhibited by overexpressed Bcl-xL when compared to vector expressing cells (MCF-7(FV)) (Figure 2D) no reduction in DICE was observed 10 days after infection. These data suggest that DICE can be induced in cancer cells irrespective of the presence of a number of oncogenic lesions frequently found in human cancers, which raises the question of whether it affects normal cells. Isolated primary human ovarian fibroblasts (NOFs) were much more resistant to DICE induced by knockdown of either CD95 or CD95L when compared to NOFs immortalized by expressing human hTERT (Figure 2E) suggesting that DICE preferentially affects cancer cells.
DICE Is Independent of Caspases, RIPK1, and MLKL.
To facilitate analysis of the cell death pathway(s) that underlie DICE, we generated a panel of single cell clones from CT26L-pTIP-L3 cells by limited dilution cloning that died completely when CD95L was knocked down (Figure S4A-C and data not shown). Cells of clone 18 (CT26L-pTIP-L3/18) died 5 days after addition of Dox (Figure S4C and Movie S1). In contrast, death of clone 22 cells (CT26L-pTIP-L3/22) was delayed (Figure S4C), consistent with the delayed knockdown of CD95L after addition of Dox in these cells (Figure S4B).
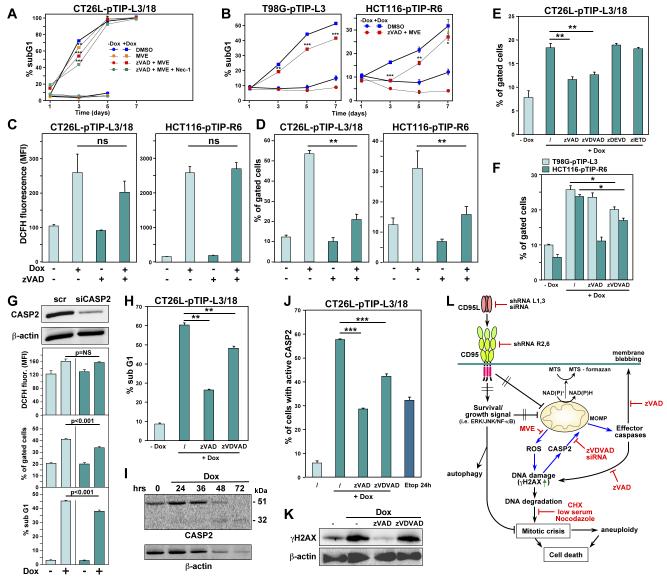
When CT26L-pTIP-L3/18 cells were studied by time-lapse video microscopy after addition of Dox, clear signs of membrane blebbing were observed just before the cells died (Movie S2), suggesting that caspase activation contributed to DICE. To test whether cells died of canonical caspase-dependent apoptosis or necroptosis mediated by the RIP1 kinase activity, a number of cell lines including CT26L-pTIP-L3/18, CAKI-1-pTIP-R6, HCT116-pTIP-R6, and T98G-pTIG-L3 were pretreated with either the panspecific caspase inhibitor zVAD-fmk (zVAD), the RIP1 kinase inhibitor Necrostatin-1 (Nec-1), or both (Figure 3A and B). When CT26L-pTIP-L3/18 cells were observed in the presence of zVAD, none of the cells showed the typical apoptotic morphology (inset in Figure 3A, Figure S5A, and Movie S3). Neither Nec-1 alone nor Nec-1 in combination with zVAD-fmk reduced cell death in any of the cell lines. While treatment of cells other then CT26L cells with zVAD slowed down DICE (induced by either knockdown of CD95 or CD95L), it did not rescue cells from death (Figure 3C). The full activity of both zVAD and Nec-1 was established using appropriate cell death systems (Figure S5B and S5C). Using HT-29 cells recently described with knocked down MLKL, a critical component of necroptosis pathway (Zhao et al., 2012), we found that DICE does not depend on MLKL for its execution (Figure S5D). Canonical necroptosis induced in these cells by treatment with TNFα, a Smac mimetic, and zVAD was significantly blocked in the absence of MLKL. These data suggested that during DICE, although caspases were activated and caused membrane blebbing, cells eventually died by a mechanism independent of apoptosis or RIP1/MLKL-dependent necroptosis.
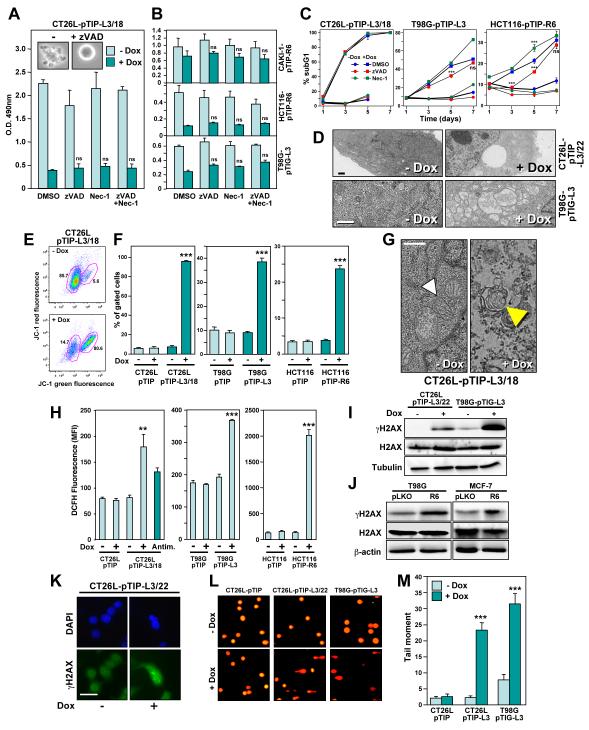
Figure 3. DICE Represents a Form of Necrosis Independent of RIP1 that Leads to Mitochondrial ROS Production and DNA Double Strand Breaks.
(A) CT26L-pTIP-L3/18 cells or (B) CAKI-1-pTIP-R6, HCT116-pTIP-R6, or T98G-pTIG-L3 cells were incubated with or without Dox after pretreatment with 20 μM zVAD-fmk, 20 μM Necrostatin-1, a combination of both inhibitors, or DMSO for 4 days (CT26L cells) or 5 days (other three cell lines). Cell survival was quantified by MTS assay.
