Heterochromatin protein 1 (HP1a) plays conserved roles in gene silencing and heterochromatin and is also implicated in transcription, DNA replication, and repair. Using BioTAP-XL mass spectrometry and sequencing across multiple life stages of Drosophila, Alekseyenko et al. identify HP1a chromatin-associated protein and RNA interactions. They discover 13 novel candidates among the top interactions. Furthermore, HP1a selectively associates with a broad set of RNAs transcribed from repetitive regions. The validation of several novel HP1a protein interactors reveals new HP1a links to chromatin organization and function.
Keywords: HP1a, ChIP, formaldehyde cross-linking, LC-MS/MS, Bayesian analysis
Abstract
Heterochromatin protein 1 (HP1a) has conserved roles in gene silencing and heterochromatin and is also implicated in transcription, DNA replication, and repair. Here we identify chromatin-associated protein and RNA interactions of HP1a by BioTAP-XL mass spectrometry and sequencing from Drosophila S2 cells, embryos, larvae, and adults. Our results reveal an extensive list of known and novel HP1a-interacting proteins, of which we selected three for validation. A strong novel interactor, dADD1 (Drosophila ADD1) (CG8290), is highly enriched in heterochromatin, harbors an ADD domain similar to human ATRX, displays selective binding to H3K9me2 and H3K9me3, and is a classic genetic suppressor of position-effect variegation. Unexpectedly, a second hit, HIPP1 (HP1 and insulator partner protein-1) (CG3680), is strongly connected to CP190-related complexes localized at putative insulator sequences throughout the genome in addition to its colocalization with HP1a in heterochromatin. A third interactor, the histone methyltransferase MES-4, is also enriched in heterochromatin. In addition to these protein–protein interactions, we found that HP1a selectively associated with a broad set of RNAs transcribed from repetitive regions. We propose that this rich network of previously undiscovered interactions will define how HP1a complexes perform their diverse functions in cells and developing organisms.
HP1a (heterochromatin protein 1) protein was first described by James and Elgin (1986) as a prominent component of heterochromatin in Drosophila. HP1a is now known to play conserved roles in gene silencing from fission yeast to humans through heterochromatin formation and maintenance (for review, see Eissenberg and Elgin 2000; Eissenberg and Elgin 2014). HP1a has also been linked to processes such as gene transcription, DNA replication, and repair (for review, see Fanti and Pimpinelli 2008; Soria et al. 2012; Elgin and Reuter 2013). HP1a may play these diverse roles in the context of distinct multicomponent complexes within varied chromatin environments. Thus, how HP1a and other epigenetic factors interact with appropriate partners within their chromatin context remains an important question.
ChIP–chip (chromatin immunoprecipitation [ChIP] coupled with microarray analysis) and ChIP-seq (ChIP combined with deep sequencing) experiments demonstrate that HP1a is indeed highly enriched over pericentric heterochromatin but in addition binds active genes in the euchromatin and heterochromatin (Piacentini et al. 2003; de Wit et al. 2007). HP1a has two conserved domains, an N-terminal chromodomain and a C-terminal chromoshadow domain, separated by a hinge region. The HP1a chromodomain binds specifically to dimethylated and trimethylated H3K9 (H3K9me2 and H3K9me3) (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002). Structural studies suggest that the chromoshadow domain is required for HP1a dimerization and interaction with proteins that possess a conserved motif, PXVXL (Thiru et al. 2004).
HP1a interactors have been identified previously using yeast two-hybrid screens (Shaffer et al. 2002; Giot et al. 2003), and HP1a has been recovered from pull-downs of several chromatin-associated protein complexes (Badugu et al. 2003; Lin et al. 2008; Emelyanov et al. 2010; Nozawa et al. 2010). HP1a also has been analyzed by inducible expression, affinity purification, and mass spectrometry (Lin et al. 2008; Ryu et al. 2014). However, a comprehensive picture based on HP1a expressed at endogenous levels within the organism has been lacking. In addition to protein–protein interactions, HP1a complexes may also interact with RNAs associated with silencing, based on the roles for dsRNA in gene silencing in Schizosaccharomyces pombe and plants (for review, see Grewal and Moazed 2003) and the association of HP1 with pericentric heterochromatic transcripts in mouse cells (Maison et al. 2011). To begin to address the many outstanding questions regarding HP1a targeting and function, we took a cross-linking approach termed BioTAP-XL that preserves protein:protein and protein:RNA interactions, which otherwise might be disrupted during typical biochemical procedures (Alekseyenko et al. 2014). Using BioTAP-XL affinity purification across multiple life stages of Drosophila, including the biochemically challenging larval and adult stages, we identified the majority of previously described HP1a partners and discovered 13 novel candidates among the top interactions. Furthermore, we also identified repetitive RNAs associated with HP1a. Our validation of several novel HP1a protein interactors encompasses biochemical, genetic, and genomic evidence for new HP1a links to chromatin organization and function.
Results
HP1a-BioTAP displays heterochromatin targeting and proper function in vivo
To investigate the composition of HP1a interactions across the diverse life stages of Drosophila, we constructed an HP1a-BioTAP transgene containing the HP1a promoter and flanking regions to preserve its native expression pattern. In parallel, we created an analogous construct of MSL3-BioTAP, an X-chromosome-specific dosage compensation regulator that provided important contrasting data sets to assess the specificity of our HP1a results. While a biotin tag-based, one-step affinity purification was previously sufficient to define MSL3 protein interactions in S2 tissue culture cells (Wang et al. 2013), we found that BioTAP two-step purification (Alekseyenko et al. 2014) yielded superior quality and is essential for analysis of more complex samples such as larvae and adult flies (Fig. 1A; see below). To test the BioTAP fusion proteins for function, we created transgenic flies and stable transfected cell lines. The HP1a-BioTAP transgene rescued the HP1a lethal mutant phenotype (50%–70% rescue), and the MSL3-BioTAP transgene rescued the msl3 male lethal phenotype (80%) (see the Supplemental Material). Furthermore, both proteins displayed their correct size and subnuclear localization in transfected S2 cells (Supplemental Fig. S1) and, in larvae, displayed the expected genomic localization on polytene chromosomes, with HP1a-BioTAP in pericentric heterochromatin (Fig. 1B), and MSL3-BioTAP on the male X chromosome (Fig. 1C). Correct targeting of HP1a-BioTAP and MSL3-BioTAP was detected in the presence of the endogenous, untagged versions of each protein, demonstrating that the tagged versions compete well with their respective native proteins.
Figure 1.
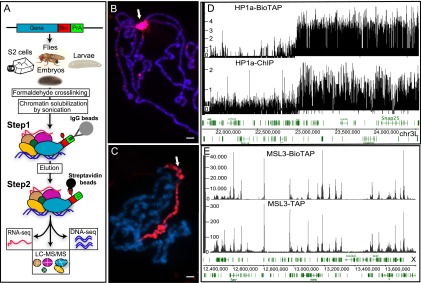
Overview of the BioTAP-XL purification strategy and validation of MSL3-BioTAP- and HP1a-BioTAP-tagged proteins. (A) The BioTAP tag includes two epitopes: protein A and Bio, a 75-amino-acid sequence that is biotinylated in vivo. To preserve endogenous expression patterns, genomic BioTAP-tagged transgenes (generated by BAC recombineering) were introduced into flies or S2 cells. Crude nuclear extracts from cells and from different life stages of flies were cross-linked using formaldehyde, sonicated, and subjected to TAP, first with rabbit IgG agarose beads eluted under denaturing conditions and subsequently using streptavidin agarose beads. The resulting DNA and RNA were analyzed by high-throughput sequencing. Peptides from the protein fraction were released by direct on-bead trypsin digestion and then identified by LC-MS/MS. Immunostaining detected the specific signal on pericentric heterochromatin for HP1a-BioTAP (B) and on the male X chromosome for MSL3-BioTAP with PAP antibody (red) (C). Hoechst staining of DNA is shown in blue. Bars, 10 μm. (D) Representative ChIP-seq profiles of HP1a-BioTAP on the heterochromatin–euchromatin border of chromosome 3L correspond well with ChIP-seq data (HP1a-ChIP) obtained from the modENCODE consortium (Kharchenko et al. 2011). (E) Representative ChIP-seq profiles of MSL3-BioTAP in S2 cells on the X chromosome. The results correlated well with previous MSL3-TAP ChIP-seq (Alekseyenko et al. 2008).
