Abstract
Based on hemagglutinin (HA) and neuraminidase (NA), influenza A virus is divided into 18 different HA (H1 to H18) and 11 NA types (N1 to N11), opening the possibility for reassortment between the HA and NA genes to generate new HxNy subtypes (where x could be any HA and y is any NA, possibly). In recent four years, since 2010, highly pathogenic avian influenza (HPAI) viruses of H5N1 subtype (HPAI A/H5N1) have become highly enzootic and dynamically evolved to form multiple H5 HA clades, particularly in China, Vietnam, Indonesia, Egypt, Cambodia, and Bangladesh. So far, after more than 10 years emerged in Vietnam (since late 2003), HPAI A/H5N1 is still posing a potential risk of causing outbreaks in poultry, with high frequency of annual endemics. Intragenic variation (referred to as antigenic drift) in HA (e.g., H5) has given rise to form numerous clades, typically marking the major timelines of the evolutionary status and vaccine application in each period. The dominance of genetically and antigenically diversified clade 2.3.2.1 (of subgroups a, b, c), clade 1.1 (1.1.1/1.1.2) and re-emergence of clade 7.1/7.2 at present, has urged Vietnam to the need for dynamically applied antigenicity-matching vaccines, i.e., the plan of importing Re-6 vaccine for use in 2014, in parallel use of Re-1/Re-5 since 2006. In this review, we summarize evolutionary features of HPAI A/H5N1 viruses and clade formation during recent 10 years (2004-2014). Dynamic of vaccine implementation in Vienam is also remarked.
Keywords: Orthomyxoviridae, Subtypes, HPAI A/H5N1, Genotypes, Clades, Reassortment, Vaccines, Vietnam
Introduction
Avian influenza is an acute infectious disease that occurs and spreads very fast in poultry, waterfowls, wild birds, animals, and able to zoonotically be transmitted to humans. Avian influenza is listed as one of the most dangerous diseases by World Organisation for Animal Health (OIE; http://www.oie.int/eng/normes/mmanual/A_summry.htm) that a special attention is needed for control. Influenza A virus belongs to the family of Orthomyxoviridae, of which the envelope is embedded by two surface glycoproteins, e.g., hemagglutinin (HA) and neuraminidase (NA). The virus has a spherical appearance with a diameter of 80-120 nm, or polymorphological shape, sometimes is elongated, like fibers and arranged in a sequence. Virus particles (virions) have a simple structure, which includes envelope, capsid and a core containing genetic material (viral genome) with molecular weight of about 250 million daltons [1]. Influenza A virus genomes contain a single-stranded negative RNA ([-]ssRNA) of 8 segments (polymerase basic 2 [PB2], polymerase basic 1 [PB1], polymerase acid [PA], HA, nucleoprotein [NP], NA, matrix [M], non-structural polypeptides [NS]) [1,2], encoding for 15 proteins in overall, including PB2, PB1, PA, HA, NP, NA, M1, M2, NS1, NS2, and supplementary proteins recently identified with little or unknown functions [3], i.e., PB1-F2 [4], PB1-N40 [5], PA-X [6], PA-N155 and PA-N182 [7]. Up-to-date, there are 18 HA and 11 NA types for influenza A viruses to be known, based on the composition and activity of the HA (from H1 to H18) and the NA (from N1 to N11) genes, especially, by recent addition of two new HA (ie., H17 and H18) and two new NA types (N10 and N11) detected in bats [8,9]. The first 15 HA and 9 NA types are originally found in aquatic fowles which served as a source for subsequent reassortment [3]. The numerous types of HA and NA enable the components going through genetic reassortments between HA and NA for generation of new subtypes [10,11,12].
Major polypeptides encoded by 8 segments of the genome of the influenza A virus are as follows:
The polymerase complex (PB2, PB1, and PA): The polymerase enzymes encoded by segments 1, 2, and 3, respectively, are responsible for transcription and replication of viral RNA [13]. Additionally, the recently identified polypeptides of PB1-F2, PB1-N40, PA-X, PA-N155, and PA-N182 encoded within segments 2 and 3, respectively, have confirmed the influenza virus as a top of the most complicated viral agents by multi-disciplinary utilization of the genome and the functional polypeptides [3].
The surface antigenic proteins (HA and NA): HA assists virus in attachment to the cell receptors; and by being cleaved by protease, facilitating intrusion of viral genetic material into the cell. NA functions in promotion of the assembly and the release of the virus progenies from the cell receptor [14]. The HA and NA antigenic glycoproteins determine the specificity of different subtypes of influenza A viruses and NA is a target for antiviral treatment [15]. Moreover, HA and NA also play a pivotal role in determining the antigenicity of vaccine production [11,14,16].
The proteins encoded by internal genes: They include NP, M (M1 and M2), and NS (comprising NS1 and NS2): Products of these genes play important roles in packaging of viral genomic RNA during replication, in intracellular transporting pathways and in shipping mechanism through the channel components [17,18]. M1 protein sustains the virion structure, playing multiple role in the replication, assembly, and release of viruses [1]. The viral ribonucleoprotein complexes (vRNPs) contain viral RNA, viral polymerases and viral NPs which are essentially formed during viral infection and replication [19,20]. NS2 product also known as a nuclear export protein or NEP is essential for transportation of the vRNPs to cytoplasm [1].
After more than 10 years of highly pathogenic avian influenza (HPAI) A/H5N1 emergence in Asia, the reassortments of HAs and NAs have always occured to generate new subtypes [21,22]. Just in a short time of recent 5 years, more lethal, pandemic candidate and/or pandemic subtypes of influenza A viruses have been reassorted and emerged in wide geographical localities [3]. These are represented by, namely, the H5N1 variants of highly differentiated clades, the pandemic A/H1N1-2009, the new subtypes of H7N9 and H10N8 in China that posed a high risk across different nations [23]. The rapid emergence of the antigenic drifted HPAI H5N1 and the newly reassorted H7N9 and H10N8 viruses raise the urgent needs to definitely update vaccine development from antigenicity-matching strains for actual application [15,24,25,26]. Fortunately, a great technology, reverse genetics has contributed at a great speed, to solve the vaccine development in the shortest time, that new vaccine(s) against most recently emerging subtypes, with high immunity and warranted protection, including the H5N1 and H7N9 vaccines have been generated [25,26,27,28,29,30].
