Transposable elements (TEs) occupy a large fraction of metazoan genomes and pose constant threats to genomic integrity. Small noncoding piwi-interacting RNAs (piRNAs) recognize and silence a diverse set of TEs in germ cells. Pezic et al. show the piRNA pathway is required to maintain a high level of the repressive H3K9me3 histone modification on long interspersed nuclear elements (LINEs) in mammalian germ cells. The analyses reveal that the piRNA pathway targets full-length elements of actively transposing LINE families but not the copious small fragments present throughout the genome.
Keywords: H3K9me3, piRNA, transposon
Abstract
Transposable elements (TEs) occupy a large fraction of metazoan genomes and pose a constant threat to genomic integrity. This threat is particularly critical in germ cells, as changes in the genome that are induced by TEs will be transmitted to the next generation. Small noncoding piwi-interacting RNAs (piRNAs) recognize and silence a diverse set of TEs in germ cells. In mice, piRNA-guided transposon repression correlates with establishment of CpG DNA methylation on their sequences, yet the mechanism and the spectrum of genomic targets of piRNA silencing are unknown. Here we show that in addition to DNA methylation, the piRNA pathway is required to maintain a high level of the repressive H3K9me3 histone modification on long interspersed nuclear elements (LINEs) in germ cells. piRNA-dependent chromatin repression targets exclusively full-length elements of actively transposing LINE families, demonstrating the remarkable ability of the piRNA pathway to recognize active elements among the large number of genomic transposon fragments.
Repetitive elements occupy a large portion of mammalian genomes (∼50% in humans and ∼40% in mice) and play important roles in large-scale chromatin structure, expression of adjacent protein-coding genes, and evolution of genomes (Mouse Genome Sequencing Consortium 2002; Slotkin and Martienssen 2007; Goodier and Kazazian 2008). Several types of short repeats—so-called satellite sequences—occupy vast domains of centromeric and pericentric regions of mammalian chromosomes and are required for chromosomal structure and segregation during cell division. Other repeats are scattered throughout the genome intermingled with genes. Many of these repeats are derived from transposable elements (TEs) that have the capacity to replicate and insert into new genomic locations, leading to their expansion in the genome. Several classes of TEs are abundant and active in the murine genome, including the long interspersed nuclear elements (LINEs) that are also active in the human genome and the long terminal repeat (LTR) elements that are related to retroviral sequences.
TEs represent a large threat to genomic integrity due to their capacity to induce dsDNA breaks, insertional mutations, and chromosome rearrangements (Goodier and Kazazian 2008). This threat is particularly critical in germ cells, as genome changes induced by TEs will be transmitted to the next generation. Many species have evolved efficient strategies to control TE activity. In germ cells of flies and mice, TE silencing is orchestrated by a specific class of small RNAs called piRNAs (piwi-interacting RNAs), which are associated with members of the Piwi clade of Argonaute proteins. One of the Piwi proteins in mice, MIWI2, localizes to the nucleus and is required for establishment of the CpG DNA methylation mark on genomic sequences of transposons in germ cells (Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). In Drosophila, piRNAs also repress TEs in the nucleus. Flies, however, lack genomic DNA methylation, and instead, the piRNA pathway has been linked to repressive histone modifications (Klenov et al. 2011; Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013).
Epigenetic mechanisms such as DNA methylation and post-translational modifications of histones control patterns of gene expression in each cell type of multicellular organisms (Reik 2007). Modifications of N-terminal histone tails play major roles in both activation and repression of genes as well as in regulation of large-scale chromatin structure. Specifically, trimethylation of histone 3 at Lys9 (H3K9me3) is generally associated with sequences that are not expressed, including genes that are repressed in particular tissues as well as permanently silenced heterochromatic regions (Barski et al. 2007; Mikkelsen et al. 2007; Reik 2007). Chromatin immunoprecipitation (ChIP) coupled to massively parallel sequencing technology (ChIP-seq) has enabled genome-wide studies of histone modifications (Mikkelsen et al. 2007); however, most studies have focused on single-copy sequence elements such as genes and their regulatory regions. Here, we studied the H3K9me3 mark on repetitive elements in different tissues in mice and explored the link between this mark and the piRNA pathway. We found that several LTR and LINE families have a high level of H3K9me3 in both somatic and germ cells. This repressive histone modification is further elevated on LINE1 elements in germ cells, and this increase depends on the piRNA pathway. The spreading of H3K9me3 into the genomic environment surrounding LINE1 elements allowed us to accurately determine the genomic targets of piRNA-guided repression. Our analysis revealed that the piRNA pathway targets a subset of LINE1 loci that contain full-length retrotransposon insertions but not the copious small fragments present throughout the genome.
Results
Distribution of H3K9me3 on TEs in somatic and germ cells
To understand the genome-wide distribution of the H3K9me3 mark on TEs in somatic and germ cells, we performed ChIP-seq on liver cells, somatic cells of the testis, and premeiotic male germ cells (spermatogonia) isolated from 10-d-old (10 d post-partum [dpp]) mice. Two independent methods were used for purification of germ and somatic cells from testis: fluorescent-activated cell sorting (FACS) based on the GFP signal from the GFP-Mili transgene expressed exclusively in germ cells (Aravin et al. 2008) and magnetic-activated cell sorting (MACS) using an antibody against the cell surface marker EpCAM, expressed only on spermatogonia (Anderson et al. 1999; Kanatsu-Shinohara et al. 2011). Isolation of germ cells by FACS resulted in >95% germ cell purity, as compared with ∼50% in the MACS-sorted samples. For each cell type, we generated and sequenced H3K9me3 ChIP and control input libraries from several independent biological samples (Supplemental Table S1).
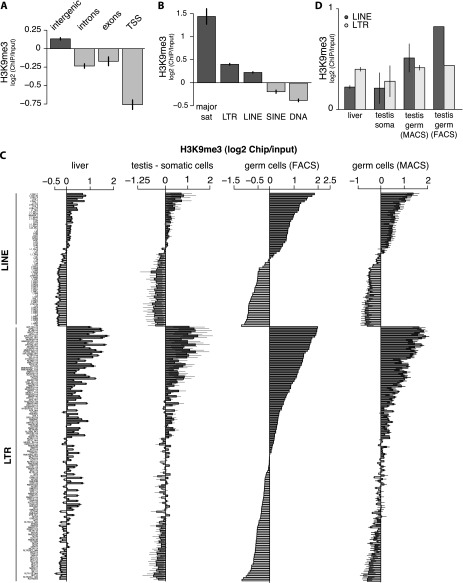
We analyzed the density of the H3K9me3 mark over different genomic features. In all three cell types, the H3K9me3 signal was strongly reduced at transcription start sites (TSSs) of protein-coding genes compared with its average genomic density (Fig. 1A; Supplemental Fig. S1A). Exons and introns also showed moderate depletion of the H3K9me3 mark. In contrast, the H3K9me3 mark was slightly enriched in intergenic regions of the genome.
Figure 1.
Distribution of the H3K9me3 mark along the genome of mouse somatic and male germ cells. (A) Distribution of the H3K9me3 mark in liver cells of 10-dpp animals on four types of genomic partitions (TSSs, exons, introns, and intergenic space). Values of four ChIP-seq replicas were normalized to the respective input samples and averaged. Error bars show standard deviations. (B) Distribution of the H3K9me3 mark in liver cells of 10-dpp animals over five major TE classes (LINEs, SINEs, LTRs, DNA, and satellites). Values of four ChIP-seq replicas were normalized to the respective input samples and averaged. (C) Enrichment of the H3K9me3 mark over TE families within the LTR and the LINE classes in liver cells, testicular somatic cells, and FACS- or MACS-sorted male germ cells (spermatogonia) of 10-dpp animals. Only families with at least 5000 mapped reads in the library were considered. Families were sorted according to H3K9me3 levels in germ cells. For liver and testicular somatic cells, values of four and two ChIP-seq replicas were averaged, respectively. Three independent biological replicas were used to measure H3K9me3 signal in spermatogonia: One was obtained by FACS-sorting using GFP-Mili, and two were obtained by MACS-sorting with the EpCAM cell surface marker. (D) Distribution of the H3K9me3 mark on all families of LINE and LTR classes combined. For somatic testicular cells and liver cells, values from two and four ChIP-seq libraries were averaged, respectively. For MACS-sorted germ cells, two ChIP-seq libraries were averaged. The error bars show standard error.
Intergenic regions and introns are composed of unique sequences and several types of repetitive elements. Repetitive sequences, particularly TEs, represent significant challenges for ChIP-seq analysis, as the majority of sequencing reads derived from these regions map to multiple positions in the genome, preventing unambiguous conclusions about their origins (Day et al. 2010). In standard ChIP-seq analysis pipelines, such multimapping reads are discarded. The commonly used alternative for analyzing repetitive sequences is mapping to a set of “consensus” TE sequences (Mikkelsen et al. 2007). However, this approach leads to information loss, as many reads that diverge from the consensus sequence are discarded. To circumvent this problem, we considered all reads that mapped to the genome without mismatches and calculated enrichment of the H3K9me3 chromatin mark over different TE families (see the Materials and Methods and Supplemental Table S2 for detailed description). Our approach generated a reliable measurement of histone mark enrichment over aggregated copies of TE families in the genome.
In agreement with previous observations (Mikkelsen et al. 2007; Matsui et al. 2010), we found that different types of repeats have distinct H3K9me3 signatures. Three classes—namely, satellite repeats, LTRs, and LINEs—were enriched in the H3K9me3 mark in all three cell types, whereas SINE (short interspersed nuclear element) and DNA transposons were depleted (Fig. 1B; Supplemental Fig. S1B). Next, we analyzed the distribution of the H3K9me3 mark on specific TE families and found large differences between families of each repeat class that were largely consistent in all three cell types (Fig. 1C). The results obtained on FACS-sorted spermatogonia and two independent samples of MACS-sorted spermatogonia were consistent and showed a high level of correlation between the two methods (Fig. 1C; Supplemental Fig. S1C). Among both LTR and LINE, several families showed strong enrichment in the H3K9me3 mark, while other families showed no enrichment or even depletion. Interestingly, in both classes, families that are known to be transcriptionally and tranpositionally active, such as the LTR families IAPEz, IAPEy, and MaLR (Smit 1993; Baust et al. 2003; Svoboda et al. 2004; Zhang et al. 2008) and the LINE families L1-Gf, T, and A (DeBerardinis et al. 1998; Naas et al. 1998; Mears and Hutchison 2001), had high levels of H3K9me3. A highly active but nonautonomous LTR family, ETn, was also enriched for the H3K9me3 mark, as was L1-F, which is transcribed at high levels but is thought to be incapable of transposition due to mutations in the ORF (Adey et al. 1991).
Three classes of repeats (satellites, LTRs, and LINEs) had elevated levels of H3K9me3 in each of the three cell types; however, notable differences between tissues were apparent. The most apparent difference was a large increase of the H3K9me3 signal on LINEs in germ cells compared with either type of somatic cells (Fig. 1D). In somatic cells, LTR elements showed higher enrichment of the H3K9me3 mark than LINEs, while in FACS-sorted germ cells, enrichment of the mark over LINE sequences was almost twice as high as on LTRs (Fig. 1D). The high level of the H3K9me3 mark on LINEs was also observed in MACS-sorted germ cells, although the difference was less pronounced compared with FACS-sorted cells, likely due to the inferior purity of MACS-sorted germ cells.