The correct localization of HP1a-BioTAP and MSL3-BioTAP was further confirmed by BioTAP-XL tandem affinity purification (TAP) from the corresponding transformed S2 cell lines (see the Materials and Methods) followed by DNA sequencing. Our results demonstrate the specific targeting of HP1a to the heterchromatic regions and MSL3 to the X chromosome, respectively, and fully recapitulate the detailed ChIP-seq enrichment profiles previously published for these proteins (Fig. 1D,E). Furthermore, the enrichment (immunoprecipitation/input ratio) magnitudes achieved by the BioTAP-XL approach were notably higher than those observed in previous TAP- or antibody-based ChIP-seq experiments (e.g., ∼6.5 times higher than anti-HP1a immunoprecipitation and ∼2.5 times higher than MSL3 TAP). While cross-linking might be expected to result in high background interactions, the stringent conditions used in the two-step BioTAP-XL affinity purification actually result in virtually no background signal when pull-downs are compared with untagged negative controls by gel electrophoresis and colloidal blue staining (Supplemental Fig. S2).
Application of a statistical method, Bayesian analysis of protein enrichment (Bamse), to analyze HP1a interactions revealed by BioTAP-XL
To examine the protein composition of HP1a complexes, we prepared cross-linked chromatin from HP1a-BioTAP isolated from S2 tissue culture cells, embryos, larvae, and adults (see the Materials and Methods). The analysis of larvae and adults, two challenging life stages for typical purification procedures, served as a new test for the effectiveness of BioTAP-XL. For protein analysis after affinity purification of HP1a-BioTAP complexes, we used on-bead trypsin digestion and analyzed the recovered peptides using liquid chromatography-tandem mass spectrometry (LC-MS/MS). In parallel with the HP1a experiments, we characterized the protein composition of MSL3-BioTAP and untagged mock pull-downs to control for interactors that are isolated in a bait-independent manner. Importantly, we also analyzed the input chromatin from each stage to determine enrichment after pull-down, a measurement we previously found to provide strong evidence for specificity after affinity purification (Wang et al. 2013; Alekseyenko et al. 2014).
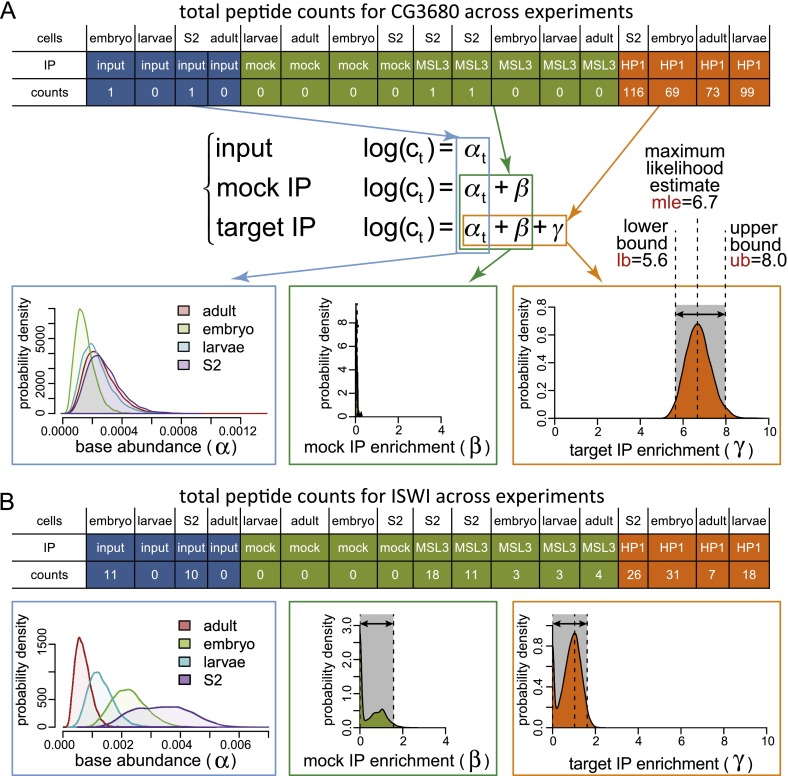
To evaluate enrichment of specific BioTAP-XL pull-downs over the multiple types of controls, we developed a statistical method (Bamse). The approach controls for multiple sources of bias, including biological replicate variability, differential base-level abundance of proteins in different cell types, variability in peptide ionization or detection rates, and nonspecific effects of pull-down procedures (see the Materials and Methods). Briefly, in evaluating whether a given protein shows specific enrichment (in log2 scale), the method examines the likelihood of possible values for three key parameters (Fig. 2): base-level abundance of that protein in each cell type (αt), the extent to which the protein is enriched due to mock or nonspecific pull-down (β), and the extent to which the protein is enriched by the specific (i.e., HP1a) pull-down (γ). The probability that a given protein shows enrichment magnitude γ in the HP1a-BioTAP pull-down given the observed LC-MS/MS data is summarized by the posterior probability distribution of γ. Figure 2 provides contrasting examples of results for CG3680, a strongly enriched candidate interactor (see below), and ISWI, an abundant chromatin component showing significantly less enrichment.
Figure 2.
Bamse. (A) Our analysis method aims to identify proteins exhibiting statistically significant enrichment in target pull-downs relative to controls based on an agglomeration of data from different cell types. The peptide counts observed for CG3680 in different mass spectrometry samples (top table) are modeled as variables distributed according to a negative binomial distribution, with the mean rate described by a log-linear model (middle formula). The log-linear model separates contributions of base-level protein abundance in the cell (αt), enrichment in pull-downs due to nonspecific effects (β), and enrichment due to specific association with the target protein (γ). (Bottom plots) A Bayesian approach was used to infer the distributions of likely values for these parameters (i.e., posterior distributions), and the range of statistically likely values of the target-specific fold enrichment magnitude γ was used to evaluate the significance of the association with the target. For CG3680, the 95% confidence interval of the target-specific enrichment magnitude γ lies within the range of 5.6–8.0 (log2 scale). (B) The posterior distributions are shown for the ISWI protein, which is detected in the HP1a BioTAP pull-downs but shows only small enrichment magnitude (<1.7 on log2 scale), partly due to nonspecific pull-down abundance β (with MSL3 control).