In this review, we focus on the recently-introduced HA evolution, preperably, on H5 HA of HPAI A/H5N1 strains and the evolutionary features of clade formation in Vietnam, by 1) summarizing the antigenic drift of H5 HA and the emergence of its clade/subclades in the past eleven years, since 2003; 2) analyzing phylogenetic relationships of different HA clades between Vietnam and neighbouring countries; 3) looking at spatial-temporal transmission of clades and dynamic vaccine implementation in Vietnam. Literature has been taken for use mainly from the international sources but in some cases, from some of local publications with actual citations.
Evolutionary Features for Emergence of Genotypes and Clades of H5N1 in Vietnam
It is noted that the precursor highly pathogenic A/H5N1 avian virus was first time formed in 1996, as a result of the genetic reassortment of the H5 HA and the N1 NA types in wild geese in Guangdong, China (A/Goose/Guangdong/1/96[H5N1]); and the reassortants subsequently derived from this ancestral source, have since then spread over the world causing numerous epidemics [3,14,31]. Although the more ancient strain of A/H5N1 was, indeed, recorded in 1959, indicated by the low pathogenic strain of A/chicken/Scotland/1959(H5N1) (GenBank: CY015081, GU052518 for HA genes), nearly 20 years in the past, particularly in the last 10 years, due to the actual emergence of HPAI influenza A viruses, the HPAI A/H5N1 subtypes have caused enormous loss to poultry, wild birds and humans [3]. Although in history, the evolutionary pressure has posed substantial impact on the dynamics of the genetic variations of all the 8 genomic segments in order to diversify the pathogenicity/antigenicity and the host adaptability of the influenza A viruses, most of which have frequently occured in the HA and NA genes, resulting in global impact by generation of epizootic outbreaks and pandemics [12,16].
Antigenic HA polypeptide (i.e., HA, for H5N1, referred to as H5) is encoded by segment 4, functionally characteristic to the specific binding process to the surface receptor on the cell membrane [14,15,16,32]. HA has the ability to create genetic mutations in different sites (especially, substituting the amino acids at the protease cleaving site, at the point between HA1 and HA2) or to reassort to generate variants, altering surface antigenic properties leading to the correlative changes in immune response [14,31,33]. NA antigenic polypeptide of H5N1 (i.e., N1) encoded by segment 6, is an enzymatic protein, cutting off virus-cell receptor molecules linked to sialic acid (N-acetylneuramic acid) to timely release virus progenies from the infected cells during viral infection [14,34].
Since the first time appearance in 1996 in Guangdong (Southern China), there have been overall 12 genotypes formed from the ancestral A/Gs/Gd1/1996 H5N1 strain, including GD, A, B, C, D, E, X(X0-X3), V, Y, W, Z(Z+) and G [35,36]. In a short time after the first emergence in Guangdong, the original genotypes (e.g., GD, A, C, D, E) of the Guangdong lineage have entirely disappeared, giving way to the next genotypes/subgenotypes (V, W, X1, X2, X3, Y, Z, and Z+) evolutionarily emerged [37]. The appearance of the Z genotype with strains of higher pathogenicity in Southeast Asian countries, was an evidence for the "antigenic drift" in genetic constellation of the influenza A/H5N1 genome [35,38]. Genotype Z is characterized by the deletions in NA and NS segments. However, some viruses isolated in Southern China (Guangdong, Guangxi, and Hunan provinces) have substantially diversified to form further genotypes, i.e., Z, V, W, G; and the genotype G was the result of reassortant between W and Z [35,39]. In Vietnam, genotypes Z and G are dominant and long last since 2005 [40,41,42]. The H5N1 strains isolated in Vietnam during 2004-2006, were of the Guangdong sublineage, belonging to the genotype Z. Recently, the additional genotype G has emerged during 2007-2008; and after 2010, both of Z and G genotypes have coexisted up to date [40,43]. Two major groups of subgenotypes have been found to exist in Vietnam, subgenotype N (North) predominant in the North and S (South) in the South [36,41]. Between 2007 and 2010, it was noted that the circulating strains were of mixed sublineages, i.e., Guangdong-like, Fujian-like and Qinghai-like co-existed [41,42,43].
In the recent 10 years, the H5 antigenic gene has high tendency to drift, generating complicated intragenic changes. As results, the newly emerged variants have enhanced their adaptability to multi-host transmissions, helping the HPAI A/H5N1 viruses to spread throughout the world; hence the level of pathogenicity has been exacerbated [32,34]. Since 1996, during 8 years by 2004, H5N1 has diversified to generate 10 distinct clades (0-9) due to the antigenic mutational drift in the H5 gene, all of which have always originally first time formed in China [3]. The emergence of the HPAI A/H5N1 viruses in early 2000s has accelerated the H5 gene undergoing onto dynamic evolution in a high speed, generating numerous new clades/subclades of the second-, third-, and forth-order subclades, mostly within clades 1 and 2 [3,44]. Not all the above mentioned clades have been introduced into Vietnam (Fig. 1) [41,42,43,44,45,46], but largely, the rooting clades have further diversified to form new serial-order subclades, heavily affecting the avian influenza occurences in poultry and wild birds, and complicating the epidemiologic situation in Vietnam [40,42,44,47,48,49].
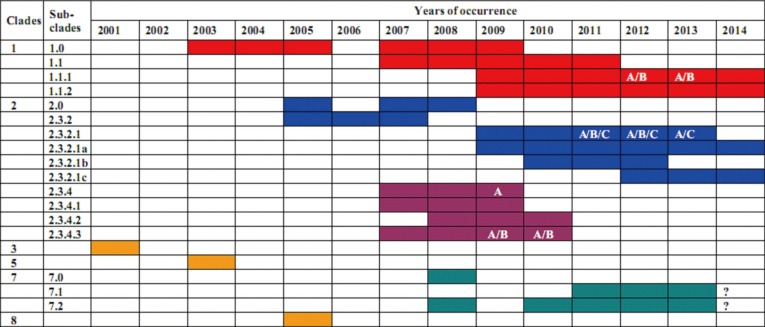
Fig. 1.
Emergence timelines of hemagglutinin-based clades of A/H5N1 in Vietnam (only those clades that emerged and identified in Vietnam are presented). Data used for making timelines of H5N1 occurence were collected from the following published sources: Nguyen et al. [41,42], Creanga et al. [43], World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group [44], Inui [45]; A/B/C marks for clades were taken from the above listed publications; question mark (?) indicates upcoming confirmation of clades in 2014.