High levels of H3K9me3 on several LTR and LINE families in both somatic and germ cells indicate that these families are effectively recognized by chromatin-based repression mechanisms present in all cell types. However, the increase in H3K9me3 on LINE families in germ cells indicates that these cells use an additional mechanism to establish the repressive mark on these elements, which is either absent or less active in somatic cells.
Spreading of H3K9me3 into TE flanking sequences
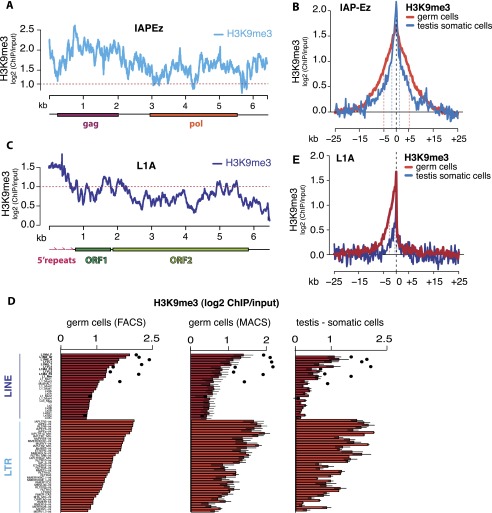
The high level of H3K9me3 mark found on several LTR and LINE families prompted us to analyze the distribution of the H3K9me3 mark within the bodies of these elements and its spreading into their genomic environment. We found that the H3K9me3 signal is evenly distributed along the bodies of LTR elements, as represented by IAPEz (Fig. 2A). Furthermore, the strong H3K9me3 signal spread both upstream of and downstream from LTR elements. Metaplot analysis of H3K9me3 spreading showed that the level of H3K9me3 decreased twofold at ∼1.5 kb from the element. The mark spread even further in germ cells (Fig. 2B).
Figure 2.
Profiles of the H3K9me3 mark along retrotransposon bodies and flanking sequences. (A) Distribution of normalized level of the H3K9me3 mark along an LTR IAPEz element consensus sequence in liver cells of 10-dpp animals. The Y-axis shows enrichment (log2) of H3K9me3 signal in ChIP compared with input DNA. Signals above the red dashed line indicate at least twofold enrichment. (B) Metaplot of input-normalized H3K9me3 signal in regions flanking all IAPEz insertions in the genome of testicular somatic cells and spermatogonia. Only uniquely mapped reads were considered. Red and blue dashed lines show the distance at which the signal dropped twofold from the peak value. (C) Distribution of the normalized level of the H3K9me3 mark along a LINE1 element (L1-A) consensus sequence in liver cells of 10-dpp animals. The Y-axis shows enrichment (log2) of H3K9me3 signal in ChIP compared with input. Signals above the red dashed line indicate at least twofold enrichment. (D) Distribution of the H3K9me3 mark on LTR and LINE families in liver cells, testicular somatic cells, and spermatogonia of 10-dpp animals. The black dots show enrichment on L1 regions corresponding to the first 500 bp of the consensus (the 5′ repeats). Only families that are enriched in the H3K9me3 mark are shown. (E) Metaplot of input-normalized H3K9me3 signal in genomic regions flanking all genomic L1-A insertions in testicular somatic cells and spermatogonia. Only uniquely mapped reads were considered. Red and blue dashed lines show the distance at which the signal dropped twofold from the peak value.
In contrast to LTR elements, several LINE families showed asymmetric distribution of H3K9me3 along the bodies of the elements. H3K9me3 was high at the 5′ ends of these elements, in the region that corresponds to several copies of the ∼200-base-pair (bp) monomers—also referred to as 5′ repeats—that function as the promoter, but dropped toward the middle and 3′ end of the L1 (Fig. 2C). Indeed, in L1 families Gf, F, T, and A, the H3K9me3 signal over the first 500 bp of the element was, on average, 1.5 times higher than the average over all genomic L1 sequences (Fig. 2D). In somatic cells, the level of H3K9me3 on the 5′ end of L1 is close to the level of this mark over the most enriched LTR elements, while in germ cells, H3K9me3 signal on L1 5′ ends exceeds the amount detected on LTRs (Fig. 2D). The H3K9me3 mark was also enriched in the genomic environment of L1 insertions; however, in contrast to LTRs, this effect was restricted to the region upstream of the L1 sequence (Fig. 2E). Therefore, asymmetry in spreading of the H3K9me3 mark outside of the L1 insertion correlates with the H3K9me3 profile on the L1 body, which shows high signal on the 5′ portion and low signal on the 3′ portion of the element. In accordance with elevated H3K9me3 levels on bodies of L1 elements in spermatogonia compared with somatic cells, the level of the mark upstream of L1 was also higher in germ cells than in somatic tissues (Fig. 2E). This result further supports the existence of a germline-specific mechanism that targets L1 elements for H3K9 trimethylation. The piRNA pathway is a good candidate for such a mechanism, as it targets TEs and operates exclusively in male germ cells.
The role of the piRNA pathway in TE repression in germ cells
To investigate whether the piRNA pathway is responsible for the increased level of H3K9me3 on TEs in the germline, we profiled this mark in germ cells of Miwi2 knockout and heterozygous (control) mice using ChIP-seq. MIWI2 is the only nuclear Piwi protein in mice, and it is required for DNA methylation of TE sequences in germ cells (Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). Although MIWI2 expression ceases soon after birth, previous studies have shown that the effects of Miwi2 deficiency on TE expression and DNA methylation persist in post-embryonic germ cells (Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008). Accordingly, we chose 10-dpp animals to analyze the H3K9me3 profiles to ensure obtaining a sufficient amount of material for ChIP-seq.
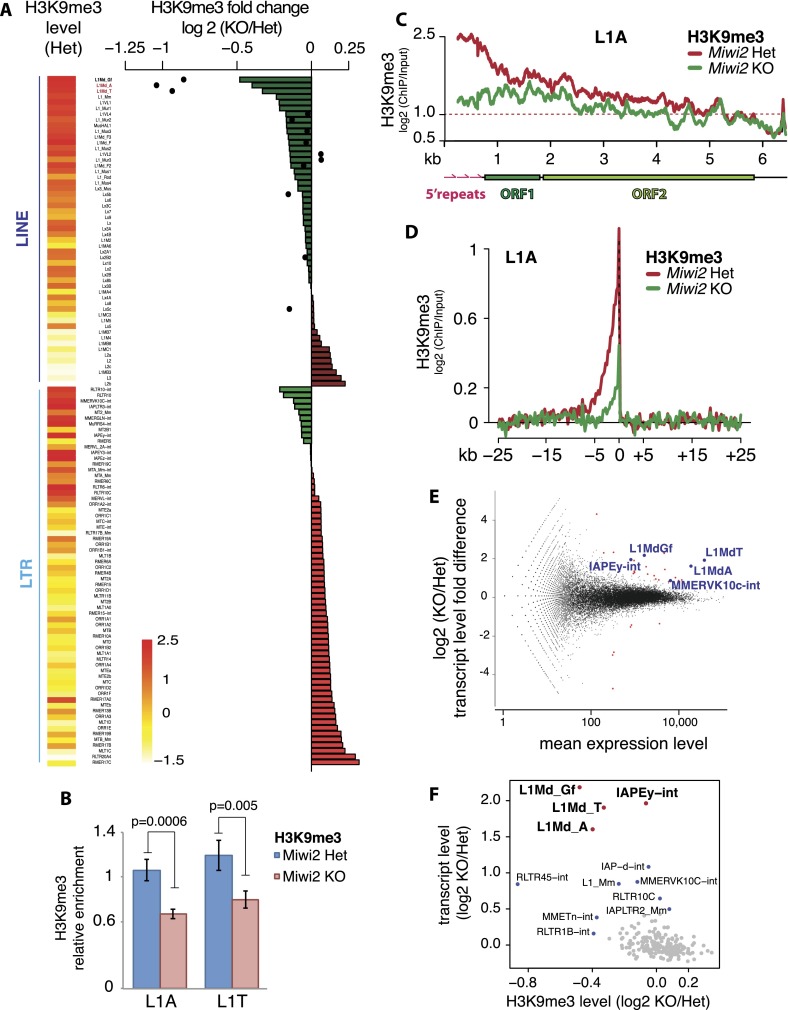
The dysfunction of the piRNA pathway in Miwi2 knockout animals did not lead to dramatic changes in the H3K9me3 signal over TSSs, exons, introns, or intergenic sequences (Supplemental Fig. S2A). Furthermore, the histone methylation of aggregated LTRs and LINEs did not change (Supplemental Fig. S2B). However, several TE families showed decreases in the H3K9me3 mark in Miwi2 knockout compared with control mice (Fig. 3A). This difference was not a consequence of a global loss of nucleosomes, as there was no difference in H3 occupancy on these TE families in Miwi2 knockout (Supplemental Fig. S2C). Importantly, the L1 families A, T, and Gf, which have the highest levels of H3K9me3 among all LINEs in wild-type germ cells, all showed a decrease in H3K9me3 in the Miwi2 mutant (Fig. 3A,B). The effect was particularly pronounced on the 5′ portions of these L1 family elements (Fig. 3A,C; Supplemental Fig. S2D–F). The loss of H3K9me3 over L1 bodies correlated with the loss of signal upstream of L1 insertions (Fig. 3D). The decrease of H3K9me3 mark on L1-A, T, and Gf was consistent in ChIP-seq experiments performed on both FACS- and MACS-sorted germ cells (Supplemental Fig. S2D,G) and could be observed using several different normalization strategies (Supplemental Fig. S3), indicating that this result is not caused by the technical variability between the samples. In addition, we confirmed the decrease in H3K9me3 mark on L1-A and L1-T families in spermatogonia of Miwi2 knockout mice using ChIP-qPCR performed on several independent biological samples (Fig. 3B).
Figure 3.
Effect of Miwi2 mutation on the H3K9me3 mark and TE expression in spermatogonia. (A) Differences in H3K9me3 levels over LINE and LTR families in spermatogonia of 10-dpp Miwi2 knockout (KO) animals relative to those in their heterozygous littermates. Levels of H3K9me3 on 5′ repeats of selected LINEs are displayed as black dots. Heat map shows H3K9me3 levels in LINE and LTR families in germ cells from Miwi2 heterozygous animals; the scale bar at the bottom shows the log2 (ChIP/input) values of these levels. Two L1 families, L1-A and L1-T, analyzed by independent ChIP-qPCR experiments shown on B, are marked in red. (B) ChIP-qPCR on L1-T and L1-A elements confirms a decrease in the H3K9me3 mark in spermatogonia of Miwi2 knockout animals. Spermatogonia were sorted from testes of 10-dpp animals by MACS using the EpCAM cell surface marker. The signal was normalized to respective ChIP input and internally normalized to signal over IAPLTR1a. Two independent ChIP experiments were performed, and error bars show standard deviation. P-values were calculated using t-test based on two sets of qPCR triplicates for each ChIP. (C) Distribution of the H3K9me3 mark along the consensus sequence of L1-A in spermatogonia of Miwi2 knockout and control animals. The Y-axis shows enrichment (log2) of H3K9me3 signal in ChIP-seq compared with input. Signals above the red dashed line indicate at least twofold enrichment. (D) Metaplot of the input-normalized level of the H3K9me3 mark in genomic regions flanking all L1-A insertions in spermatogonia of Miwi2 knockout and control animals. Only uniquely mapped reads were considered. Tandem L1-A genomic insertions were excluded from the analysis. (E) Differential expression of genes and TEs in testes of 10-dpp Miwi2 knockout and control animals as measured by RNA-seq. The fold change in expression upon Miwi2 knockout (Y-axis) is plotted against the average expression level (X-axis). Each dot corresponds to a gene or TE family. Genes (red) and TEs (blue) that passed the multiple testing-adjusted P-value < 0.01 threshold are shown. (F) Correlation between the changes in H3K9me3 and expression levels of TE families upon Miwi2 knockout. The X-axis shows the fold difference of H3K9me3 signal in TE families in spermatogonia of Miwi2 knockout and control animals (negative values indicate the loss of H3K9me3 signal in Miwi2 knockout animals). The Y-axis shows the fold difference in abundance of TE transcripts in Miwi2 knockout compared with control mice. LINE and LTR families with at least 5000 mapped reads were considered.
