The FOXO transcription factors suppress cell proliferation and cell survival by transcriptionally activating specific gene targets linked to diverse cancer regulatory pathways. Here Zheng et al. identify FOXO3a as an EglN2 prolyl hydroxylase substrate. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by displacing the deubiquitinase USP9x. Consequently, loss of EglN2 leads to the accumulation of FOXO3a, which suppresses Cyclin D1 expression. These findings provide key insights into post-transcriptional control of FOXO3a and provide a new avenue for pharmacologically altering Cyclin D1 activity.
Keywords: EglN2, FOXO3a, USP9x, prolyl hydroxylation, Cyclin D1, breast cancer
Abstract
The three EglN prolyl hydroxylases (EglN1, EglN2, and EglN3) regulate the stability of the HIF transcription factor. We recently showed that loss of EglN2, however, also leads to down-regulation of Cyclin D1 and decreased cell proliferation in a HIF-independent manner. Here we report that EglN2 can hydroxylate FOXO3a on two specific prolyl residues in vitro and in vivo. Hydroxylation of these sites prevents the binding of USP9x deubiquitinase, thereby promoting the proteasomal degradation of FOXO3a. FOXO transcription factors can repress Cyclin D1 transcription. Failure to hydroxylate FOXO3a promotes its accumulation in cells, which in turn suppresses Cyclin D1 expression. These findings provide new insights into post-transcriptional control of FOXO3a and provide a new avenue for pharmacologically altering Cyclin D1 activity.
The HIF transcription factor, which consists of a labile α subunit and a stable β subunit, is a master regulator of genes that promote adaptation to hypoxia, including genes linked to metabolism, angiogenesis, and proliferation (Semenza 2013). HIF provided the first example of the use of prolyl hydroxylation as an intracellular signal. Specifically, HIF1α is hydroxylated on prolyl residues 402 and 564 by members of the EglN prolyl hydroxylase (also called PHD prolyl hydroxylase) family when oxygen is plentiful (Ivan et al. 2001; Jaakkola et al. 2001; Masson et al. 2001; Kaelin and Ratcliffe 2008). Prolyl-hydroxylated HIFα binds to the von Hippel-Lindau (VHL) E3 ligase complex, which polyubiquitylates HIFα and targets it for proteasomal degradation (Kaelin and Ratcliffe 2008).
There are three EglN family members in humans and mice (EglN1, EglN2, and EglN3). Their enzymatic activity requires oxygen, ascorbic acid, iron, and α-ketoglutarate (α-KG). Under hypoxic conditions, EglNs lose their activity and fail to hydroxylate HIFα, which leads to HIFα stabilization (Kaelin 2005; Kaelin and Ratcliffe 2008). HIFα then forms a complex with its binding partner, HIF1β (ARNT); translocates into the nucleus; binds to target genes containing hypoxia response elements (HREs); and activates their transcription (Kaelin 2005; Kaelin and Ratcliffe 2008). Notably, the oxygen Km values for the EglN prolyl hydroxylases are slightly above atmospheric oxygen (Epstein et al. 2001; Hirsila et al. 2003; Kaelin and Ratcliffe 2008). As a result, they are poised to act as oxygen sensors, coupling changes in oxygen availability to changes in HIF-dependent transcription.
EglN1 is clearly the primary HIF prolyl hydroxylase under normal conditions based on studies in cell culture and with genetically engineered mice (Berra et al. 2003; To and Huang 2005; Takeda et al. 2006; Minamishima et al. 2008). Perhaps for this reason, EglN1 is required for embryonic development, whereas EglN2−/− and EglN3−/− mice are viable (Takeda et al. 2006, 2007; Minamishima et al. 2008). EglN2 and EglN3, however, contribute to HIFα regulation under certain conditions, such as following EglN1 inactivation (Minamishima et al. 2009).
There is also mounting indirect evidence that EglN2 and EglN3 have HIF-independent functions. For example, EglN2 hydroxylase activity regulates Cyclin D1 accumulation and proliferation in a HIF-independent manner, and EglN3 can promote apoptosis in a HIF-independent manner (Lee et al. 2005; Bishop et al. 2008; Zhang et al. 2009; Tennant and Gottlieb 2010). However, the EglN2 and EglN3 hydroxylation targets responsible for these two phenotypes have remained elusive. For example, EglN2 appears to control Cyclin D1 at the mRNA level, and there is no evidence that EglN2 hydroxylates Cyclin D1 directly (Zhang et al. 2009).
A number of groups have attempted to identify novel EglN targets, including EglN2 and EglN3 targets. Taylor and coworkers (Cummins et al. 2006) provided indirect evidence that IκBKB is hydroxylated by EglN2, which could potentially contribute to negative regulation of NFkB by EglN2. Stamler and colleagues (Xie et al. 2009) discovered that β2-adrenergic receptor could be hydroxylated by EglN3 and subsequently ubiquitylated by pVHL. Semenza and colleagues (Luo et al. 2011) reported that PKM2 hydroxylation by EglN3 promotes its binding to HIF1 and enhances the transactivation of HIF1 target genes. For many of these and other putative EglN substrates, it has been difficult to demonstrate hydroxylation both in vitro and in vivo, possibly due to technical factors.
The FOXO transcription factors suppress cell proliferation and cell survival by transcriptionally activating specific gene targets that are linked to diverse cancer regulatory pathways (Greer and Brunet 2005; Huang and Tindall 2007). Activation of PI3K by extracellular growth signals leads to FOXO phosphorylation at three conserved Ser/Thr sites by AKT, whereupon the FOXOs are translocated to the cytoplasm and degraded (Greer and Brunet 2005; Huang and Tindall 2007). The role of the FOXOs in cancer has recently received increasing support from genetic studies in mice and human tumors (Paik et al. 2007; Cancer Genome Atlas Research Network 2008).
Identification of EglN substrates by unbiased mass spectrometry methods has so far proved challenging. This might relate to low abundance of the substrates, low affinities of the enzyme–substrate interactions, and the fact that both hydroxylation and spontaneous oxidation lead to the same change in mass (+16). We adapted a previously reported 96-well decarboxylation assay to screen for proteins that can be hydroxylated by EglN2 in vitro (Zhang et al. 1999). We focused on 1000 proteins previously linked to breast cancer because EglN2−/− mice exhibit mammary gland hypoproliferation and because loss of EglN2 inhibits breast cancer growth (Witt et al. 2006; Zhang et al. 2009). We identified FOXO3a as an EglN2 prolyl hydroxylase substrate. Prolyl hydroxylation by EglN2 destabilizes FOXO3a by displacing the deubiquitinase USP9x. Consequently, loss of EglN2 leads to the accumulation of FOXO3a, which suppresses Cyclin D1.
Results
Screen for novel EglN2 substrates
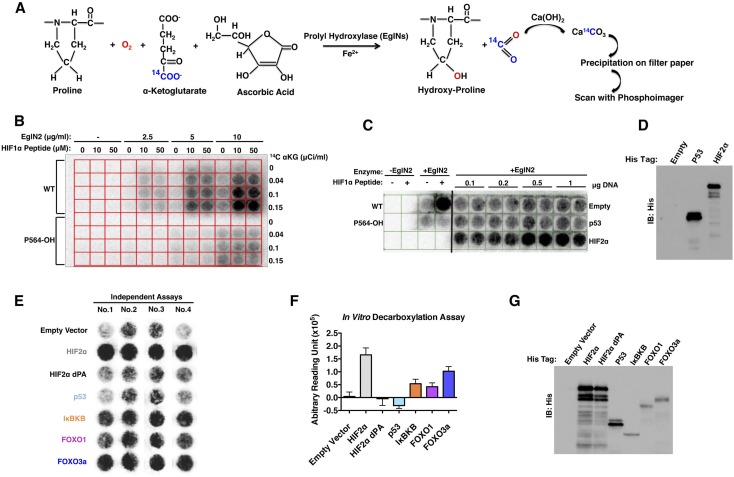
To screen for EglN2 substrates, we modified a previously published in vitro hydroxylation assay that can be used in a 96-well plate format (Zhang et al. 1999). This assay is based on the knowledge that hydroxylation by α-KG-dependent dioxygenases results in the decarboxylation of α-KG and the release of CO2. Hydroxylation in the presence of α-KG radiolabeled with 14C at the α carbon position leads to the release of radioactive CO2, which can then be captured with filters that are presaturated with Ca(OH)2 and tightly clamped to each plate. CO2 release, quantified with a phosphoimager, provides a measure of hydroxylation in each well (Fig. 1A).
Figure 1.
Screen for EglN2 substrates. (A) Biochemical basis for screen. Hydroxylation of a substrate by EglN2 is coupled to release and capture of 14C-labeled CO2. (B) Hydroxylation assays performed with varying amounts of recombinant EglN2, 14C-αKG, and synthetic HIF1α (556–575) peptides (wild-type or containing hydroxyproline at 564 position). (C) Hydroxylation assays using unprogrammed wheat germ lysate (Empty) or wheat germ lysate programmed to produce His-tagged p53 or HIF2α. Hydroxylation assays with no EglN2 (first two columns) or with EglN2 in the absence or presence of synthetic HIF1α peptides (columns 3,4) were included as controls. Increasing amounts of plasmid DNA were used for in vitro transcription/translation, as indicated. (D) Immunoblot analysis of in vitro translation products used in C. (E) Representative images from four independent hydroxylation assays. (F) Quantification of signal intensity in E (arbitrary units) after subtracting signal intensity from the empty vector well. (G) Immunoblot analysis of in vitro translation products as in E.
All three EglN family members can hydroxylate synthetic HIF1α peptides in vitro (Bruick and McKnight 2001; Epstein et al. 2001; Ivan et al. 2001). In pilot experiments, we confirmed that this 96-well plate assay could detect hydroxylation of a wild-type HIF1α peptide (corresponding to HIF1α residues 556–575) over a range of peptide, enzyme, and α-KG concentrations (Fig. 1B). Conversely, a synthetically prehydroxylated analog of this HIF1α peptide (Pro564-OH) served as a negative control in these experiments (Fig. 1B).
