Mobile elements are important evolutionary forces that challenge genomic integrity. Long interspersed element-1 (L1/LINE-1) is the only autonomous transposon still active in the human genome. Here, Castro-Diaz et al. show that early embryonic control of L1 is an evolutionarily dynamic process. The data indicate that two transcriptional repression systems are engaged in repressing L1 in human embryonic stem cells. A model is proposed in which newly emerged L1 lineages are first suppressed by DNA methylation-inducing small RNA-based mechanisms before sequence-specific protein repressors are selected.
Keywords: DNA methylation, evolution, KAP1, KRAB-ZFPs, LINE1, embryonic stem cells
Abstract
Mobile elements are important evolutionary forces that challenge genomic integrity. Long interspersed element-1 (L1, also known as LINE-1) is the only autonomous transposon still active in the human genome. It displays an unusual pattern of evolution, with, at any given time, a single active L1 lineage amplifying to thousands of copies before getting replaced by a new lineage, likely under pressure of host restriction factors, which act notably by silencing L1 expression during early embryogenesis. Here, we demonstrate that in human embryonic stem (hES) cells, KAP1 (KRAB [Krüppel-associated box domain]-associated protein 1), the master cofactor of KRAB-containing zinc finger proteins (KRAB-ZFPs) previously implicated in the restriction of endogenous retroviruses, represses a discrete subset of L1 lineages predicted to have entered the ancestral genome between 26.8 million and 7.6 million years ago. In mice, we documented a similar chronologically conditioned pattern, albeit with a much contracted time scale. We could further identify an L1-binding KRAB-ZFP, suggesting that this rapidly evolving protein family is more globally responsible for L1 recognition. KAP1 knockdown in hES cells induced the expression of KAP1-bound L1 elements, but their younger, human-specific counterparts (L1Hs) were unaffected. Instead, they were stimulated by depleting DNA methyltransferases, consistent with recent evidence demonstrating that the PIWI–piRNA (PIWI-interacting RNA) pathway regulates L1Hs in hES cells. Altogether, these data indicate that the early embryonic control of L1 is an evolutionarily dynamic process and support a model in which newly emerged lineages are first suppressed by DNA methylation-inducing small RNA-based mechanisms before KAP1-recruiting protein repressors are selected.
More than half of the human genome is derived from mobile elements, most of which are retrotransposons spreading by reverse transcription of an RNA intermediate and integration of the resulting DNA product (Cordaux and Batzer 2009). These endogenous retroelements (EREs) represent essential evolutionary forces but also threats to genomic integrity and, as such, are subjected to transcriptional repression from the earliest stages of embryogenesis. Reciprocal selective pressures are exerted between EREs and host defenses engaged in their control, which can often be traced through phylogenetic studies (Furano and Boissinot 2008).
Long interspersed element-1 (L1, also known as LINE-1) is the only autonomous transposon still active in humans. About 500,000 copies of L1 are present in the human genome, amounting to some 20% of its DNA content. Many L1 integrants are 5′-truncated owing to the abortive tendency of the target-primed reverse transcription mechanism used by this class of retroelements. Nevertheless, the human genome contains some 100 retrotransposition-competent L1 elements, >40 of which are highly active (Brouha et al. 2003; Beck et al. 2011). Furthermore, L1 provides the trans-acting functions required for the transposition of nonautonomous retroelements such as SINEs (short interspersed nuclear elements, which include Alu repeats in humans) and SVAs (SINE–VNTR–Alu, a composite hominoid-restricted ERE) (Dewannieux et al. 2003; Finnegan 2012). The 6- to 7-kb-long genome of a full-length L1 comprises a 5′ untranslated region (UTR) promoter region; two ORFs encoding, respectively, a nucleic acid-binding protein and a product endowed with endonuclease and reverse transcriptase activity; and a 3′ UTR ending with a poly(A) tail (Babushok and Kazazian 2007; Rosser and An 2012). As other EREs, L1 shapes transcriptional networks, for instance, through L1-initiated cellular transcripts or L1-contained enhancers or insulators (Speek 2001; Nigumann et al. 2002; Matlik et al. 2006; Slotkin and Martienssen 2007; Faulkner et al. 2009). L1 elements present in the human or mouse genomes can be subdivided into subfamilies based on nucleotide substitutions, insertions, and/or deletions. Furthermore, phylogenetic studies interestingly indicate that this class of retroelements displays an unusual pattern of evolution in which a single L1 lineage at a time is generally active within the genome of a species and amplifies to thousands of copies before its replacement by a new lineage, likely under selective pressures exerted by host defense mechanisms (Cordaux and Batzer 2009).
EREs are silenced during early embryogenesis by histone methylation, histone deacetylation, and DNA methylation through sequence-specific mechanisms that counter the wave of epigenetic modifications—mainly DNA demethylation—required for the reprogramming typical of this developmental period (Rowe and Trono 2011). For endogenous retroviruses (ERVs), key mediators of this process are the DNA-binding Krüppel-associated box domain-containing zinc finger proteins (KRAB-ZFPs) and their cofactor, KAP1 (KRAB-associated protein 1), also known as TRIM28 (tripartite motif protein 28) (Wolf and Goff 2007, 2009; Matsui et al. 2010; Rowe et al. 2010). In human embryonic stem (hES) and mouse ES (mES) cells, the KRAB-ZFP-mediated docking of KAP1 at EREs triggers the formation of heterochromatin through the recruitment of the SETDB1 (also known as ESET) histone methyltransferase, responsible for trimethylating histone 3 at Lys9; histone deacetylases; and HP1 (heterochromatin protein 1), which collectively induce transcriptional repression (Schultz et al. 2002; Ivanov et al. 2007). The further recruitment of DNA methyltransferases (DNMTs) results in permanent silencing marks, which are subsequently maintained throughout development without need for persistent expression of sequence-specific ERE-recognizing repressors (Quenneville et al. 2012; Rowe et al. 2013a).