In contrast to LINEs, the LTR elements with high levels of H3K9me3, such as IAPEz and IAPEy families, were not affected by the piRNA deficiency. The mark was also preserved in both flanks of LTR (IAPEz) elements in Miwi2 knockout mice (Supplemental Fig. S4A). As expected, there was no difference in H3K9me3 levels on L1 and LTR elements between heterozygous and knockout samples in somatic cells of testis and in liver cells (Supplemental Fig. S4B,C).
As an alternative approach to examine the effect of piRNA deficiency, we identified all genomic regions that showed change in H3K9me3 enrichment in Miwi2 knockout germ cells compared with control cells. We assessed ChIP signal in 1-kb windows in the libraries from heterozygous and knockout animals and then ranked the windows by the degree of change in H3K9me3 signal in knockout animals. Out of 6866 windows that showed a decrease in the H3K9me3 mark in Miwi2 knockout, 53.24% contained at least one fragment of a LINE retrotransposon. Moreover, when we examined the windows with reduced levels of H3K9me3 signal together with their flanking regions (10 kb in total), we found that LINEs occupied 35.21% of these regions, compared with 21.15% in the entire genome. Therefore, regions of the genome with less H3K9me3 signal in Miwi2 knockout cells compared with control cells are significantly enriched in LINEs (permutation test P < 10−6).
To investigate how loss of the H3K9me3 mark observed in the Miwi2 knockout mice affects expression of TEs, we profiled the transcriptome of whole testis isolated from 10-dpp Miwi2 knockout and heterozygous mice (Supplemental Table S3). Expression levels of the majority of LTR and LINE families did not change in the Miwi2 mutant (Fig. 3E) despite the fact that they are targeted by abundant piRNAs (Supplemental Fig. S5). However, in agreement with previous results (Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; Shoji et al. 2009), we found that expression of several LINE and LTR families was increased in germ cells of Miwi2-deficient mice (Fig. 3E). Among LTR elements, only two families had significantly increased levels of expression in the mutant: IAPEy was up-regulated by 3.9-fold (P < 5.76 × 10−11, adjusted for multiple testing), and MMERVK10C was up-regulated by 1.8-fold (P < 0.02, adjusted for multiple testing). Neither family had decreased H3K9me3 signal in the Miwi2 mutant, indicating that their derepression in Miwi2 knockout is independent of this mark. Miwi2 deficiency also resulted in significant derepression of several LINE families: L1-Gf, L1-T, and L1-A (4.6-fold, 3.8-fold, and 3.1-fold activation, respectively, with P-values < 3.28 × 10−15, adjusted for multiple testing). Up-regulation of transcript levels of L1-T elements was confirmed by RT-qPCR on two independent biological samples (Supplemental Fig. S6A). Importantly, all three L1 families that are derepressed in the Miwi2 mutant had increased levels of H3K9me3 in germ cells compared with somatic cells (Supplemental Fig. S6B) and showed loss of H3K9me3 in the Miwi2 mutant (Fig. 3A,B,F). This result suggests that the piRNA pathway is responsible for the higher levels of H3K9me3 on these L1 elements in germ cells.
MIWI2 targets only full-length L1 copies for chromatin repression
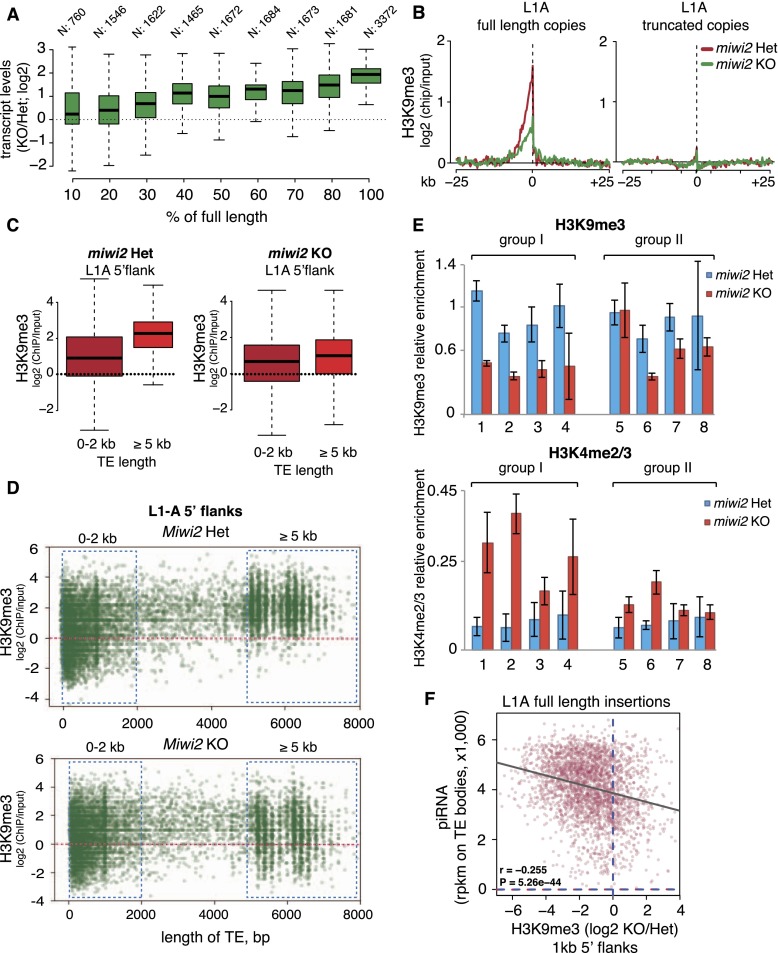
The decrease in levels of the H3K9me3 mark in Miwi2 mutant observed on aggregated genomic sequences of L1-A, L1-T, and L1-Gf is rather small. Furthermore, genome-wide analysis reveals that the number of regions that lose H3K9me3 (6866) is small compared with the number of L1 fragments in the genome (>900,000 according to the RepeatMasker annotation). Both observations imply the possibility that the piRNA pathway might affect chromatin marks of a small set of L1 genomic copies. Most copies of L1 are truncated and lack the 5′ region that functions as the promoter (DeBerardinis and Kazazian 1999; Goodier et al. 2001; Mouse Genome Sequencing Consortium 2002), so individual insertions differ in length (with full-length elements being ∼6 kb long). They also differ in similarity to consensus sequence, as there are many old insertions that diverged over time. We found that derepression of L1 in Miwi2 knockout spermatogonia positively correlated with both the length of the element and similarity to consensus (Fig. 4A; Supplemental Fig. S6C). These results indicate that a deficiency in the piRNA pathway affects a specific subset of L1 elements that are full-length and close to consensus; however, this analysis did not allow us to identify the individual copies that are targeted.
Figure 4.
Distribution of the H3K9me3 mark on individual L1 copies. (A) Correlation between the length of L1-A insertions and their derepression in testes of Miwi2 knockout (KO) mice. All genomic copies of L1-A were binned in groups based on their length. The number of loci in each category is indicated above the boxes. The Y-axis shows fold change in the transcript abundance in testes of 10-dpp Miwi2 knockout animals relative to their heterozygous littermates measured by RNA-seq. Boxes correspond to the 25th and 75th percentiles, and the lines inside the boxes are the medians. The whiskers spread to either 1.5 of IQR (interquartile range) or the farthest outlier if the outlier was within the 1.5 IQR distance. (B) Metaplot of the input-normalized level of the H3K9me3 mark in genomic regions flanking L1-A insertions in spermatogonia of 10-dpp Miwi2 knockout and control mice. All genomic L1-A copies were separated into full-length (with the preserved 500 bp of the 5′ end) and truncated copies. Only reads uniquely aligned to flanks of stand-alone L1-A insertions were considered for this analysis. (C) Distribution of the H3K9me3 mark in 1-kb flanks upstream of full-length (>5 kb) and truncated (<2 kb) L1-A insertions in spermatogonia of Miwi2 knockout and control animals. Only uniquely mapped ChIP-seq reads were considered. Boxes and whiskers show percentiles and IQR. (D) Scatter plot representation of the results shown in C. The plot shows the input-normalized level of the H3K9me3 mark in the 1-kb upstream flanks of individual L1-A copies (Y-axis) in relation to the length of each insertion (X-axis) in spermatogonia of Miwi2 knockout and control mice. The dots correspond to individual L1 copies that had at least one read mapped to their flanks in both ChIP and input libraries (9855 insertions in control and 10,072 in knockout). (E) ChIP-qPCR analysis of H3K9me3 and H3K4me2/3 levels on eight individual full-length L1-A insertions in Miwi2 knockout and control spermatogonia. The four loci in group I had a strong decrease in the H3K9me3 mark upon Miwi2 deficiency according to ChIP-seq shown on D, while the four loci in group II showed mild or no decrease in ChIP-seq. Germ cells were sorted by MACS from 10-dpp animals. H3K9me3 and H3K4me2/3 ChIPs were performed on the same material. H3K9me3 signal was normalized to input and a control region with high H3K9me3 (RMER1B). H3K4me2/3 signal was normalized to input and H3K4me2/3 level on the promoter of the gene encoding RNA polymerase II (Pol II). Error bars show standard deviation. Genomic loci and P-values for analyzed L1-A loci are listed in Supplemental Table S5. P-values were calculated using the t-test. (F) The correlation between the number of piRNAs that are able to target individual full-length L1-A loci and their repression as measured by the drop in H3K9me3 signal in Miwi2 knockout animals compared with the control. The X-axis shows the fold difference of H3K9me3 signal in 1-kb flanks of individual L1-A insertions in spermatogonia of Miwi2 knockout and control animals (negative values indicate the loss of H3K9me3 signal in Miwi2 knockout animals). The Y-axis shows the abundance of MIWI2-associated piRNAs that target the corresponding L1-A loci in wild-type embryonic (embryonic day 16.5 [E16.5]) testes. The number of piRNAs was normalized to lengths of L1-A loci (reads per kilobase of sequence per million mapped reads [RPKM]). An unlimited number of perfect (i.e., no mismatches) genomic alignments per piRNA read was allowed for this analysis.
To investigate the effect of Miwi2 deficiency on individual copies of L1, we took advantage of the fact that the H3K9me3 mark spreads upstream of the L1 insertion into the genomic environment. This allowed us to unambiguously compare individual L1 copies by measuring H3K9me3 signal upstream of individual insertions, as the upstream reads could be uniquely mapped. First, we found a high level of H3K9me3 upstream of aggregated full-length L1-A insertions in spermatogonia of control mice, while truncated copies have significantly lower levels of the mark (Fig. 4B,C). This result is in agreement with previous observations that most genomic L1-A copies are truncated from the 5′ end and lack the 5′ portion that has a high level of H3K9me3. Furthermore, levels of the H3K9me3 mark were decreased in Miwi2 knockout spermatogonia on flanking sequences of full-length L1-A copies, but there was little difference around truncated copies (Fig. 4C,D). As expected, Miwi2 deficiency does not affect the H3K9me3 mark on 3′ flanking sequences of L1-A elements or on flanking regions of L1-F elements (Supplemental Figs. S7, S8).