Next, we confirmed that this assay could detect hydroxylation of full-length HIF2α produced using wheat germ extracts. In these experiments, increasing amounts of plasmid DNA suitable for in vitro transcription and translation were added to fixed amounts of wheat germ extract, which was then used in the hydroxylation assays. Wheat germ extract programmed to produce HIF2α, but not p53, produced hydroxylation signals above background over a range of plasmid concentrations (0.1–1.0 μg) (Fig. 1C,D).
We next tested the in vitro translation products of ∼1000 cDNAs previously linked to breast cancer (Witt et al. 2006). Each cDNA was tested in quadruplicate in separate plates (one cDNA per well per plate × four plates). Wild-type HIF2α as well as a HIF2α variant in which its two prolyl hydroxylation sites have been changed to alanine (HIF2α dPA) (Kim et al. 2006) were included as positive and negative controls, respectively, in each plate. FOXO3a consistently produced a hydroxylation signal despite being produced at lower levels than HIF2α (Fig. 1E–G). Another FOXO family member, FOXO1, as well as IκBKB also produced hydroxylation signals (Fig. 1E–G.). We focused on FOXO3a because it consistently produced stronger hydroxylation signals than did either FOXO1 or IκBKB and because FOXO3a suppresses the proliferation of a variety of cell lineages, including mammary cells, which are known to be affected by EglN2 loss (Hu et al. 2004; Yang et al. 2008; Lin et al. 2011).
Hydroxylation of FOXO3a by EglN2
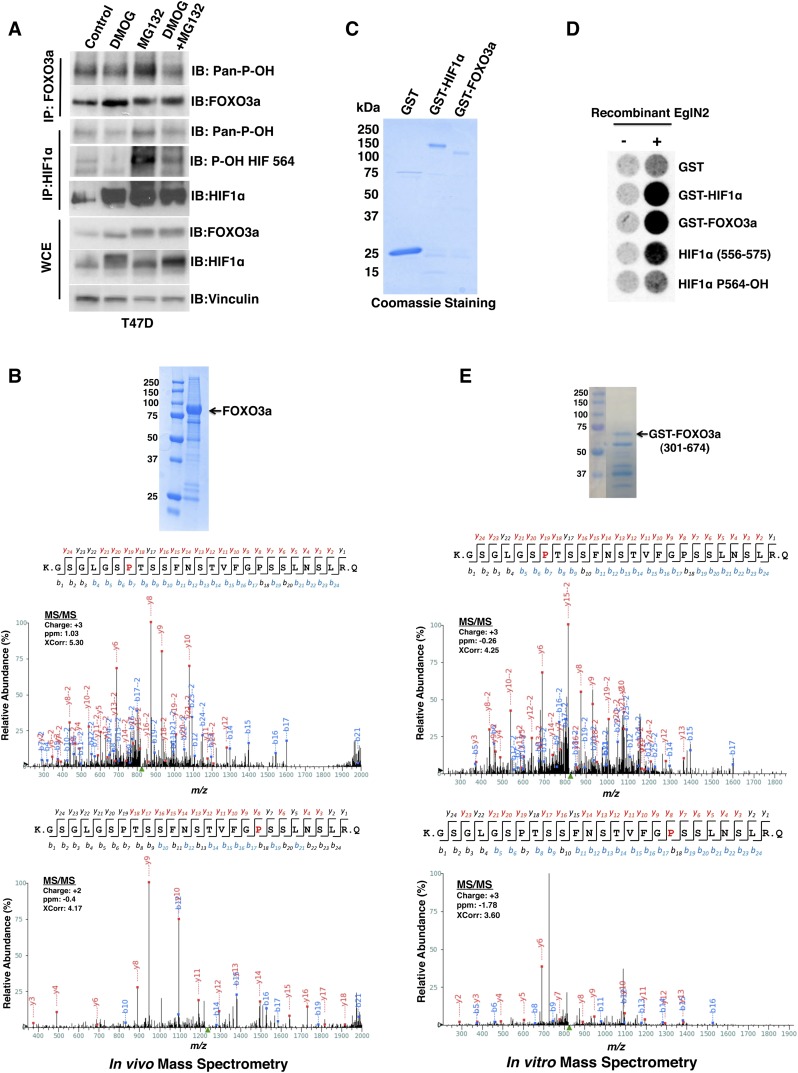
To determine whether FOXO3a can be hydroxylated by EglN2 in vivo, we treated T47D breast cancer cells with the prolyl hydroxylase inhibitor DMOG, the proteasome inhibitor MG132, both, or neither (vehicle alone). Following FOXO3a and HIF1α immunoprecipitation, Western blots were performed to examine FOXO3a and HIF1α hydroxylation using an anti-hydroxyproline antibody (Pan-P-OH). In multiple independent experiments, we detected increased HIF1α and FOXO3a prolyl hydroxylation signals upon MG132 treatment, which was abrogated by concurrent treatment with DMOG (Fig. 2A). The superior signal to noise ratio for HIF1α hydroxylation using an antibody that specifically recognizes hydroxylation of HIF1α Pro564 (P-OH HIF 564) relative to the Pan-P-OH antibody presumably reflects the higher affinity of the former antibody relative to the latter (Fig. 2A). The increased HIF1α prolyl hydroxylation signal with MG132 treatment was expected because HIF1α is rapidly degraded following prolyl hydroxylation. The fact that FOXO3a behaved similarly suggested that prolyl hydroxylation likewise destabilizes FOXO3a.
Figure 2.
EglN2 prolyl hydroxylates FOXO3a in vivo and in vitro. (A) Immunoblot (IB) assays of whole-cell extracts (WCEs) and immunoprecipitates (IPs) of T47D cells treated with either vehicle control, 1 mM DMOG, 10 μM MG132, or both 1 mM DMOG and 10 μM MG132. (B) 293FT cells were cotransfected to produce HA-FOXO3a and Flag-EglN2 and then treated with MG132. (Inset) HA-FOXO3a was purified by anti-HA immunoprecipitation followed by SDS–polyacrylamide gel electrophoresis and Coomassie blue staining. Shown are liquid chromatography-tandem mass spectrometry (LC-MS/MS) data for the excised FOXO3a band corresponding to FOXO3a peptides hydroxylated at Pro426 or Pro437. The red “P” indicates a hydroxylated proline residue. (C,D) Coomassie blue staining (C) and in vitro hydroxylation assays (D) with the indicated GST fusion proteins. Synthetic HIF1α peptides corresponding to residues 556–575 were included as controls in D. P564-OH indicates 556–575 peptide hydroxylated at Pro564. (E) GST-FOXO3a (301–674) was incubated with recombinant EglN2, resolved by SDS–polyacrylamide gel electrophoresis, and stained with Coomassie blue. Shown are MS/MS data for the excised FOXO3a band corresponding to FOXO3a peptides hydroxylated at Pro426 or Pro437. The red “P” indicates a hydroxylated proline residue. The peak heights are the relative abundances of the corresponding fragment ions, with the annotation of the identified matched N terminus-containing ions (b ions) in blue and C terminus-containing ions (y ions) in red.
Next, we used mass spectrometry to identify the potential FOXO3a prolyl hydroxylation sites. Toward this end, 293FT embryonic kidney cells were cotransfected to produce either HA-HIF1α or HA-FOXO3a together with Flag-EglN2 and treated with MG132. The HA-tagged proteins were recovered by immunoprecipitation and resolved by SDS–polyacrylamide gel electrophoresis. The bands of interest were identified by Coomassie blue staining, excised, and analyzed by mass spectrometry (Fig. 2B; Supplemental Fig. S1A). Using this approach, we identified hydroxylation of HIF1α Pro564, as expected, as well as hydroxylation of FOXO3a prolyl residues 426 and 437, which are conserved across different species (Supplemental Fig. S1B).
To determine whether EglN2 can hydroxylate these sites in vitro, we first used our radioactive decarboxylation assay to confirm that EglN2 can hydroxylate a GST-FOXO3a fusion protein produced in Escherichia coli. GST-HIF1α full-length and GST were included as positive and negative controls, respectively (Fig. 2C,D). Next, we performed in vitro hydroxylation assays with recombinant EglN2 and either GST-HIF1α or a GST-FOXO3a fusion protein containing FOXO3a residues 301–674 as substrates. Mass spectrometry analysis confirmed hydroxylation of the same sites identified in vivo; namely, prolyl residue 564 on HIF1α and prolyl residues 426 and 437 on FOXO3a (Fig. 2E; Supplemental Fig. S1C,D). Hydroxylation of GST-FOXO3a was not detected when EglN2 was omitted and was greatly diminished when prolyl residues 426 and 437 were mutated to alanines (Supplemental Fig. S1E,F). We were unable to reproducibly detect hydroxylation of synthetic FOXO3a peptides spanning residues 400–440 or 420–444, possibly because additional FOXO3a residues contact EglN2 or because hydroxylation of FOXO3a by EglN2 requires a FOXO3 conformational determinant that is not assumed by these peptides. A similar situation has been described for the lysyl hydroxylation of eRF1 by Jmjd4 (Feng et al. 2014).