By 2014, for example, clade 2 has diversified into different subclades and expanded over the world. The 2.3.4 clade alone evolved to the forth order subclades of 2.3.4.1, 2.3.4.2, 2.3.4.3, 2.3.4.4, 2.3.4.5, 2.3.4.6 over China [50] of which the subclades 2.3.4.1, 2.3.4.2, 2.3.4.3 have emerged and existed in Vietnam for a period of 4 years, during 2007-2010 (Fig. 1). The newly emerged clade 2.3.2 appeared more expandable in Vietnam, Southeast Asia, North Asia and many countries in Eastern Europe from 2009 to 2014, widely detected in wild birds, then poultry in China (Qinghai lake region), Mongolia, Russia, Japan, Korea, Laos, Hong Kong, Vietnam, Japan, Bangladesh, India, Bulgaria and recently, in Indonesia [42,51,52,53,54,55,56,57,58]. These strains are now classified into forth and fifth-order clades with an additional letter of the right-most digit implemented [44].
Major clades, such as 2.1.3, 2.2.1, 2.3.2, have also undergone substantial evolution in HA gene, resulting in a deep diversification, even within clade 2.3.2.1, there have been novel endemic, dynamically differentiated variants of a, b, c subgroups, during 2010-2014 in Vietnam. These 2.3.2.1a/b/c viruses, are preferably circulating in waterfowles, sometimes with no symptoms, thus are confusingly affecting the vaccination programs as well as complicating the epidemiological situation in a number of countries in the region [42,43,58,59]. Although six clades have been found among the A/H5N1 strains isolated since 2003 in Vietnam, including 1, 2, 3, 5, 7, 8, only three (clades 1, 2 and 7) have rooted to spatially/temporally last and to monophyletically diversify into discrete subclades of one-, two-, three- or forth-order designations (Fig. 1). These clades of 1, 2, 7 and their digit number-order subclades seemed to undergo on in-depth H5 based-drifting for formation of the further fifth-order groups, e.g., in case of clade 2.3.2.1a/b/c, officially nomenclatured by adding alphabetical implementation [44]. The derivative strains of these clades are firmly now circulating in multiple species of gallinaceous poultry throughout Vietnam [42,43,48,60].
Diversification of the Predominant Clades (1, 1.1, 2.3.4, 2.3.2.1 and 7, 7.1, 7.2) in Vietnam
Approaching 18 years from the time of first generation, the Gs/Guangdong lineage-H5N1 avian influenza virus reassortants have proved to be the most persistent subtypes expressing the highest pathogenicity in wild birds and domestic poultry [3]. The further descent lineage-viruses spread as largely as reaching overall 63 countries with deaths of 250 million of gallinaceous poultry and wild birds; and hundreds of human victims [25]. Vietnam was among one of the first countries outside of China affected by this HPAI H5N1 subtype and become enzootic with a number of persistent outbreaks annually.
Numerous clades have been generated during last ten years indicated by major timelines as below:
During 2003-2005: It was the first time in late 2003, that the HPAI A/H5N1 viruses have been introduced into Vietnam; and during this period, viruses of clade 1 were predominantly circulating in all provinces of the country [40,41].
In 2006: Vietnam has applied nationwide vaccination program (Re-1, clade 0 vaccine), and there was no report of any outbreak this year.
Between 2007 and 2009: This period was geographically marked by the major presence of clade 2.3.4 of the Fujian sublineage (and its further subclades 2.3.4.1, 2.3.4.2, and 2.3.4.3) in the North; then these (sub)/clades have completely disappeared from the country; no report of clade 2.3.4 was made after 2009. Viruses of clade 1 (and its clade 1.1) were dominating Mekong Delta provinces in the South, and starting evolving into clades 1.1.1 and 1.1.2 [41,42]. Re-1 and Re-5 vaccines (clade 2.3.4 origin) imported from China were in use and a national project of testing NIBRG-14 masterseed (rgA/Vietnam/1194/2004, clade 1, obtained from National Institute for Biological Standards and Control [NIBSC, UK]), was launched for vaccine production [61,62]. In this period, the first detection of a newly designated clade 7 at ports of entry in Lang Son province was published [60,63].
In 2010: This year was the time for the new clades emerged, and alternatively, for the appearance of periodically existed viruses [42]. The emergence of newly introduced clade 2.3.2 and further evolution of clade 2.3.2.1 into 2.3.2.1a/b was noted in the North; and the continual evolution of clade 1.1 of the Cambodian lineage into 1.1.1 and 1.1.2 occured in the South [42,43,47,64]. Re-1 and Re-5 vaccines were in use.
During 2011-2012: The dynamic formation of a, b, and particularly, c subgroups of clade 2.3.2.1 was in progress in whole country [43,65]; and because of antigenicity-matching failure for clade 2.3.2.1b, the Re-1/Re-5 vaccination was ceased in some provinces in the North [66]. NIBRG-14 based vaccine (namely, NAVET-Vifluvac) was successfully tested, indicating reliable efficacy against viruses of the current clades, particularly when used with immunomodulating adjuvants like β-glucan [61,62,67]. Clades 1.1.1/1.1.2 were in complete dominance in the South and Re-1 vaccine was still in use [47,49].
During 2013-2014: There was a wide circulation of clade 2.3.2.1a,c in whole country and no report of clade 2.3.2.1b was made anymore. Concurrently, clades 1.1.1/1.1.2 were heading from the Mekong Delta region to Central Vietnam. This time, the re-recognition of clade 7.1/7.2 evolving from clade 7 was reported. Due to decline in protection and wide spread of clade 2.3.2.1c, Re-6 vaccine is planned to import for use in 2014 [49]; according to the Vietnam authority, Department of Animal Health, 2014 (http://www.cucthuy.gov.vn/).