Despite the lower overall level of H3K9m3 on truncated insertions (<2 kb), we detected a high level of the mark on a number of these sequences (Fig. 4D). This might be a consequence of the genomic location of these insertions; namely, TE fragments present in heterochromatic regions would have higher H3K9me3 levels as a consequence of their chromatin environment. In agreement with this hypothesis, we found that truncated copies that have high H3K9me3 are twice as likely to be located in intergenic space and not in introns of protein-coding genes compared with copies that have a low level of the H3K9me3 mark (Supplemental Table S4).
Individual full-length L1 copies show significant variability in degree of the decrease of the H3K9me3 mark upon Miwi2 deficiency (Fig. 4D). This might reflect a genuine biological difference between individual copies located in different chromatin environments or be caused by an inaccuracy in measurement of the H3K9me3 mark on individual copies due to the low number of ChIP-seq reads per copy. To address this question, we selected eight full-length L1-A insertions that have a high level of H3K9me3 mark in upstream flanks as measured by ChIP-seq (Supplemental Table S5). Half of these copies (group I) had a strong decrease in the H3K9me3 mark upon Miwi2 deficiency, while the other half (group II) showed mild or no decrease in the ChIP-seq experiment (Supplemental Table S5). Next, we measured the H3K9me3 signal on both groups in independent biological samples using ChIP-qPCR with primers designed to detect individual copies. The same material was also used to profile H3K4me2/3, a mark that strongly correlates with transcriptional activity, in promoter regions of the same individual L1-A insertions. This allowed us to understand whether the drop in the H3K9me3 signal on individual L1 copies correlates with their transcriptional activation. All four L1-A copies that show a decrease in the H3K9me3 mark upon Miwi2 deficiency by ChIP-seq (group I) also showed a significant drop in signal as measured by ChIP-qPCR on independent biological samples (Fig. 4E; Supplemental Table S5). Importantly, these copies also showed a twofold to fourfold increase in the H3K4me2/3 mark on their promoters, indicating that the decrease in the H3K9me3 mark correlates with transcriptional activation of individual L1-A copies (Fig. 4E). Profiling of chromatin marks on group II L1-A copies showed that while two of them do not lose the H3K9me3 mark as was expected, two others showed a significant decrease in H3K9me3 in Miwi2 mutants, although the magnitude of change was smaller compared with group I copies (see Supplemental Table S5). Interestingly, change (or the absence of it) in the H3K9me3 mark for group II copies also correlated with the change in the H3K4me2/3 mark. These results indicate that ChIP-seq overall is quite accurate in detecting the level of H3K9me3 on individual L1 copies, although it is not perfect, likely due to the low number of reads recovered per copy. Importantly, our data indicate that loss of the H3K9me3 mark on individual L1 copies correlates with transcriptional activation of these copies, as reflected by an increase in the H3K4me2/3 mark on their promoter regions.
To understand whether the difference in repression of individual full-length L1 copies depends on the ability of piRNAs to target individual retrotransposon loci, we counted embryonic piRNAs mapped to all full-length L1-A copies in the genome. We found significant correlation between the number of piRNAs that are able to target each L1-A locus and the level of piRNA-dependent repression (as measured by a change in the H3K9me3 mark in flanking sequence in Miwi2 knockout vs. heterozygous animals) (Fig. 4F). This result indicates that even full-length L1 copies are repressed to a different degree that is directly proportional to the number of piRNAs that target each element.
Taken together, our results indicate that deficiency in the piRNA pathway exclusively affects full-length copies of transpositionally active L1 families, and the strength of repression is proportional to the number of piRNAs targeting each full-length copy. This result explains the moderate effect of Miwi2 deficiency on the level of the H3K9me3 mark on L1 elements considered in bulk, as full-length copies represent a minor fraction of all genomic copies of these retrotransposons. These results are also in agreement with the proposed model that piRNAs recognize nascent transcripts to induce chromatin changes, as only full-length copies are transcribed and therefore can be targeted.
The effect of L1 silencing on expression of adjacent genes
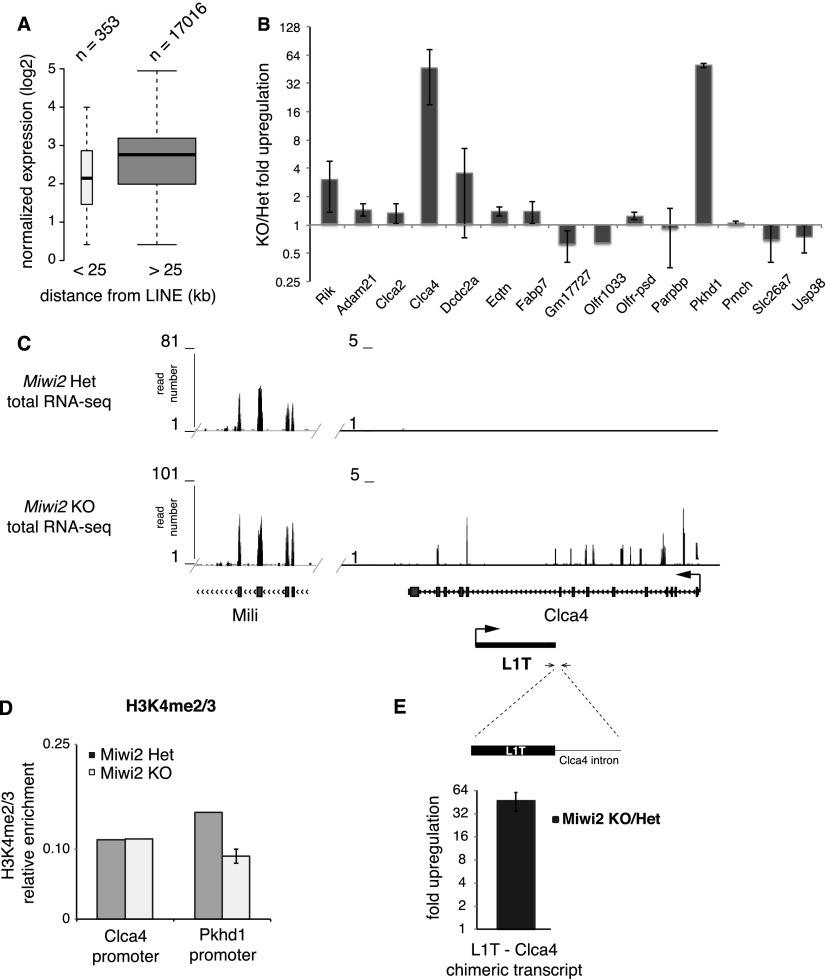
The spreading of repressive chromatin marks such as H3K9me3 into the genomic environment might regulate expression of nearby genes, as demonstrated by the phenomenon of position effect variegation (Henikoff 1990; Schotta et al. 2003; Slotkin and Martienssen 2007). To investigate the role of TEs in expression of host genes, we first analyzed genomic positions of L1 elements relative to genes. In general, full-length L1 insertions (≥6 kb) of active families (T, A, and Gf) are located far from the TSS of any gene. The median distance between an L1 and a TSS is >500 kb, which is at least 20 times greater than the distance that the H3K9me3 mark can spread from the L1. Indeed, of >17,000 genes expressed in testis, only 353 TSSs are located within 25 kb of a full-length LINE. Interestingly, these genes are expressed, on average, at significantly lower levels compared with the rest of the genes (P < 1.63 × 10−22) (Fig. 5A). This implies that spreading of the H3K9me3 mark might affect expression of the nearby genes.
Figure 5.
The influence of L1 on the expression of adjacent genes. (A) Relationship between the gene expression level in testis and the distance between the gene promoter and the neighboring L1 insertion. Genes with TSSs within 25 kb of full-length (≥6000 bp) L1-A, L1-T, and L1-Gf elements were considered. The Y-axis shows normalized expression as determined by RNA-seq averaged between testes of Miwi2 knockout (KO) and heterozygous 10-dpp animals. Boxes and whiskers show percentiles and IQR as in Figure 3E. (B) RT-qPCR analysis of expression of 15 genes with TSSs <25 kb from a full-length L1 insertion in testes of Miwi2 knockout and heterozygous 10-dpp animals. Results of two biological replicates were averaged; error bars correspond to standard deviations. (C) Genomic environment of the L1-T insertion in the intron of the Clca4 gene (chr3: 144,796,386–144,849,612). Shown are RNA-seq (polyA-selected total RNA) tracks from 10-dpp Miwi2 heterozygous (Het) and knockout testis. Only unique mappers are shown. Arrows designate TSSs and direction of the transcription for Clca4 and L1-T. A portion of the Mili gene is shown for comparison with the Clca4 locus. The Y-axis shows the number of reads. Note that the majority of the RNA-seq reads detected in Miwi2 knockout mapped to intronic sequences of the Clca4 locus, while almost all reads from the Mili gene correspond to exons. (D) ChIP-qPCR analysis of H3K4me2/3 enrichment on the promoters of Clca4 and Pkhd1 in MACS-sorted spermatogonia from 10-dpp Miwi2 knockout and control littermates. Signal was normalized to input and to the H3K4me2/3 level on the promoter of the gene encoding RNA Pol II. Error bars indicate standard deviation. (E) RT-qPCR analysis of the L1-T/Clca4 chimeric transcript in testes of 10-dpp Miwi2 knockout and control animals. Forward primer is in the L1-T insertion, and the reverse primer is in the intronic region of Clca4. RNA was isolated from total testis of two independent sets of Miwi2 heterozygous and knockout littermates. RNA levels were normalized to actin mRNA levels. Error bars indicate standard deviation.
To directly test whether the presence of an L1 represses expression of adjacent protein-coding genes, we looked at changes in gene expression in Miwi2 knockout testis compared with controls using RNA sequencing (RNA-seq). This analysis revealed that, of the genes with TSSs within 25 kb of L1 elements, expression of three genes (Dcdc2a, Olfr856-ps1, and Pkhd1) changed significantly (P < 0.0002 at a false discovery rate [FDR] < 10%). Importantly, all three genes were up-regulated in the Miwi2 mutant (Supplemental Table S6). Independent RT-qPCR analysis of the expression of 15 genes that have TSSs within 25 kb of a full-length L1 insertion revealed that expression of only two genes, Clca4 and Pkhd1, significantly increased in the Miwi2 mutant compared with control cells (Fig. 5B). Bioinformatic analysis revealed that genes that are derepressed in the Miwi2 mutant do not have significant complementarity to known piRNAs and thus are not targeted directly by piRNAs (data not shown).
To understand whether up-regulation of genes adjacent to L1 insertions in Miwi2 mutants is caused by transcriptional activation due to change in chromatin structure, we measured the H3K9me3 and the H3K42/3 marks on promoters of Clca4 and Pkhd1. ChIP-qPCR failed to detect significant change in either mark on gene promoters upon Miwi2 deficiency (Fig. 5D; data not shown). Therefore, we carefully examined RNA-seq reads derived from the Clca4 locus and found that the majority of reads in this locus detected in Miwi2 mutant are mapped to introns of Clca4, indicating that they are derived from unprocessed transcript (Fig. 5C). Furthermore, we noticed that the primers we used to detect up-regulation of Clca4 transcript by qRT-PCR (Fig. 5B) do not span an exon–exon junction; another set of primers designed to detect spliced Clca4 mRNA failed to detect up-regulation in testes of Miwi2 knockout and control animals (data not shown). Together, these results suggest that the RNA transcribed from the Clca4 locus in Miwi2 knockout animals does not correspond to properly processed Clca4 mRNA but rather to an aberrant transcript. We propose that transcription initiated by the L1-T element might not terminate properly and extends through the Clca4 locus. Indeed, RT–PCR with primers designed to detect the L1-T readthrough chimeric transcript detected an ∼60-fold increase in the amount of such RNA in Miwi2 knockout (Fig. 5E).