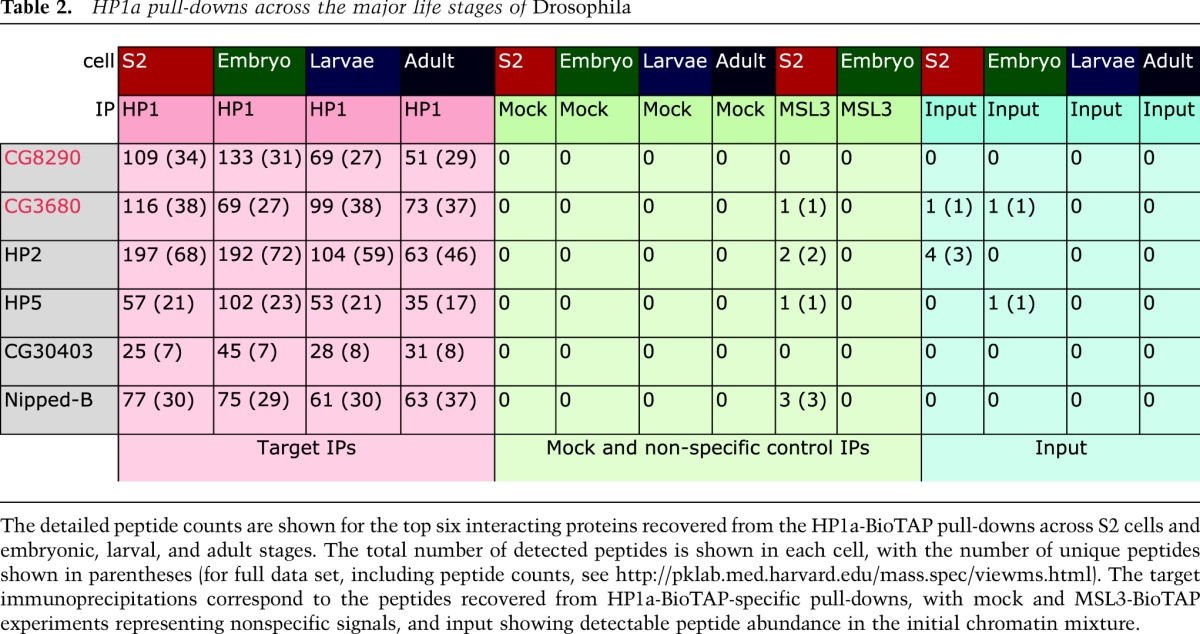
Ranking HP1a-interacting proteins by their enrichment magnitude using Bamse, we recovered candidates known to function in heterochromatin, transcription, replication, DNA repair, general chromatin organization, and cell division with high reproducibility among different samples. The top 32 interacting proteins are shown with representative peptide counts in Table 1 (for full data set, including peptide counts, see http://pklab.med.harvard.edu/mass.spec/viewms.html). A more detailed view of the top six interactors across the Drosophila life cycle is shown in Table 2. The protein lists enriched in parallel by purification of MSL3-BioTAP in the same Drosophila life stages (Supplemental Table S1) provided negative control data sets for the HP1 results and also strong validations for the proteins discovered to interact with MSL3 in S2 cells (Wang et al. 2013).
Table 1.
HP1a protein interactors ranked by confidence score
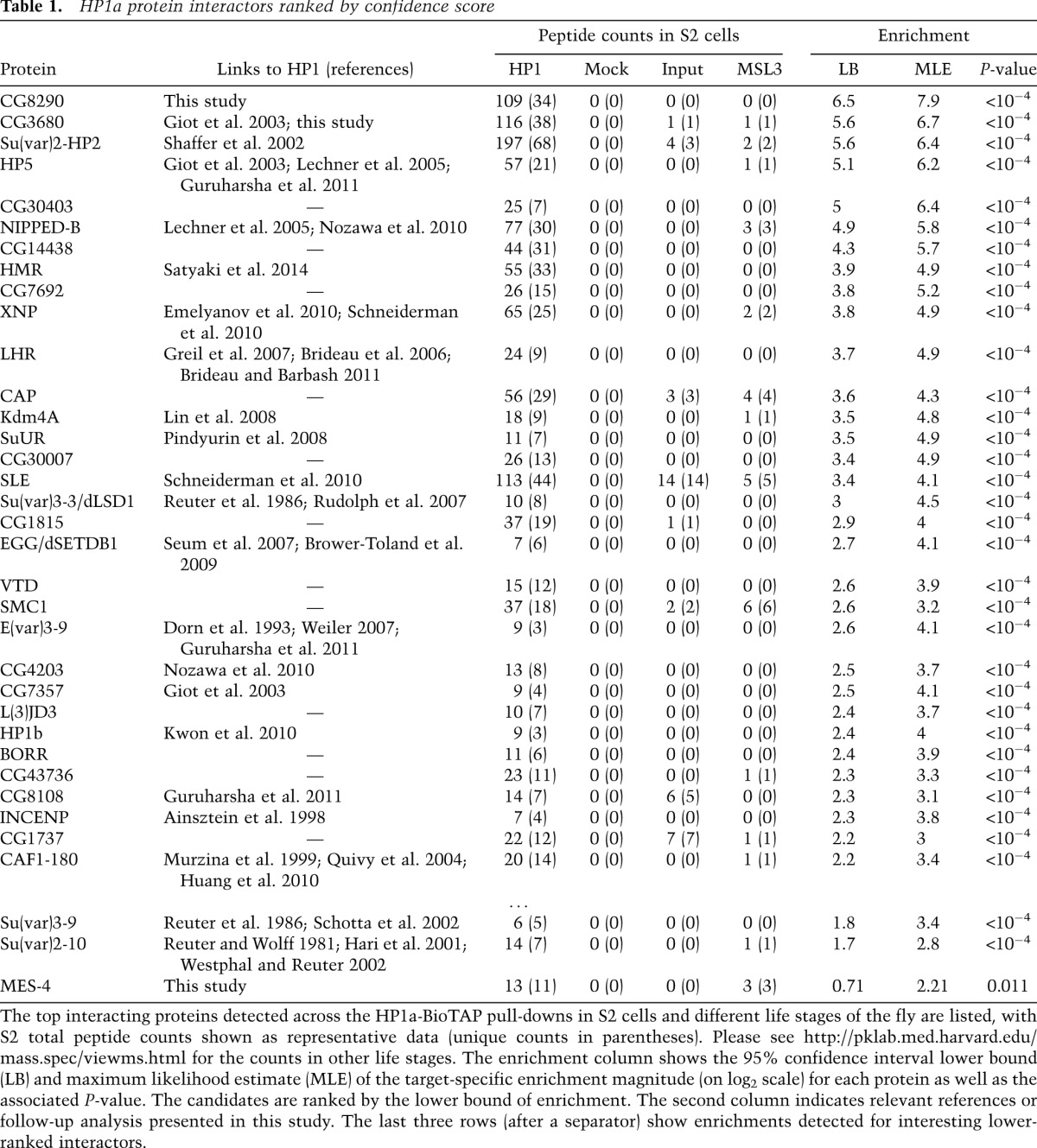
Table 2.
HP1a pull-downs across the major life stages of Drosophila
The top HP1a-interacting proteins could be divided into three categories. The first group comprises the proteins for which association with Drosophila HP1a has been previously identified through genetic studies of position-effect variegation (PEV) {Suppressor of variegation 3-9 [SU(VAR)3-9], SU(VAR)2-10, SU(VAR)3-3, XNP, SLE, dSETDB1, E(VAR)3-9, and CAF1-180} and/or through molecular analysis (yeast two-hybrid or protein pull-downs) (HP2, HP3, HP5, CG3680, KDM4A, SUUR, HMR, CG7357, and HP1b). The second group encompasses interactors for which, to our knowledge, the connection to HP1a has not been described in Drosophila but was reported for the homologous proteins in different organisms (CG4203, Nipped-B/hNIPBL, and INCENP/hINCENP). The third category includes potential novel HP1a interactors (CG8290, CG30403, CG14438, CG7692, CG30007, CAP, CG1815, VTD, CG4203, l(3)j2D3, BORR, CG1737, SMC1, and MES-4).