EglN2 regulates FOXO3a abundance
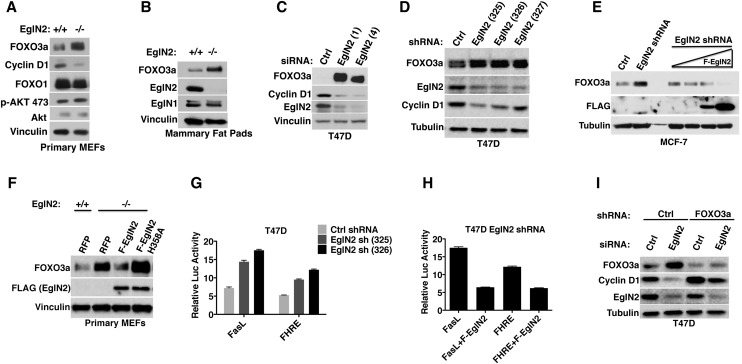
Prolyl hydroxylation regulates HIF1α stability, and the FOXO3a prolyl hydroxylation signal, like that of the HIF1α prolyl hydroxylation signal, was increased by proteasomal blockade in Figure 2A, suggesting that prolyl hydroxylation regulates FOXO3a abundance. Consistent with this idea, FOXO3a protein levels were increased in primary mouse embryonic fibroblasts (MEFs), immortalized MEFs, and mammary fat pads derived from EglN2−/− female mice compared with littermate EglN2+/+ controls, and these differences were abolished by exposure to DMOG or hypoxia (Fig. 3A,B; Supplemental Fig. S2A). The regulation of FOXO3a by EglN2 appeared to be post-transcriptional because Foxo3a mRNA levels were similar in EglN2+/+ and EglN2−/− MEFs (Supplemental Fig. S2B). Notably, AKT activity, which regulates FOXO3a localization and degradation (Brunet et al. 1999; Huang and Tindall 2007), was not altered by EglN2 loss, as determined by AKT phosphorylation at Ser473 (Fig. 3A). This suggests that regulation of FOXO3a by EglN2 and regulation of FOXO3a by AKT are distinct from one another. EglN1 loss, in contrast to EglN2 loss, did not induce FOXO3a in MEFs and mammary fat pads despite modestly up-regulated Foxo3a mRNA expression, indicating that regulation of FOXO3a is specific to EglN2 (Supplemental Fig. S2C–E).
Figure 3.
FOXO3a protein abundance is regulated by EglN2 and links EglN2 to Cyclin D1. (A,B) Immunoblot analysis of primary MEFs (A) and mammary fat pads (B) derived from EglN2+/+ and EglN2−/− littermates. (C,D) Immunoblot analysis of T47D cells after transfection with EglN2 siRNA (C) or infection with lentiviruses encoding EglN2 shRNAs. Unrelated nontargeting siRNA and shRNA sequences were used as controls (Ctrl). (E) Immunoblot analysis of MCF-7 cells that were infected with a lentivirus encoding EglN2 shRNA or Ctrl shRNA and, after drug selection, superinfected with a lentivirus encoding Flag-EglN2 (F-EglN2) under the control of a doxycycline-inducible promoter. Increasing amounts of doxycycline were added to the cells 48 h before lysis as indicated by the triangle. (F) Immunoblot analysis of primary MEF cells that were infected with a lentivirus encoding red fluorescent protein (RFP) or Flag-tagged EglN2 (wild-type or H358A). (G,H) Luciferase reporter assay of T47D cells that were infected with a lentivirus encoding EglN2 shRNA (325 or 326) or Control (Ctrl) shRNA followed by transfection with either FasL or FHRE reporter plasmid in the absence or presence of shRNA-resistant Flag-EglN2 (F-EglN2) with TK-Renilla as an internal control. (I) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding FOXO3a shRNA or control (Ctrl) shRNA and then transfected with either EglN2 siRNA or Ctrl siRNA.
Consistent with these results, down-regulation of EglN2 with two independent siRNAs and three independent shRNAs up-regulated FOXO3a in T47D breast carcinoma cells (Fig. 3C,D; Supplemental Fig. S2F). Similar results were obtained in MCF-7 cells (Supplemental Fig. S2G). As expected from the analysis of EglN1−/− MEFs and mammary fat pads, depletion of EglN1 by siRNA in T47D and MCF-7 cells did not affect FOXO3a protein (Supplemental Fig. S2F,G). Importantly, up-regulation of FOXO3a by EglN2 RNAi was reversed by coexpression of an shRNA/siRNA-resistant version of wild-type EglN2, but not by a catalytically dead (H358A) EglN2 variant (Fig. 3E,F; Supplemental Fig. S2H). EglN2 depletion activated FOXO3a-responsive promoters (FasL or FHRE) in luciferase-based reporter assays (Fig. 3G). The increased FOXO3a reporter activity could be reversed by cotransfection with an shRNA-resistant version of Flag-EglN2 (Fig. 3H).
We next asked whether up-regulation of FOXO3a could account for our earlier observation that EglN2 loss causes loss of Cyclin D1 (Zhang et al. 2009). To address this, we stably infected T47D breast carcinoma cells to express an effective FOXO3a shRNA or a control shRNA and then transiently transfected them with siRNAs against EglN2 or control siRNA (Ctrl). Down-regulation of FOXO3a blunted the effect of the EglN2 siRNA on Cyclin D1 levels (Fig. 3I), suggesting that EglN2 loss leads to FOXO3a accumulation, which then suppresses Cyclin D1. This finding is consistent with previous reports that FOXO3a suppresses Cyclin D1 (Ramaswamy et al. 2002; Schmidt et al. 2002).
Regulation of FOXO3a by EglN2 is HIF-independent
Treatment of T47D cells with various pharmacological prolyl hydroxylase inhibitors promoted the accumulation of FOXO3a in EglN2-proficient cells but did not further induce FOXO3a in cells already expressing high levels of FOXO3a by virtue of shRNA-mediated EglN2 depletion (Fig. 4A). Cells treated with MG132, which should induce FOXO3a, were included as an additional control for comparison. These results further support that control of FOXO3a by EglN2 is linked to EglN2 catalytic activity (see Fig. 3F; Supplemental Fig. S2H). They left open, however, the possibility that regulation of FOXO3a abundance by EglN2 is an indirect consequence of activating HIF, which is the canonical EglN substrate, instead of, or in addition to, changes in FOXO3a hydroxylation. A link between HIF and hypoxic induction of FOXO3a has been suggested before (Bakker et al. 2007; Jensen et al. 2011). To address this, we infected T47D or BT474 breast cancer cells with shRNAs against ARNT, the critical binding partner for HIFα’s transcriptional activity. Down-regulation of EglN2 with siRNA in these cells still induced FOXO3a (Fig. 4B; Supplemental Fig. S3A). Moreover, we did not observe induction of HIF1α protein levels in breast cancer cells after EglN2 loss alone (Fig. 4A), consistent with EglN1 being the primary regulator of HIF1 under most conditions (Berra et al. 2003). We did note that one of the two ARNT shRNAs depressed basal FOXO3a levels in BT474 breast cancer cells (Supplemental Fig. S3A). The significance of this finding is unclear. Collectively, these results suggest that control of FOXO3a by EglN2 is HIF-independent.
Figure 4.
Regulation of FOXO3a by EglN2 is post-transcriptional and HIF-independent. (A) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding EglN2 shRNA or control shRNA (Ctrl) and then treated with 200 μM CoCl2, 1 mM DMOG, 200 μM DFO, 10 μM MG132, or DMSO (Control) for 16 h. (B) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding either ARNT shRNA (3146) or Ctrl shRNA and, after drug selection, transfected with either EglN2 siRNA or Ctrl siRNA or treated with 1 mM DMOG. (C) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding either EglN2 shRNA or Ctrl shRNA and then treated with 100 μg/mL cycloheximide (CHX) for the indicated amount of time. (D) Quantitative RT–PCR (qRT–PCR) analysis of mRNA from T47D cells that were transfected with the indicated siRNAs. Error bars represent one SEM. (E,F) Immunoblot analysis of MCF-7 cells that were infected with a lentivirus encoding HA-FOXO3a and, after hygromycin selection, treated with either 10 μM MG132, 200 μM CoCl2, 200 μM DFO, 1 mM DMOG, or DMSO control for 16 h. (F) The cells were superinfected with a lentivirus encoding EglN2 shRNA (327) or control shRNA after the initial hygromycin selection. The cells were then placed under both hygromycin and puromycin selection and treated with DMOG, MG132, or vehicle control.
Regulation of FOXO3 by EglN2 is post-transcriptional
To further understand how EglN2 regulates FOXO3a protein levels, we measured FOXO3a protein stability and mRNA levels in cells depleted of EglN2. Loss of EglN2 prolonged FOXO3a protein stability, as determined by cycloheximide chase experiments (Fig. 4C), but did not alter Foxo3a mRNA abundance (Fig. 4D; Supplemental Fig. S2B), suggesting that the control of FOXO3a by EglN2 is post-transcriptional.
In support of this conclusion, we next infected two different breast cancer cell lines (MCF-7 and T47D) with a lentivirus encoding HA-tagged FOXO3a under the control of a weak heterologous promoter (UBC). Treatment of these cells with either prolyl hydroxylase inhibitors or a virus expressing an EglN2 shRNA promoted the accumulation of HA-FOXO3a (Fig. 4E,F; Supplemental Fig. S3B). These interventions did not affect other HA-tagged proteins expressed from this same lentiviral vector (data not shown). In Figure 4F, treatment of MCF-7 cells expressing the EglN2 shRNA with either DMOG or MG132 led to further accumulation of HA-FOXO3a, perhaps because down-regulation of EglN2 with the shRNA was incomplete.