Taken together, our results indicate that spreading of the repressive H3K9me3 mark from adjacent L1 elements does not have a major effect on expression of host genes in germ cells. In line with our data, a recent study in mouse embryonic stem (ES) cells also found the spreading of heterochromatin from endogenous retroviruses (ERVs) into adjacent genes to be rare (Rebollo et al. 2011). Furthermore, we showed that in at least one case, what at first glance looked like L1-induced regulation of an adjacent host gene is in fact caused by readthrough transcription of a TE through the gene locus rather than genuine activation of the expression of the host gene. Similarly, a recent study showed that activation of ERVs in ES cells caused by deficiency in the SetDB1 histone methyltransferase also lead to formation of chimeric transcripts between retrotransposons and host genes (Karimi et al. 2011). The readthrough transcription of a host gene locus initiated by a retrotransposon promoter might represent a first step in a process of transposon domestication that culminates with transposon-derived sequences used as regulatory elements for host gene expression (Duhl et al. 1994; Peaston et al. 2004; Romanish et al. 2007; Wang et al. 2007; Feschotte 2008; Li et al. 2012).
The link between the piRNA pathway, the H3K9me3 mark, and DNA methylation
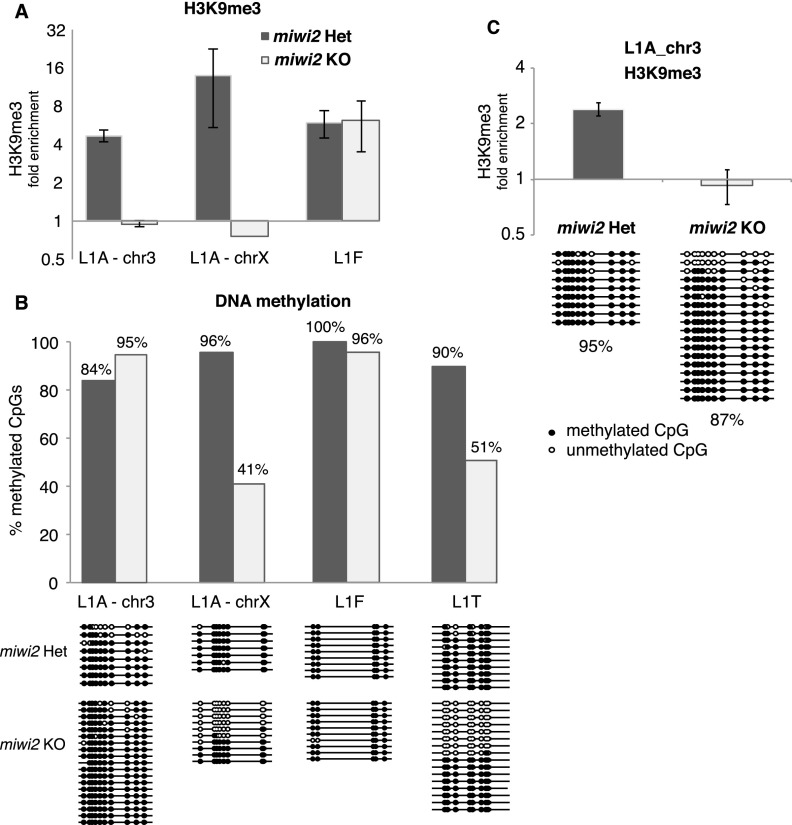
Previous studies showed that failure of the piRNA pathway leads to loss of DNA methylation on L1 copies, supporting the hypothesis that piRNAs might guide establishment of DNA methylation patterns in germ cells (Carmell et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008). To understand the relationship between the piRNA pathway, H3K9me3, and DNA methylation, we analyzed two genomic regions in which the level of the H3K9me3 mark upstream of the L1-A element is decreased in Miwi2 knockout cells compared with levels in controls (Fig. 4E, insertions 1 and 2; Supplemental Table S5) and a control L1-F region with no change in the Miwi2 mutant. First, we performed independent ChIP-qPCR to measure H3K9me3 signals on the same loci using two different biological samples. ChIP-qPCR confirmed loss of the H3K9me3 mark in the Miwi2 mutant for both L1-A loci, indicating complete penetrance of Miwi2 deficiency (Fig. 6A). Next, we analyzed DNA methylation of these regions and an additional L1T insertion using bisulfate conversion followed by sequencing. Like the H3K9me3 signal, DNA methylation at the L1-F locus was not affected by Miwi2 deficiency. One of the L1-A loci and the L1-T locus showed significant loss of CpG methylation in the Miwi2 mutant in addition to the decrease in the H3K9me3 mark (Fig. 6B). The other L1-A locus did not show a change in DNA methylation despite the decrease in H3K9me3 signal (Fig. 6B). To exclude the possibility that the observed differences are due to differences between samples, we confirmed that there was a loss of H3K9me3 signal without an accompanying change in DNA methylation status at this locus by performing both ChIP-qPCR and bisulfite sequencing on the same starting material (Fig. 6C). This result indicates that the H3K9me3 mark is deposited on L1 elements in a piRNA-dependent fashion, which does not always correlate with CpG DNA methylation.
Figure 6.
H3K9me3 and DNA methylation marks on individual L1 loci. (A) Level of the H3K9me3 mark on three individual L1 loci in spermatogonia of 10-dpp Miwi2 knockout (KO) and control animals. Spermatogonia were purified by MACS, and the H3K9me3 signal was measured by ChIP-qPCR. Means for two qPCRs on ChIP samples from two Miwi2 heterozygous animals and one knockout animal are shown. Error bars show standard deviation. An L1-F insertion served as a control for ChIP efficiency. All ChIP signals are normalized to input and a negative control region. (B) DNA methylation analysis of the same three loci shown in A and an additional L1-T locus. The plot shows the percentage of methylated CpGs on each locus. The actual sequenced clones are shown below the graph. (C) Analysis of H3K9me3 and DNA methylation on the L1-A locus on chr3 shown in A and B performed on the same starting material.
Discussion
Distribution of the H3K9me3 mark on different repeat classes
Our study focused on the profile of the H3K9me3 mark on the repetitive portion of the genome in mouse somatic and germ cells. Although TEs and other repetitive regions comprise almost half of the genome, they have received surprisingly little attention in the majority of genomic and epigenetic studies. Several recent studies established the importance of the H3K9me3 mark for silencing LTR ERV families in ES cells (Matsui et al. 2010; Rowe et al. 2010). Histone modifications on LINEs have not been extensively analyzed genome-wide, and there is conflicting evidence on whether they are targets for H3K9me3 deposition (Martens et al. 2005; Mikkelsen et al. 2007; Fadloun et al. 2013).
We found high levels of H3K9me3 signal on three repeat classes (satellites, LTRs, and LINEs) in all three cell types studied (liver cells, testicular somatic cells, and spermatogonia). Our study also revealed substantial variability in the levels of H3K9me3 between repetitive element families within each class (Fig. 1C). Interestingly, LINE and LTR families that are active in the murine genome (Ostertag and Kazazian 2000) had high levels of H3K9me3, while the majority of ancient, inactive families had low levels of the mark. In somatic tissues, the enrichment of H3K9me3 on LTR elements and particularly on the families with the highest level of the mark was stronger than on LINEs (Fig. 1B–D). However, careful examination of H3K9me3 patterns on LTRs and LINEs revealed a more complex picture: We found that the distribution of the mark along the body of the element differed for LTRs and LINEs. Whereas the mark was high along the entire length of the LTR element, 5′ regions of L1 elements, composed of short repeats that function as promoters, had significantly higher H3K9me3 signal than the rest of the element body (Fig. 2A,C). This pattern was also reflected in the spreading of the H3K9me3 mark into the genomic environment: H3K9me3 signal spread symmetrically (both upstream and downstream) from LTRs, as observed previously (Rebollo et al. 2011), and asymmetrically (only upstream) from L1 elements (Fig. 2B,E).
Most L1 copies are truncated from the 5′ end due to a nonprocessive reverse transcriptase activity during retrotransposition (Luan et al. 1993; Ostertag and Kazazian 2000; Eickbush and Jamburuthugoda 2008). Therefore, any analysis that considers all genomic copies of a particular LINE family together results in a gross underestimation of the level of H3K9me3 signals on the 5′ portions of full-length L1 elements. Indeed, when we calculated the H3K9me3 signal on the 5′ repeats of L1-F, L1-Gf, L1-A, and L1-T families, levels were similar to those observed on the most enriched LTR elements (Fig. 2D). Overall, our results indicate that active LTR and LINE families are recognized as targets for repressive histone marks and also explain why enrichment of H3K9me3 was not detected on L1 previously.
The role of piRNAs in establishing the H3K9me3 mark on TEs in germ cells
How LTRs and LINEs are recognized as targets for H3K9 trimethylation is not well understood, although, in ES cells, sequences of several LTR families are bound by a complex that contains sequence-specific KRAB-ZFP (Krüppel-associated box domain-zinc finger) proteins. These sequence-specific DNA-binding proteins recruit TRIM28/KAP1 and its binding partner, the SetDB1/ESET histone methyltransferase, resulting in silencing of targeted LTRs (Wolf and Goff 2009; Matsui et al. 2010; Rowe et al. 2010). No similar complex was detected for LINEs.
The elevated level of the H3K9me3 mark on L1 in spermatogonia suggests the existence of a pathway that targets these elements for chromatin repression specifically in male germ cells. The piRNA pathway is a good candidate for such a mechanism, as it operates exclusively in male germ cells and is able to target TEs using sequence-specific piRNA guides. The piRNA-guided repression of TEs requires expression of MIWI2, which enters the nucleus of germ cells in late embryogenesis (Aravin et al. 2008). We addressed the role of the piRNA pathway in establishing the H3K9me3 mark on TEs by comparing the profiles of this mark in somatic and germ cells of Miwi2 mutants and their heterozygous littermates. We found that a deficiency in the piRNA pathway did not affect levels of the H3K9me3 mark in somatic cells (Supplemental Fig. S4). This result is expected from previously reported expression patterns of Piwi proteins and piRNAs that seem to be restricted to germ cells (Kuramochi-Miyagawa et al. 2004, 2008; Carmell et al. 2007), although other reports proposed that the piRNA pathway might also repress TEs in other cell types (Marchetto et al. 2013). In contrast to somatic cells, we found significant differences in the H3K9me3 signal in spermatogonia of Miwi2-deficient mice and heterozygous mice: All three L1 families that were transcriptionally derepressed in Miwi2 mutants (L1-A, L1-T, and L1-Gf) also had decreased levels of the H3K9me3 mark, with the effect most pronounced on their 5′ regions (Fig. 3; Supplemental Fig. S2). Loss of the mark on the body of L1 elements correlated with a decreased H3K9me3 signal upstream of L1 elements (Fig. 3D). In contrast to L1-A, L1-T, and L1-Gf elements, we found that L1-F elements, which are significantly older and have lost the ability to retrotranspose, are not targeted by the piRNA pathway. One possibility is that the majority of L1-Fs are accumulated in heterochromatin, explaining the high level of H3K9me3 mark on their sequences independently of Miwi2. Indeed, analysis of H3K9me3 levels downstream from L1-F insertions (which reflects the genomic environment of the element, since there is no spreading of the H3K9me3 mark downstream from the insertion) revealed a weak trend, supporting this hypothesis (data not shown). Taken together, our results indicate that the piRNA pathway targets active L1 families for chromatin modifications.