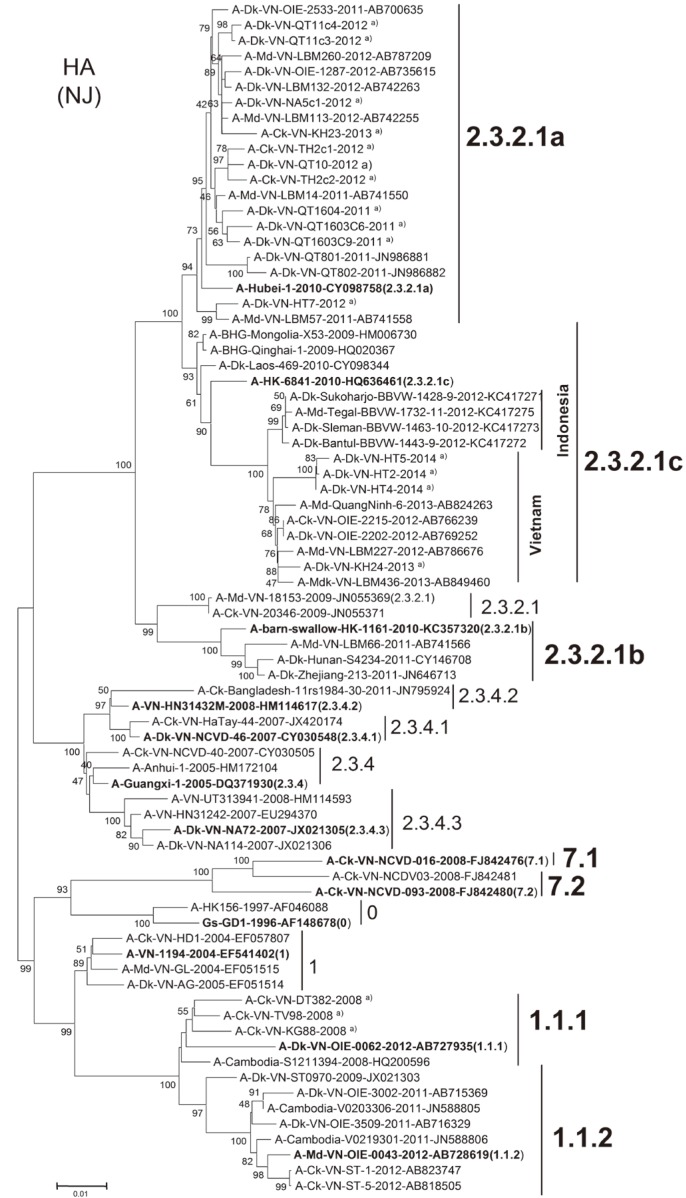
Tracing back the original source of H5N1, it is indicated that all viruses of the pre-existing clades 2.3.4, 2.3.2, 1.1, 7 before 2010; and the current circulating 2.3.2.1a/(b)/c, clades 1.1.1/1.1.2 and clades 7.1/7.2 in Vietnam have been introduced from Southern provinces of China during 2003-2014 [43]. It should also be noted that the very virulent clade 1.1.1/1.1.2 viruses currently dominating the South of Vietnam have evolved from a Cambodian sublineage of clade 1.1 [3]. To examine the clustering of viruses in different clades using World Health Organization (WHO)/OIE/Food and Agriculture Organization of the United Nations (FAO)-2014-nomenclaturing system, we include those previously listed as references for clading source and the current sequences obtained by our study from samples collected during 2008-2014. The phylogenetic tree resulting from H5 nucleotide data (using 77 sequences comprising of representative clades) is shown in Fig. 2. It clearly shows that most recent 2009-after collected isolates of Vietnam are completely clustered with the H5N1 HPAI strains of newly described clade 2.3.2.1 (and of its clade 2.3.2.1a/b/c) emerged after 2009, closely linked to A/Hubei/1/2010 (clade 2.3.2.1a), to A/barn-swallow/Hong Kong/1161/2010 (clade 2.3.2.1b) and A/Hong Kong/6841/2010 (clade 2.3.2.1c), respectively.
Fig. 2.
Phylogenetic tree based on the hemagglutinin H5 sequences (1,588 nucleotides) of representative A/H5N1 viruses including those of predominant clades in Vietnam since 2004. Topology was constructed by MEGA6.06 using the neighbor-joining analysis of the boostrap method with 1,000 replications (values <40 were not shown) [72]. Clades were nomenclatured based on the criteria and the reference strains determined in World Health Organization (WHO)/World Organisation for Animal Health (OIE)/Food and Agriculture Organization of the United Nations (FAO)-2014 system [44]. Major clades of H5N1 are shown as groups of monophyletic branches. Regarding to clade 2.3.2.1, viruses are divided into clade 2.3.2.1a (A/Hubei/1/2010-like); 2.3.2.1b (A/barn-swallow/Hong Kong/1161/2010-like); and 2.3.2.1c (A/Hong Kong/6841/2010-like). Note: Clades/subclades for prototype reference strains are bolded and parenthesized for indication. VN, Vietnam; HK, Hong Kong; KR, South Korea; KH, Cambodia. a)Viruses from our own study, marked at end of each strain. Scale bar at bottom indicates the number of nucleotide substitutions per site; the corresponding GenBank accession number are denoted at end of each sequence where applicable.
Strains forming clade 2.3.2.1a and clade 2.3.2.1b, comprised HA sequences (majority are of our study, be included in analysis) which were closely linked to A/Hubei/1/2010 and A/barn-swallow/Hong Kong/1161/2010, respectively. Clade 2.3.2.1c viruses, all clustered together, around the reference A/Hong Kong/6841/2010 strain, are of some typical characteristics of taxonomic grouping. It is indicated that the old strains, originally isolated in 2009 at Qinghai lake (China), or those before 2009 including the ancestral A/BHG/Qinghai/1/2009 (accession No. HQ020367) [68] formed a basal branch, formerly named as clade 2.3.2, now classified as clade 2.3.2.1c (Fig. 2). The tree shown in Fig. 2 supported a precise topology of clade 2.3.2.1c strains, with the Vietnamese (of those isolated in 2014 of our study as well) and Indonesian viruses [58] in two separate groups. Distinct from clade 1 they have evolved from, all HA sequences of clades 1.1.1/1.1.2 performed a monophyletically close group, regardless of where they were identified from isolates, in Vietnam or Cambodia (Fig. 2).
Antigenic Characteristics of A/H5N1 Viruses in Vietnam
Since their re-emergence in 2003, HPAI A/H5N1 viruses have become enzootic in some countries and continue causing outbreaks in poultry as well as sporadic human infections. The A/H5N1 viruses have diversified both genetically and antigenically leading to the need for multiple vaccine development. Evolution of the HA gene of influenza A/H5N1 viruses emerged in Vietnam has marked major antigenic changes, affecting immune response in the vaccinated poultry [42,47].
Amino acid analysis of H5 HA of the emerging strains isolated during 2010-2013 in Vietnam has revealed that the polybasic cleavage site (amino acid 338-345, H5 numbering) of these viruses, was similar to the H5 HAs belonging to viruses of clade 2.3.2.1 and its subgroups (2.3.2.1a,b,c) recently reported in China, South Korea, Japan, Mongolia, Nepal, Laos, Russia, Bangladesh and Indonesia [3,58,69,70]; and similar to those of clade 2.3.2 origin, but different from clade 2.3.4 and clade 1 (and 1.1) (Table 1) [3,42,43]. The strains found in South of Vietnam have evolved from clade 1, resulting in subclades 1.1.1 and 1.1.2, similar to that of the Cambodian clade 1.1 [3,44,70]. However, amino acids of receptor binding site of strains (QSG, position 238-240) in all clades in Vietnam and elsewhere were conserved (Table 1), keeping the ability of binding specificity stable throughout the evolution of HA [14,59].