(C) DNA fragmentation quantified in the indicated cells after treatment with Dox for up to 7 days in the presence of either zVAD-fmk (20 μM in CT26L and T98G, 10 μM in HCT116 cells) or Necrostatin-1 (20 μM in T98G, 10 μM in CT26L and HCT116 cells). The day 7 data points for CT26L cells are not shown because cells started to die due to overconfluency. P-values were calculated by ANOVA.
(D) Transmission electron micrographs of CT26L-pTIP-L3/22 and T98G-pTIG-L3 cells 4 days after incubation with or without Dox. Size bars, 1 μm.
(E) Representative FACS plot of CT26L-pTIP-L3/18 cells after incubation with and without Dox for 3 days after staining the cells with JC-1. Left gate: Intact cells, right gate: cells with loss of mitochondrial transmembrane membrane potential.
(F) Events (percent) in right gate of the indicated cell lines treated as in (E) for 3 days (CT26L) or 4 days (T98G, HCT116) (data are mean ± s.d.). P-values were calculated by ANOVA.
(G) Electron micrographs of CT26L-pTIP-L3/18 cells treated or not with Dox for 3 days. White arrow head, intact mitochondrion; yellow arrow head, onion shaped structure found instead of mitochondria. Size bar, 500 nm.
(H) Quantification of ROS production by DCFH fluorescence in cells treated with or without Dox for 2 days (CT26L), 4 days (T98G), 5 days (HCT116), or CT26L cells treated with antimycin for 24 hours. P-values were calculated by ANOVA.
(I, J) Western blot analysis for γH2AX and H2AX in CT26L-pTIP-L3/22 and T98G-pTIG-L3 cells, 2 days and 4 days after Dox treatment, respectively (I). T98G and MCF-7 cells were infected with pLKO control virus or pLKO-R6 (J), and Western blot analysis was performed after 5 days.
(K) Immunofluorescence microscopy of CT26L-pTIP-L3/22 cells treated or not with Dox for 2 days. Cells were fixed with paraformaldehyde and stained for γH2AX (green) and DAPI (blue). Scale bar = 100 μm.
(L) CT26L-pTIP, CT26L-pTIP-L3/22 (2 days Dox), and T98G-pTIG-L3 (4 days Dox) cells were subjected to an alkaline comet assay. Representative fluorescence photomicrographs of comet assay showing DNA migration pattern in single cell gel electrophoresis stained with SYBRGreen-1.
(M) Quantification of tail moment of cells in (L) expressed in arbitrary units. Values represent the average of all cells within a given sample population. P-values were calculated using t-test.
** = p<0.001; *** = p<0.0001; ns = not significant.
One of the characteristics of cells dying by DICE was that they displayed elongated odd shapes and increased in size (Figure S5E, S5F, and S5G). This cell size increase was cell autonomous because when HepG2 cells infected with the GFP expressing version of the inducible lentiviral vector pTIG-L3 were mixed 1:1 with cells infected with control virus, only the GFP positive cells showed an increase in cell size after addition of Dox (Figure S5H). An increase in cell size caused by influx of water is a classic sign of necrosis (Festjens et al., 2006). To determine the effect of DICE on subcellular structures, we subjected CT26L-pTIP-L3/22 and T98G-pTIG-L3 cells to transmission electron microscopy analysis 4 days after addition of Dox (Figure 3D). Cells showed partially condensed nuclei and contained large dilated vesicles, consistent with ongoing necrosis. Thus, while DICE has some features of apoptotic cell death, it is at its core a form of necrosis.
While the experiments with cells overexpressing Bcl-xL (Figure 2D) suggested that the proapoptotic activity of mitochondria is dispensable for the execution of DICE, mitochondria may become dysfunctional during DICE. Indeed, by using the dyes JC-1 (Figure 3E) and DiOC6 (data not shown), DICE in CT26L-pTIP-L3/18, T98G-pTIP-L3 and HCT116-pTIP-R6 cells was accompanied by a drop in mitochondrial transmembrane potential (Δψm), a measure of mitochondrial outer membrane permeabilization (MOMP). These data suggested that CD95 and CD95L are required to maintain mitochondrial integrity in cancer cells. This was confirmed by transmission electron microscopy. Figure 3G shows CT26L-pTIP-L3/18 cells 3 days after knockdown of CD95L. Functional mitochondria were visible in these cells prior to addition of Dox (white arrow head in Figure 3G), but were not detectable after addition of Dox. Instead, onion shaped membrane structures (yellow arrow head in Figure 3G), which were the remnants of mitochondria, were detected.
DICE Involves ROS Production and DNA Damage
Necrotic cell death is often accompanied by production of reactive oxygen species (ROS) (Festjens et al., 2006). Knockdown of CD95L in CT26L-pTIP-L3/18 or in T98G cells or knockdown of CD95 in HCT116 cells resulted in a significant increase in intracellular ROS (Figure 3H). Production of high amounts of ROS can cause DNA double strand breaks (DSBs), leading to DNA damage, resulting in cell death. Inducible knockdown of CD95L resulted in an increase in phosphorylated H2AX (γH2AX), a marker for DNA DSBs (Figure 3I). Knockdown of CD95 using shRNA lentiviruses expressing R6 also induced γH2AX (Figure 3J). In CT26L-pTIP-L3/22 cells undergoing DICE, γH2AX was detected as foci in the nucleus (Figure 3K). To confirm that DSBs had occurred, cells were subjected to a Comet assay. Both CT26L/22 and T98G cells undergoing DICE showed a pronounced increase in cells with degraded DNA (Figure 3L and 3M).
To determine whether the production of ROS during DICE was a key element that induces DSBs and death, cells were treated with mitochondrially targeted vitamin E (MVE). In CT26L cells, MVE significantly reduced ROS generated during DICE (Figure S6A), resulting in a strong reduction in γH2AX formation, which was accompanied by a substantial reduction in DNA fragmentation and a reduction in cell death as monitored by MTS assay and phase contrast microscopy. In T98G cells undergoing DICE, addition of MVE completely blocked ROS production and reduced γH2AX. This, however, had no effect on cell viability and only a minimal effect on DNA degradation (Figure S6B).