Previous studies detected a modest up-regulation of L1 in KAP1- or SETDB1-deleted mES cells (Matsui et al. 2010; Rowe et al. 2010), suggesting that this class of ERE is regulated by alternative pathways. In line with this hypothesis, recent data pointed to the importance of small RNA-based repression in the control of L1 expression in human pluripotent stem cells (Ciaudo et al. 2013; Fadloun et al. 2013; Heras et al. 2013; Marchetto et al. 2013). The present study reveals that the KRAB/KAP1 pathway and DNA methylation, the known output of small RNA-based mechanisms, are both engaged in restricting L1 in ES cells but act on evolutionarily distinct sets of elements, which provides a remarkable illustration of the reciprocal selective pressures exerted between EREs and the host mechanisms responsible for their control.
Results
KAP1 associates with full-length L1 in hES cells
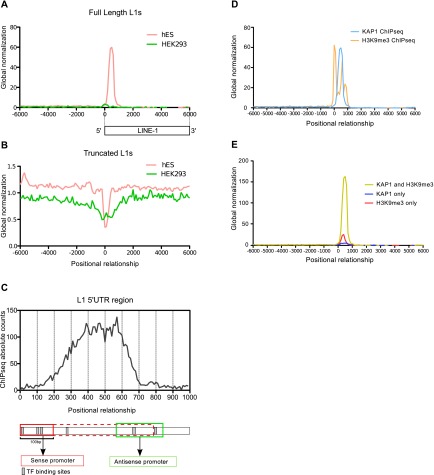
In order to investigate a possible role for KAP1 in the control of L1, we performed chromatin immunoprecipitation (ChIP) followed by deep sequencing (ChIP-seq) in H1 hES cells. We found that ∼8% of the total of L1-derived sequences annotated in the human genome somehow overlapped with KAP1 peaks in these cells. As most of the L1 sequences are 5′ truncated, we reasoned that only L1 copies endowed with a 5′ UTR would require transcriptional control; hence, we focused our analysis on L1 sequences >5 kb, assuming that they corresponded in their majority to full-length integrants. Fulfilling this prediction, 52% of these L1 sequences harbored a KAP1 peak, usually over their first 1000 base pairs (bp), contrasting with only 2% of elements <5 kb (Fig. 1A,B; Supplemental Fig. S1). Furthermore, while KAP1 was present on full-length L1s in hES cells, it was not significantly enriched at any L1-derived sequence in the differentiated human cell line HEK293 (Fig. 1A,B; Supplemental Fig. S1). A more detailed mapping of the ChIP-seq tags indicated that most of the KAP1 peaks targeted the middle region of the 5′ UTR, encompassing L1 nucleotides +300 to +600 (Fig. 1C). H3K9me3-specific ChIP-seq analyses confirmed a strong coincidence between KAP1 peaks and deposition of this repressive mark at the 5′ end of full-length L1 elements (Fig. 1D), with only a small minority of L1 bearing only KAP1 and with H3K9me3 seldom detected without the corepressor (Fig. 1E). Extending these findings, we could document the accumulation of KAP1 and H3K9me3 at the 5′ end of full-length L1 elements by performing the same type of analysis in mES cells (Supplemental Fig. S2).
Figure 1.
KAP1 coincides with H3K9me3 at the 5′ end of full-length L1 in hES cells. Distribution of ChIP-seq KAP1 peaks relative to the 5′ end of full-length elements (A) or the center of truncated L1 elements (B) in hES and HEK293 cells. The profiles were normalized to the total number of ChIP-seq peaks for each cell line. (C) KAP1 ChIP-seq peak distribution over the first kilobase of L1. The L1 5′ UTR is schematized below, with sense and antisense promoters as red and green boxes, respectively. Sense promoter is diversely depicted as mainly located in the first 100 bp or extending up to 700 bp. (D) Overlap of KAP1 and H3K9me3 ChIP-seq tags relative to the 5′ end of full-length L1 elements. (E) Relative frequency of KAP1+H3K9me3, KAP1-only, and H3K9me3-only peaks at this location.
KAP1-bound (KB) L1 sequences can act as cis-repressors in hES cells
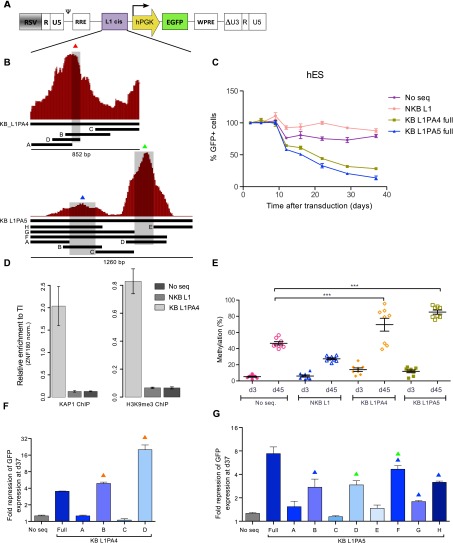
In order to assess the functional consequences of KAP1 recruitment at L1 sequences, we cloned KB regions (as defined by ChIP-seq in hES cells) from an L1PA4 and an L1PA5 element upstream of a PGK-GFP reporter cassette within the context of lentiviral vectors using corresponding non-KB (NKB) L1 fragments as negative controls (Fig. 2A,B). We then monitored GFP fluorescence in hES and 293T cells transduced with these vectors (Fig. 2C; Supplemental Fig. S3A). Expression from vectors containing full-length KB L1 fragments was progressively repressed in hES cells but not in 293T cells. In contrast, GFP fluorescence induced by the empty vector or harboring NKB L1 fragments remained strong over time in both cell types. ChIP followed by quantitative PCR (ChIP-qPCR) with PGK-specific primers confirmed that repression correlated with KAP1 enrichment and deposition of H3K9me3 (Fig. 2D; Supplemental Fig. S3B). Furthermore, L1-mediated KAP1 recruitment strongly stimulated the CpG methylation of the adjacent PGK promoter in hES cells (Fig. 2E). In order to define further the L1PA4- and L1PA5-derived KAP1-recruiting elements identified through this assay, we cut these ∼1-kb-long sequences into subfragments of ∼200 bp (Fig. 2B). This revealed that the cis-repressors contained in these retrotransposons coincided with the top of the corresponding KAP1 ChIP-seq peak (Fig. 2B; Supplemental Figs. 2FG, S3C,D). Of note, the KAP1-binding L1PA4 D subfragment induced faster and stronger repression than its full-length parent, suggesting that the latter contains elements with conflicting influences. In addition, while the tested L1-PA4 leader contained one KAP1-responsive cis-repressor, its L1PA5 counterpart harbored two such elements. Collectively, these data support a model in which the KAP1 corepressor is tethered to the 5′ end of subfamilies of L1 elements in hES cells, triggering their epigenetic silencing. Of note, our attempt to abrogate L1-induced KAP1-mediated repression of the PGK promoter by shRNA-mediated KAP1 depletion failed, probably because sufficient levels of KAP1 knockdown could not be maintained over time (data not shown).