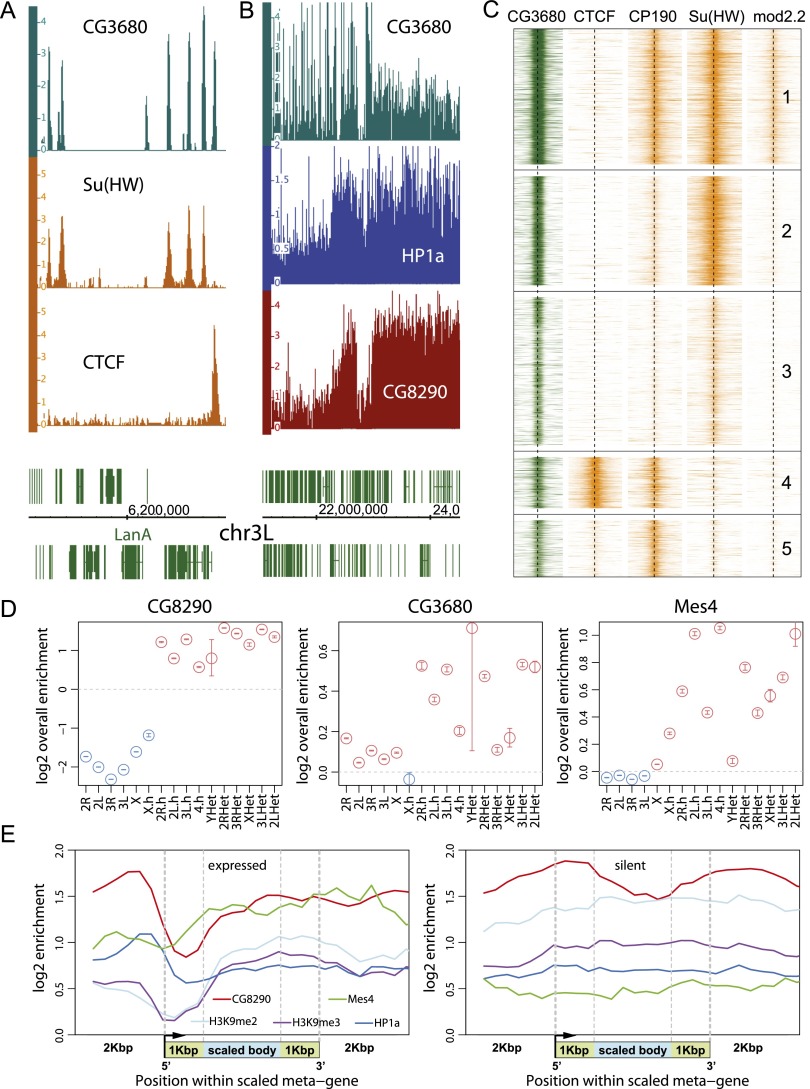
We had prior interest in MES-4 as a putative histone H3K36 methyltransferase. To determine its chromosomal localization, we produced anti-MES-4 antibodies through the modEncode project (Kharchenko et al. 2011) and transgenic flies expressing MES-4 tagged at its N terminus with protein A. We performed immunostaining of polytene chromosomes in transgenic larvae and found that both endogenous MES-4, detected by anti-MES-4 antibodies, and tagged MES-4, detected by PAP antibodies, were localized in the pericentric heterochromatin regions and on the fourth chromosome, which are also the classical targets for HP1a (Supplemental Fig. S3). We further confirmed and refined MES-4 localization to heterochromatin at high resolution by ChIP–chip in S2 cells using anti-MES-4 antibodies. (Fig. 3D; Supplemental Fig. S4). While broadly enriched with HP1a across heterochromatin (Supplemental Fig. S4), MES-4 shows higher average enrichment in transcriptionally active compared with transcriptionally silent heterochromatic genes (Fig. 3E; Supplemental Fig. S5).
Figure 3.
Genomic distribution of CG3680, CG8290, and MES-4 proteins. (A) Within the euchromatic portion of the Drosophila genome, CG3680 exhibits prominent binding positions coinciding with those of several insulator proteins, including CTCF and Su(Hw). The plots show enrichment of the DNA fraction of the CG3680 BioTAP pull-down for a euchromatic region of chromosome 3L along with ChIP-seq enrichment for the insulator proteins in S2 cells. (B) A region of chromosome 3L in S2 cells containing the euchromatic–heterochromatic boundary is shown. Both CG3680 and CG8290 show a broad pattern of enrichment in the heterochromatin, resembling that of HP1a. However, the enrichment magnitude of CG3680 is lower, particularly in contrast to its insulator-associated euchromatic binding positions. (C) CG3680 euchromatic binding positions coincide with several different classes of insulators. The heat maps show ChIP-seq enrichments of Drosophila insulator proteins around all euchromatic CG3680 positions in S2 cells (from top to bottom). Most of the euchromatic CG3680 sites can be classified into those associated with gypsy-like insulator combinations [Su(Hw) + CP190 + mod2.2; cluster 1], standalone Su(Hw) (clusters 2 and 3), or CTCF + CP190 (cluster 4 insulators). (D) CG3680, CG8290, and MES-4 show statistically significant enrichment in the heterochromatic portions of the Drosophila genome. The maximum likelihood estimate (circles) over the overall enrichment levels ([red] enriched; [blue] depleted) and the 95% confidence intervals (whiskers) are shown for the euchromatic portions of the assembled chromosome arms (e.g., 2L), assembled pericentromeric heterochromatin regions (e.g., 2L.h), and unassembled heterochromatic contigs (e.g,. 2LHet). (E) Metagene profiles of CG8290 and MES-4 in S2 cells with comparison with HP1 and H3K9me2/3 modification (Kharchenko et al. 2011) around expressed and silent heterochromatic genes.
We further selected two top-scoring, previously uncharacterized hits for validation: CG8290, enriched 239-fold in our HP1a-BioTAP-XL pull-down, and CG3680, enriched 104-fold (Table 1). We created stable S2 cell lines expressing genomic clones of CG8290 and CG3680 tagged with BioTAP and performed affinity purification under stringent, non-cross-linked conditions (500 mM salt). Western blot analysis demonstrated that HP1a interacts with both CG8290-BioTAP and CG3680-BioTAP but not with MSL3-BioTAP or untagged extract, directly confirming the interactions detected by mass spectrometry (Supplemental Fig. S6).