The Final Phase of DICE Resembles Mitotic Catastrophe
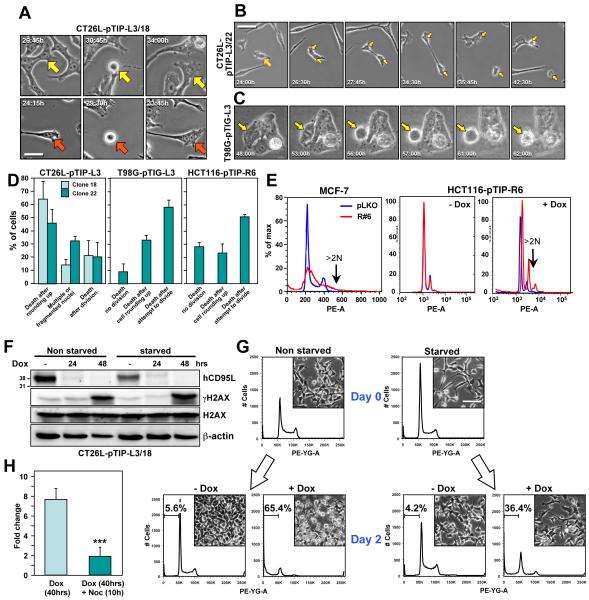
We sought to determine the consequences of DNA damage observed in cells that die by DICE. We performed single cell analyses of CT26L pTIP-L3 clones 18 and 22 using time-lapse video microscopy. Following addition of Dox, a significant proportion of cells attempted to divide, but failed, resulting in cells with multiple or fragmented nuclei (Figure 4A and 4D and Movie S4). Another fraction of cells rounded up and then either died immediately or died following cytokinesis (Figure 4B and Movie S5). When stained with DAPI, numerous cells with anaphase bridges, evidence of failed cytokinesis, were detected (Figure S7A). In T98G-pTIP-L3 and HCT116-pTIP-R6 cells, almost all cells that died had attempted to divide as evidenced by the appearance of a metaphase plate just before rounding up and dying (Figure 4C and 4D, Movie S6 and data not shown). When stained with DAPI, multiple cells were detected with massively fragmented nuclei (Figure S7B). In addition, PI staining and flow cytometry indicated that some of the cancer cells showed signs of aneuploidy after induction of DICE (Figure 4E). These data suggested that for cells to die by DICE, they have to enter mitosis. If true, then blocking entry into the cell cycle should prevent death. To test this directly, we arrested CT26L-pTIP-L3/18 cells in G0 by culturing them for two days in medium without FCS and then added Dox. Serum starvation did not prevent CD95L knockdown (Figure 4F); however, it resulted in a reduction in the percentage of dead cells from 65% to 36% (Figure 4G). Serum starvation did not, however, reduce the accumulation of DNA DSBs as indicated by the appearance of γH2AX (Figure 4F). Finally, we induced CD95L knockdown in CT26L-pTIP-L3/18 cells by addition of Dox and after 40 hours arrested cells in G2/M by treating them for 10 hours with the microtubule inhibitor nocodazole. This treatment also significantly reduced DICE (Figure 4H). Two additional compounds were found to slow down DICE presumably due to their activity to arrest cells in the cell cycle (Supplementary Results and Figure S7C-G).
Figure 4. DICE Occurs when Cells Enter Mitosis.
(A-C) Single shots from a time-lapse video photographic sequence of (A) CT26L-pTIP-L3/18, (B) CT26L-pTIP-L3/22, and (C) T98G-pTIG-L3 cells, after addition of Dox (see Movies S4-S6). Arrows in A indicate cells with multiple nuclei; arrow in B labels a dividing cell; arrow in C marks a cell attempting to divide. Scale bar = 100 μm.
(D) Quantification of cell behavior after addition of Dox to the indicated cells.
(E) FACS plots showing cell cycle profiles of MCF-7 cells 7 days after infection with pLKO or pLKO-R6 (left panel) or HCT116-pTIP-R6 treated or not with Dox for 5 days (middle and right panels).
(F) Western blot analysis of hCD95L, γH2AX, H2AX, and Actin in CT26L-pTIP-L3/18 cells after serum starvation (0.1% FCS) for 2 days, followed by treatment with Dox for the indicated times.
(G) FACS plots and representative micrographs of CT26L-pTIP-L3/18 cells cultured in serum (10% FCS) and under starvation conditions (0.1% FCS) for 2 days and then treated with Dox for an additional 2 days. Gate shows the sub G1 fraction. Size bar = 100 μm.
(H) Specific cell death (fold change in sub G1 peak) in CT26L-pTIP-L3/18 cells 40 hrs after addition of Dox when compared to the cells with or without treatment with 50 nM nocodazole during the last 10 hours of incubation. P-values were calculated by ANOVA. *** = p<0.0001. See also Figure S7, Movie S4, S5, and S6.
DICE Engages a Complex Cell Death Program that Is Difficult to Block
None of the tested inhibitors of apoptosis, necrosis, mitochondrial integrity, ROS, or their combination completely blocked DICE in any of the cell lines. In addition, a large number of compounds targeting various cancer relevant pathways were found to have no effect on the execution of DICE in CT26L-pTIP-L3/18 cells. Furthermore, four commonly used chemotherapeutic drugs also did not inhibit or promote the execution of DICE in either HCT116-pTIP-R6 or CAKI-1-pTIP-R6 cells after addition of Dox (data not shown, tested compounds listed in Table S3).