Figure 2.
KAP1-binding L1 fragments can induce repression and DNA methylation of a heterologous promoter in hES. (A) KB (KB L1PA4 and KB L1PA5) and NKB (NKB L1PA4) L1 sequences were cloned in depicted lentiviral vector upstream of a PGK-EGFP expression cassette. The resulting vectors were transduced in hES, and EGFP expression was monitored over time by FACS. (B) Schematic representation of the KAP1 ChIP peaks mapped on the L1PA4 and L1PA5 5′ end, with indication of derived fragments and subfragments cloned in the vector depicted in A. (C) Monitoring of GFP expression in hES cells transduced with the indicated vectors. (No seq) Lentiviral vector with no ERE-derived fragment upstream of the expression cassette. The figure shows the mean and SD of two biological replicates. (D) KAP1 and H3K9me3 recruitment to indicated lentiviral vectors in hES, assessed 35 d after transduction by ChIP-qPCR using PGK-specific primers. The figure illustrates the mean and SD of technical replicates. This experiment was performed twice with similar results (see Supplemental Fig. S3). Relative enrichment was determined by normalizing to a known positive (ZNF180 3′ UTR) control. (E) Influence of the L1 cis-acting sequences on the methylation of the nearby PGK promoter. Methylation of eight CpG positions was evaluated by pyrosequencing at days 4 and 35 after transduction of hES cells with the PGK-GFP lentiviral vectors. Mean and standard error mean (SEM) of two biological replicates is shown. Statistical differences were determined by one-way ANOVA test using the Bonferroni multiple test adjustment. (***) P ≤ 0.001. (F,G) Fold repression of the indicated vectors containing L1 subfragments described in B, assessed 37 d after transduction (respect to day 5). Overtime fold repression is presented in Supplemental Figure S3. Colored triangles indicate the presence of L1 sequences overlapping with the summits of the respective KAP1 ChIP-seq peaks as depicted in B.
The KRAB–KAP1 system recognizes evolutionarily discrete subfamilies of human and mouse L1
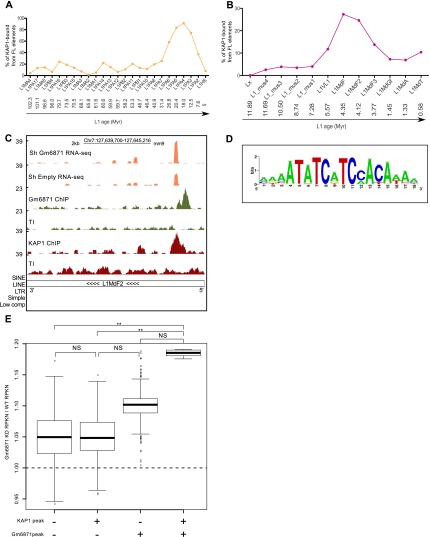
L1 displays an unusual pattern of evolution in mammals, with a single active lineage at any given time (Smit et al. 1995; Khan 2005; Sookdeo et al. 2013). This allows the approximate aging of L1 integrants in the genome of higher species and their sequence-based grouping in chronologically ordered subfamilies. By exploiting this feature, we could determine that KAP1 associated with only a small percentage of full-length human L1 belonging to lineages older than 26.8 million years (L1MA4 to L1PA7 subfamilies) and was practically absent from L1Hs; that is, L1 elements that invaded the human ancestral genome after the human–chimpanzee divergence some 7.6 million years ago. In contrast, KAP1 was recruited to a high fraction of L1PA6 to L1PA3 elements, peaking at >80% for the L1PA5, L1PA4, and L1PA3 subfamilies (Fig. 3A). Furthermore, H3K9me3 enrichment over full-length L1s from different subfamilies matched their KAP1-binding pattern, with this histone mark absent from very old L1s, highly enriched on the KAP1-recruiting L1PA5/PA4 and the rare KB L1HS, and present, albeit at much lower levels, on KAP1-devoid L1Hs (Supplemental Fig. S4). Remarkably, a similarly chronological pattern of KAP1 recruitment was recorded in mES cells, with KAP1 enrichment the highest on L1MdF and L1MdF2, estimated to be between 7.3 million and 3.8 million years old, and much lower on both older and younger L1 integrants (Fig. 3B).
Figure 3.
Evolutionally dynamic and KRAB-ZFP-mediated KAP1–L1 interaction. Percentage of KB full-length (FL) L1 elements per subfamily in hES (A) and mES (B) cells, arranged from the oldest to the youngest subfamily using ages obtained from previously published divergence analysis studies (Khan 2005; Sookdeo et al. 2013). (Myr) Million years. (C) Screenshot of a representative L1MdF2 element, illustrating RNA-seq coverage plots from control (shEmpty) and Gm6871 knockdown mES cells as well as gm6871 and Kap1 ChIP-seq tracks. (D) Putative gm6871 DNA-binding motif identified by computing gm6871 ChIP-seq peaks with the RSAT software (Thomas-Chollier et al. 2012). (E) Relative change in the expression (RPKN [ normalized reads per kilobase]) of murine full-length L1s bound or not bound by KAP1 and/or Gm6871 between Gm6871 knockdown and wild-type mES cells. The raw data were bootstrapped 1000 times with a resampling size of 100 for the plot design. The statistical analyses were calculated on the entire raw data by Wilcoxon nonparametric test. (NS) P > 0.05; (**) P ≤ 0.01.