Table 1.
Amino acid residues at sites of protease cleavage, receptor binding, epitopes of antigenic sites and a site of variable glycosylation of the H5 hemagglutinin polypeptide
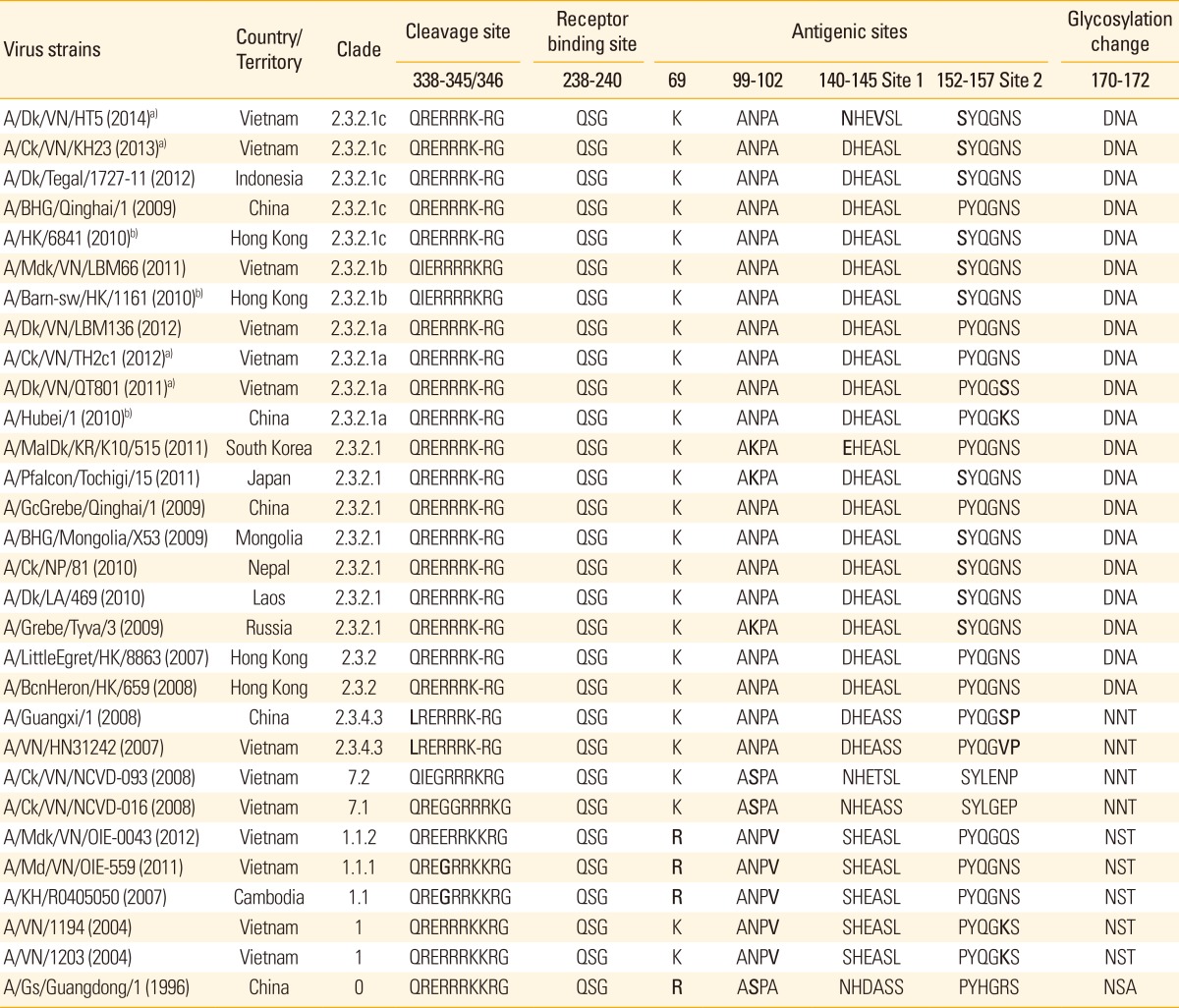
Bold letter of amino acid indicates a change in comparison to other strains of same clade.
Dk, duck; Ck, chicken; BHG, black headed gull; HK, Hong Kong; Mdk, muscovy duck; VN, Vietnam; sw, swallow; MalDk, mallard duck; KR, South Korea; KH, Cambodia; Gs, goose.
a)Viruses from our own study.
b)Prototype reference strains for clade 2.3.2.1a/b/c.
Analysis of amino acid identity at the antigenic sites (Table 1) revealed that all strains of clade 2.3.2.1a/b/c have amino acid K at antigenic position 69; ANPA at 99-102 (for some strains, N was replaced by K or S); and a highly conserved sequence, DHEASL, at 140-145 (antigenic site 1) except for the Vietnamese clade 2.3.2.1c strains of 2014 who possess NHEVSL. A/Hubei/1/2010 and A/Barn-sw/HK/1161(2010), the prototype vaccine strains [30,44], share close identity to the viruses within clade 2.3.2.1 at most of major sites, except at antigenic site 2, where strains of clades 2.3.2.1b,c were noticed making the sequence PYQGNS to become SYQGNS (Table 1). Whether these amino acid substitutions, alternatively, at the antigenic site 2, i.e., S152P and K156S/N, between strains of clades 2.3.2.1b,c and 2.3.2.1a, have caused antigenicity-mismatching immune response or not, still a question to be clarified.
Glycosylation at amino acid 170 in HA1 (position 170-172) is completely lost by changing from NST (as seen in clade 1 and 1.1 strains) and NNT (clade 2.3.4) to amino acids DNA in all strains of clades 2.3.2 origin and current 2.3.2.1(a,b,c) of Vietnam, China, Bangladesh, Indonesia and other countries [43,59,71], including those strains collected in 2014 of our current study (Table 1).
Spatial-Temporal Transmission of H5N1 Clades and Dynamic of Vaccine Application in Vietnam
Clade 2.3.2.1 and clade 1.1 (now 1.1.1/1.1.2) have revealed a great diversification due to their complex evolution of HA and endemically spatial transmission after 2010 in Vietnam. Clades 7 and 7.1/7.2 were temporarily detected before and now re-emerged in some places.