In an attempt to identify compounds that would block DICE, we performed a small molecule screen using the 1120 FDA approved compounds of the Preswick library (www.prestwickchemical.com/index.php?pa=26), but did not find a compound that prevented DICE at 10 μM (data not shown). To identify components of the DICE signaling pathway, we performed a rescue screen using 77,720 lentiviral based shRNAs targeting 16,143 unique genes. Since using the shRNA library involved a puromycin selection step, we generated a clone of CT26L-pTIG-L3 cells by first sorting for GFP positivity followed by limited dilution cloning. Similar to CT26L-pTIP-L3 or CT26L-pTIP-L3/22 cells, these cells also died quantitatively after addition of Dox (Figure S8A). Not a single clone grew out after repeated infection with the shRNA library, suggesting that knockdown of a single gene cannot rescue cells from DICE (data not shown). In order to identify genes that could at least modulate DICE, we combined the genome-wide shRNA screen with next generation sequencing (Figure S8B-S8D, Supplementary Results). As a result, 169 death attenuators (prosurvival genes) were identified as well as 907 death promoters (prodeath genes). Both groups of genes were subjected to a gene ontology analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Figure S8E). The 83 prosurvival genes contained a cluster of genes known to be negative regulators of cell death. Interestingly, the most highly enriched cluster of genes among the prosurvival genes were connected to glutathione metabolism, which is consistent with ROS being involved in the execution of DICE. In contrast, the 433 prodeath genes were enriched in genes known to be positive regulators of cell death. Consistent with our observation of DNA damaging stress in these cells, a highly enriched gene cluster among the prodeath genes was involved in the stress response and DNA repair. A number of these shRNAs that target genes that promote DICE were tested individually, but none of them blocked DICE (data not shown). This again suggested that DICE cannot be inhibited by targeting individual genes and likely involves activation of more than one death pathway.
Caspase-2 Contributes to the Execution of DICE
One of the inhibitors that slowed down DICE was the caspase inhibitor zVAD-fmk (Figure 3C). Even in CT26L-pTIP-L3/18 cells, zVAD-fmk reduced cell death early after DICE induction at around 40 hours (data not shown). When combined with the ROS inhibitor MVE, zVAD slowed down DICE in different cells at early stages (Figure 5A and B). Interestingly, by combining more of the inhibitors, early attenuation of DICE in CT26L-pTIP-L3/18 cells was enhanced stepwise (Figure 5A). This suggests that DICE comprises multiple subpathways that contribute to the initiation of DICE, and we sought to determine the order of these pathways. Treatment with zVAD-fmk alone did not significantly affect the production of ROS in CT26L-pTIP-L3/18 cells after knockdown of CD95L or in HCT116-pTIP-R6 cells after knockdown of CD95 (Figure 5C), suggesting that ROS production occurred upstream of activation of caspases in the DICE pathway. Interestingly, MOMP was dependent on the activation of caspases (Figure 5D). To determine which caspase regulated MOMP, a set of different caspase inhibitors was tested. Of the tested inhibitors, inhibitors of caspase-3/7 and caspase-8 had no effect (Figure 5E). In contrast, the caspase-2 inhibitor zVDVAD-fmk reduced MOMP in CT26L cells almost as effectively as zVAD-fmk. This inhibitor also reduced MOMP in T98G and HCT116 cells undergoing DICE (Figure 5F), and this finding was validated by knocking down caspase-2 in CT26L-pTIP-L3/18 cells (Figure 5G). Caspase-2 is known to be activated by DNA damage (Castedo et al., 2004). Consequently, inhibition of caspase-2 significantly reduced DNA degradation in cells undergoing DICE (Figure 5H), suggesting that caspase-2 is upstream of DNA degradation, but downstream of DNA damage (DNA DSBs). Activation of caspase-2 during DICE was detected by Western blotting (Figure 5I) and by a caspase-2 specific in situ activity assay (Figure 5J). To place caspase-2 activation in the DICE signaling pathway, we treated cells undergoing DICE with caspase inhibitors and monitored the formation of γH2AX (Figure 5K). Consistent with caspase-2 being downstream of DSBs, treatment of cells with the caspase-2 inhibitor did not affect formation of γH2AX. In contrast, zVAD-fmk prevented DSBs, suggesting that another caspase dependent pathway independent of ROS must be contributing to DSBs that are seen during DICE. The data suggest that loss of either CD95 or CD95L in cancer cells causes metabolic stress, loss of mitochondria, production of mitochondrial ROS, and DNA damage, followed by an inability of cells to undergo proper cytokinesis. In summary, DICE is reminiscent of a necrotic form of mitotic catastrophe with signs of apoptosis.
Figure 5. During DICE, DNA DSBs Cause Activation of Caspase-2, Resulting in MOMP.
(A, B) Cell death quantified in the indicated cells after treatment with Dox for up to 7 days in the presence of the indicated inhibitors (zVAD and Nec-1 as in Figure 5C; MVE: 1 μM in CT26L, 0.25 μM in T98G, and 0.2 μM in HCT116 cells). The day 7 data points for CT26L cells are not shown because cells started to die due to overconfluency. P-values were calculated by ANOVA.
(C, D) Quantification of ROS produced (C) and changes in MOMP (D) in cells after knockdown of either CD95L or CD95. Analyses were performed 2 days (CT26L) and 4 days (HCT116) after addition of Dox. P-values were calculated by ANOVA.
(E) Quantification of MOMP in CT26L cells 40 hours after knockdown of CD95L. Cells were pretreated for 1 hour with different caspase inhibitors as indicated. P-values were calculated using t-test.
(F) Quantification of MOMP in cells 3 days (T98G) or 4 days (HCT116) after knockdown of CD95L. Cells were pretreated for 1 hour with different caspase inhibitors as indicated.
(G) From the bottom, DNA degradation, MOMP quantification and ROS staining of CT26L-pTIP-L3 cells transfected with either scrambled (scr) siRNA or a siRNA targeting mouse caspase-2 (100 nM) and treated with Dox for 40 hours. The top panel shows a Western blot for caspase-2 and actin (loading control).
(H) Quantification of DNA degradation in CT26L-pTIP-L3/18 cells 40 hours after knockdown of CD95L. Cells were pretreated for 1 hour with different caspase inhibitors as indicated.
(I) Western blot analysis of caspase-2 and actin in CT26L-pTIP-L3 cells after addition of Dox.
(J) Caspase-2 activity assay of CT26L-pTIP-L3/18 cells 48 hours after knockdown of CD95L. Cells were pretreated with caspase inhibitors where indicated. Etoposide treatment for 24 hours was used as a positive control. P-value was calculated using t-test.