Previous studies show that dysfunction of the piRNA pathway leads to loss of CpG methylation on LTR and LINE1 in germ cells. This result led to the hypothesis that piRNAs might guide the DNA methyltransferase complexes to their chromatin targets. Our findings that piRNAs also affect the H3K9me3 mark raise the question of the relative contribution of the two mechanisms, DNA methylation and histone modification, to piRNA-dependent TE repression. Mice deficient in Dnmt3L, a protein necessary for DNA methylation of TE sequences in germ cells, show strong overexpression of TEs, indicating that maintenance of DNA methylation is critical for efficient silencing (Bourc’his and Bestor 2004). Nevertheless, a direct association between DNA methyltransferases and the nuclear MIWI2 complex was not detected. It is possible that DNA methylation is parallel to or downstream from a different chromatin-modifying activity, such as piRNA-guided H3K9 trimethylation. Indeed, recent studies in Drosophila demonstrated an ability of piRNAs to establish repressive H3K9me3 marks on TE targets in the absence of DNA methylation in this organism (Klenov et al. 2011; Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). Furthermore, studies in murine ES cells showed the importance of H3K9 methylation for repression of LTR and LINE retrotransposons (Matsui et al. 2010; Rowe et al. 2010). A high level of both DNA methylation and H3K9me3 marks can be found on LTR ERV elements in ES cells; however, SetDB1-mediated deposition of H3K9me3 seems to be the primary silencing mechanism (Matsui et al. 2010). Even though LINEs were not affected by SetDB1 deficiency, the same study found up-regulation of LINEs in ES cells deficient in another H3K9 methyltransferase, Suv39h. Suv39h-deficient mice have defects in spermatogenesis similar to the ones observed in Dnmt3L and Miwi2 mutants (Peters et al. 2001; Bourc’his and Bestor 2004; Carmell et al. 2007), suggesting that these might be caused by TE activation, although the expression of retrotransposons was not analyzed in these studies. Taken together, these results suggest a role of H3K9me3 in silencing of LINEs. Other chromatin marks might also play a role in LINE repression: A recent study found a correlation between the global decrease in the H3K9me2 mark and transcriptional activation of LINEs during spermatogenesis (Di Giacomo et al. 2013).
Our data suggest that the ability of piRNAs to establish the repressive H3K9me3 marks was conserved during metazoan evolution, in contrast to DNA methylation. Future studies should reveal the interaction between piRNA-dependent DNA and histone modifications and determine the relative contribution of these two processes to TE repression.
The piRNA pathway selectively represses full-length LINE1 elements
We compared the list of TE families that are targeted by piRNAs with elements that are derepressed and/or had altered levels of H3K9me3 in the piRNA-deficient mutant. Interestingly, the three sets of TEs do not completely overlap. Most striking is that, whereas piRNAs are derived from the majority of LTR and LINE families, only a few families had increased expression in the Miwi2 mutant (Fig. 3E,F). There are several nonmutually exclusive explanations for this fact: First, many transposon families present in the murine genome are remnants of ancestral invasions, and these elements may no longer have the capacity for transposition or even to be expressed. Obviously, TEs lacking functional promoters cannot be activated even when a repressive mechanism such as the piRNA pathway is removed. Second, it is plausible that some TEs are subject to several independent mechanisms of repression, and, due to this redundancy, a deficiency in only the piRNA pathway may not be sufficient to release silencing. For example, the IAPEz family of LTR retrotransposons that is active in mice is targeted by piRNAs but did not have increased levels of expression in the Miwi2 mutant. There was a high H3K9me3 signal on this element in both somatic and germ cells that was independent of the piRNA pathway, suggesting that a non-piRNA mechanism induces chromatin silencing of IAPEz. For example, histone methyltransferase activity recruited via KRAB-ZFP DNA-binding proteins and TRIM28 might be responsible for repression in the absence of functional piRNAs. Finally, it is possible that some TE families are active during a developmental window that we did not examine.
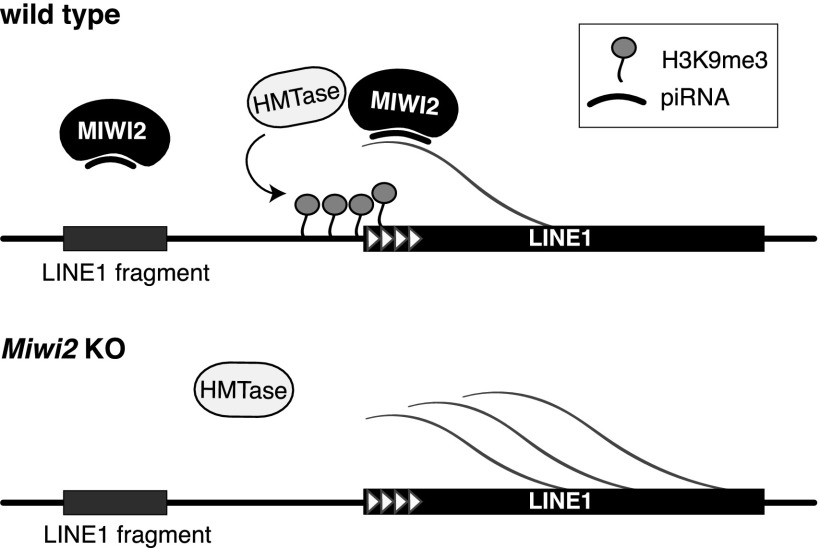
Previous studies of TEs have treated all copies that belong to a family together due to the fact that the bulk of transcriptome and genome reads derived from TEs cannot be unambiguously assigned to a particular TE copy in the genome. As a result, the average level of signal for all genomic copies of a particular TE was calculated, and possible differences between individual copies were not resolved. Here we studied individual genomic copies using unique sequence reads derived from the genomic environment of each retrotransposon insertion. A similar approach was recently used by Rebollo et al. (2011) to analyze individual copies of LTR elements in ES cells. The glimpse into the world of individual TEs revealed a remarkable picture: We found that distinct copies within the same transposon family had different levels of H3K9me3 and responded differently to defects in the piRNA pathway (Fig. 4C,D). Specifically, we observed that in contrast to the majority of truncated L1 copies, full-length L1 had high H3K9me3 signal and that these signals were decreased in the piRNA pathway mutant. Furthermore, we found that even distinct full-length L1 copies are different in the magnitude of their repression by the piRNA pathway, and the level of suppression is directly proportional to the number of piRNAs that target each individual locus. The loss of H3K9me3 mark on full-length L1 copies resulted in their transcriptional activation, as evidenced by the increase of H3K4me2/3 mark on their promoter regions (Fig. 4E). In contrast to full-length L1 copies, abundant truncated L1 copies had lower levels of H3K9me3, which did not change upon Miwi2 mutation (Fig. 4B–D). This observation explains the rather moderate effect of Miwi2 mutation on H3K9me3 marks of L1 elements considered in bulk, as the majority of genomic L1 copies are 5′-truncated. More importantly, this result sheds light on the mode of transposon recognition by the piRNA pathway. Previously, we observed that piRNAs are generated along the whole body of L1, which suggests that even truncated genomic copies might be recognized (Aravin et al. 2008). This conundrum can be resolved by a model in which TE-derived piRNAs in complex with MIWI2 target nascent transcripts, resulting in establishment of a repressive chromatin structure on these target loci (Fig. 7). As 5′-truncated L1 copies lack functional promoters, they are not transcribed and therefore are not targeted by the piRNA machinery. This mechanism makes biological sense, as it allows the piRNA pathway to ignore ancient, already inactive insertions and concentrate on silencing active, potentially dangerous TE copies.
Figure 7.
Model for piRNA-induced establishment of the H3K9me3 mark on L1 elements. Transcripts from a full-length LINE in the nucleus of embryonic prospermatogonia are recognized by a MIWI2–piRNA complex, which recruits a histone methyltransferase (HMTase). This results in deposition of the H3K9me3 mark on LINE 5′ repeats and in the adjacent upstream region. The piRNA associates only with transcripts from actively transcribed copies of TEs. Truncated copies, which are not transcribed, are not targeted by piRNAs.
Materials and methods
Animals
The Miwi2 knockout mouse model is described in Kuramochi-Miyagawa et al. (2008). For FACS, Miwi2 mice were crossed to a mouse strain expressing the GFP-Mili transgene (Aravin et al. 2008).
All experiments were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the California Institute of Technology. Animals were group-housed, unless otherwise mentioned, at 22°C –24°C with ad libitum access to food and water in a 13-h/11-h light/dark cycle in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited housing facility. Mice were euthanized according to IACUC-approved procedures.
Isolation of testicular germ and somatic cells
Testes were dissected, tunica were removed, and tubules were gently untangled using fine-tip forceps. Tubules were put in 1 mL of DMEM containing 1% fetal bovine serum (FBS) and 100 U/mL collagenase type I (Invitrogen, 17100-017PBS) and incubated with agitation for 30 min at room temperature with occasional resuspending. Cells were filtered through a 0.45-μm strainer and collected by centrifuging at 250 rcf for 5 min at room temperature. About 1 million cells are recovered from one pair of 10-dpp mouse testes. Cells were washed once in 750 μL of 1× PBS and fixed in 1% paraformaldehyde (PFA) in PBS (100 μL of PFA in 700 μL of PBS) for 5 min at room temperature. Fixation was quenched with 90 μL of 1.25 M glycine for 5 min at room temperature. Cells were pelleted at 5000 rcf for 5 min at 4°C and washed twice in 750 μL of ice-cold PBS. For FACS, cells were washed one more time in FACS buffer (1× HBSS with 10 mM HEPES, 2.5 mg/mL BSA, 25 mM MgCl2, 50μg/mL DNase I), resuspended in 1 mL of FACS buffer, and filtered through a 0.45-μm filter into FACS tubes.
Germ cells were collected, based on GFP fluorescence, from testes of mice expressing the GFP-Mili transgene. GFP fluorescence was not impaired by fixation. For MACS, cells were washed once in MACS buffer (0.5% BSA, 2 mM EDTA in 1× PBS). Cells were then resuspended in 500 μL of MACS buffer and incubated with anti-EpCAM antibody (rat monoclonal, G8.8, Santa Cruz Biotechnology, sc-53532; amount adjusted to number of cells as per the manufacturer’s instructions) for 30 min at 4°C with agitation. Cells were then washed once in 750 μL of MACS buffer and resuspended in 80 μL of MACS buffer, and 20 μL of anti-rat IgG microbeads (Miltenyi Biotec, 130-048-502) was added. Cells were incubated with beads for 15 min at 4°C without agitation, resuspended in 400 μL of MACS buffer, and applied to MS MACS separation columns (Miltenyi Biotec, 130-042-201). Cells were separated on a Mini-MACS separator (Miltenyi Biotec, 130-042-102) according to the manufacturer’s instructions. Flow-through was collected as testicular somatic cells, and the bound fraction containing germ cells was eluted. No germ cells were lost in the flow-through, and the bound fraction contained ∼50% of germ cells, compared with 10%–15% in full testis.