These evolutions and transmissions are characterized by the next descriptions, as follows:
Viruses of clade 2.3.2.1a (phylogenetically designated as A/Hubei/1/2010[H5N1]-like), emerged first time in mid-2010 in some provinces of the North of Vietnam. During the past four years since then, HA 2.3.2.1a viruses spatially spread over the coastal provinces along Central Vietnam, reaching Mekong Delta Basin, and currently (early 2014) predominantly are present in whole country [42,44]. For vaccination of poultry, Re-5, an imported vaccine from China for use in Vietnam, also known as Harbin Re-5 (reverse genetics-based A/duck/Anhui/1/2006 of HA clade 2.3.4 antigens), produced by the Harbin Veterinary Research Institute, Heilongjiang province, China, is still in nationwide use, showing reasonable efficacy for protection against clade 2.3.2.1a viruses [49]. The fact is that, circulating viruses of this 2.3.2.1a clade have not caused great loss to poultry and humans in Vietnam. Both Re-5 and Re-1 vaccines (Re-1, implemented since 2006; of rgA/Goose/Guangdong/1996 of HA clade 0, generated by reverse genetics) conferred a high level of protection (over 90%) in chickens against clade 2.3.2.1a by challenge test in laboratory; and about 70% seropositivity (by HA inhibition test) in immunized poulty in the field. However, protection level of Re-1/Re-5 vaccines, by challenging with strains of novel clade 2.3.2.1 (a, b, c), was much lower in ducks and muscovy ducks [47,49,65, 67,69,73].
Viruses of clade 2.3.2.1b (A/barn-swallow/HK/1161/2010 [H5N1]-like), were first time identified in early 2011 in some localities in the North Vietnam such as Quang Ninh, Bac Ninh, Bac Kan, Thai Nguyen, Phu Tho, Nam Dinh, Thai Binh, Son La, Ninh Binh, Nghe An [42,43,64,66]. Up-to-date, viruses of this clade 2.3.2.1b circulated restrictly in China, Hong Kong SAR and Vietnam [44]. In Vietnam, clade 2.3.2.1b viruses seemed to limit their spatial and temporal transmission in the North of Vietnam. Nghe An province was the last geographic locality where the latest clade 2.3.2.1b was detected in 2012. In 2013, no further report has been made about the presence of HA clade 2.3.2.1b viruses, assuming that they have completely disappeared from the circulating A/H5N1 population in the country [44].
Viruses of clade 2.3.2.1c (the reference strain therein is A/Hong Kong/6841/2010[H5N1]), was first time identified in 2012, from outbreaks of chickens and ducks in several boundary northern provinces, then spread southbound to Quang Ngai, Khanh Hoa provinces of Central Vietnam. These viruses are now shifting towards the provinces in Mekong Delta of Southern Vietnam, and predominantly present in the whole country [43,66,67,70]. Thus, clade 2.3.2.1c is of a great challenge to HPAI A/H5N1 control in Vietnam, since the current in-use-vaccines showed rapid decline of protection against these viruses; and of course, presenting a high risk to public health [49]. Regarding to nationwide immunization of poultry, the current-in use Re-1 and Re-5 vaccines do not have sufficient efficacy to prevent the 2.3.2.1c clade viruses [49], requesting a new vaccine with higher matching of antigenicity to be implemented [43]. The Re-6 vaccine (rgA/dk/Guangdong/S1322/2010[H5N1] of clade 2.3.2), developed by the Harbin Veterinary Research Institute (China), has been imported for use in 2014, targeting the prevailing clade 2.3.2.1 in Vietnam.
Currently, clade 2.3.2.1 tends to spread in spatial distance from North to South of Vietnam, warning the vertical transmission along the country. Concurrently, strains of more virulent clade 1.1, now evolved into 1.1.1 and 1.1.2, are predominantly present in Mekong Delta provinces, which have been phylogenetically revealed to share close evolution of the Cambodian clade 1 progenitors [42,44,73]. The presence of clade 1.1 and its subclades 1.1.1/1.1.2 in several provinces of Central Vietnam in 2013, indicated that now these clades are heading towards North Vietnam from Mekong Delta Basin. The overlapping region for spatial transmission of clades 1.1/1.1.1/1.1.2 and 2.3.2.1a/b/c is lower Central Vietnam provinces; and interestingly, contrary orientation of HPAI A/H5N1 infections is likely of a consequence of free poultry trade due to North-South/South-North transportation, between provinces within Vietnam and between Vietnam and Cambodia [43,47].
Clade 7, 7.1/7.2 viruses (A/chicken/Shanxi/10/2006-like), representing an interesting evolution of clades in Vietnam, were on site detected in chickens seized at ports of entry in 2008 [60] and subsequently, viruses of these subclades (7.1/7.2) were identified in some places of Northern Vietnam [42,43,63]. The early introduced A/chicken/Vietnam/NCVD-016/2008 (No. FJ842476) was considered as an ancestral entry strain for diversification of the recently recognized clade 7.1 and 7.2 viruses.
Conclusion
From 2009 to date, in recent 5 years, a number of new antigenic subgroups of H5N1 have been formed mainly from two sources in Vietnam: newly introduced clades (2.3.2.1) from Southern region of China, and clade 1.1 localized in the Mekong Delta evolved from Cambodian lineages, heading to move in the opposite direction between the North and South Vietnam. Meanwhile, the in-depth diversified clade 2.3.2.1 viruses (with a, b and c monophyletic groups) have complicated the epidemiologic and vaccination status in the whole country. Because of failure in immune response in poultry vaccinated with Re-1 vaccine (clade 0) and Re-5 (clade 2.3.4) to viruses of the currently circulating clade 2.3.2.1b/c, the Vietnamese government has announced a temporary suspension of the vaccination program with these vaccines in some of the northern provinces. Significant amino acid substitutions in the strains of clade 2.3.2.1c emerged since 2011, were the main cause of the protective decline of the available vaccines currently used nationwide. The biased efficacy of the Re-1 and Re-5 imported vaccines has forced Vietnam to import Re-6 vaccine (clade 2.3.2) currently being used in 2014. The transmission tendency of two predominant clades, i.e., 2.3.2.1 (a,b,c) and 1.1/1.1.1/1.1.2, needs special works for active management of the endemics under control and containing enforcement of the virus spread.
Footnotes
This work was supported by the Institutional Basic Project (to T.H.L.) from the Institute of Biotechnology (IBT) and the Vietnam Academy of Science and Technology (VAST). We extend our thanks to Prof. Chong-Woo Bae for his encouragement for writing this review.
No potential conflict of interest relevant to this article was reported.