(K) Western blot analysis of γH2AX and actin in CT26L-pTIP-L3/18 cells 40 hours after knockdown of CD95L. Some cells were pretreated with the indicated caspase inhibitors.
(L) Schematic illustrating the DICE subpathway in CT26L cells that results in activation of caspase-2 (blue arrows), which causes MOMP. Cancer cells are protected by the expression of both CD95 and CD95L. For details, see text.
* = p<0.01; ** = p<0.001; *** = p<0.0001; ns = not significant.
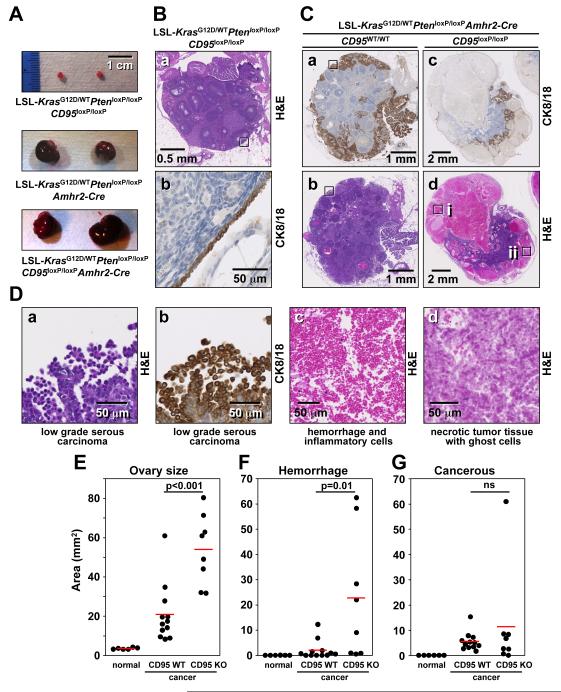
Evidence for Necrosis Induction after Knockout of CD95 in a Mouse Model of Ovarian Cancer
We previously demonstrated that in a model of endometrioid cancer, deletion of CD95 in the ovarian surface epithelium of LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxP mice severely reduced tumor formation (Chen et al., 2010). However, the reason for the failure of tumors to develop remained unclear. Recently, a spontaneous mouse model of ovarian cancer involving the same genetic lesions, but with a different delivery of Cre, was described (Mullany et al., 2011). In this model, LSL-KrasG12D/wtPtenloxP/loxP mice crossed with mice expressing the Cre recombinase under the control of the anti-Muellerian hormone receptor 2 (Amhr2-Cre) develop a low-grade form of serous epithelial ovarian cancer at 5-10 weeks of age (Mullany et al., 2011). In contrast to the model of endometrioid cancer used in our previous study in which a rapid and acute expression of Cre was achieved by injecting adenoviral Cre into mouse ovaries, Amhr2 driven Cre is expressed gradually beginning at embryonic day 3 and continuing until 7 months of age (Fong and Kakar, 2009). To determine the contribution of CD95 to this form of ovarian cancer, we crossed CD95loxP/loxP into the LSL-KrasG12D/wtPtenloxP/loxPAmhr2Cre/wt mice (Figure S9A). In 40 or 80 day old mice not expressing Amhr2-Cre, we did not detect a significant difference in the size of the ovaries in mice expressing wild-type CD95, CD95+/loxP, or CD95loxP/loxP (Figure 6A and data not shown). In contrast, all the ovaries from LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt mice were substantially larger than the ovaries from overall wild-type (normal) mice (Figure 6A, 6E, S9C). Unexpectedly, the ovaries from LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt mice were significantly larger than their CD95 wild-type counterparts (Figure 6A, 6E, S9C). A histological analysis showed that in wild-type mice, only the single layer of ovarian surface epithelial cells (OSE) was positive for cytokeratin 8/18 (CK8/18) (Figure 6B-b). In contrast, ovaries from approximately 80 day old mutant mice expressing CD95 contained cytokeratin 8/18 positive low-grade serous ovarian cancer cells (Figure 6C-a, 6C-b, 6D-a and 6D-b). The cancer developed predominantly on the surface of the ovaries (see Figure S9C). In contrast, in the ovaries from LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt mice, extended areas of the cancerous ovaries were necrotic with large numbers of infiltrating immune cells (Figure 6C-c, 6D-c, 6F and S9C). The large degree of hemorrhage which was found in the majority of CD95 deficient ovaries, but not in cancerous CD95 expressing ovaries, likely accounted for the overall size difference between cancerous ovaries expressing CD95 and lacking CD95 expression. Areas of cancerous tissue were also found in the CD95 mutant mice and they were mostly in foci in the interior of what was left of the ovaries (Figure 6C-c and S9C). In one CD95 mutant mouse, we detected classical necrotic tumor tissue with the presence of ghost cells (Figure 6C-dii and 6D-d). This necrotic tissue was not in the center of the tumor where necrosis often occurs due to hypoxic conditions, but rather at the periphery of the ovary where ovarian cancer was found in most CD95 wild-type cancerous mice. This finding is consistent with tumor cells lacking CD95 expression undergoing necrosis.
Figure 6. Detection of Necrosis in Cancerous Ovaries from CD95 Deficient KrasG12D/wt Pten−/− Mice.
(A) Representative images of ovaries from 80 day old KrasG12D/wtPten−/− mice expressing either wild-type CD95 (center) or lacking CD95 expression (bottom). Normal ovaries from an age matched LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxP mouse (without Amhr2-Cre expression) are shown as control (top).
(B) a: H&E stained normal ovary from a 83 day old LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxP mouse (#1046). b: CK8/18 staining of the boxed area of an adjacent section in a.
(C) a-d: Low power images of H&E and CK8/18 stained ovaries from LSL-KrasG12D/wtPtenloxP/loxPAmhr2-Cre mouse expressing wild-type CD95 (a, b; #1317) or lacking CD95 expression (c, d; #1254).
(D) a and b: High power images of boxed regions in C-a and C-b, respectively. c and d: High power images of box i and ii in C-d, respectively.