Isolation of liver cells
One liver lobe was dissected and immediately put in ice-cold PBS with protease inhibitor cocktail (Roche Complete Mini, EDTA-Free, catalog no. 11 836 170 001). Tissue was chopped finely with a blade and collected in 1.4 mL of PBS. For fixation, 200 μL of 8% PFA was added. Cells were fixed for 12 min at room temperature, and fixation was quenched with 180 μL of 1.25 M glycine for 10 min at room temperature, both with agitation. Tissue pieces were pelleted by centrifuging at 120 rcf for 5 min at 4°C and transferred to a glass douncer in 2 mL of ice-cold PBS. The tissue was dounced for 20–30 strokes with a loose pestle to yield a single-cell suspension. Cells were filtered through a 0.45-μm strainer, pelleted at 250 rcf for 5 min, resuspended in 1 mL of ice-cold PBS, and filtered again through Miracloth. One million cells were used for each ChIP experiment.
ChIP
Both testicular and liver fixed cells were pelleted by centrifugation at 5000 rcf for 5 min at 4°C. Up to 1 million cells were used for each ChIP experiment. Cells were resuspended in 200 μL of ChIP lysis buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 0.7% SDS, 0.1 mM AEBSF) and sonicated in a Bioruptor (Diagenode) three times at 10 cycles (30 sec sonication, 30 sec recovery) on high setting in the cold room. Sonicated chromatin was cleared by centrifuging at 20,000 rcf for 10 min at 4°C and was stored either on ice for immediate use or at −80°C. For immunoprecipitation, 50 μL of Protein G DynaBeads (Life Technologies, catalog no. 10004) slurry per sample was used, and 10 μL was used for chromatin preclearing. Beads were washed two times in 3 vol of RIPA/BSA buffer (5 mg/mL BSA, 20 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 10 mM NaF, 0.2 mM Na3VO4, and protease inhibitor cocktail. Antibody (anti-H3K9me3, Abcam, ab8898; anti-H3K4me2/3, Abcam, ab6000; anti-H3, Active Motif, #61277) was conjugated to the beads in a total volume of 500 μL of BSA/RIPA for 1.5–2 h at 4°C with agitation; 5 μL per 1 million cells was used. Beads were washed twice in BSA/RIPA. Chromatin was precleared by incubating with Protein G DynaBeads for 0.5 h at 4°C with agitation in 550 μL of BSA/RIPA; 50 μL was set aside as chromatin input material, and 150 μL of TE was added. The rest of the sample was transferred to antibody-conjugated DynaBeads and incubated for 2–3 h at 4°C with agitation. Beads were washed three times in ice-cold LiCl wash buffer (10 mM Tris-HCl at pH 7.4, 500 mM LiCl, 1% NP40, 1% Na-deoxycholate) and once in ice-cold 1× TE. Washed beads were resuspended in 200 μL of TE, and 200 μL of 2× Proteinase K buffer (200 mM Tris-HCl at pH 7.4, 25 mM EDTA, 300 mM NaCl, 2% SDS) with 10 μL of 20 mg/mL Proteinase K (New England Biolabs, P8102) was added to both ChIP and input samples. Samples were incubated for 3 h at 55°C followed by overnight incubation at 65°C to reverse the cross-links. Salt concentration was adjusted to 300 mM NaCl, and final sample volume was adjusted to 500 μL. DNA was extracted using phenol/chloroform, and chloroform and was precipitated in 2 vol of absolute ethanol for 2 h at −80°C. After centrifuging at 20,000 rcf for 30 min, pelleted DNA was washed in 500 μL of 70% ethanol, dried, and resuspended in 12 μL of water. Typically, ∼500 ng of DNA was recovered from input, and ∼50 ng of DNA was recovered from ChIP.
RNA isolation, reverse transcription, and polyA+ selection
Total RNA was isolated from total testis using Ribozol (Amresco, N580). RNA was DNase-treated using Turbo DNA-free kit (Ambion, AM1907). DNase-treated RNA (5 µg) was reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen, 18080-044) in 40 μL. After reverse transcription, 80 μL of water was added to the cDNA reaction. For library cloning, 1 μg of DNase-treated RNA was polyA+-selected using the DynaBeads mRNA purification kit (Invitrogen, 610.06).
Genomic DNA isolation and bisulfite conversion
Cells were isolated from testes as described above, resuspended in 100 μL of TE, and lysed in 1 mL of genomic DNA lysis buffer (10 mM Tris-HCl at pH 8.0, 100 mM EDTA at pH 8.0, 0.5% SDS, supplemented with 250 U of RNase I [Ambion, AM2295]). Lysate was incubated for 1 h at 37°C, transferred to a glass douncer, and homogenized further by 15 strokes with a tight pestle. Proteinase K (New England Biolabs) was added to a final concentration of 100 μg/mL and incubated for 3 h at 50°C. After cooling to room temperature, an equal volume of phenol (pH 8.0) was added, mixed well, and incubated for 10 min at room temperature with agitation. The water phase was recovered after centrifugation, and the phenol extraction was repeated one more time followed by phenol/chloroform and chloroform extraction. The water phase was then mixed with 2 vol of absolute ethanol, and 5 M NaCl was added to a final concentration of 300 mM. DNA was precipitated for 2 h at −80°C or overnight at −20°C. Pelleted DNA was washed twice with 70% EtOH, dried, and resuspended in water. A260/280 and A260/230 ratios were measured to check purity. For bisulfite conversion, 500 ng of genomic DNA was bisulfite-treated using the Active Motif MethylDetector kit (#55001) as per the manufacturer’s protocol.
DNA methylation analysis
Each locus was amplified by two successive PCR reactions using Taq polymerase (GenScript, E00007). For outer PCR, 4 μL of converted DNA was amplified using the following program: 3 min at 94°C, 30 sec at 94°C, 30 sec at 50°C, 30 sec at 72°C for 20 cycles, and 4 min at 72°C. For nested PCR, 4 μL of outer PCR reaction was used as input and amplified for 30 cycles using the same program with extension of the final amplification time to 10 min. Primer sequences are listed in Supplemental Table S8. PCR products were resolved on 1.2% agarose gel, purified, and cloned into pGEM vector (pGEM-T Vector System I, Promega, A3600). Individual clones were Sanger-sequenced and analyzed for CpG methylation status using MethTools 2.0 (http://194.167.139.26/methtools/MethTools2_submit.html).
qPCR
qPCRs were performed using 2× MasterMix (MyTaq HS mix 2×, Bioline, BIO-25046) supplemented with 0.2 μL of SYBR Green (; Molecular Probes, S7563; 1:6000 dilution in DMSO) per 20-μL reaction. For analysis of gene expression, 1 μL of cDNA was used as input. Primer sequences for qPCR analysis of gene expression are listed in Supplemental Table S9. For ChIP-qPCR using primers against TE consensuses, input DNA was diluted 1:1000, ChIP DNA was diluted 1:100, and 1 μL was used. For ChIP-qPCR on individual genomic loci, input DNA was diluted 1:100, ChIP DNA was diluted 1:10, and 1 μL was used. Primer sequences for ChIP-qPCR are listed in Supplemental Table S7.
Cloning ChIP-seq and RNA-seq libraries
Total RNA libraries were cloned using either Illumina TruSeq RNA sample prep kit (version 2, 48 rxns, RS-122-2001) or NEBNext Ultra RNA library prep kit for Illumina (New England Biolabs, E7530). ChIP libraries were cloned using NEBNext ChIP-seq library prep master kit (New England Biolabs, E6240) and amplified using NEBNext Multiplex Oligos for Illumina (New England Biolabs, E7335). Final ChIP libraries were resolved on a 1.2% agarose gel, the region in the 200- to 450-nt range was excised, and DNA was purified.
Analysis of ChIP-seq signal within genomic partitions, repeat classes, and TE families
To assess ChIP signal distribution genome-wide and on TE bodies, we considered all reads that mapped to the genome (mm10) with Bowtie 0.12.7 (Langmead et al. 2009) of up to 10,000 positions, allowing for zero mismatches. This cutoff was used to speed up the computation without sacrificing relevant information: It eliminates, on average, only 0.25% of distinct sequences (0.3%–0.6% reads), of which about a third are derived from simple repeats (Supplemental Table S2). For each read, we assigned the multiplicity score (NH) equal to the total number of valid alignments of that read in the genome. Next, we divided the genome into four genomic partitions: exons, introns, and TSSs of genes and the intergenic space. Exons were defined as the region of the genome covered by any of the exons annotated by RefSeq (Pruitt et al. 2009) (RefSeq gene table was downloaded from the University of California at Santa Cruz [UCSC] genome browser [Meyer et al. 2013]); introns were defined as the regions between exonic partitions within the boundaries of a region covered by RefSeq transcripts belonging to any gene. Regions that were between exonic partitions but outside of the gene boundaries were defined as the intergenic space. The TSS of a given gene was defined as the 1-kb region around the 5′ end of the most outstanding transcript annotated to that gene (±500 bp).
After the genomic partitions were defined, we counted reads that were mapped within partitions. Each read count was weighed by the multiplicity score (NH) associated with the read; thus, each read i incremented the sum of reads by 1/NHi. If a read overlapped with more than one partition, then the read was assigned to the partition that contained the read’s 5′ nucleotide. With the weighted counts, we calculated how many reads were mapped to each individual partition. Next, for each of the four partition types, counts of reads in individual partitions were totaled. Finally, for each of the four partition types, the total counts of reads were converted into reads per kilobase of sequence per million mapped reads (RPKM) values in order to account for differences in library depths and differences in lengths of partitions.
ChIP signal in TE families and classes was defined in the manner analogous to the one described above for genomic partitions. We considered regions annotated by RepeatMasker (http://www.repeatmasker.org) in the UCSC genome browser for the mm10 genome (Meyer et al. 2013) as belonging to a particular family of TEs to comprise one type of genomic partition. Similarly, partition types were defined for repeat classes such as LTRs, LINEs, and satellites. We then calculated how many weighted reads were mapped to each individual partition and subsequently totaled the counts for partitions of each type. Finally, for each partition type, the counts were converted into RPKM values. ChIP enrichment was defined as the ratio of RPKMs between ChIP and input libraries.
Normalization strategies for ChIP-seq experiments
A ChIP experiment relies on precipitation of a cross-linked complex of target protein with DNA, and it is conceivable that differences in precipitation efficiency between experiments might bias estimation of ChIP enrichment. To make sure that our results were not caused by the variability in efficacy of ChIP between samples, we tested two different strategies for internal normalization of ChIP-seq samples. The first strategy used normalization to H3K9me3 enrichment on major satellite repeats that have a very high level of this mark in all cell types. The second strategy used normalization to average H3K9me3 levels in all 100-kb genomic windows that have a high level of the mark. To compile a list of 100-kb windows that have a high H3K9me3 mark, we selected all windows from the control experiment (ChIP-seq on FACS-sorted spermatogonia from 10-dpp Miwi2 heterozygous mice) that had input-normalized H3K9me3 enrichment of more than twofold. In addition, we discarded windows that had <10 reads uniquely mapped to the window. We identified 3358 such windows. Next, we assessed the H3K9me3 enrichment (ChIP/Input signal) in these windows in all of our experiments and normalized by this amount the enrichment on LINE and LTR transposon families. Both normalization strategies showed that the H3K9me3 mark is depleted from 5′ ends of L1-Gf, L1-T and L1-A elements in Miwi2 knockout spermatogonia (Supplemental Fig. S3).