In the context of a screen based on ChIP-seq of mES cells with HA-tagged KRAB-ZFPs, we identified Gm6871 as a L1 ligand with 104 full-length elements bound by both KAP1 and this mouse-specific KRAB-ZFP (Fig. 3C). The majority of them belonged to the L1MdF2 (64%) and L1MdF3 (13%) subfamilies, and a search performed on all Gm6871-recruiting sequences identified a putative Gm6871 DNA-binding motif (Fig. 3D) present in 95% of these L1 elements, contrasting with only 0.2% of elements from the younger L1MdA and L1MdT subfamilies. Gm6871 was so far only a predicted gene, but we could detect its expression in both pluripotent and differentiated cells, albeit with higher levels in mES cells compared with fibroblasts (Supplemental Fig. S5), as previously described for many KRAB-ZFPs (Corsinotti et al. 2013). We also could document the nuclear localization of a HA-tagged derivative of Gm6871 expressed in mES cells by lentivector-mediated transduction (Supplemental Fig. S6A) and demonstrate an interaction between Gm6871 and KAP1 by coimmunoprecipitation of extracts from Gm6871-HA-expressing mES and 293T cells and by KAP1-GST pull-down assay (Supplemental Fig. S6B,C). To ascertain the functional relevance of this interaction, we depleted endogenous Gm6871 in mES cells by lentivector-mediated RNAi and evaluated L1 mRNA expression by RNA deep sequencing (RNA-seq). Consistent with the ChIP-seq results, upon Gm6871 knockdown, we observed a significant increase in the levels of L1 sequences identified in control cells as binding either KAP1+Gm6871 or Gm6871 alone but not of L1s bound only by KAP1 or associated with neither protein (Fig. 3C,E). In order to demonstrate further the implication of Gm6871 in the control of specific L1s, we performed a KAP1 ChIP in mES cells transduced with a control or Gm6871-directed shRNA-expressing lentiviral vector followed by qPCR with primers specific for three KAP1- and Gm6871-associated L1 elements. As controls, we included ICR (imprinting control region) sequences, known to recruit KAP1 independently of Gm6871 (Quenneville et al. 2011), and another genomic locus highly enriched for KAP1 and Gm6871 in our ChIP-seqs. The results revealed a mild but reproducible reduction of KAP1 enrichment at the tested Gm6871-recruiting loci upon Gm6871 knockdown, while association of the corepressor with ICRs was unaffected (Supplemental Fig. S7). Finally, RT-qPCR performed in mES cells confirmed that these L1 elements were up-regulated upon removal of SETDB1 (Supplemental Fig. S8), the histone methyltransferase responsible for H3K9me3 induction by the KRAB–KAP1 complex (Schultz et al. 2002; Iyengar and Farnham 2011). Altogether, these results demonstrate that Gm6871 tethers KAP1 and associated chromatin modifiers to a specific subset of murine L1s and strongly suggest that the KRAB-ZFP family at large is involved in the sequence-specific repression of LINEs in higher vertebrates.
The KB subset of L1 is activated by KAP1 depletion in hES cells
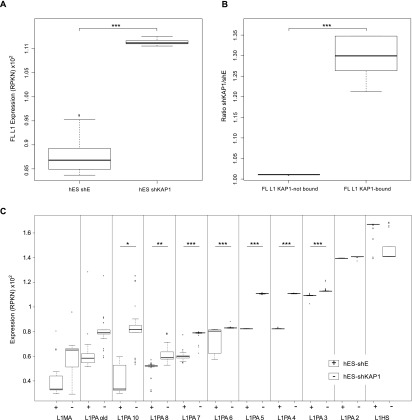
To probe the impact of KAP1 on the transcriptional control of L1, we used lentivector-mediated RNAi coupled with RNA-seq in hES cells. Global expression of full-length L1 was increased in KAP1-depleted compared with control ES cells (Fig. 4A; Supplemental Fig. S9), but this difference came only from KB elements (Fig. 4B). Analyzing levels of L1 transcripts for the various subfamilies (Fig. 4C) further revealed that ancient, infrequently KB elements were lowly expressed at baseline and were not or were only moderately affected by knocking down the corepressor. In comparison, members of the highly KAP1-enriched L1PA4 and L1PA5 subfamilies were more strongly expressed in control cells and were significantly up-regulated in KAP1-depleted cells. Of note, the fold change in L1PA4 and L1PA5 expression levels between control and KAP1-depleted cells was not only statistically significant but also the strongest among all evaluated subfamilies. Finally, expression of youngest elements (L1PA2 and L1Hs) was highest at baseline and unchanged upon KAP1 knockdown.
Figure 4.
KAP1 depletion leads to up-regulation of KB L1 in hES cells. (A) Comparative expression of full-length L1 elements in hES cells transduced with control (shE) or Kap1 knockdown (shKAP1) lentiviral vectors. (B) Relative change in the expression of full-length L1 elements bound or not by KAP1, comparing Kap1 knockdown and control hES cells. (C) Comparative expression of full-length L1 in control versus Kap1 knockdown hES cells, examining each L1 subfamily separately, arranged from the oldest to the youngest one. The “L1MA” category corresponds to the families L1MA1 to L1MA9. The “L1PA old” category corresponds to the families L1PA11 to L1PA17. The raw data have been bootstrapped 1000 times for the plot design. Expression corresponds to RPKN values (see the Material and Methods), with P-values ([*] P ≤ 0.05; [**] P ≤ 0.01; [***] P ≤ 0.001) calculated on the raw data by Wilcoxon nonparametric test.