References
- 1.Shaw M, Palese P. Orthomyxoviridae. In: Knipe DM, Howley P, editors. Field virology. 6th ed. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1151–1185. [Google Scholar]
- 2.Rabadan R, Levine AJ, Robins H. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J Virol. 2006;80:11887–11891. doi: 10.1128/JVI.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178:63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumbholz A, Philipps A, Oehring H, et al. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol. 2011;200:69–75. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- 5.Wise HM, Foeglein A, Sun J, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013;87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Types of influenza viruses [Internet] Atlanta: Centers for Disease Control and Prevention; 2013. [cited 2013 Apr 19]. Available from: http://www.cdc.gov/flu/about/viruses/types.htm. [Google Scholar]
- 11.ElHefnawi M, Sherif FF. Accurate classification and hemagglutinin amino acid signatures for influenza A virus host-origin association and subtyping. Virology. 2014;449:328–338. doi: 10.1016/j.virol.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Lu L, Lycett SJ, Leigh Brown AJ. Reassortment patterns of avian influenza virus internal segments among different subtypes. BMC Evol Biol. 2014;14:16. doi: 10.1186/1471-2148-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mair CM, Ludwig K, Herrmann A, Sieben C. Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim Biophys Acta. 2014;1838:1153–1168. doi: 10.1016/j.bbamem.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Shi Y, Gao F, et al. Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an alpha-helix structure in its cleavage site with inefficient HA1/HA2 cleavage. J Virol. 2012;86:12861–12870. doi: 10.1128/JVI.01606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tharakaraman K, Raman R, Viswanathan K, et al. Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell. 2013;153:1475–1485. doi: 10.1016/j.cell.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett. 2013;587:1206–1214. doi: 10.1016/j.febslet.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson EC, Fodor E. Transport of the influenza virus genome from nucleus to nucleus. Viruses. 2013;5:2424–2446. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Q, Chou YY, Doganay S, Vafabakhsh R, Ha T, Palese P. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J Virol. 2012;86:7043–7051. doi: 10.1128/JVI.00662-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Zhang Y, Wu B, et al. Evolutionary characterization of the pandemic H1N1/2009 influenza virus in humans based on non-structural genes. PLoS One. 2013;8:e56201. doi: 10.1371/journal.pone.0056201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei F, Shi W. Prospective of genomics in revealing transmission, reassortment and evolution of wildlife-borne Avian Influenza A (H5N1) viruses. Curr Genomics. 2011;12:466–474. doi: 10.2174/138920211797904052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runstadler J, Hill N, Hussein IT, Puryear W, Keogh M. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol. 2013;17:162–187. doi: 10.1016/j.meegid.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.To KK, Tsang AK, Chan JF, Cheng VC, Chen H, Yuen KY. Emergence in China of human disease due to avian influenza A(H10N8): cause for concern? J Infect. 2014;68:205–215. doi: 10.1016/j.jinf.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Baz M, Luke CJ, Cheng X, Jin H, Subbarao K. H5N1 vaccines in humans. Virus Res. 2013;178:78–98. doi: 10.1016/j.virusres.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spackman E, Swayne DE. Vaccination of gallinaceous poultry for H5N1 highly pathogenic avian influenza: current questions and new technology. Virus Res. 2013;178:121–132. doi: 10.1016/j.virusres.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Bu Z, Chen H. Avian influenza vaccines against H5N1 'bird flu'. Trends Biotechnol. 2014;32:147–156. doi: 10.1016/j.tibtech.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Subbarao K, Katz JM. Influenza vaccines generated by reverse genetics. Curr Top Microbiol Immunol. 2004;283:313–342. doi: 10.1007/978-3-662-06099-5_9. [DOI] [PubMed] [Google Scholar]
- 28.Kreibich A, Stech J, Mettenleiter TC, Stech O. Simultaneous one-tube full-length amplification of the NA, NP, M, and NS genes of influenza A viruses for reverse genetics. J Virol Methods. 2009;159:308–310. doi: 10.1016/j.jviromet.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Krammer F, Cox RJ. The emergence of H7N9 viruses: a chance to redefine correlates of protection for influenza virus vaccines. Expert Rev Vaccines. 2013;12:1369–1372. doi: 10.1586/14760584.2013.850036. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness [Internet] Geneva: World Health Organization; 2014. [cited 2014 Feb 20]. Available from: http://www.who.int/influenza/vaccines/virus/201402_h5h7h9h10_vaccinevirusupdate.pdf?ua=1. [Google Scholar]
- 31.Velkov T, Ong C, Baker MA, et al. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol. 2013;56:705–719. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013;9:e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013;178:99–113. doi: 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li KS, Guan Y, Wang J, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 36.Smith GJ, Naipospos TS, Nguyen TD, et al. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology. 2006;350:258–268. doi: 10.1016/j.virol.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 37.Macken CA, Webby RJ, Bruno WJ. Genotype turnover by reassortment of replication complex genes from avian influenza A virus. J Gen Virol. 2006;87(Pt 10):2803–2815. doi: 10.1099/vir.0.81454-0. [DOI] [PubMed] [Google Scholar]
- 38.Wu WL, Chen Y, Wang P, et al. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. J Virol. 2008;82:1798–1807. doi: 10.1128/JVI.02256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan L, Bahl J, Smith GJ, et al. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology. 2008;380:243–254. doi: 10.1016/j.virol.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan XF, Nguyen T, Davis CT, et al. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One. 2008;3:e3462. doi: 10.1371/journal.pone.0003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen TD, Nguyen TV, Vijaykrishna D, et al. Multiple sublineages of influenza A virus (H5N1), Vietnam, 2005-2007. Emerg Infect Dis. 2008;14:632–636. doi: 10.3201/eid1404.071343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen T, Rivailler P, Davis CT, et al. Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology. 2012;432:405–416. doi: 10.1016/j.virol.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Creanga A, Thi Nguyen D, Gerloff N, et al. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology. 2013;444:12–20. doi: 10.1016/j.virol.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 44.World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses. 2014;8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inui K. Epi-zone approach for HPAI H5N1 surveillance and control [Internet] [cited 2014 May 1]. Available from: http://www.rr-asia.oie.int/fileadmin/Regional_Representation/Programme/H_HPAI/JTF/2012_5th_ExpMeeting/2-1_Dr_Inui.pdf.