CG3680 binds pericentric heterochromatin and gypsy-like insulator-binding groups
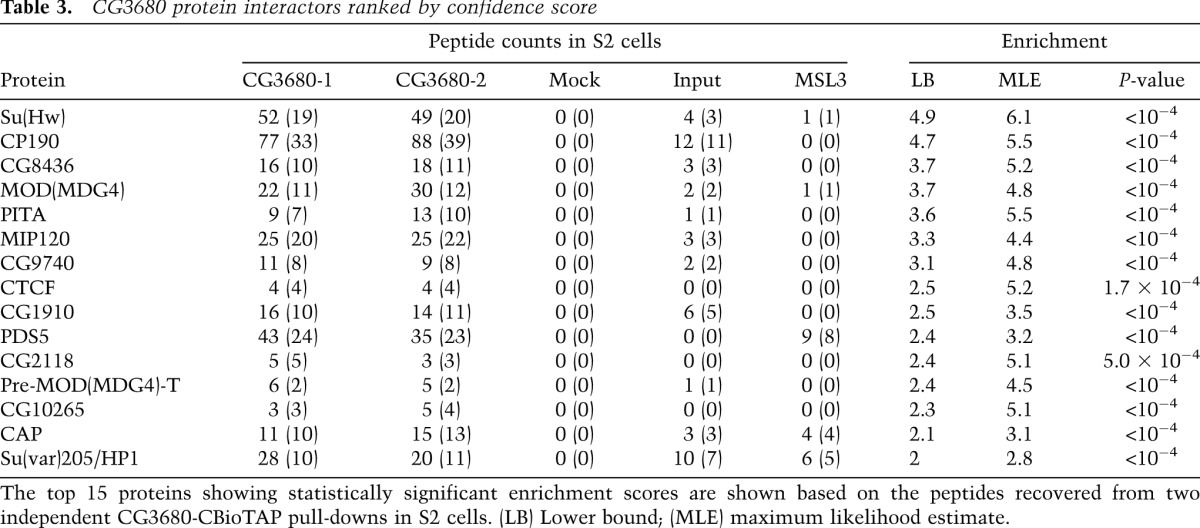
CG3680 was previously identified as a potential interactor of HP1a in a high-throughput yeast two-hybrid assay (Giot et al. 2003). The protein has a crotonase-fold domain, which in the context of human CDY-like proteins has been implicated in transfer of acetyl groups to and from chromatin (Lahn et al. 2002; Caron et al. 2003) as well as in protein multimerization (Franz et al. 2009). To further characterize the CG3680-HP1a interaction, we performed a CG3680 BioTAP-XL pull-down in S2 cells, in which HP1a was among the top 15 protein hits (Table 3). Surprisingly, the top candidates included SU(HW), MOD(MDG4), and CP190 proteins, the components of the gypsy insulator known for its effect on enhancer–promoter interactions (Georgiev and Gerasimova 1989; Pai et al. 2004). ChIP-seq analysis of the recovered DNA also indicated that CG3680 might have a dual function in the nucleus. Consistent with our LC-MS/MS results, we found that CG3680 shows numerous (>5000) narrow binding sites within euchromatic regions, nearly all of which coincide with insulator-binding sites (Fig. 3A,C; Supplemental Fig. S7). These include gypsy-like insulator sites (22%) and stand-alone SU(HW)-binding sites (57%) as well as unrelated CTCF+CP190 (11%) or stand-alone CP190-binding positions (10%). It is important to note that CG3680 was not present at all sites associated with each insulator class, suggesting that it might introduce further distinctions within these classes (Negre et al. 2010; Schwartz et al. 2012). Furthermore, regions of HP1a-CG3680 interaction (see below) appear to be distinct from the CG3680 insulator-associated binding peaks (Fig. 3B; Supplemental Fig. S7).
Table 3.
CG3680 protein interactors ranked by confidence score
Similar to HP1a, CG3680 also exhibits a broad pattern of binding in heterochromatin. Enrichment of CG3680-BioTAP in heterochromatin is statistically significant (Fig. 3D) but lower in magnitude than that observed for HP1a-BioTAP (∼1.5-fold vs. eightfold) and covers only a subset (∼35%) of HP1a-enriched regions. Metagene profiles reveal no general enrichment of CG3680 over genes and a relative depletion within transcriptionally active genes (Supplemental Fig. S5). Taken together, the reciprocal CG3680-BioTAP mass spectrometry and ChIP-seq mapping data establish CG3680 as a strong HP1a interactor in heterochromatin, with a potentially separate function in chromatin insulators. Based on these characteristics, we propose HIPP1 (HP1 and insulator partner protein-1) as an appropriate name for CG3680.
Genetic analysis of CG8290, a novel heterochromatin protein, reveals a Su(var) phenotype
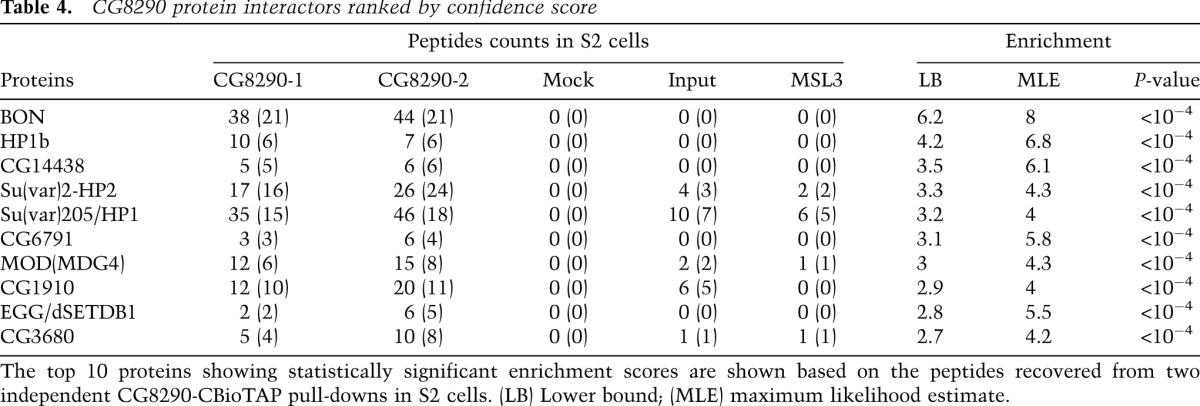
CG8290 ranked as a top interacting protein in our LC-MS/MS analysis of the HP1a pull-downs (Tables 1, 2). It was greatly enriched in all samples and was absent from all input and control BioTAP-XL experiments. Surprisingly, we found no prior reports suggesting a link between CG8290 and HP1a. In reciprocal pull-downs using CG8290-BioTAP in S2 cells, we identified HP1a among the top five proteins recovered by the LC-MS/MS analysis (following Bonus [BON], HP1b, CG14438, and HP2) (Table 4). Both BON and HP2 have been implicated in HP1a-mediated repression (Shaffer et al. 2002; Beckstead et al. 2005). The results of ChIP-seq analysis of the CG8290-associated DNA showed that pericentric heterochromatin was the main target of the CG8290-BioTAP protein, similar to HP1a (Fig. 3B,D). We also created CG8290-BioTAP transgenic flies and confirmed the heterochromatic localization of CG8290 by immunofluorescence studies in larval polytene chromosomes (Supplemental Fig. S3). Detailed analysis of the CG8290 ChIP-seq data shows that CG8290 enrichment in heterochromatic genes closely resembles that of HP1a, including pronounced depletion at active transcription start sites, suggesting strong association between these proteins and likely involvement of CG8290 in heterochromatin gene regulation (Fig. 3E; Supplemental Fig. S5).
Table 4.
CG8290 protein interactors ranked by confidence score
To ask what might be the functional consequences of the HP1a–CG8290 interaction discovered via BioTAP-XL, we induced excision of a P element inserted in the 5′ untranslated region (UTR) of cg8290 to create a series of cg8290 deletion alleles and selected cg82901 and cg82902 for further analysis. cg82902 eliminates most of the coding region without affecting the neighboring PI31 gene (Fig. 4D). We found that homozygous deletion flies are viable (data not shown) but display a moderately strong dose-dependent Su(var) phenotype, consistent with a function in heterochromatic silencing (Fig. 4E).
Figure 4.