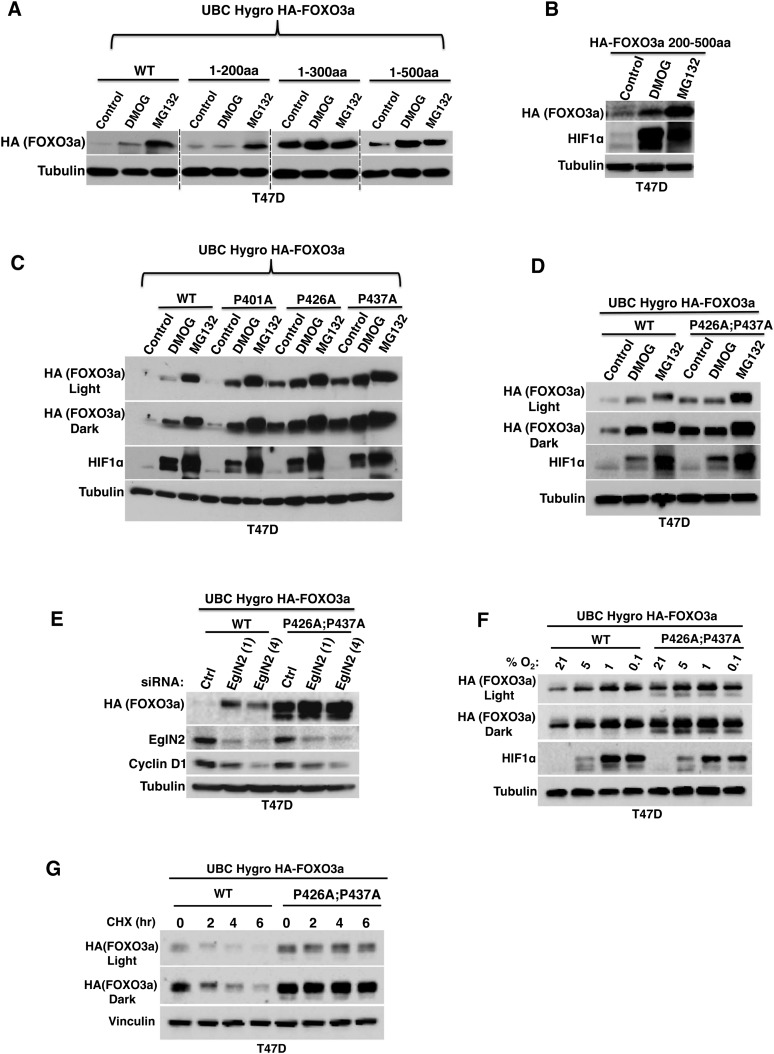
FOXO3a stability is regulated by prolyl hydroxylation
In order to narrow down the region of FOXO3a that is responsible for its regulation by EglN2, we generated different FOXO3a truncation mutants and tested their responsiveness to DMOG and MG132. As shown in Figure 5A, the region of FOXO3a corresponding to residues 1–500 was sufficient for induction by DMOG and MG132, while FOXO3a polypeptides corresponding to residues 1–200 and 1–300 were not induced by DMOG. FOXO3a contains a nuclear localization signal (NLS) between amino acids 242 and 259 that is important for regulating its subcellular localization and protein stability (Brunet et al. 1999; Tsai et al. 2007). Therefore, we then tested an HA-FOXO3a variant corresponding to FOXO3a residues 200–500 and found that this region was sufficient to confer sensitivity to DMOG (Fig. 5B).
Figure 5.
FOXO3a prolyl hydroxylation sites govern FOXO3a stability. (A–E) Immunoblot analysis of T47D cells that were infected with lentiviruses encoding the indicated HA-FOXO3a variants and then treated with either 1 mM DMOG, 10 μM MG132, or DMSO control for 16 h (A–D) or transfected with EglN2 siRNA (1 or 4) or control siRNA (E). (WT) Wild-type. (C,D) Light or dark refers to different exposure times. (F,G) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding either wild-type FOXO3a or P426A;P437A followed by hygromycin selection and treatment with either hypoxia for 16 h (F) or 100 μg/mL cycloheximide (CHX) for the indicated amount of time (G). Light or dark refers to different exposure times.
Since we determined that EglN2 hydroxylates FOXO3a on Pro426 and Pro437 in vitro and in vivo, we next generated full-length HA-FOXO3a variants in which Pro426, Pro437, or both were changed to alanine. Mutation of either Pro426 or Pro437 led to partial stabilization of FOXO3a, which was further enhanced by DMOG treatment (Fig. 5C). This effect was specific because mutating another prolyl residue located nearby, Pro401, did not alter FOXO3a abundance in these assays (Fig. 5C). Importantly, mutating both Pro426 and Pro437 to alanine (P426A;P437A) rendered FOXO3a insensitive to DMOG treatment (Fig. 5D), EglN2 depletion (Fig. 5E), and hypoxia (Fig. 5F), indicating that these two sites are necessary and that either is sufficient for regulation of FOXO3a protein stability by the EglN2 prolyl hydroxylase. Consistent with these findings, mutations of both prolines (P426A;P437A) significantly prolonged FOXO3a protein stability compared with wild-type FOXO3a, as determined by cycloheximide chase experiments (Fig. 5G).
Prolyl hydroxylation of FOXO3a disrupts its deubiquitylation by USP9x
Prolyl hydroxylation affects the stability of both HIF1α and FOXO3a, the former because prolyl hydroxylation promotes the binding of the pVHL ubiquitin ligase (Ivan et al. 2001; Jaakkola et al. 2001). A number of FOXO3a ubiquitin ligases have been reported, including β-TRCP and MDM2 (Yang et al. 2008; Tsai et al. 2010). In pilot studies, however, we found no evidence that hydroxylation promotes the binding of these two ligases to FOXO3a (Supplemental Fig. S4A).
In an attempt to identify the molecular mechanism by which FOXO3a stability is regulated by the EglN2 prolyl hydroxylase, we decided to identify protein whose binding to FOXO3a could be influenced by FOXO3a hydroxylation. We treated T47D cells stably expressing HA-FOXO3a under the control of the weak UBC promoter with either DMOG or vehicle and then recovered the HA-FOXO3a by immunoprecipitation. Mass spectrometry analysis of the immunoprecipitates from the DMOG-treated samples (but not the vehicle control) revealed peptides derived from FOXO3a, as expected, as well as two peptides derived from the USP9x deubiquitinase (Supplemental Fig. S4B). Recovery of USP9x in such immunoprecipitates was confirmed by Western blot analysis (Supplemental Fig. S4C) but was substoichiometric, perhaps reflecting a low affinity or fast off rate typical of many enzyme–substrate interactions.
USP9x has been reported to stabilize various proteins (Mcl-1, SMAD4, SMURF1, and Itch) involved in cancer by preventing their degradation through the removal of conjugated ubiquitin (Mouchantaf et al. 2006; Dupont et al. 2009; Schwickart et al. 2010; Xie et al. 2013). Due to the large size of USP9x, four different USP9x fragments, corresponding to N1 (residues 1–674), N2 (residues 675–1217), C1 (residues 1218–2106), and C2 (residues 2107–2570), were produced by in vitro translation and used in FOXO3a-binding assays. The C2 fragment, but not the other fragments, bound to GST-FOXO3 in vitro (Supplemental Fig. S4D). Next, these same four USP9x fragments were produced as GST fusion proteins and used to capture full-length FOXO3a produce by in vitro translation. GST-C2, but not the other GST-USP9x, bound FOXO3a (Supplemental Fig. S4E,F). These results are consistent with an earlier study that implicated the C2 region of USP9x in substrate binding (Xie et al. 2013).
To determine whether USP9x regulates FOXO3a stability, we infected T47D and MCF7 breast cancer cells with five different hairpins against USP9x and observed that all five decreased both endogenous and exogenous FOXO3a protein levels (Fig. 6A,B; Supplemental Fig. S4G). Similar results were obtained after transient transfection with USP9x shRNA/siRNAs in MCF-7, T47D, and 293T cells (Fig. 6C; Supplemental Fig. S4H,I). Moreover, we could rescue FOXO3a levels in cells expressing an effective USP9x shRNA by coexpressing an shRNA-resistant version of USP9x (Fig. 6C). As expected, down-regulation of FOXO3a in cells after shRNA-mediated USP9x depletion reflected enhanced FOXO3a ubiquitylation and decreased FOXO3a stability (Fig. 6D,E). In a complementary set of experiments, we found that exogenous expression of wild-type USP9x, but not a previously reported catalytically inactive USP9x variant (C1556S) (Schwickart et al. 2010; Xie et al. 2013), promoted the accumulation of FOXO3a (Fig. 6F). Collectively, these results suggest that USP9x promotes FOXO3a stability.
Figure 6.
Differential binding of USP9x underlies control of FOXO3a stability by EglN2. (A) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding the indicated USP9x shRNAs or Ctrl shRNA. (B) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding HA-FOXO3a and then superinfected with a lentivirus encoding USP9x shRNA (364 or 064) or Ctrl shRNA. (C) Immunoblot analysis of 293T cells that were infected with a lentivirus encoding USP9x shRNA (364) or Ctrl shRNA and then, after drug selection, transfected with a plasmid encoding V5-tagged mouse full-length USP9x or empty vector (Ctrl). (D) Immunoblot (IB) assays of whole-cell extracts (WCE) and immunoprecipitates (IP) of T47D cells that were infected with a lentivirus encoding USP9x shRNA (sh128 or sh364) or Ctrl shRNA and then, after drug selection, transiently transfected with plasmids encoding Flag-ubiquitin and HA-FOXO3a followed by treatment with 10 μM MG132 for 16 h. (E) Immunoblot analysis of MCF-7 cells that were infected with a lentivirus encoding shRNA against either USP9x (364) or Ctrl shRNA followed by 100 μg/mL cycloheximide for the indicated duration. (F) Immunoblot analysis of 293T cells that were cotransfected with plasmids encoding V5-tagged USP9x (wild-type or C1556S) and HA-FOXO3a. (G) Immunoblot (IB) assays of whole-cell extracts (WCE) and immunoprecipitates (IP) of T47D cells that were transfected with a plasmid encoding HA-FOXO3a (wild-type or P426A;P437A) followed by treatment with 10 μM MG132 for 16 h. (H) Immunoblot analysis of bound USP9x recovered from lysed T47D cells that were incubated with 20 μL of Neutravidin-Sepharose preloaded with the indicated FOXO3a peptides. (I,J) Immunoblot analysis of T47D cells that were infected with a lentivirus encoding USP9x shRNA (364) or Ctrl shRNA and then, after drug selection, treated with the indicated drugs or siRNAs.