Analysis of ChIP-seq signal in 1-kb regions flanking TE insertions
For profiling ChIP signal in regions flanking TE insertion, we mapped the library reads to the mm10 genome with Bowtie 0.12.7 (Langmead et al. 2009), allowing for zero mismatches, and reads with more than one valid alignment were discarded (i.e., we only considered uniquely mapping reads for this analysis). The upstream and downstream 1-kb flanks of regions annotated by RepeatMasker as a TE were analyzed using the same procedure as described above for TE sequences with the exception that here we counted only reads uniquely mapped to TE flanking regions. The conceptual difference between the flanking region approach versus body approach was that analysis of flanking regions generally allows for unambiguous allocation of sequencing reads, whereas mapping to repetitive elements was frequently ambiguous. Therefore, the analysis of the flanking regions allowed the association of upstream and downstream RPKM values with each individual copy of a TE.
Meta-analysis of ChIP-seq signal in 25-kb flanks of TE insertions
Similar to the analysis of regions flanking TE insertions, we considered only those reads that mapped to the mm10 genome uniquely with zero mismatches (Langmead et al. 2009). Next, we took 25-kb regions on the 5′ and 3′ sides of selected TE families defined according to RepeatMasker track in the UCSC browser and divided them into 250-bp bins. TEs closer than 25 kb to the end of the chromosome were excluded from the analysis. The ChIP-seq reads that overlapped with each individual bin were counted. If a read overlapped with two bins, then the bin containing the 5′ nucleotide of the read was assigned the count. Reads were totaled for all copies of TEs to produce per-bin summary counts. The per-bin summary counts were divided by the total number of uniquely mapped reads in the library in order to account for the library depth. Finally, we divided the normalized per-bin summary counts in ChIP libraries by those in the input libraries, which produced a metaprofile of ChIP enrichment.
We noticed that in all ChIP-seq libraries, despite the ChIP enrichment near the edges of 25-kb regions flanking TEs being flattened out, the baseline of the enrichment on the edges was >1. We hypothesize that this is due to global biases in ChIP-seq read distributions; for example, it was suggested that ChIP is biased toward open regions of chromatin (Chen et al. 2012). Therefore, the high ChIP enrichment baseline may reflect the fact that only a subset of the genome is “ChIP-able,” whereas the whole genome is represented in the input library. To account for such differences between ChIP and input libraries, besides the input normalization of ChIP signal, we introduced an additional normalization step. Judging from the shapes of the metaprofile enrichment curves, we concluded that the distal 5 kb of sequence of 25-kb regions flanking TEs represents the true unbiased baseline of ChIP enrichment. Therefore, we computed the average ChIP enrichment between 5-kb upstream and 5-kb downstream flanking regions and normalized the metaprofile by this amount.
Analysis of ChIP-seq signal on TE consensus sequences
To assess ChIP signal distribution genome-wide and on TE bodies, we mapped the library reads to all RepBase TE consensus sequences with Bowtie 0.12.7 (Langmead et al. 2009), allowing for three mismatches and allowing an unlimited number of valid alignments for each read. Next, with BEDTools (Quinlan and Hall 2010), we evaluated coverage of each base in the consensus sequence. To account for differences in library depths, the consensus coverage was normalized by the total number of mapped reads. Rather than normalizing by the total number of reads mapped to the consensus, we normalized by the total number of reads mapped to the mm10 genome (zero mismatch policy, allowing for 10,000 valid alignments per read). We reasoned that normalizing by reads mapping to the consensus would artificially inflate estimates for input libraries because input reads are relatively randomly distributed. Disregarding the fraction of the library mapping outside TEs would artificially increase input coverage of TEs. Ideally, normalization should be performed by the number of reads mapped to mm10 with an unlimited number of valid alignments per read. However, allowing unlimited multimapping for mm10 alignment was not computationally feasible, and the error associated with restriction of multimapping to 10,000 was minimal (Supplemental Table S1). To estimate ChIP signal on TE consensus sequences, the per-base coverage in ChIP libraries was divided by the coverage in corresponding input libraries.
RNA-seq profiling of differential expression of genes and TEs
To facilitate transcriptome analysis, RNA-seq libraries were first computationally depleted of rRNA-derived reads by subtracting those reads that map to rRNA sequences (GenBank identifiers: 18S, NR_003278.3; 28S, NR_003279.1; 5S, D14832.1; and 5.8S, K01367.1) with Bowtie 0.12.7 with up to three mismatches (Langmead et al. 2009). Next, we aligned the libraries to RefSeq mouse transcriptome (the RefSeq gene table was downloaded from the UCSC Genome Browser), allowing for up to three mismatches and an unlimited number of valid alignments per read. The alignments were then unambiguously assigned to individual transcripts using eXpress (Roberts and Pachter 2013). The output of eXpress contains per-transcript RPKM and per-transcript read count estimates. Per-transcript RPKM values were contrasted directly for testis versus liver gene expression comparison (Supplemental Fig. S3). For other types of analyses, per-transcript read counts were summed for transcripts belonging to each gene to produce per-gene read counts. We assumed that genes that received zero read counts in either RNA-seq library were not detectably expressed and removed these genes from the analysis.
To estimate RNA-seq read counts and RPKMs for TEs and other repetitive sequences, we aligned the in silico rRNA-depleted libraries to the mm10 genome with Bowtie 0.12.7 (Langmead et al. 2009), allowing for zero mismatches and 10,000 valid alignments per read. In order to count reads mapped to individual TE families and compute corresponding RPKM values, we proceeded in the same fashion as described above for ChIP-seq libraries.
To estimate differential expression of genes, TEs, and other repetitive elements, we assumed that the technical error in RNA-seq estimation of TE expression is the same as for coding gene transcript expression. Therefore, to increase the power of the analysis, we combined TE and gene read counts and then modeled dispersion (i.e., noise) and estimated differential expression using DESeq (Anders and Huber 2010). DESeq was implemented via the R statistical environment (version 3.0.1) as described in the DESeq documentation accompanying the R interface of the software.
Differential occupancy of H3K9me3 across 1-kb genomic windows
We aligned reads to the mm10 genome with Bowtie 0.12.7 (Langmead et al. 2009), allowing for zero mismatches. Only uniquely mapping reads were considered for this analysis. Next, we broke the mm10 genome into nonoverlapping 1-kb windows and counted the number of reads mapped to each window in ChIP-seq libraries from FACS-sorted germ cells of Miwi2 knockout and control mice. If a read overlapped with two windows, then the window with the 5′ nucleotide of the read received the count for that read. In the end, we received the list of windows with counts of overlapping reads from two ChIP-seq libraries. Only windows that had at least one read in both knockout and control libraries were considered.
For differential occupancy analysis, the per-window counts from knockout and control ChIP-seq libraries were compared with DESeq (Anders and Huber 2010). Normalization, modeling of dispersion, and assignment of fold changes and P-values were done as described in the DESeq documentation accompanying the R interface of the software. We identified 6866 windows in which the levels of the H3K9me3 mark in Miwi2 knockout were decreased relative to the levels in the control samples (P < 0.05). With meta-analysis of ChIP signal in regions flanking TE insertions, we established that the mark spreads from TEs into the environment. Therefore, we extended each of the genomic windows that show differential H3K9me3 signal by 4.5 kb on both sides to produce 10-kb windows. As a control, we took all 1-kb genomic windows, extended each 10 kb, and, for each window, estimated the percentage of bases covered by LINE or LTR transposons. The average TE content in genomic windows that contained at least one ChIP-seq read in knockout and control libraries comprised the expected background TE content. We next calculated the average TE content in the differential occupancy windows and compared it with the control. In order to assess the statistical significance of the comparison of TE content in the identified 6866 regions that had decreases in the H3K9me3 mark, we sampled the control windows at random 6866 times and calculated the average TE content in the sample. The procedure was repeated 1,000,000 times, enabling us to directly estimate the empirical P-value associated with the comparisons of TE content.
Accession numbers
RNA-seq, small RNA, and small DNA data were deposited in the Gene Expression Omnibus database under accession number GSE58332.
Acknowledgments
We thank Katalin Fejes Tóth and members of the Aravin laboratory for helpful discussion and comments on the manuscript. We thank Rochelle Diamond and Diana Perez (California Institute of Technology) for invaluable help with cell sorting. We thank Alyssa Maskell (California Institute of Technology) for assistance with mouse work. We thank the Baltimore laboratory (California Institute of Technology) for lending us their magnetic cell-sorting equipment. We thank Sailakshmi Subramanian (Mount Sinai), Stijn van Dongen (EMBL-EBI), Georgi Marinov, Henry Amrhein, and Diane Trout (all California Institute of Technology) for help with bioinformatic and statistical analysis, and Igor Antoshechkin (California Institute of Technology) for help with RNA-seq. This work was supported by grants from the National Institutes of Health (R00 HD057233, R01 GM097363, and DP2 OD007371A), the Searle Scholar Award, and the Packard Fellowship to A.A.A.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.240895.114.
References
- Adey NB, Comer MB, Edgell MH, Hutchison CA III 1991. Nucleotide sequence of a mouse full-length F-type L1 element. Nucleic Acids Res 19: 2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Schaible K, Heasman J, Wylie C 1999. Expression of the homophilic adhesion molecule, Ep-CAM, in the mammalian germ line. J Reprod Fertil 116: 379–384 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Baust C, Gagnier L, Baillie GJ, Harris MJ, Juriloff DM, Mager DL 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J Virol 77: 11448–11458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chen Y, Negre N, Li Q, Mieczkowska JO, Slattery M, Liu T, Zhang Y, Kim TK, He HH, Zieba J, et al. 2012. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods 9: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day DS, Luquette LJ, Park PJ, Kharchenko PV 2010. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 11: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Kazazian HH Jr 1999. Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics 56: 317–323 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH Jr 1998. Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat Genet 20: 288–290 [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O’Carroll D 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608 [DOI] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS 1994. Neomorphic agouti mutations in obese yellow mice. Nat Genet 8: 59–65 [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Jamburuthugoda VK 2008. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134: 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla MEE 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338 [DOI] [PubMed] [Google Scholar]
- Feschotte C 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH Jr 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S 1990. Position-effect variegation after 60 years. Trends Genet 6: 422–426 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Takashima S, Ishii K, Shinohara T 2011. Dynamic changes in EPCAM expression during spermatogonial stem cell differentiation in the mouse testis. PLoS ONE 6: e23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci 108: 18760–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Akagi K, Hu Y, Trivett AL, Hlynialuk CJW, Swing DA, Volfovsky N, Morgan TC, Golubeva Y, Stephens RM, et al. 2012. Mouse endogenous retroviruses can trigger premature transcriptional termination at a distance. Genome Res 22: 870–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Marchetto MCN, Narvaisa I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Mears ML, Hutchison CA III 2001. The evolution of modern lineages of mouse L1 elements. J Mol Evol 52: 51–62 [DOI] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2013. The UCSC genome browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH 1998. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr 2000. Biology of mammalian L1 retrotransposons. Annu Rev Genet 35: 501–538 [DOI] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR 2009. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res 37: D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. 2011. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet 7: e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Roberts A, Pachter L 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods 10: 71–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL 2007. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet 3: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G 2003. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14: 67–75 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9–MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Smit AFA 1993. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res 21: 1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285 [DOI] [PubMed] [Google Scholar]
- Wang SH, Elgin SCR 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci 104: 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL 2008. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet 4: e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]