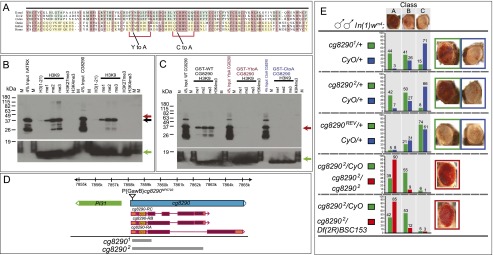
CG8290 recognizes H3K9me2/3 through its ADD domain and functions in heterochromatin silencing. (A) Alignment of the ADD domains from dipteran CG8290 (Drosophila melanogaster, Drosophila virilis, and Culex pipiens) and vertebrate ATRX (Danio rerio, Gallus gallus, and Homo sapiens). Residues conserved among dipterans or vertebrates are shaded blue and yellow, respectively. Residues shared by at least five of the six species shown here are highlighted in pink. Mutations Y101A and C117A at conserved positions implicated in H3K9me recognition and used as controls in H3K9 peptide pull-downs are depicted below. Red brackets denote the residues forming a composite H3K9me3 pocket (Iwase et al. 2011). (B) Anti-GST Western blot analysis showing recovery of the CG8290 GST-ADD domain after pull-down with biotinylated histone H3 tail peptides methylated at H3K9 but not at H3K4 or H3K27 residues, consistent with the published human ATRX ADD data (positive control). Mobilities of the GST-tagged CG8290 and ATRX ADD domains are slightly different and are marked by red and black arrows. A green arrow indicates biotinylated histone H3 peptides used in pull-downs, detected with streptavidin-HRP. (C) Western blot analysis of the CG8290 GST-ADD domain showing that binding to methylated histone H3K9 peptides is abrogated by point mutations Y101A and C117A. Positions of wild-type and mutant CG8290 ADD are indicated by the red arrow. The green arrow indicates the signal from biotinylated histone H3 peptides used in pull-downs. (D) Organization of the cg8290 locus. cg8290 (blue) produces three types of transcripts encoding three CG8290 isoforms. Coding sequences and UTRs are colored purple and orange, respectively, with the position of the ADD domain indicated. Insertion of P{GawB}cg8290NP0793 is indicated by a triangle. Two null alleles, cg82901 and cg82902, are deletions of the cg8290 coding sequence, not affecting the upstream PI31 gene. The deletions are indicated by gray boxes. (E) cg8290 alleles display a dose-dependent PEV phenotype and act as Su(var)s in the white-mottled genetic background. A dominant Su(var) phenotype is observed in wm4h/Y; cg82901 or 2/+ flies, which is further enhanced in homozygotes (wm4h/Y; cg82902/cg82902) and trans-heterozygotes (wm4h/Y; cg82902/Df(2R)BSC153). cg8290REV is a precise excision of P{GawB} and serves as a genetic background control. Df(2R)BSC153 is a 341-kb deletion removing cg8290 and other genes; it was obtained on a background distinct from that of cg82902, so its use in combination with cg8290 helps exclude the possible contribution of linked PEV modifiers on the cg82902 chromosome. Distribution of eye color classes (A, B, or C, depicted above) is distinct between cg82901 or 2/+ and cg8290REV/+ or CyO/+. Cg8290-null animals (cg82902/cg82902 and cg82902/Df(2R)BSC153) display a further increase in eye pigmentation.
CG8290 exhibits affinity for H3K9me2/3 through an ATRX-like ADD domain
The CG8290 gene encodes several predicted protein isoforms (Fig. 4D). All CG8290 isoforms share a common N-terminal ADD domain (amino acids 54–192). The ADD domain has been reported to confer binding to histone tails and is present in the well-studied mammalian proteins ATRX and DNMT3a (Dhayalan et al. 2011; Eustermann et al. 2011; Iwase et al. 2011). Recently, it was shown that in the context of human ATRX, the ADD domain binds the histone H3 tail, specifically the H3K4me0K9me2/3 modification. This recognition was enhanced by interaction with HP1a, which also recognizes the same epitope, although likely on a neighboring nucleosome (Dhayalan et al. 2011; Iwase et al. 2011). Given that the fly homolog of ATRX, XNP, lacks an ADD domain (Bassett et al. 2008; Valadez-Graham et al. 2012), we wanted to explore the possible link between the CG8290 ADD, HP1a, and H3K9me2/3 recognition.
The alignment of insect CG8290-derived and vertebrate ATRX-derived ADD domains (Fig. 4A) indicates that the homology is essentially limited to cysteine residues, comprising the basic zinc finger fold of this domain; however, several residues previously implicated in pathogenesis in ATRX mutant patients and shown to abrogate H3K9me2 binding (Iwase et al. 2011) are also conserved. Therefore, we expressed wild-type and mutant versions of the CG8290 ADD domain (155 amino acids) as GST fusions and tested their binding to various biotinylated histone H3 tail peptides. The data presented in Figure 4B indicate that, like the ADD from human ATRX (positive control), the CG8290 ADD specifically binds H3K9me peptides, with pronounced preference for H3K9me2 and H3K9me3. Next, we created individual point mutations in two conservative parts of the composite H3K9me3-binding pocket in the ADD domain. Consistent with the human ADD structural data (Iwase et al. 2011), Y101A and C117A mutants in CG8290 displayed severely reduced binding to histone peptides irrespective of their methylation status (Fig. 4C). Based on these characteristics, we propose dADD1 (Drosophila ADD-1) as an appropriate name for CG8290.
Identification of HP1a-associated RNAs
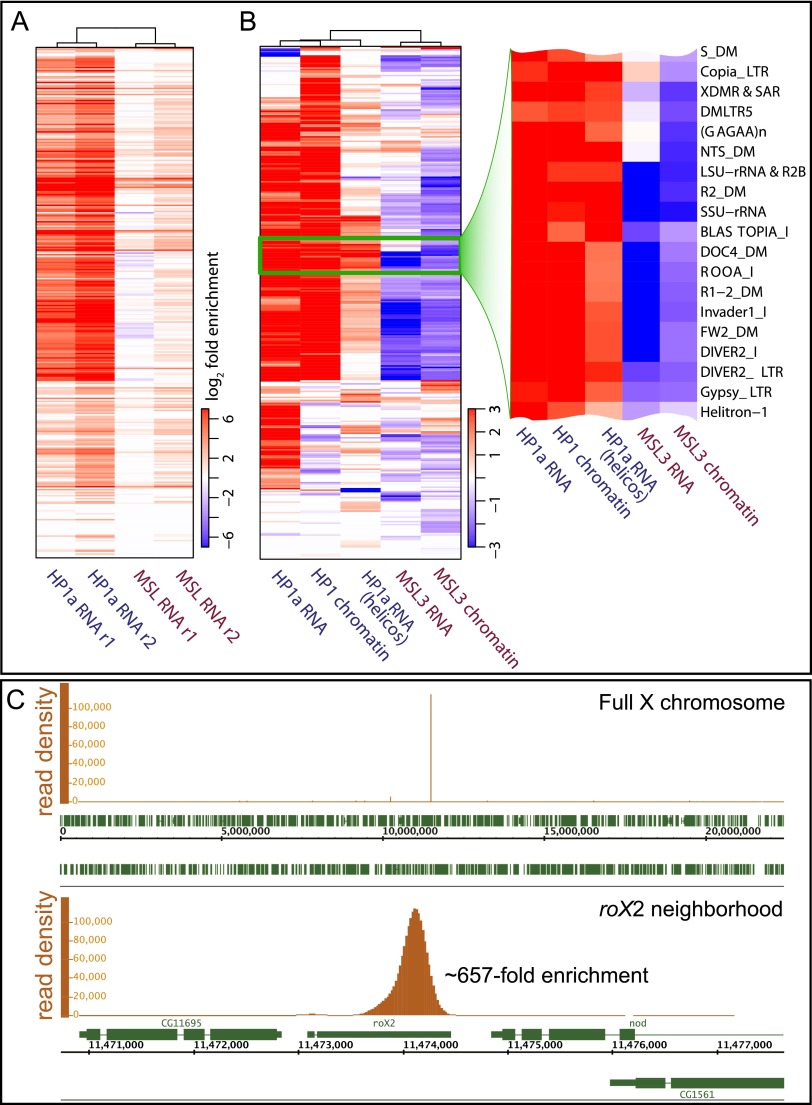
How heterochromatin in Drosophila is initially formed is still an open question. To investigate whether chromatin-associated RNAs might contribute and determine whether the BioTAP approach would be effective in detecting RNA components in addition to the protein and DNA components of HP1a-associated chromatin, we also purified the RNA fractions from HP1a-BioTAP and MSL3-BioTAP pull-downs. The MSL complex provided a very important proof of principle, as the two noncoding roX RNAs are known to be major components (Amrein and Axel 1997; Meller et al. 1997). In S2 cells, roX2 RNA is the major noncoding RNA (ncRNA) component, as roX1 is expressed at very low levels (Smith et al. 2000). When examined by random-primed, strand-specific RNA sequencing (RNA-seq), we observed strong enrichment for roX2 RNA in the MSL3-BioTAP pull-down from S2 cells (77-fold and 57-fold in the two replicates, respectively). In the HP1a pull-down from S2 cells, we observed strong enrichment for RNA matching repetitive regions of the genome (Fig. 5A).