Although these results indicated that USP9x could protect FOXO3a from ubiquitin-mediated proteolysis, they did not address the importance of FOXO3a hydroxylation in this process. To address this question, we next transiently transfected T47D cells to produce either wild-type HA-FOXO3a or the HA-FOXO3a variant in which prolyl residues 426 and 437 were converted to alanine (P426A;P437A) and treated them with MG132 in an attempt to equalize the amount of exogenous FOXO3a. We repeatedly detected increased coimmunoprecipitation of USP9x with the P426A;P437A mutant relative to wild-type FOXO3a (Fig. 6G). In a complementary set of experiments, synthetic, biotinylated FOXO3a peptides corresponding to FOXO3a residues 420–444 were used to capture USP9x from T47D extracts, as determined by immunoblot analysis. The 420–444 peptide readily captured USP9x unless either or both prolyl sites were synthetically hydroxylated (Fig. 6H). Moreover, inhibition of EglN2 with prolyl hydroxylase inhibitors (DFO and DMOG) (Fig. 6I) or an effective siRNA (Fig. 6J) did not increase FOXO3a protein levels in T47D breast cancer cells in which USP9x was depleted. The effects were specific because loss of USP9x did not prevent the induction of FOXO3a by MG132 (Fig. 6J). Collectively, these results suggest that prolyl hydroxylation by EglN2 diminishes the binding of USP9x to FOXO3a, leading to FOXO3a destabilization.
Discussion
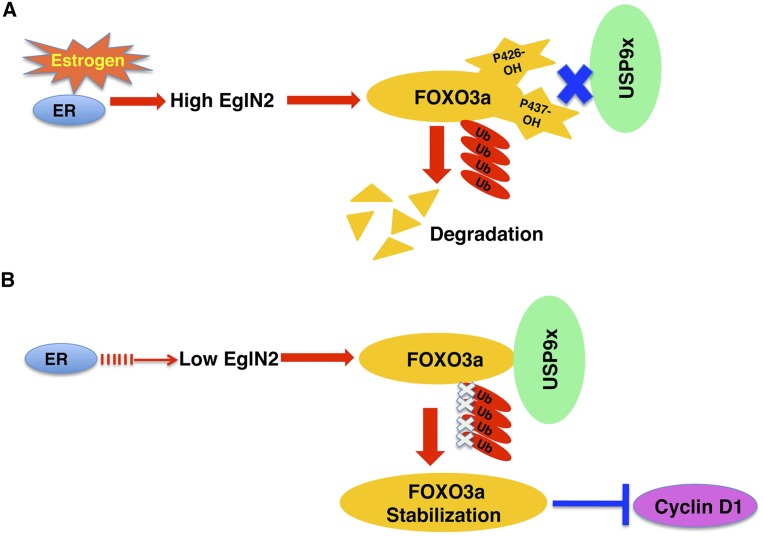
We developed an in vitro hydroxylation screen that identified FOXO3a as a potential EglN2 substrate and found that EglN2 can hydroxylate FOXO3a on prolyl 426 and 437 sites in vitro and in vivo. FOXO3a hydroxylation disrupts its interaction with the USP9x deubiquitinase. The dissociation from USP9x promotes FOXO3a ubiquitylation and proteasomal degradation (Fig. 7A). Loss of EglN2 promotes the accumulation of FOXO3a as a result of enhanced FOXO3a protein stability, which suppresses Cyclin D1 expression (Fig. 7B).
Figure 7.
Proposed models for FOXO3a regulation by EglN2. (A) Estrogen binding with ER transcriptionally induces EglN2, which triggers FOXO3a hydroxylation on prolyl residue 426 and 327 sites. Hydroxylation of these sites dissociates FOXO3a from USP9x deubiquitinase, thereby facilitating FOXO3a ubiquitylation and degradation. (B) Depletion of EglN2 by either lack of ER activity or EglN2 siRNAs/shRNAs prevents FOXO3a from being hydroxylated. As a result, USP9x binds with FOXO3a and prevents FOXO3a from being ubiquitylated and degraded. Stabilization of FOXO3a decreases Cyclin D1 expression.
Regulation of FOXO3a by EglN2 appears to be important in signaling by estrogens. Activation of the estrogen receptor α (ERα) by estrogens induces EglN2 mRNA (Seth et al. 2002; Appelhoff et al. 2004; Zhang et al. 2009), and EglN2 promotes mammary proliferation (Seth et al. 2002; Zhang et al. 2009). Conversely, loss of EglN2 decreases mammary gland proliferation during lactation and decreases the proliferation of estrogen-responsive breast cancer cells in vitro and in vivo (Zhang et al. 2009). Our findings suggest that these phenotypes are due at least partly to EglN2’s role in the regulation of FOXO3a and its downstream target, Cyclin D1, which plays a critical role in mammary proliferation. Interestingly, FOXO3a has been reported to control the transcription of ERα and, through physical association, affect ERα-dependent transcription (Guo and Sonenshein 2004). If true, this would suggest that induction of EglN2 and down-regulation of FOXO3a constitute a feedback loop with ERα.
Genetic studies in mice indicate that members of the FOXO family, including FOXO3a, can act as tumor suppressor genes. Somatic deletion of all three FOXOs (FOXO1, FOXO3, and FOXO4) engenders a cancer-prone condition in mice, further strengthening its tumor suppressor role in vivo (Paik et al. 2007). Consistent with this, decreased expression of FOXO3a associated with poor outcomes in breast cancer and neuroblastoma (Sunayama et al. 2011; Jiang et al. 2013; Santo et al. 2013). Conversely, FOXO3a nuclear localization is associated with good prognosis in luminal-like breast cancer (Habashy et al. 2011). Moreover, exogenous expression of FOXO3a inhibits tumor growth in vitro and in vivo in breast cancer (Hu et al. 2004; Yang et al. 2008; Lin et al. 2011; Sisci et al. 2013).
It is well established that FOXO3a is regulated by phosphorylation on T32 and S253 by AKT/SGK kinase, which creates binding sites for chaperone protein 14-3-3 (Brunet et al. 1999, 2001, 2002; Huang and Tindall 2007). Binding to 14-3-3 promotes the translocation of FOXO3a from nucleus to cytoplasm, where it undergoes proteasomal degradation (Brunet et al. 2002). Similarly, it is also known that phosphorylation of FOXO3a at S294, S344, and S425 by ERK promotes the binding of MDM2 to FOXO3a, which likewise promotes its proteasomal degradation (Yang et al. 2008). Finally, phosphorylation of FOXO3a by IκB kinase on S644 was reported to affect FOXO3a stability by promoting the binding of β-TRCP E3 ligase (Hu et al. 2004; Tsai et al. 2010). Our findings reveal another layer to the regulation of FOXO3a stability, in this case mediated by changes in deubiquitylation rather than ubiquitylation.
USP9x appears to be capable of either enhancing or suppressing tumor growth in a context-dependent manner. For example, USP9x has been reported to stabilize the anti-apoptosis protein Mcl-1 and promote tumor cell survival in follicular lymphomas and diffuse large B-cell lymphomas (Schwickart et al. 2010). On the other hand, by using transposon-mediated insertional mutagenesis, Tuveson and colleagues (Perez-Mancera et al. 2012) identified USP9x as a gene that, when inactivated, cooperates with oncogenic Kras to accelerate pancreatic ductal adenocarcinoma (PDA). Our studies show that loss of USP9x leads to loss of FOXO3a. This would be predicted to promote tumor growth in certain settings for the reasons outlined above.
As an EglN2 substrate, FOXO3a is stabilized by hypoxia and hypoxia mimetics. In addition, FOXO3a appears to be a direct HIF target (Bakker et al. 2007; Jensen et al. 2011). FOXO3a clearly plays a role in adaptation to hypoxic stress (Scott et al. 2002; Miyamoto et al. 2007; Tothova and Gilliland 2007; Mabon et al. 2009; Jensen et al. 2011). Induction of FOXO3a by hypoxia might help conserve ATP by restricting cell proliferation as well as by reprogramming cell metabolism. In this regard, FOXO3a was reported to regulate oxygen consumption and reactive oxygen species (ROS) production by inhibiting mitochondrial gene expression (Jensen et al. 2011; Ferber et al. 2012). FOXO3a activation by hypoxia could decrease mitochondrial gene expression and mitochondria DNA copy number, which could shift cellular metabolism from oxidative phosphorylation to glycolysis and reduce oxidative stress. In this way, FOXO3a would, like HIF, serve as another important regulator mediating cellular adaptation to hypoxia. It is also possible that FOXO3a tunes the HIF response in certain settings, as suggested before (Bakker et al. 2007; Jensen et al. 2011). Notably, EglN2−/− mice display decreased oxygen consumption and increased hypoxia tolerance associated with increased expression of pyruvate dehydrogenase 4 (PDK4), which is responsive to FOXO3a (Kwon et al. 2004; Aragones et al. 2008).
It remains to be determined whether EglN2 has additional substrates, especially as our screen was restricted to 1000 cDNAs. Moreover, we have not yet done secondary analyses of other potential positives from our screen, such as the FOXO3a paralog FOXO1 and IκBKB. With regard to IκBKB, Taylor and coworkers (Cummins et al. 2006) showed that EglN2 loss as well as hydroxylase inhibitors activate NFκB and provided indirect evidence that IκBKB was hydroxylated, including the presence of a peptide sequence similar to the sequence that is prolyl-hydroxylated in HIFα. However, they were unable to confirm that EglN2 hydroxylates IκBKB. Our data suggest that this question should now be revisited.