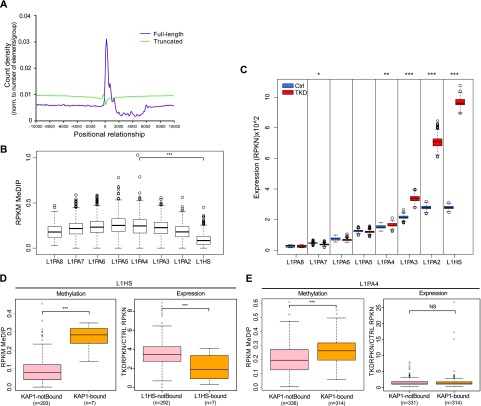
Youngest human L1 are up-regulated upon depletion of DNMTs
DNA methylation is involved in the long-term transcriptional control of EREs, including L1. Correspondingly, our analysis of MeDIP-seq data from the Epigenomics Mapping Consortium (Bernstein et al. 2010) indicated that DNA methylation is enriched at the 5′ region of mappable full-length L1 integrants in human H1 ES cells (Fig. 5A). It also revealed L1PA4 and L1PA5 as the most methylated and L1Hs as the least methylated subfamilies (Fig. 5B). Therefore, we investigated the relative impact of KAP1-mediated and DNA methylation-mediated mechanisms in the control of L1. For this, we generated hES cell populations depleted for the de novo (DNMT3A and DNMT3B) and maintenance (DNMT1) DNMTs by lentivector-mediated RNAi. DNMT3A and DNMT3B could be stably knocked down for >22 d, whereas DNMT1 expression was partially recovered at that point, suggesting a growth disadvantage in the absence of this enzyme (data not shown). Still, cells in which all three DNMTs were strongly depleted (Supplemental Fig. S9A,B) could be readily obtained and kept in culture for the time of our study, as reflected by their complete loss of DNA methylation at the GRB10 ICR after 5 or 9 d of triple knockdown (Supplemental Fig. S9C). Most interestingly, comparing the expression of individual L1 elements revealed that, in DNMT-depleted cells, it was the members of the youngest, KAP1-unbound L1 subfamilies (L1PA2 and L1Hs) that were the most up-regulated at that point, whereas older elements, whether KAP1-controlled or not, were not or were very modestly affected (Fig. 5C). In an attempt to explore further the interplay between KAP1-mediated and DNA methylation-mediated repression of L1, we separated L1PA4 and L1Hs family members in KB and KAP1-devoid elements. Next, we looked at their methylation levels at baseline and at their expression upon DNMT triple knockdown. For L1Hs, we found that the KAP1-devoid elements, which were the overwhelming majority within this group, were significantly less methylated at baseline than their rare KB counterparts and that they alone were induced upon DNMT knockdown (Fig. 5D). Within the L1PA4 subfamily, baseline DNA methylation was globally higher and more homogeneous, with only slightly lower levels for KAP1-free members. Furthermore, expression of all L1PA4 elements was comparable in control and triple knockdown cells. Of note, depleting KAP1 in DNMT knockdown cells was highly toxic, precluding the further exploration of potential synergies between the two L1 repression pathways.
Figure 5.
DNMT depletion induces up-regulation of younger L1 subfamilies. (A) Comparative expression of full-length L1 in control and DNMT knockdown (triple knockdown [TKD]) hES cells, analyzing each subfamily separately as described in Figure 4C. (B) Distribution of MeDIP-seq reads relative to the 5′ end of full-length (pink line) or truncated (yellow line) L1, normalizing profiles to the total number of elements per group. (C) DNA methylation levels on full-length L1 elements separated by subfamilies. (D) DNA methylation levels in full-length L1PA4 and L1Hs bound or not by KAP1 in hES cells, based on the numbers of MeDIP-seq reads per million base pairs per kilobase of L1 (RPKM). (E) Relative change in the expression of the same L1 elements, comparing triple knockdown and control hES cells. P-values ([NS] P > 0.05; [*] P ≤ 0.05; [**] P ≤ 0.01; [***] P ≤ 0.001) were calculated with Wilcoxon nonparametric test.
Discussion
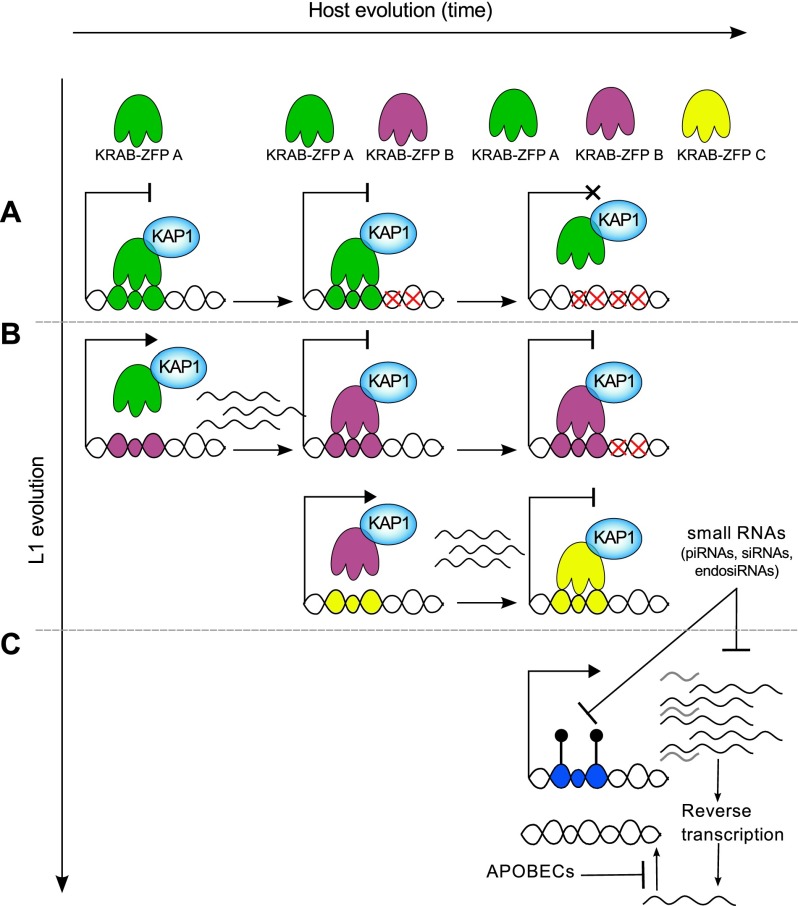
The transcriptional silencing of EREs is essential to protect genomic integrity, particularly during the vulnerable phases of developmental reprograming that occurs in ES and germ cells. Previous studies have revealed the roles of KRAB-ZFPs and their cofactor, KAP1, in the early embryonic repression of ERVs (Wolf and Goff 2009; Rowe et al. 2010; Tan et al. 2013), whereas small RNA-based mechanisms have been thought to prevail for the silencing of L1 elements, as initially discovered in germ cells (Yang and Kazazian 2006; Aravin et al. 2007; Carmell et al. 2007; Beck et al. 2011). The present study actually establishes that L1 expression is also controlled by the KRAB–KAP1 system. Furthermore, our data, coupled with the recent demonstration that PIWI partakes in the regulation of L1Hs elements in human pluripotent cells (Marchetto et al. 2013), strongly support an evolutionary model in which the transcription of newly emerged L1 lineages is first repressed by small RNA-induced DNA methylation before KAP1-mediated silencing takes over through the selection of KRAB-ZFPs capable of tethering the master corepressor to their sequence (Fig. 6).