Figure 5.
RNA-seq analysis of BioTAP-XL pull-downs. (A) Enrichment of repeat-derived RNA in HP1a-BioTAP cross-linked complexes from S2 cells compared with MSL3-BioTAP complexes from S2 cells detected using a random-priming approach for cDNA synthesis and Illumina RNA-seq. The plot shows log2 average enrichment across all annotated repeat types in Drosophila (each row corresponds to a repeat type), estimated relative to a corresponding input sample (total chromatin-associated RNA). Only statistically significant enrichment/depletion levels are shown in color. (B) Enrichment of the same spectrum of repeat-derived RNAs (rows are ordered as in A) assessed using HP1a-BioTAP embryos and either an on-bead ligation method for cDNA priming (HP1a RNA) or direct RNA-seq (HP1a RNA; Helicos). The enrichment was calculated compared with unrelated BioTAP-XL pull-down samples from embryos (see the Supplemental Material). The enrichment levels are also shown for the DNA fraction of the BioTAP pull-downs, illustrating the abundance of HP1a binding to the repetitive regions of the genome (HP1a chromatin) as well as the lack of such binding in the case of the control MSL3 complex (MSL3 chromatin). The zoomed-in version shows selected repeat details, with the full list in Supplemental Data File 1. Repeat enrichment was not observed for MSL3-associated RNA from S2 cells using the on-bead ligation approach (MSL3 RNA). (C) Analysis of the RNA recovered from the MSL3-BioTAP pull-down (on-bead method, Illumina platform) confirms the presence of roX2 RNA as the main RNA component of the MSL complex in S2 cells.
We wished to confirm these results in embryos and also explore additional methods, as we postulated that there might be biases in the repetitive RNA representation described above due to the random-priming step. Therefore, we examined the HP1a RNA pull-down profiles from embryos using two additional protocols. The first protocol used on-bead RNA adapter ligation, a modified version of the CRAC technique (Granneman et al. 2009) followed by RNA-seq on the Illumina platform. The second protocol used a simple reverse-cross-linking RNA recovery step followed by Helicos Direct RNA-seq, bypassing reverse transcription and PCR amplification (see the Materials and Methods; Ozsolak et al. 2009). Each of these approaches provided potential advantages. In the case of the CRAC technique, these include the use of a specific RNA adapter for reverse transcription and the retention of the RNAs of interest on the bead, allowing multiple rounds of washing. In the case of direct RNA-seq, omission of ligation and amplification steps might be expected to decrease biases in final sequence representation.
All three methods resulted in enrichment of repetitive RNAs after HP1 pull-down but to varying degrees (Fig. 5B; Supplemental Data File 1). The on-bead ligation method may be superior when considering roX2 RNA recovery from MSL3-BioTAP pull-downs as a positive control (Fig. 5C). In summary, all three methods showed enrichment of repetitive RNAs in HP1a pull-downs; thus, it is possible that transcripts originating from repetitive sequences may help guide or facilitate the function of HP1a complexes, analogous to the function of centromeric repetitive RNAs in fission yeast. However, a direct test for the function of repetitive RNA, separate from its DNA coding sequences within heterochromatin, remains technically challenging.
Discussion
HP1a and its interactors participate in multiple chromatin-associated complexes
In this study, we identified HP1a interactors using a chromatin-based biochemical approach (BioTAP-XL). We found connections between HP1a and factors responsible for chromatin organization, gene transcription, replication, and DNA repair in agreement with previously reported results for Drosophila and human HP1. Surprisingly, among the top interactors, we discovered multiple, previously unstudied proteins. Most of the new proteins were present in all developmental stages of the fly and fell into two categories. The first category was interactors for which the connection to HP1a was not known for Drosophila but was reported for the homologous proteins in different organisms; for example, Nipped-B (hNIPBL) and INCENP (hINCENP) (Ainsztein et al. 1998; Lechner et al. 2005). The second group includes novel HP1a interactors such as CG8290, CG30403, CG14438, and MES-4.
We validated the two top candidates, CG8290 and CG3680, and proposed naming them dADD1 and HIPP1, respectively. Analysis of our dADD1-BioTAP ChIP-seq results together with immunofluorescence localization studies strongly suggests that the main target of this protein is pericentric heterochromatin, correlating with the primary localization of HP1a. LC-MS/MS analysis of proteins associated with dADD1 showed that besides HP1a and HP2, the top candidate interactor is BON. BON is the only Drosophila homolog of the TIF1 family, and human TIF1 members interact with and phosphorylate HP1 (Le Douarin et al. 1998; Nielsen et al. 1999; Ryan et al. 1999). No direct interactions were observed between BON and HP1a in flies despite the observation that bonus could act as both an enhancer and suppressor of PEV (Beckstead et al. 2005). Therefore, we propose that dADD1 could act as a bridge between HP1a and BON.
The most interesting finding regarding dADD1 is its relationship to the human ATRX protein through its ADD domain. ATR-X (α-thalassemia/mental retardation, X-linked) syndrome is a human congenital disorder that causes severe intellectual disabilities. Mutations in the ATRX gene, which encodes an ATP-dependent chromatin remodeler, are responsible for the syndrome. Approximately half of the missense mutations in affected individuals are clustered in a cysteine-rich domain termed ADD, and the other half cluster in the SNF2-type ATP-dependent chromatin-remodeling domain (Iwase et al. 2011). The ADD domain was shown to bind the H3K9me3 chromatin mark and recruit ATRX to pericentric heterochromatin. In flies, the reported ortholog of ATRX is XNP (Bassett et al. 2008). XNP has the SNF2-type ATP-dependent chromatin-remodeling domain but is missing the ADD domain. It was shown previously as well as in this study that XNP interacts physically and functionally with HP1a (Emelyanov et al. 2010). At the same time, it was reported that XNP is not a general component of heterochromatin. Instead, XNP localizes to active genes and to a major focus near the heterochromatin of the X chromosome, corresponding to an unusual, decondensed block of satellite DNA (Schneiderman et al. 2009). We speculate that in flies, the SNF2 and ADD domains of human ATRX are divided between two proteins, XNP and dADD1. Interestingly, both proteins strongly interact with HP1a but are rather weak interactors with each other based on our results from the dADD1-BioTAP pull-down.
HIPP1/CG3680 was the second top candidate in our validation analysis. It was previously scored as a potential partner of HP1a in a high-throughput yeast two-hybrid assay (Giot et al. 2003), but to our knowledge, this interaction was not pursued. Here we discover association between HP1a and HIPP1 using multiple biochemical approaches. The ChIP-seq together with the LC-MS/MS results of the HIPP1-BioTAP pull-down suggest that the protein associates with HP1a within heterochromatin and also plays an HP1a-indpendent role within insulator complexes in euchromatin. The association with insulators is further strengthened by the recent identification of CG8436 and CG9740 (Table 3) as novel CP190-interacting proteins in S2 cells (Cuartero et al. 2014). Overall, our analyses of HIPP1 suggest separate roles in the euchromatic and heterochromatic portions of the genome, but we cannot exclude the possibility that the presence of HIPP1 simultaneously in both locations has some common purpose.
HP1a chromatin interaction networks
Our study provides an initial, unbiased view of HP1a-associated complexes on chromatin across the life cycle of Drosophila melanogaster. We were able to expand the BioTAP-XL approach to biochemically challenging life stages and identify protein–RNA associations in addition to protein–DNA and protein–protein interactions from a single chromatin preparation. A strength of BioTAP-XL is that it requires no prior knowledge of the biochemical properties of a given complex beyond the ability to assess whether the BioTAP-tagged bait protein retains its wild-type function.