Strategies to induce FOXO3a signaling in cancer might have therapeutic potential, especially in cancers such as breast cancer in which expression of FOXO3a or loss of Cyclin D1 inhibits cell proliferation. It has been argued that certain cytotoxic drugs, such as Paclitaxel and Imatinib, induce FOXO3a protein by indirectly promoting FOXO3a dephosphorylation (Sunters et al. 2003, 2006; Essafi et al. 2005). Targeted agents that block PI3K/AKT signaling also induce FOXO3a (Sunters et al. 2006). Our study opens another avenue for manipulating FOXO3a activity. In particular, our work predicts that EglN2 inhibitors would promote the accumulation of FOXO3a and in turn down-regulate Cyclin D1, leading to impaired cancer cell proliferation. In this regard, it has already been established that EglN family members can be inhibited with drug-like small organic molecules in humans (Nwogu et al. 2001; Ivan et al. 2002; Fraisl et al. 2009; Beuck et al. 2012). It will be important to determine whether small molecule EglN2 inhibitors display anti-cancer activity either alone or when combined with other agents such as cytotoxics, Cdk4 inhibitors, or PI3K/AKT inhibitors.
Materials and methods
Cell culture
MEF, 293FT, and MCF-7 cells were cultured in DMEM containing 10% fetal bovine serum (FBS) plus 1% penicillin and streptomycin (Pen Strep). BT474 and T47D cells were maintained in RPMI medium containing 10% FBS plus 1% Pen Strep. Following lentiviral infection, cells were maintained in the presence of 200 μg/mL hygromycin or 2 μg/mL puromycin, depending on the vector. All cells were maintained at 37°C in 5% CO2.
Plasmids
Full-length HA-tagged FOXO3a was amplified by PCR with a 5′ primer that introduced a NotI site and an HA tag and a 3′ primer that introduced a BamHI site. The PCR product was digested with BamHI and NotI and cloned into pcDNA3.1(+) (Invitrogen) vector cut with these two enzymes. FOXO3a P426A, P437A, and P426A;P437A cDNAs were generated by site-directed mutagenesis. The FOXO3a insert was cut with BamHI and NotI and ligated into pGEX-4T-2 (Amersham) vector cut with these two enzymes to make the full-length GST-FOXO3a plasmid. The FOXO3a cDNA encoding residues 301–674 was amplified by PCR with a 5′ BamHI and a 3′ NotI site and ligated into pGEX-4T-2 vector cut with these two enzymes to make the GST-FOXO3a (301–674) plasmid.
Full-length Flag-tagged EglN2 was amplified by PCR with a 5′ primer that introduced an EcoRI site and a Flag tag and a 3′ primer that introduced a BamHI site. The PCR product was digested with EcoRI and BamHI and cloned into pcDNA3.1(−) vector cut with these two enzymes. Full-length human EglN2 was amplified by PCR with a 5′ BamHI and a 3′ EcoRI site. The EglN2 PCR product was digested with BamHI and EcoRI and ligated into pGEX-4T-2 vector to make the GST-EglN2 plasmid. pLenti6-Flag-EglN2 was described previously (Koivunen et al. 2012). pLenti6-Flag-EglN2 (H358A) was generated by using site-directed mutagenesis. The inducible expression plasmid for Flag-tagged EglN2 was generated by PCR of EglN2 cDNA in pcDNA-HA-EglN2 plasmid with a 5′ primer that introduced an XmaI and a Flag tag and a 3′ primer that introduced a ClaI site. The PCR product was digested with XmaI and ClaI and cloned into pTripZ vector (Thermo Scientific) cut with AgeI and ClaI. In order to generate shRNA-resistant EglN2 cDNAs, translationally silent mutations were introduced into EglN2 shRNA recognition sites by site-directed mutagenesis as indicated previously (Zhang et al. 2009).
The UBC Hygro HA-FOXO3a plasmids encoding full-length FOXO3a, FOXO3a (1–200), FOXO3a (1–300), FOXO3a (1–500), and FOXO3a (200–500) were made by PCR-amplifying a FOXO3a cDNA with a 5′ primer that introduced a BamHI site and an HA tag and a 3′ primer that introduced a NotI site. The PCR product was digested with BamHI and NotI and ligated into pLenti-UBC-pGK-Hyg (a modified version of pLL3.7, Addgene) vector cut with these two enzymes. FasL and FHRE plasmids were obtained from Addgene.
V5-tagged mouse USP9x, catalytic-dead USP9x C1556S, and GST-tagged human USP9x (N1, N2, C1, and C2) plasmids were generously provided by Dr. Feng Cong (Novartis). Human USP9x cDNA fragments (N1, N2, C1, and C2) were subcloned into pcDNA3.1 vector with an N-terminal Flag tag.
Plasmid pSP64polyA (Promega no. P1241) was modified to allow for recombination subcloning of cDNAs from pDNR1r plasmids (Clontech) using the Cre-loxP system and gain an N-terminal RGS-6xHis tag; the final plasmid was intended to allow for easy/effective in vitro transcription/translation of His-tagged proteins using the SP6 promoter. All plasmids were confirmed by DNA sequencing.
Generation of cDNA library in PSP64 PolyA vector
Glycerol stocks for ∼1000 gateway-compatible donor plasmids containing ORF cDNA previously linked to breast cancer were obtained from the Harvard Institute for Proteomics (Witt et al. 2006). One-hundred nanograms of Miniprep DNA for these clones was mixed with the destination vector pSP64PolyA RGS-6HisLoxP (modified pSP64PolyA [Promega], which allows for in vitro transcription and introduces an N-terminal RGS-6His tag), 1 μL of CRE recombinase (New England Biolabs), and CRE reaction buffer (33 mM NaCl, 50 mM Tris-HCl, and 10 mM MgCl2) in a 10-μL reaction. The recombination reaction was performed for 30 min at 37°C followed by 10 min of heat inactivation at 70°C in a 96-well format. Five microliters of each recombination reaction was then incubated with 20 μL of competent E. coli followed by plating onto plates compatible with ampicillin, chloramphenicol, and sucrose triple selection. Miniprep DNA was isolated from surviving colonies and authenticated by colony PCR.
Generation of recombinant EglN2
Full-length human EglN2 with C-terminal Flag and His tags was expressed using a baculovirus in H5 insect cells and immunopurified with anti-Flag antibody, as previously described (Hirsila et al. 2005).
In vitro decarboxylation assay
In vitro decarboxylation assay was modified from a previously published in vitro hydroxylation assay (Zhang et al. 1999). In brief, peptides or in vitro translated protein were supplemented with 50 mM HEPES (pH 7.4), 1500 U/mL catalase, 100 μM FeSO4, 1 mM ascorbic acid, 0.15 μCi/mL [1-14C] α-KG (Perkin Elmer), and 1 μg of purified recombinant EglN2 in a 100-μL reaction volume in a 96-well format. Radiolabeled CO2 was captured with glass fiber filter paper (catalog no. IH-201-A, Inotech Biosystems International) soaked with 30 mM saturated Ca(OH)2. A plastic adhesive microtiter plate sealer film was applied to the one side of the filter paper that was not in contact with the plate. The plate and filter paper with plastic sealer were sandwiched between two custom-made aluminum plates. The upper plate had a protective rubber layer that contacted the plastic-coated filter paper. The plate was clamped together by hand-tightening four thumbscrews. The clamped plate was transferred to a 37°C oven and allowed to incubate for 2 h, after which the apparatus was disassembled. The filter papers were dried under a heat lamp for 20 min and exposed to storage phosphorscreens overnight. Images were recorded using a Typhoon 8600 imager (Amersham Biosciences). Arbitrary signal intensity was calculated by ImageQuant software, with the background signal (empty vector control) being subtracted.
Immunoprecipitation
Cells were lysed in in EBC lysis buffer supplemented with complete protease inhibitors (Roche Applied Bioscience). The lysates were clarified by centrifugation and then mixed with primary antibodies or 3F10 anti-HA-conjugated beads (Roche Applied Bioscience) overnight at 4°C. For primary antibodies, protein G Sepharose was added for an additional 2 h (Roche Applied Bioscience). The bound complexes were washed with NETN buffer eight times and were eluted by boiling in SDS loading buffer. Bound proteins were resolved in SDS-PAGE followed by immunoblot analysis.
Peptide synthesis
All peptides were synthesized at the W.M. Keck Biotechnology Resource Center at Yale University. Each contained an N-terminal biotin and free C terminus and was synthesized in 25 μM scale. The sequences were as follows: HIF1α (556–575), DLDLEMLAPYIPMDDDFQLR; HIF1α P564-OH, DLDLEMLAP#YIPMDDDFQLR (# denotes hydroxylation); FOXO3a (400–440), QPSPTGGLMQRSSSFPYTTKGSGLGSPTSSFNSTVFGPSSL; FOXO3a (420–444), GSGLGSPTSSFNSTVFGPSSLNSLR; P426-OH, GSGLGSP#TSSFNSTVFGPSSLNSLR (# denotes hydroxylation); P437-OH, GSGLGSPTSSFNSTVFGP#SSLNSLR (# denotes hydroxylation); and P426-OH;P437-OH, GSGLGSP#TSSFNSTVFGP#SSLNSLR (# denotes hydroxylation).
GST protein purification and GST pull-down
GST plasmids were transformed with BL21-competent cells. Single colonies were picked and cultured overnight in 50 mL of LB medium containing ampicillin. Five milliliters of overnight culture was diluted into 500 mL of fresh LB medium with ampicillin and grown for 2–3 h at 37°C with shaking until OD600 reached 0.8–1.0, at which point 0.2 mM IPTG was added to induce GST protein production. Four hours later, the bacteria were pelleted by centrifugation and disrupted by a nanodebee homogenizer. Cleared bacteria lysates were purified using Glutathione-Sepharose 4B. Twenty microliters of Glutathione-Sepharose 4B preloaded with ∼2 μg of recombinant protein of GST fusion proteins was incubated with either in vitro translated protein in 500 μL of NETN buffer or EBC cell lysates. After overnight incubation, bound complexes were washed eight times with NETN buffer times followed by boiling in SDS loading buffer and SDS-PAGE.