Figure 6.
Model for the evolutionally dynamic control of L1. (A) Very ancient L1s (shown in the top row) may have been once recognized by the KRAB/KAP1 system but have since then accumulated mutations (red crosses) abrogating binding by cognate KRAB-ZFPs but also transcription ability. (B) More recent subfamilies recruit KAP1 through sequence-specific KRAB-ZFPs but also may have some mutations taming their baseline expression. (C) The youngest L1 elements are highly transcribed and are not yet recognized by any KRAB-ZFP but produce small RNAs such as piRNAs, which in turn down-regulate their expression via DNA methylation and see their retrotransposition further blocked by proteins such as APOBEC family members.
In both hES and mES cells, we found that KAP1 regulates L1 but that this control is restricted to lineages that have entered the corresponding ancestral genomes during the periods 31 million to 7.6 million years ago and 5.6 million to 3.8 million years ago, respectively. We identified a novel KRAB-ZFP responsible for tethering KAP1 to and controlling the expression of a subset of murine L1, strongly suggesting that these DNA-binding proteins are collectively involved in recognizing this class of retroelements, as previously observed for other EREs (Tan et al. 2013), and that in return, L1 has contributed to the species-specific diversification of the KRAB-ZFP gene family. However, we also determined that younger L1 lineages are generally not subjected to KRAB/KAP1-mediated regulation, whether in humans or mice. We found that the human-specific L1Hs, most of which neither recruit KAP1 nor are activated by KAP1 depletion, were instead induced upon depletion of DNMTs in hES cells. This observation fits well with the recent discovery that the PIWI-interacting RNAs (piRNAs)–PIWI system partakes in the early embryonic control of youngest L1 lineages in humans and apes (Marchetto et al. 2013). PIWI-mediated control, which was initially thought to be relevant only in germ cells, is indeed triggered by the recognition of L1-proximal sequences by a complex encompassing a member of the PIWI subclade of Argonaute proteins and L1-derived piRNAs, which leads to L1 transcriptional inhibition via DNA methylation (Aravin et al. 2007; Carmell et al. 2007; De Fazio et al. 2011). Whether other small RNA-based mechanisms reported to partake in the early embryonic control of L1 (Ciaudo et al. 2013; Fadloun et al. 2013; Heras et al. 2013) also act in a lineage-specific fashion remains to be determined.
Our finding that KAP1 binds a significant subset of L1s in ES cells but only exceptionally in HEK293T cells fits with the establishment of permanent silencing marks on EREs, including LINEs, during the early embryonic period. However, that it still is found on some L1 integrants in the differentiated cells suggests that particular L1s and their control mechanisms have been coopted to fulfil some roles in adult somatic tissues.
The presence of two KAP1-repressed DNA elements in a L1PA5-derived sequence (Fig. 2) and the weak effect of Gm6871 knockdown on L1 transcription raise the possibility of some redundancy in the KRAB-ZFP-mediated control of L1s. Furthermore, although several mechanisms of L1 restriction have been described, their inactivation never results in spectacular up-regulation of these elements (nothing comparable, for instance, with the several hundred-fold induction undergone by some ERVs when KAP1 is deleted in mES cells) (Rowe et al. 2010). While this suggests that L1s are subjected to several layers of control, KAP1-restricted L1s belong to subfamilies more ancient and less active than human L1Hs and may have accumulated, over time, mutations that attenuate their transcriptional potential, dampening their up-regulation upon KAP1 removal. As for more ancient L1 lineages, their lack of KAP1 binding, coupled with their low level of baseline expression and inertia upon either KAP1 or DNMT depletion, is likely explained by the accumulation of inactivating mutations, alleviating the need for any sort of transcriptional control.
That KAP1-regulated elements are unaffected by depleting DNMTs, whereas the KAP1 recruitment at ERVs ultimately leads to their DNA methylation, is not surprising. The KRAB/KAP1 system indeed represses transcription of EREs primarily via histone deacetylation, H3K9 trimethylation, and HP1 recruitment, with DNA methylation occurring only secondarily to ensure the permanence of the silencing process (Quenneville et al. 2012; Rowe et al. 2013a). Our observation that KB murine L1 elements are up-regulated upon Setdb1 knockout in mES cells confirms the primary importance of histone methylation-based mechanisms in their control. Some rare L1Hs were found to be KB and accordingly were barely induced upon DNMT knockdown, in contrast to their far more prevalent KAP1-devoid counterparts. For L1PA4 subfamily members, we did not see any induction in DNMT triple knockdown cells whether they bore KAP1 or not. However, these elements were globally highly methylated, which may explain their resistance to the DNMT knockdown. It could also be that other epigenetic modifications, some of which may be lasting consequences of earlier KAP1 recruitment, partake in their repression.
Retrotransposons are mutagenic yet harbor cis-acting activities, many of which contribute to shaping transcriptional networks, including in ES cells (Bourque et al. 2008; Kunarso et al. 2010; Jacques et al. 2013; Rowe et al. 2013b; Ward et al. 2013). They thus have both a detrimental and an evolutionarily beneficial potential, which requires that they be very delicately controlled. We propose that this is accomplished, at least for the youngest, most active L1 elements, via autoregulation of piRNA production, a repression mechanism that is in part self-imposed. For elements that escape this process, additional restrictions are exerted at the post-transcriptional level, for instance, through lethal editing of reverse transcripts by the APOBEC3B cytidine deaminase (Bogerd et al. 2006; Chiu and Greene 2008; Wissing et al. 2011; Marchetto et al. 2013). After some time, KAP1-induced restriction, which appears more stringent, takes over through the selection of L1-recognizing KRAB-ZFPs. It will be interesting to ask whether a similar level of complexity prevails to the control of this class of retroelements in germ cells, where the reprogramming of epigenetic marks opens another window for their activation.