(E-G) Morphometric analysis of ovary size (E), area of tissue hemorrhage (F), and area of cancerous tissues as assessed through CK8/18 staining (G). Both ovaries of the 6 LSL-KrasG12D/wtPtenloxP/loxP CD95loxP/loxP mice (normal) and the 4 LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2-Cre mice (cancer) were analyzed. P-values were calculated using t-test. ns = not significant.
See also Figure S9.
Lack of Cancer Formation in Mice Deficient for CD95 in the Ovaries
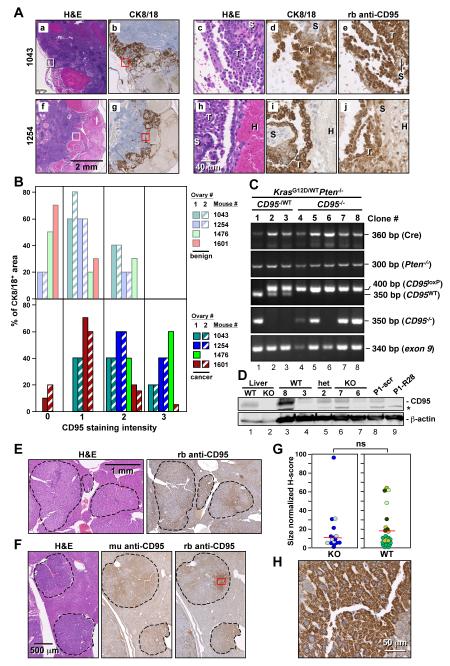
Given the inefficiency of Amhr2-Cre to recombine target genes (Fong and Kakar, 2009), we wondered whether the tumors that were detected in the KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt mice had fully recombined CD95 alleles or whether they still expressed CD95 protein. We therefore stained the ovaries from the four mice with the LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt genotype, which should lack CD95 expression, with an anti-mouse CD95 antibody and analyzed the expression of CD95 in the CK8/18 positive tumor regions (Figure 7A and data not shown). Compared to the CK8/18 negative benign ovarian tissue, CD95 expression was elevated in all tumor areas when compared to benign tissue (Figure 7B). Examples for two of the CD95 mutant mice are shown in Figure 7A. In both cases, strong cytosolic staining was detected for both CK8/18 and CD95 only in the tumor cells, but not in the stromal or infiltrating cells (Figure 7A-d, 7A-e, 7A-i, and 7A-j). The immunohistochemistry data suggested that almost all cancer cells that formed in the ovaries with a CD95 deficient genotype in fact expressed CD95. To confirm this finding, we isolated cancer cells from the ascites of the CD95 mutant mouse #1043 and a CD95 heterozygous mouse by long term culture (15 passages) and then set up single cell cloning at 0.5 cells per well. Only clones that were derived from a single cell were selected for genotyping, effectively excluding contamination by noncancer cells (Figure 7C). Three clones derived from a LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/wtAmhr2Cre/wt mouse as well as five clones derived from a LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2Cre/wt mouse were randomly selected for analysis. In all eight clones, both Pten alleles were deleted (Figure 7C) and the LSL-KrasG12D gene was recombined (data not shown). In contrast, of the five CD95 knockout clones, four had recombined only one CD95 locus and one (clone 6) retained both CD95 alleles in wild-type configuration (Figure 7C, lanes 4-8). Of the three clones heterozygous for CD95, only one had recombined the CD95loxP/loxP and two clones had both a wild-type CD95 locus and an intact CD95loxP allele (Figure 7C, lanes 1-3). Remarkably, all eight clones had at least one intact CD95 allele as evidenced by the presence of exon 9 (Figure 7C). Finally, we performed Western blot analysis for CD95. While the expression of CD95 varied greatly between the isolated clones (even among the clones isolated from CD95 wild-type mice), all tested clones expressed detectable CD95, confirming the PCR analysis and the immunohistochemistry. Interestingly, clone 7 isolated from the CD95 knockout mouse expressed CD95 protein at a level similar to what was expressed in the heterozygous clone 2 (Figure 7D, lanes 6 and 5). These data suggest that CD95 is critical for the survival of the cells in this type of cancer, and the necrosis observed in ovaries with deleted CD95 was likely caused by the elimination of CD95. It appeared that cells could only become cancerous if Cre had selectively recombined LSL-KrasG12D and deleted Pten without affecting the CD95 locus. In order to test whether cancer cells from these mice with ovarian cancer are also dependent on CD95 stimulation by CD95L for their survival, we sequentially transfected a cell line derived from these mice which expresses CD95 (P1 in Figure 7D, lane 8) with two siRNAs targeting mouse CD95L (Figure S9E). This induced both significant growth reduction and cell death. This data support a crucial role for CD95 in the survival of ovarian cancer cells.
Figure 7. Cancerous Cells Developing from LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2-Cre and DEN Treated CD95loxP/loxPAlb-Cre Mice Express CD95.
(A) Immunohistochemistry analysis of sections of ovaries from two LSL-KrasG12D/wtPtenloxP/loxPCD95loxP/loxP mice. a, b, f, and g: larger areas of two ovaries. c, d, e: higher magnifications of H&E, CK8/18 and CD95 staining of the areas boxed in a and b. h, i, j: higher magnifications of H&E, CK8/18 and CD95 staining of the areas boxed in f and g. T, tumor; H, hemorrhage; S, stroma.
(B) Quantification of CD95 staining intensity in ovaries of 4 KrasG12D/wtPten−/− CD95 mutant mice in benign (top) and CK8/18 positive cancerous areas (bottom). Ovary #2 in mouse #1476 had so much infiltration that neither benign nor tumor tissue could be detected. Ovary #2 in mouse #1601 did not contain any benign tissue.
(C) PCR analysis of cloned cell lines derived from mice with ovarian cancer heterozygous or homozygous for CD95loxP. Primers used in the third panel detect the right loxP site in the CD95 gene.
(D) Western blot analysis of single cell clones derived from mice with a genotype of either wild-type, heterozygous, or homozygous deletion of CD95. As a positive control, extracts of a cell line (P1) derived from KrasG12D/wtPten−/− mice (Mullany et al., 2011) are shown infected with a lentiviral shRNA targeting mouse CD95 (R28) or a nontargeting shRNA (scr) as well as liver extracts from wild-type (wt) and CD95loxP/loxPAlb-Cre (ko) mice. *= unspecific band.