Peptide-binding assay
Peptides (50 μg) were incubated with 20 μL of Neutravidin-Sepharose (Pierce) and 500 μg of EBC extracts prepared from T47D parental cells or T47D cells expressing Flag-EglN2 in a total volume of 500 μL overnight at 4°C. The Sepharose was washed eight times with NETN buffer. Bound proteins were then eluted by boiling in SDS loading buffer, resolved by SDS-PAGE, and detected by immunoblot analysis.
Mice
EglN2−/− mice were generously provided by Regeneron Pharmaceuticals. MEFs were isolated from embryonic day 13.5 (E13.5) as described previously (Zhang et al. 2009). EglN1Flox/Flox (F/F) mice were described previously (Minamishima et al. 2008; Moslehi et al. 2010). All experiments related to mice complied with National Institutes of Health guidelines and were approved by the Animal Care and Use Committees at either the Dana-Farber Cancer Institute or University of North Carolina at Chapel Hill.
Western blot analysis and antibodies
EBC buffer (50 mM Tris-HCl at pH 8.0, 120 mM NaCl, 0.5% NP40, 0.1 mM EDTA, and 10% glycerol) supplemented with complete protease inhibitor (Roche Applied Biosciences) was used to harvest whole-cell lysates. Mouse mammary glands and tumor tissues were lysed in 8 M urea containing 40 mM Tris-HCl (pH 7.6) and 2.5 mM EDTA. Cell lysate concentrations were measured by Bradford assay. Equal amounts of cell lysates were resolved by SDS-PAGE. Rabbit anti-FOXO3a (no. 2497), P-OH HIF564 (no. 3434), FOXO1 (no. 2880), EglN1 (no. 4835), p-AKT473 (no. 9271), and AKT (no. 9272) antibodies were obtained from Cell Signaling. Rabbit anti-EglN2 (no. NB100-310) and HIF1α (no. NB100-479) antibodies were obtained from Novus Biological. Rabbit anti-Cyclin D1 antibody (no. RB-010-P0) was obtained from Neomarker. Mouse anti-ARNT (no. 611079) and HIF1α (no. 610958) antibodies were obtained from BD Bioscience. Anti-HA antibody (no. MMS-101P) was obtained from Covance. Anti-vinculin (no. V9131), tubulin (no. T9026), and Flag (M2, no. F3165) antibodies were obtained from Sigma. Anti-V5 antibody (no. R960-25) was obtained from Invitrogen. Anti-USP9x (no. A301-350) and anti-ubiquitin (no. A300-317A) antibodies were obtained from Bethyl Laboratory. Anti-GST (no. SC-138) antibody was obtained from Santa Cruz Biotechnology. The pan-hydroxylated proline antibody was obtained from Advanced Targeting Systems (no. AB-T044). Peroxidase-conjugated goat anti-mouse secondary antibody (no. 31430) and peroxidase-conjugated goat anti-rabbit secondary antibody (no. 31460) were obtained from Thermo Scientific.
In vivo and in vitro mass spectrometry
For in vitro mass spectrometry, 2 μg of GST-HIF1α or GST-FOXO3a (301–674) was incubated in 50 mM Tris-HCl (pH 7.8), 100 μM dithiothreitol (DTT), 1500 U/mL catalase, 10 μM FeSO4, 1 mM ascorbic acid, 0.2 mM α-KG, and 2 μg recombinant EglN2 enzyme in 100-μL reactions. The reaction tubes were incubated for 1 h at 37°C. Samples were resolved by SDS-PAGE followed by Coomassie staining. Bands corresponding to GST-HIF1α and GST-FOXO3a were excised for mass spectrometry.
For in vivo mass spectrometry, eight p100 dishes of 293FT cells were cotransfected with plasmids encoding Flag-EglN2 and either HA-HIF1α or HA-FOXO3a. Thirty-six hours post-transfection, cells were treated with 10 μM MG132 for 16 h before harvesting. Cleared cell lysates were incubated with anti-HA-agarose (3F10, Roche Applied Science) followed by four to five washes with NETN lysis buffer (120 mM NaCl, 20 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 0.5% [v/v] Nonidet P-40 [NP-40]). Proteins were eluted by boiling in SDS-containing sample buffer, resolved by SDS-PAGE, and identified by Coomassie staining. HIF1α and FOXO3a bands were excised for mass spectrometry.
The bands containing HIF1α or FOXO3a were reduced with 10 mM DTT for 30 min, alkylated with 55 mM iodoacetamide for 45 min, and in-gel-digested with trypsin enzymes. The resulting peptides were extracted from the gel and analyzed by nanoscale-microcapillary reversed-phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (Villen et al. 2008). All peptide matches were filtered based on mass deviation, tryptic state, XCorr, and dCn and confirmed by manual validation.
siRNAs and lentiviral shRNA vectors
Nontargeting control siRNA number 2 was obtained from Dharmacon (catalog no. D0012100220). FOXO3a siRNA was obtained from Cell Signaling Technology (catalog no. 6302). EglN2 (1) and EglN2 (4) siRNAs were described previously (Zhang et al. 2009): USP9x si1 target sequence, CGCCTGATTCTTCCAATGAAA; and USP9x si2 target sequence, GTACGACGATGTATTCTCA. EglN2 shRNA in pCCLsin.PPT.hPGK.GFP.Wpre lentiviral vector was described previously (Zhang et al. 2009). Lentiviral EglN2 shRNAs, ARNT shRNAs, and USP9x shRNAs were obtained from Broad Institute TRC shRNA library. Target sequences are listed as follows: Ctrl shRNA, CAACAAGATGAAGAGCACCAA; EglN2 (325), GCTGCATCACCTGTATCTATT; EglN2 (326), GCCACTCTTTGACCGGTTGCT; EglN2 (327), ACTGGGACGTTAAGGTGCATG; ARNT (1770), GAGAAGTCAGATGGTTTATTT; ARNT (3146), CCTTTGTCTTTCTGTGTACTT; USP9x (064), GGTCGTTACAGCTAGTATTTA; USP9x (362), CGATTCTTCAAAGCTGTGAAT; USP9x (363), CGACCCTAAACGTAGACATTA; USP9x (364), CGCCTGATTCTTCCAATGAAA; USP9x (128), GAGAGTTTATTCACTGTCTTA; and FOXO3a shRNA, CATGTTCAATGGGAGCTTGGA.
Virus production and infection
Lentiviral infection was carried out essentially as previously described (Moffat et al. 2006). Briefly, 293T cells were transfected with Lipofectamine 2000, and viruses were collected 48 and 72 h later. After passing through a 0.45-μM filter, virus supernatants were used to infect target cells in the presence of 8 μg/mL polybrene.
Cell culture luciferase assays
T47D cells and their derivatives were plated on a 24-well plate and transfected with 150 ng of either FasL or FHRE reporter plasmid (Addgene), 10 ng of the TK-Renilla luciferase expression plasmid (Promega), and 500 ng of pcDNA or pcDNA Flag-EglN2 (EglN2 shRNA resistant) with Lipofectamine 2000 (Invitrogen) reagent according to the manufacturer’s instructions. Thirty-six hours later, luciferase activity of cells extracts was measured with the Dual Luciferase assay system (Promega) according to the manufacturer’s instructions and a Berthold Technologies luminometer.
Cycloheximide pulse chase assay
For cycloheximide assays, cells were treated with 100 μg/mL cycloheximide (Sigma) in appropriate growth medium. At various times thereafter, cells were lysed and subjected to immunoblot analysis.
FOXO3a ubiquitylation assay
Twenty-four hours after transfection T47D cells were treated with 5 μM MG132 overnight and then lysed in TBS plus 1% SDS. Cell lysates were boiled for 10 min followed by addition of 1 mL of EBC lysis buffer (with protease inhibitor cocktail) and sonication. Samples were chilled for 20 min on ice before anti-HA conjugated Sepharose (3F10, Roche Applied Bioscience) was added into each sample. Following incubation for 4 h at 4°C with gentle mixing, the Sepharose was washed three times with 1 mL of prelysis buffer (20 mM Tris-HCl at pH 7.4, 650 mM NaCl, 1 mM EDTA, 1% Triton X-100). The proteins were eluted by boiling in SDS-containing sample buffer and resolved by SDS-PAGE prior to immunoblot analysis.
Acknowledgments
We thank Regeneron Pharmaceutical for the EglN2−/− mice, Dr. Silvia Giordano for the pCCLsin.PPT.hPGK.GFP.Wpre lentiviral vector, Dr. Feng Cong for USP9x plasmids, Dr. Charles M Perou for suggestions, and members of Zhang and Kaelin laboratory for helpful discussions. Q.Z. was supported by Friends Award from Dana-Farber Cancer Institute, Terri Brodeur Breast Cancer Foundation, and Ruth L. Kirschstein National Research Service Award post-doctoral fellowship awards. This work has been supported by K99/R00 award from NIH (to Q.Z.), University Cancer Research Fund (to Q.Z.), and grants from National Institutes of Health (2R01CA68490-16, 1K99CA160351-01, and 1F32CA139929-01), Howard Hughes Medical Institute, and Breast Cancer Research Foundation to W.G.K.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242131.114.