Materials and methods
Plasmids and lentiviral vectors
pLKO.1.puro shRNA vectors were used for KAP1, Gm6871, and DNMT1 knockdown. shRNAs against DNMT3A and DNMT3B were cloned into the pLVTHM vector, which was further modified to express neomycin, hygromycin, or blasticidine resistance genes instead of GFP. For each shRNA vector, an empty version (without shRNA) was cloned as a control. The shRNA targeting sequences were obtained through the RNAi Consortium (http://www.broadinstitute.org/rnai/public) and are listed in the Supplemental Material (Supplemental Table 2). L1 cis-acting sequences (see “Genomic Coordinates” in the Supplemental Material) were cloned into the pENTR/D/TOPO vector an then into an in-house cloned gateway destination vector by LR recombination (pRRL.R1-R2.PGK.GFP). Codon-optimized Gm6871 was synthesized and introduced by Gateway cloning in a puromycin selectable lentivector under a tetracyclin-inducible TRE promoter to obtain an HA-tagged protein (pSIN-TRE-Gm6871-3xHA, Addgene). LV production protocols are detailed at http://tronolab.epfl.ch. LV backbones are available at Addgene (http://www.addgene.org).
ES cell culture and transduction
The H1 hES cell line (WA01, WiCell) was cultured in mTeSR1 medium (Stem Cell Technologies) on hES-qualified Matrigel (BD Biosciences) and in the presence of ROCK inhibitor (Y-27632). mES (ES3 and J1) cell lines were cultured as previously described (Rowe et al. 2013b). J1 cells culture was further supplemented with 1 μM PD0325901 and 3 μM CHIR99021. mES cells were grown on 0.1% gelatin-coated (48723-500G-F, Sigma) plates. Transductions were done at a multiplicity of infection (MOI; determined in HCT116 or 3T3 cells) of 0.25–50. Whenever required, cells were selected with 100 μg/mL hygromycin, 10 μg/mL blasticidin, 0.25 μg/mL or 1.0 μg/mL puromycin, or 200 μg/mL neomycin. Pluripotency was monitored by FACS using a human pluripotent stem cell transcription factor analysis kit (BD Biosciences) or mouse anti SSEA-1 PE-conjugated antibody (560142, BD Pharmingen).
RT-qPCR and RNA-seq
Total RNA was extracted and DNAse I-treated using a spin column-based RNA purification kit (Macherey Nagel). cDNA was synthesized starting from 500 ng of RNA and using random hexamers and SuperScript II (Invitrogen). Primers (Supplemental Table 1) were used for SYBR Green qPCR (Applied Biosystems), and their specificity was confirmed with dissociation curves. RT-qPCR reactions were performed in triplicate for each cDNA sample. hES RNA-seq was generated with RNA extracted 14 d after KAP1 depletion (Turelli et al. 2014) or 9 d after triple DNMT depletion. mES RNA-seq was done in ES3 cells 4 d after the sh-Gm6871 knockdown vector transduction (MOI50), in duplicate (independent transductions). Knockdown levels were of 0.87 and 0.88 by qPCR. The 76- or 100-bp single-end reads from the Illumina HiSeq sequencing instrument were mapped using the Bowtie short read aligner (Langmead et al. 2009) to the annotated sequence of individual full-length L1 (minimum 5 kb in length) (lists provided in the Supplemental Material). The annotation and genomic coordinates of full-length L1 elements were obtained from the University of California at Santa Cruz genome browser. Reads mapping to multiple locations were evenly distributed across those locations, and a maximum of three mismatches was allowed. The RPKN (normalized reads per kilobase) values were calculated using an in-house R program and correspond to the read counts normalized to the length of the repeated element and to the total number of reads mapped to the transcriptome.
ChIP-seq and ChIP-qPCR
Chromatin was prepared from 1 × 107 H1 hES or J1 mES cells (for KAP1 ChIPs) and from 2 × 107 ES3 cells (for Gm6871 ChIP) as previously described (Barde et al. 2013; Rowe et al. 2013b; Turelli et al. 2014) with KAP1-specific (Tronolab, SY326768 or ab10483, Abcam), H3K9me3-specific (Diagenode), or HA-specific (Covance; MMS-101P) antibodies. For sequencing, total input (TI) and ChIP library preparation was performed as described in Santoni de Sio et al. (2012) using between 2 and 10 ng of chromatin. Sequencing was performed on an Illumina genome analyzer IIx, with each library sequenced in 80-base single-read or 100-bp reads run. The 80- to 100-bp single-end or paired-end reads generated were mapped to the human genome assembly hg19 or mouse genome assembly mm9 using the Bowtie short read aligner (Langmead et al. 2009), allowing up to two to three mismatches, and all multiple matches were discarded. The peaks were called using the MACS program (Zhang et al. 2008) and were normalized to the TI. When defining KB and NKB L1 sequences, only KAP1 peaks with a MACS score [Log10(pval)] >100 were considered. ChIP-seq data in HEK293 cells (Iyengar et al. 2011) were obtained from the ENCODE database (https://genome.ucsc.edu/ENCODE). H3K9me3 ChIP-seq data in mES (ES3) cells was previously published in Rowe et al. (2013b). Motif search was performed with RSAT (Thomas-Chollier et al. 2012) using Gm6871 called peaks as input and unbound repeated regions as background control. Correlation analysis between ChIP-seq peaks, MeDIP-seq tags, and L1 elements was done using the ChIP-cor analysis module (http://ccg.vital-it.ch/chipseq/chip_cor.php).
FACS analysis
Cells were analyzed on a FACScan machine (Becton Dickinson). Analysis was performed with FlowJo software (version 8-1.8.6, Treestar, Inc.).
DNA methylation
For quantitative bisulfite pyrosequencing, genomic DNA was converted (1–2 μg per sample) using an Epitect bisulfite kit (591014, Qiagen) and used for PCR (primers were designed on the converted antisense and sense strand, respectively, using PyroMark Assay Design 2.0 software). Purity of PCR products was verified on agarose gels for each experiment before immobilizing on 96-well plates using a vacuum prep workstation and pyrosequencing using PyroMark gold reagents (972804, Qiagen; Center for Integrative Genomics, University of Lausanne, Switzerland). Results were analyzed using Pyro Q-CpG software. Primer sequences are in Supplemental Table 1. MeDIP-seq data sets (Hs1376 and Hs1303) were downloaded from http://www.genboree.org/epigenomeatlas. COBRA methylation analysis was performed using primers for the GRB10 human ICR (see the Supplemental Material) and as previously described (Xiong and Laird 1997).