- 46.Long NT, Thanh TT, van Doorn HR, et al. Recent avian influenza virus A/H5N1 evolution in vaccinated and unvaccinated poultry from farms in Southern Vietnam, January-March 2010. Transbound Emerg Dis. 2011;58:537–543. doi: 10.1111/j.1865-1682.2011.01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrel MA, Emch M, Nguyen T, Todd Jobe R, Wan XF. Population-environment drivers of H5N1 avian influenza molecular change in Vietnam. Health Place. 2012;18:1122–1131. doi: 10.1016/j.healthplace.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cha RM, Smith D, Shepherd E, et al. Suboptimal protection against H5N1 highly pathogenic avian influenza viruses from Vietnam in ducks vaccinated with commercial poultry vaccines. Vaccine. 2013;31:4953–4960. doi: 10.1016/j.vaccine.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 49.Gu M, Zhao G, Zhao K, et al. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg Infect Dis. 2013;19:2021–2024. doi: 10.3201/eid1912.130340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang HM, Batchuluun D, Kim MC, et al. Genetic analyses of H5N1 avian influenza virus in Mongolia, 2009 and its relationship with those of eastern Asia. Vet Microbiol. 2011;147:170–175. doi: 10.1016/j.vetmic.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 51.Hu X, Liu D, Wang M, et al. Clade 2.3.2 avian influenza virus (H5N1), Qinghai Lake region, China, 2009-2010. Emerg Infect Dis. 2011;17:560–562. doi: 10.3201/eid1703.100948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert M, Jambal L, Karesh WB, et al. Highly pathogenic avian influenza virus among wild birds in Mongolia. PLoS One. 2012;7:e44097. doi: 10.1371/journal.pone.0044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchida Y, Suzuki Y, Shirakura M, et al. Genetics and infectivity of H5N1 highly pathogenic avian influenza viruses isolated from chickens and wild birds in Japan during 2010-11. Virus Res. 2012;170:109–117. doi: 10.1016/j.virusres.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Nagarajan S, Tosh C, Smith DK, et al. Avian influenza (H5N1) virus of clade 2.3.2 in domestic poultry in India. PLoS One. 2012;7:e31844. doi: 10.1371/journal.pone.0031844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Islam MR, Haque ME, Giasuddin M, et al. New introduction of clade 2.3.2.1 avian influenza virus (H5N1) into Bangladesh. Transbound Emerg Dis. 2012;59:460–463. doi: 10.1111/j.1865-1682.2011.01297.x. [DOI] [PubMed] [Google Scholar]
- 56.Marinova-Petkova A, Georgiev G, Seiler P, et al. Spread of influenza virus A (H5N1) clade 2.3.2.1 to Bulgaria in common buzzards. Emerg Infect Dis. 2012;18:1596–1602. doi: 10.3201/eid1810.120357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dharmayanti NL, Hartawan R, Pudjiatmoko, et al. Genetic characterization of clade 2.3.2.1 avian influenza A(H5N1) viruses, Indonesia, 2.0.1.2. Emerg Infect Dis. 2014;20:671–674. doi: 10.3201/eid2004.130517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu L, Bao L, Yuan J, et al. Antigenicity and transmissibility of a novel clade 2.3.2.1 avian influenza H5N1 virus. J Gen Virol. 2013;94(Pt 12):2616–2626. doi: 10.1099/vir.0.057778-0. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen T, Davis CT, Stembridge W, et al. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology. 2009;387:250–256. doi: 10.1016/j.virol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Le TH, Le KX, Cuong PV, et al. Adjuvant effects of Sophy β-glucan on H5N1 and H5N2 vaccination using a mouse model. Trop Med Health. 2010;38:23–27. [Google Scholar]
- 61.Le T, Le T, Doan TH, et al. The adjuvant effect of Sophy beta-glucan to the antibody response in poultry immunized by the avian influenza A H5N1 and H5N2 vaccines. J Microbiol Biotechnol. 2011;21:405–411. [PubMed] [Google Scholar]
- 62.Davis CT, Balish AL, O'Neill E, et al. Detection and characterization of clade 7 high pathogenicity avian influenza H5N1 viruses in chickens seized at ports of entry and live poultry markets in Vietnam. Avian Dis. 2010;54(1 Suppl):307–312. doi: 10.1637/8801-040109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 63.Tung DH, Van Quyen D, Nguyen T, Xuan HT, Nam TN, Duy KD. Molecular characterization of a H5N1 highly pathogenic influenza virus clade 2.3.2.1b circulating in Vietnam in 2011. Vet Microbiol. 2013;165:341–348. doi: 10.1016/j.vetmic.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 64.Bui VN, Dao TD, Nguyen TT, et al. Pathogenicity of an H5N1 avian influenza virus isolated in Vietnam in 2012 and reliability of conjunctival samples for diagnosis of infection. Virus Res. 2014;179:125–132. doi: 10.1016/j.virusres.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tien NN. Current situation of avian influenza, period of 2009-2012, and actual prevention solutions. Vet Sci Tech. 2013;20:82–90. [Google Scholar]
- 66.Hanh TX, Thi P, Thuy DH, et al. Testing the effect of the NAVET-Vifluvac vaccine against A/H5N1 viruses of clade 1.1 and 2.3.2.1c by challenging and direct contact of virulent strains. Vet Sci Tech. 2013;20:22–29. [Google Scholar]
- 67.Li Y, Liu L, Zhang Y, et al. New avian influenza virus (H5N1) in wild birds, Qinghai, China. Emerg Infect Dis. 2011;17:265–267. doi: 10.3201/eid1702.100732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nga NT, Le TH. Clarification of new clades 1.1 and 2.3.2.1 of avian influenza virus (H5N1) in Vietnam based on genetic and phylogenetic analysis of hemagglutinin (H5) genes of strains collected during 2004-2011. Vet Sci Tech. 2012;19:20–28. [Google Scholar]
- 69.Nga NT, Khien DV, Le TH. Confirmation of clades 2.3.2.1a and 2.3.2.1c of A/H5N1 viruses isolated in 2013 in Vietnam. Vet Sci Tech. 2013;20:16–21. [Google Scholar]
- 70.Gerloff NA, Khan SU, Balish A, et al. Multiple reassortment events among highly pathogenic avian influenza A(H5N1) viruses detected in Bangladesh. Virology. 2014;450-451:297–307. doi: 10.1016/j.virol.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 71.Phuong do Q, Dung NT, Jorgensen PH, Handberg KJ, Vinh NT, Christensen JP. Susceptibility of Muscovy (Cairina Moschata) and mallard ducks (Anas Platyrhynchos) to experimental infections by different genotypes of H5N1 avian influenza viruses. Vet Microbiol. 2011;148:168–174. doi: 10.1016/j.vetmic.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 72.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 60. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sorn S, Sok T, Ly S, et al. Dynamic of H5N1 virus in Cambodia and emergence of a novel endemic sub-clade. Infect Genet Evol. 2013;15:87–94. doi: 10.1016/j.meegid.2012.05.013. [DOI] [PubMed] [Google Scholar]