(E) H&E and CD95 staining of a liver section of a wild-type mouse 8 months after injection of DEN.
(F) H&E and CD95 staining of a liver section of a CD95loxP/loxPAlb-Cre mouse 8 months after injection of DEN using two different antibodies (a mouse monoclonal (mu) or a rabbit polyclonal (rb) anti-CD95 antibody).
(G) Quantification of CD95 expression in all nodules in sections from two CD95loxP/loxPAlb-Cre and 4 CD95loxP/loxP mice. A total of 15 tumor nodules were analyzed from CD95 knockout livers and 20 nodules were analyzed from wild-type livers. Liver nodules found in individual mice are shown in different colors. ns = not significant.
(H) Magnification of the CD95 stained tumor region in F boxed in red.
Lack of Cancer Formation in Mice Deficient for CD95 in the Liver
Prompted by the data on the role of CD95 in the new model of low-grade serous ovarian cancer, we revisited our previous observation that liver cancer was reduced by 50% in mice lacking CD95 expression in the liver (CD95loxP/loxPAlb-Cre mice) in a model of chemically induced liver cancer (Chen et al., 2010). The expression of Cre driven by the albumin promoter has also been reported to be quite inefficient (Postic and Magnuson, 2000). We therefore took liver sections of 4 randomly chosen wild-type and 2 CD95−/− liver knockout mice 8 months after injection with diethylnitrosamine (DEN) and stained them for CD95. In wild-type mice, numerous cancerous liver nodules were detected. We detected upregulation of CD95 in the cancerous lesions when compared to the surrounding benign liver tissue (Figure 7E and data not shown). Interestingly, a clear staining of CD95 was also found in the liver nodules that had formed in the CD95−/− mice (Figure 7F). This was established by using two independent anti-CD95 antibodies (one mouse monoclonal and one rabbit polyclonal). Quantification of CD95 staining in nodules detected in sections of each liver showed that there was no significant difference in the CD95 expression between cancer nodules that had formed in wild-type mice when compared to CD95loxP/loxPAlb-Cre mice (Figure 7G). These data suggest that similar to the ovarian cancer models, liver tumors that lack CD95 expression barely if at all formed. When magnified, it became clear that most of the CD95 staining in tumor nodules was found in intracellular vesicles (Figure 7H), suggesting that the tumor promoting activity of CD95 may occur intracellularly. In summary, our data indicates that CD95 and CD95L do not just promote tumor growth, but they are also crucial for the survival of solid cancer cells.
DISCUSSION
We report that knockdown of either CD95 and/or CD95L in 15 cancer cell lines, which represent 8 different types of solid cancers, caused substantial induction of cell death in every cell line. In addition, analysis of the data on 12 arrayed genome-scale shRNA screens identified CD95L as a gene critical for the survival of cancer cells. CD95 was also identified in the lethality screens, however, with less potency than CD95L. Finally, we provide evidence that in two genetic mouse tumor models of ovarian and liver cancer, cancer cells that form still express CD95, confirming the in vivo findings that cancer cells barely develop in the absence of CD95. Death induced by CD95R/L elimination (DICE) is characterized by cell swelling, appearance of large dilated vacuoles, generation of ROS, and DNA DSBs. This is followed by reduction in cell growth, activation of caspase-2 resulting in MOMP, and ultimately, cell death (Figure 5L). Activation of effector caspases allows the cells to bleb and likely also contributes to DNA degradation. However, inhibiting caspases did not prevent cells from undergoing DICE. In addition, blocking ROS and subsequent DSBs also did not prevent the cells from dying. Cells can die earlier, but eventually die by a form of mitotic catastrophe at the first attempt to undergo cytokinesis. DICE is independent of the initiator caspase-8. It is a form of necrosis, but does not depend on the RIP1/MLKL containing necrosome, as neither necrostatin-1 treatment nor MLKL knockdown attenuated it. DICE can kill cancer cells independently of p53 or DICER expression and is not affected by overexpression of Bcl-xL.
While DICE was initially discovered in cancer cell lines, our data from mouse tumor models of low-grade serous ovarian cancer and liver cancer suggest that CD95 may also be required for the survival of cancer cells in vivo (see Supplementary Discussion). Based on evidence from both CD95 and CD95L knockout mice that do not show signs of cell death or growth deficiencies in any tissue outside the immune system (Adachi et al., 1995; Karray et al., 2004), we predicted that DICE preferentially affects cancer cells with little effect on normal cells. This prediction was supported by the finding that hTERT immortalized fibroblasts were more susceptible to DICE induction induced by either CD95 or CD95L knockdown than nonimmortalized fibroblasts (see Figure 2E).
While inhibition of caspases did not block DICE, it significantly slowed it down at least at earlier time points. The caspase most significant for DICE was caspase-2. Consistent with its role in DICE, caspase-2 is known to be activated by DNA damage and has functionally been linked to mitotic catastrophe (Castedo et al., 2004). Caspase-2 is an attractive candidate for being a component of the DICE pathway because it plays a role in cell cycle maintenance (Dorstyn et al., 2012) and acts as a tumor suppressor in mouse models of lymphoma and breast cancer (Ho et al., 2009; Parsons et al., 2013).
In summary, DICE comprises multiple death pathways, and we did not find a single small molecule inhibitor or shRNA that can block DICE. The only compounds that could at least temporality halt the execution of DICE were compounds that cause cell cycle arrest, which is consistent with our finding that cells without CD95 or CD95L expression cannot traverse through cytokinesis. DICE may therefore be an interesting new way to kill cancer cells without them easily acquiring resistance.