Immunoblotting
Cells were washed with ice-cold PBS and resuspended in radioimmunoprecipitation (RIPA) buffer to prepare total cell extracts. Protein amount was quantified by BCA protein assay reagents (Pierce) and normalized for loading on a 10% denaturing SDS–polyacrylamide gel. Wet transfer was performed, and the primary antibodies used were anti-DNMT1 (rabbit pAb; ab87654, Abcam), anti-DNMT3A (mouse mAb; ab13888, Abcam), anti-DNMT3B (rabbit pAb; ab2851, Abcam), and β-tubulin (rabbit pAb; ab21058, Abcam).
Immunofluorescence
mES cells were transduced with Gm6871-HA, ZFP809-HA, or LacZ-HA and cultured with 5 μg/mL doxycycline. Cells were fixed in methanol for 10 min and labeled with anti-HA antibody (MMS-101P, Covance) followed by Alexa488-conjugated anti-mouse antibody. Nuclei were stained with DAPI. Images were acquired using a 63× lens on a Zeiss Axiovert 200M microscope.
Coimmunoprecipitation and GST pull-down
Gm6871-HA, ZFP809-HA, KRAB-deleted ZFP809, or LacZ-HA plasmids were used to transduce mES cells or transfect 293T cells. Cells were cultured with 5 μg/mL doxycycline for at least 48 h, harvested, and lysed with lysis buffer (50 mM Tris HCl at pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycolate) supplemented with protease inhibitors under constant agitation for 30 min. Lysate was sonicated twice for 10 sec at 30% duty cycle. Immunoprecipitation was performed overnight with HA antibody (MMS-101P, Covance) in immunoprecipitation buffer (50 mM Tris HCl at pH 8, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40) supplemented with protease inhibitors. All steps were performed at 4°C. Immunoblotting was performed with either anti-KAP1 antibody (ab10483, Abcam) followed by HRP-conjugated anti-rabbit antibody or HRP-conjugated anti-HA antibody (12013819001, Roche). Ex vivo GST pull-down assay was performed as previously described (Yahi et al. 2008).
Accession numbers
RNA-seq and ChIP-seq data were deposited in the Gene Expression Omnibus database at the NCBI under the accession numbers GSE57989 (Turelli et al. 2014) and GSE58323.
Acknowledgments
We thank E. Planet for help with data analysis, the staff of our Genomics core facility for sequencing, Vital-IT for computing, C. Raclot and S. Offner for technical help, and the members of our laboratory for stimulating discussions. This work was financed through grants from the Swiss National Science Foundation and the European Research Council to D.T (ERC 268721). N.C.-D. designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. G.E. and A.C. designed, performed, and analyzed experiments. A.K. and J.D. analyzed data. B.Y. and M.F. made intellectual contributions. S.M.J. performed experiments. P.T. designed, performed, and analyzed experiments and helped supervise the project. D.T. conceived and directed the study, analyzed and interpreted the results, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.241661.114.
References
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH 2007. Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat 28: 527–539 [DOI] [PubMed] [Google Scholar]
- Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, Offner S, Marquis J, Kapopoulou A, Vanicek J, et al. 2013. A KRAB/KAP1–miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science 340: 350–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12: 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci 103: 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol 26: 317–353 [DOI] [PubMed] [Google Scholar]
- Ciaudo C, Jay F, Okamoto I, Chen CJ, Sarazin A, Servant N, Barillot E, Heard E, Voinnet O 2013. RNAi-dependent and independent control of LINE1 accumulation and mobility in mouse embryonic stem cells. PLoS Genet 9: e1003791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cordaux R, Batzer MA 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D 2013. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS ONE 8: e56721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla M-E 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338 [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- Finnegan DJ 2012. Retrotransposons. Curr Biol 22: R432–R437 [DOI] [PubMed] [Google Scholar]
- Furano AV, Boissinot S 2008. Long interspersed nuclear elements (LINEs): evolution. eLS doi: 10.1002/9780470015902.a0005304.pub2 [Google Scholar]
- Heras SR, Macias S, Plass M, Fernandez N, Cano D, Eyras E, Garcia-Perez JL, Cáceres JF 2013. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol 20: 1173–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ 2011. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 286: 26267–26276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ 3rd, Farnham PJ 2011. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol 31: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9: e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H 2005. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlik K, Redik K, Speek M 2006. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol 2006: 71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634 [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D 2012. The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Reports 2: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser JM, An W 2012. L1 expression and regulation in humans and rodents. in Front Biosci (Elite Ed) 4: 2203–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D 2013a. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D 2013b. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res 23: 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, Genolet R, Thomas JH, Luescher IF, Pinschewer D et al. 2012. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J 26: 4561–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Smit AF, Tóth G, Riggs AD, Jurka J 1995. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246: 401–417 [DOI] [PubMed] [Google Scholar]
- Sookdeo A, Hepp CM, McClure MA, Boissinot S 2013. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV 2013. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res 11: 1045–1059 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M, Darbo E, Herrmann C, Defrance M, Thieffry D, van Helden J 2012. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc 7: 1551–1568 [DOI] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. 2014. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res doi: 10.1101/gr.172833.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MC, Wilson MD, Barbosa-Morais NL, Schmidt D, Stark R, Pan Q, Schwalie PC, Menon S, Lukk M, Watt S, et al. 2013. Latent regulatory potential of human-specific repetitive elements. Mol Cell 49: 262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC 2011. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem 286: 36427–36437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW 1997. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25: 2532–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahi H, Fritsch L, Philipot O, Guasconi V, Souidi M, Robin P, Polesskaya A, Losson R, Harel-Bellan A, Ait-Si-Ali S 2008. Differential cooperation between heterochromatin protein HP1 isoforms and MyoD in myoblasts. J Biol Chem 283: 23692–23700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kazazian HH Jr 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 13: 763–771 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]