References
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM 2004. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 279: 38458–38465 [DOI] [PubMed] [Google Scholar]
- Aragones J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, et al. 2008. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet 40: 170–180 [DOI] [PubMed] [Google Scholar]
- Bakker WJ, Harris IS, Mak TW 2007. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell 28: 941–953 [DOI] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J 22: 4082–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuck S, Schanzer W, Thevis M 2012. Hypoxia-inducible factor stabilizers and other small-molecule erythropoiesis-stimulating agents in current and preventive doping analysis. Drug Test Anal 4: 830–845 [DOI] [PubMed] [Google Scholar]
- Bishop T, Gallagher D, Pascual A, Lygate CA, de Bono JP, Nicholls LG, Ortega-Saenz P, Oster H, Wijeyekoon B, Sutherland AI, et al. 2008. Abnormal sympathoadrenal development and systemic hypotension in PHD3−/− mice. Mol Cell Biol 28: 3386–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME 2001. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a). Mol Cell Biol 21: 952–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB 2002. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol 156: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, Godson C, Nielsen JE, Moynagh P, Pouyssegur J, et al. 2006. Prolyl hydroxylase-1 negatively regulates IκB kinase-β, giving insight into hypoxia-induced NFκB activity. Proc Natl Acad Sci 103: 18154–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. 2009. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136: 123–135 [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, Medema RH, Lam EW 2005. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24: 2317–2329 [DOI] [PubMed] [Google Scholar]
- Feng T, Yamamoto A, Wilkins SE, Sokolova E, Yates LA, Munzel M, Singh P, Hopkinson RJ, Fischer R, Cockman ME, et al. 2014. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell 53: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A 2012. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ 19: 968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisl P, Aragones J, Carmeliet P 2009. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov 8: 139–152 [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24: 7410–7425 [DOI] [PubMed] [Google Scholar]
- Guo S, Sonenshein GE 2004. Forkhead box transcription factor FOXO3a regulates estrogen receptor α expression and is repressed by the Her-2/neu/phosphatidylinositol 3-kinase/Akt signaling pathway. Mol Cell Biol 24: 8681–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habashy HO, Rakha EA, Aleskandarany M, Ahmed MA, Green AR, Ellis IO, Powe DG 2011. FOXO3a nuclear localisation is associated with good prognosis in luminal-like breast cancer. Breast Cancer Res Treat 129: 11–21 [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J 2003. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem 278: 30772–30780 [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J 2005. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J 19: 1308–1310 [DOI] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. 2004. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117: 225–237 [DOI] [PubMed] [Google Scholar]
- Huang H, Tindall DJ 2007. Dynamic FoxO transcription factors. J Cell Sci 120: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Ivan M, Haberberger T, Gervasi DC, Michelson KS, Gunzler V, Kondo K, Yang H, Sorokina I, Conaway RC, Conaway JW, et al. 2002. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc Natl Acad Sci 99: 13459–13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Jensen KS, Binderup T, Jensen KT, Therkelsen I, Borup R, Nilsson E, Multhaupt H, Bouchard C, Quistorff B, Kjaer A, et al. 2011. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J 30: 4554–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zou L, Lu WQ, Zhang Y, Shen AG 2013. Foxo3a expression is a prognostic marker in breast cancer. PLoS ONE 8: e70746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG 2005. Proline hydroxylation and gene expression. Annu Rev Biochem 74: 115–128 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr, Ratcliffe PJ 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30: 393–402 [DOI] [PubMed] [Google Scholar]
- Kim WY, Safran M, Buckley MR, Ebert BL, Glickman J, Bosenberg M, Regan M, Kaelin WG Jr 2006. Failure to prolyl hydroxylate hypoxia-inducible factor α phenocopies VHL inactivation in vivo. EMBO J 25: 4650–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Huang B, Unterman TG, Harris RA 2004. Protein kinase B-α inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53: 899–910 [DOI] [PubMed] [Google Scholar]
- Lee S, Nakamura E, Yang H, Wei W, Linggi MS, Sajan MP, Farese RV, Freeman RS, Carter BD, Kaelin WG Jr, et al. 2005. Neuronal apoptosis linked to EglN3 prolyl hydroxylase and familial pheochromocytoma genes: developmental culling and cancer. Cancer Cell 8: 155–167 [DOI] [PubMed] [Google Scholar]
- Lin C, Wu Z, Lin X, Yu C, Shi T, Zeng Y, Wang X, Li J, Song L 2011. Knockdown of FLOT1 impairs cell proliferation and tumorigenicity in breast cancer through upregulation of FOXO3a. Clin Cancer Res 17: 3089–3099 [DOI] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabon ME, Scott BA, Crowder CM 2009. Divergent mechanisms controlling hypoxic sensitivity and lifespan by the DAF-2/insulin/IGF-receptor pathway. PLoS ONE 4: e7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ 2001. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J 20: 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Bardeesy N, Cullen D, Bronson RT, Kaelin WG Jr 2008. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood 111: 3236–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG Jr 2009. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol 29: 5729–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. 2007. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1: 101–112 [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Moslehi J, Minamishima YA, Shi J, Neuberg D, Charytan DM, Padera RF, Signoretti S, Liao R, Kaelin WG Jr 2010. Loss of hypoxia-inducible factor prolyl hydroxylase activity in cardiomyocytes phenocopies ischemic cardiomyopathy. Circulation 122: 1004–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A 2006. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem 281: 38738–38747 [DOI] [PubMed] [Google Scholar]
- Nwogu JI, Geenen D, Bean M, Brenner MC, Huang X, Buttrick PM 2001. Inhibition of collagen synthesis with prolyl 4-hydroxylase inhibitor improves left ventricular function and alters the pattern of left ventricular dilatation after myocardial infarction. Circulation 104: 2216–2221 [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mancera PA, Rust AG, van der Weyden L, Kristiansen G, Li A, Sarver AL, Silverstein KA, Grutzmann R, Aust D, Rummele P, et al. 2012. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2: 81–91 [DOI] [PubMed] [Google Scholar]
- Santo EE, Stroeken P, Sluis PV, Koster J, Versteeg R, Westerhout EM 2013. FOXO3a is a major target of inactivation by PI3K/AKT signaling in aggressive neuroblastoma. Cancer Res 73: 2189–2198 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH 2002. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol 22: 7842–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O’Rourke K, Bazan F, Eastham-Anderson J, et al. 2010. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463: 103–107 [DOI] [PubMed] [Google Scholar]
- Scott BA, Avidan MS, Crowder CM 2002. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296: 2388–2391 [DOI] [PubMed] [Google Scholar]
- Semenza GL 2013. Cancer-stromal cell interactions mediated by hypoxia-inducible factors promote angiogenesis, lymphangiogenesis, and metastasis. Oncogene 32: 4057–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth P, Krop I, Porter D, Polyak K 2002. Novel estrogen and tamoxifen induced genes identified by SAGE (serial analysis of gene expression). Oncogene 21: 836–843 [DOI] [PubMed] [Google Scholar]
- Sisci D, Maris P, Cesario MG, Anselmo W, Coroniti R, Trombino GE, Romeo F, Ferraro A, Lanzino M, Aquila S, et al. 2013. The estrogen receptor α is the key regulator of the bifunctional role of FoxO3a transcription factor in breast cancer motility and invasiveness. Cell Cycle 12: 3405–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunayama J, Sato A, Matsuda K, Tachibana K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Sakurada K, et al. 2011. FoxO3a functions as a key integrator of cellular signals that control glioblastoma stem-like cell differentiation and tumorigenicity. Stem Cells 29: 1327–1337 [DOI] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, Coffer PJ, Medema RH, Coombes RC, Lam EW 2003. FoxO3a transcriptional regulation of Bim controls apoptosis in Paclitaxel-treated breast cancer cell lines. J Biol Chem 278: 49795–49805 [DOI] [PubMed] [Google Scholar]
- Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ, Burgering BM, Coombes RC, Lam EW 2006. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res 66: 212–220 [DOI] [PubMed] [Google Scholar]
- Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH 2006. Placental but not heart defects are associated with elevated hypoxia-inducible factor α levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol 26: 8336–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Cowan A, Fong GH 2007. Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation 116: 774–781 [DOI] [PubMed] [Google Scholar]
- Tennant DA, Gottlieb E 2010. HIF prolyl hydroxylase-3 mediates α-ketoglutarate-induced apoptosis and tumor suppression. J Mol Med 88: 839–849 [DOI] [PubMed] [Google Scholar]
- To KK, Huang LE 2005. Suppression of hypoxia-inducible factor 1α (HIF-1α) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J Biol Chem 280: 38102–38107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Gilliland DG 2007. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell 1: 140–152 [DOI] [PubMed] [Google Scholar]
- Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD 2007. Crystal structure of the human FOXO3a–DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res 35: 6984–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WB, Chung YM, Zou Y, Park SH, Xu Z, Nakayama K, Lin SH, Hu MC 2010. Inhibition of FOXO3 tumor suppressor function by βTrCP1 through ubiquitin-mediated degradation in a tumor mouse model. PLoS ONE 5: e11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villen J, Beausoleil SA, Gygi SP 2008. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics 8: 4444–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt AE, Hines LM, Collins NL, Hu Y, Gunawardane RN, Moreira D, Raphael J, Jepson D, Koundinya M, Rolfs A, et al. 2006. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res 5: 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Xiao K, Whalen EJ, Forrester MT, Freeman RS, Fong G, Gygi SP, Lefkowitz RJ, Stamler JS 2009. Oxygen-regulated β(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci Signal 2: ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, Wilson C, Nathans R, Zhang J, Kirschner MW, et al. 2013. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem 288: 2976–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. 2008. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 10: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Qi RC, Chen T, Chung TD, Stern AM, Hollis GF, Copeland RA, Oldenburg KR 1999. Development of a carbon dioxide-capture assay in microtiter plate for aspartyl-β-hydroxylase. Anal Biochem 271: 137–142 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Gu J, Li L, Liu J, Luo B, Cheung HW, Boehm JS, Ni M, Geisen C, Root DE, et al. 2009. Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell 16: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]