EXPERIMENTAL PROCEDURES
Generation of the TetOn shRNA Expressing Cells
The pTIG lentiviral vector (pHIV7-TetR-IRES-GFP) was a kind gift from Dr. John J. Rossi (Beckman Research Institute of the City of Hope, Duarte, California) (Aagaard et al., 2007). To replace the GFP sequence with a puromycin resistance sequence, the puromycin resistance gene was excised from pUC57-puro using PmlI/BsrgI and inserted into pTIG-U6tetOshRNA. The shRNA oligonucleotides (shScr, L1, L3, and R6) were obtained from IDT, and were cloned directly into the BsaBI/SphI restriction sites in pTIG or pTIP. To generate cells stably expressing the inducible shRNA constructs, cells were either infected with pTIG vectors and sorted for GFP expression using a Beckman Coulter MoFlo high speed multilaser droplet cell sorter or were infected with pTIP vectors and maintained in puromycin containing medium unless doxycycline (Dox) was added. shRNAs were induced by adding 0.1 μg/ml Dox unless otherwise indicated.
Generation of KrasG12D/wtPtenloxP/loxPCD95loxP/loxPAmhr2-Cre Mice
To obtain female littermates with CD95loxP/loxP present or absent within the spontaneous ovarian cancer mouse model, founder mice on a genetic background previously published (Mullany et al., 2011) were bred in two steps (see Figure S9A). In the parental generations, the Amhr2-Cre+ gene was bred only through the male lineage, while the LSL-KrasG12D gene was only bred through the female lineage. The genotype of the original parents for the sire was Kraswt/wt PtenloxP/loxP CD95wt/wt Amhr2-Cre and for the dam was LSL-KrasG12D/wt PtenloxP/loxP CD95loxP/wt. In the F1 generation, the genotype of the sire was Kraswt/wt PtenloxP/loxP CD95wt/loxP Amhr2-Cre/wt, and the genotype of the dam was LSL-KrasG12D/wtKraswtPtenloxP/loxP CD95wt/loxP. In the F2 generation, both parents had the genotype CD95loxP/wt to allow for both CD95wt/wt (“CD95-wt-mice”) and CD95loxP/loxP (“CD95-ko-mice”) in the next generation. The expected ratio for each female genotype was 1:32.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. E. Lengyel and A. Mitra for providing primary omental fibroblasts. We would like to thank Drs. B. Vogelstein for providing the HCT116 p53−/− and HCT116 Dicer−/− cells, T. Li for performing the tissue stainings for CD95, and Sam Bettis for performing the small molecule screen.. We would like to thank Drs. J. Lahti, J. Rossi, and Zhen-Gang Liu for providing the NB7 cells, the Tet-inducible vector, and the MLKL knockdown cells, respectively. We like to thank Dr. J. Licht for help in performing Comet assays. The HTS Core Facility is supported by Mr. William H. Goodwin and Mrs. Alice Goodwin, the Commonwealth Foundation for Cancer Research, the ETC of MSKCC, the Lillian S. Wells Foundation, and by a NIH/NCI Cancer Center Support Grant 5 P30 CA008748-44. S.B. is supported in part by NIH/NCI training grant T32CA09560. This work was funded by the NCI (RO1 CA112240 to M.E.P.).
Footnotes
Additional experimental procedures can be found in the supplement.
References
- Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Targeted mutation in the Fas gene causes hyperplasia in peripheral lymphoid organs and liver. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- Aoki K, Kurooka M, Chen JJ, Petryniak J, Nabel EG, Nabel GJ. Extracellular matrix interacts with soluble CD95L: retention and enhancement of cytotoxicity. Nat Immunol. 2001;2:333–337. doi: 10.1038/86336. [DOI] [PubMed] [Google Scholar]
- Bhinder B, Antczak C, Ramirez CN, Shum D, Liu-Sullivan N, Radu C, Frattini MG, Djaballah H. An arrayed genome-scale lentiviral-enabled short hairpin RNA screen identifies lethal and rescuer gene candidates. Assay Drug Dev Technol. 2013;11:173–190. doi: 10.1089/adt.2012.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhinder B, Djaballah H. A simple method for analyzing actives in random RNAi screens: introducing the “H Score” for hit nomination & gene prioritization. Comb Chem High Throughput Screen. 2012;15:686–704. doi: 10.2174/138620712803519671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Valent A, Raslova H, Yakushijin K, Horne D, Feunteun J, Lenoir G, Medema R, et al. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 2004;23:4362–4370. doi: 10.1038/sj.onc.1207572. [DOI] [PubMed] [Google Scholar]
- Chen L, Park SM, Tumanov AV, Hau A, Sawada K, Feig C, Turner JR, Fu YX, Romero IL, Lengyel E, et al. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Puccini J, Wilson CH, Shalini S, Nicola M, Moore S, Kumar S. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ. 2012;19:1288–1298. doi: 10.1038/cdd.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Vandenabeele P. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta. 2006;1757:1371–1387. doi: 10.1016/j.bbabio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Fong MY, Kakar SS. Ovarian cancer mouse models: a summary of current models and their limitations. J Ovarian Res. 2009;2:12. doi: 10.1186/1757-2215-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci U S A. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karray S, Kress C, Cuvellier S, Hue-Beauvais C, Damotte D, Babinet C, Levi-Strauss M. Complete loss of Fas ligand gene causes massive lymphoproliferation and early death, indicating a residual activity of gld allele. J Immunol. 2004;172:2118–2125. doi: 10.4049/jimmunol.172.4.2118. [DOI] [PubMed] [Google Scholar]
- Martin-Villalba A, Llorens-Bobadilla E, Wollny D. CD95 in cancer: tool or target? Trends Mol Med. 2013;19:329–335. doi: 10.1016/j.molmed.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Mullany LK, Fan HY, Liu Z, White LD, Marshall A, Gunaratne P, Anderson ML, Creighton CJ, Xin L, Deavers M, et al. Molecular and functional characteristics of ovarian surface epithelial cells transformed by KrasG12D and loss of Pten in a mouse model in vivo. Oncogene. 2011;30:3522–3536. doi: 10.1038/onc.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 2013;20:1174–1182. doi: 10.1038/cdd.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME, Budd RC, Desbarats J, Hedrick SM, Hueber AO, Newell MK, Owen LB, Pope RM, Tschopp J, Wajant H, et al. The CD95 receptor: apoptosis revisited. Cell. 2007;129:447–450. doi: 10.1016/j.cell.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Katsaros D, et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA J1 - PNAS. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci U S A. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.