Our follow-up analysis by BioTAP-XL tagging of two previously uncharacterized but strong HP1a interactors revealed that they coexist with HP1a in heterochromatin but also exhibit distinct binding locations in the euchromatic portion of the genome, likely shared with distinct partners identified in their protein interaction mass spectrometry lists. We believe that this constitutes a very promising beginning for the construction of an HP1a chromatin interaction network. For example, future investigations of potential subcomplexes could be pursued by asking whether post-translational modifications might govern subsets of HP1a–protein, HP1a–DNA, or HP1a–RNA interactions. Indeed, HP1a is known to have multiple sites of phosphorylation in vivo (Eissenberg et al. 1994), and we found that we could identify phosphorylated HP1a peptides from our complex peptide mixtures in multiple life stages (Supplemental Fig. S8) that are consistent with those identified in a global phosphoproteome analysis of Drosophila embryos (Zhai et al. 2008). Site-specific mutagenesis of these sites might interfere with subsets of interactions of the HP1a-BioTAP bait protein and could be assessed in parallel for phenotypes using classical genetics, thereby revealing how subsets of key functional interactions may be regulated by post-translational modifications. Furthermore, we could also detect phosphorylation of HIPP1, dADD1, and other HP1 interactors in S2 cells, embryos, larvae, and adults (Supplemental Fig. S8; Supplemental Data File 2). Future mining and BioTAP-XL analysis of the full HP1 protein interaction list will provide ample material to further define potential HP1 subcomplexes and their nucleic acid-binding properties. In summary, despite years of prominence in the growing field of epigenetics, many critical aspects of HP1a targeting and function still remain mysterious. We propose that continuing to decipher the function of HP1a and its multiple partners will require a concerted, “chromatin-centric” approach.
Materials and methods
BioTAP-XL for Drosophila S2 cells, embryos, larvae, and adults
Fly embryo preparation
To generate 100 g of 6- to 20-h embryos, flies were grown at 25°C with 65% humidity using large embryo collection cages (Flystuff, catalog no. 59-101) containing molasses plates with yeast paste. Embryos were washed off the plate with embryo saline solution (7 g of NaCl, 300 μL of Triton X-100 per/1 L), dechorionized in 50% bleach solution for 3 min, washed twice with dH2O, and blot-dried with paper towels. Using a motorized Teflon pestle, 10 g of embryos was dounced in 40 mL of embryo nuclear extraction buffer (NEB) (10% sucrose, 20 mM HEPES at pH 7.6, 10 mM NaCl, 3 mM MgCl2, 0.2% Triton) with 0.1 mM PMSF buffer for 10 strokes. The embryonic slurry was filtered using a single layer of miracloth (Calbiochem, catalog no. 475855). The filter was washed with an additional 60 mL of cold embryo NEB with 0.1 mM PMSF. The filtered homogenate (100 mL) was immediately transferred into a 225-mL T-flask filled with 360 mL of room temperature PBS and 40 mL of 37% formaldehyde and then treated as described below for S2 cells, starting from the pelleting of cross-linked nuclei.
Larval and adult fly preparations
One-hundred grams of third instar larvae or adult flies was collected. Larvae were separated from the food by washing in 20% sucrose (see Ashburner 1989). Flies were collected 1–12 h after eclosion. Whole animals were rapidly frozen in liquid N2 and stored at −80 °C until ready for assaying. Frozen samples (four sets of 25 g) were ground to a powder with a ceramic mortar and pestle chilled with liquid N2. Powder from 100 g of starting material was carefully added to a Clarkson BB250S 1-L blender with a stainless steel container filled with 900 mL of NEB + 0.1 mM PMSF (25 °C). Warning: Avoid pouring residual N2 into the blender and wear face protection to avoid splashing! The mixture was stirred for 30 sec and then blended for 30 sec at speed 1 and 30 sec at speed 2. Crushed larval or fly extract was immediately transferred into 4-gal Nalgene wide-mouth LDPE carboys (catalog no. 73004) with 4500 mL of PBS and 500 mL of 37% formaldehyde at room temperature and then treated as described below for S2 cells, starting from the pelleting of cross-linked nuclei.
Drosophila S2 cell culture
Cells were grown in four 50-mL large T-flasks (225 cm2) in 10% FBS-supplemented Schneider’s Drosophila medium (Invitrogen, catalog no. 11720) to a density of ∼5 × 106 cells per milliliter. Cell cultures were split in half by adding an equal volume of HyClone CCM3 serum-free medium (Thermo Scientific, catalog no. SH30065) and grown in eight T-flasks (225 cm2) to a density of ∼5 × 106 cells per milliliter. Cells were transferred into four 2.8-L Fernbach glass flasks (Bellco Glass, Inc.). For each flask, 100 mL of cells was added into 400 mL of HyClone CCM3 medium, and cells were grown at 90 rpm and 26.5°C to a density of ∼1 × 107 cells per milliliter, with cell viability >94%. Cells were pelleted by spinning at 600g for 10 min at 4°C and washed two to three times in 500 mL of PBS. Between washes, cells were pelleted as above.
Formaldehyde cross-linking
Harvested S2 cells were homogenized by using a 100-mL dounce homogenizer (Bellco Glass, Inc.) and 10 strokes each of A and B pestles. For every 4–5 mL of cell pellet volume, 100 mL of NEB + 0.1 mM PMSF prechilled on ice was added. Without delay, 100 mL of cell/nuclear homogenate was poured into a T-225 flask containing a room temperature mixture of 360 mL of PBS and 40 mL of 37% formaldehyde and incubated for 30 min at 25°C on a orbital shaker platform with vigorous shaking (100 rpm). Fixed nuclei were pelleted by spinning at 4000g for 10 min at 4°C. The supernatant was carefully decanted, and the nuclear pellet was washed four times with 100 mL of ice-cold PBS with 0.1 mM PMSF. Nuclei were pelleted between washes at 4000g for 10 min at 4°C. Nuclei were resuspended in glycerol buffer and snap-frozen in liquid N2 prior to further processing.
Chromatin preparation
Frozen nuclear extracts from Drosophila S2 cells, embryos, third instar larvae, and adult flies were thawed and spun down at 4000g for 10 min at 4°C. Pellets were washed with 10–20 vol of TE buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA at pH 8.0) with 0.1 mM PMSF and spun down at 4000g for 10 min at 4°C. Pellets were again resuspended with 10 vol of TE buffer with 0.1 mM PMSF by pipetting up and down (for third instar larval or adult fly chromatin, a motorized Teflon pestle was used). SDS was added to the mixture to a final concentration of 1%. The mixture was inverted in the tube 10 times and spun down at 4000g for 10 min at 4°C. The supernatant was carefully removed (note that the pellet may be quite loose), and the pellet was resuspended with 10 vol of TE buffer with 0.1 mM PMSF by pipetting and further spun down at 4000g for 10 min at 4°C. This washing step was repeated twice. The pellet was resuspended with 1.5 vol of TE buffer with 0.1 mM PMSF by pipetting up and down (for third instar larvae or adult flies, motorized Teflon pestle was used). SDS was added to a mixture to a final concentration of 0.1%. The resulting viscous mixture was sonicated in 4.5-mL aliquots using a Misonix Sonicator 3000 with Microtip power output level 7 and total sonication processing time of 3.5 min (15-sec pulse “on” and 45-sec “off” time) to generate DNA fragments in the range of 300–2000 base pairs (bp).
Triton X-100 (1% final) and NaCl (140 mM final) were added to the sonicated samples. Samples were mixed on a rotating wheel for 5 min at 4°C and spun down at 10,000g for 10 min at 4°C. The supernatant containing the soluble chromatin was collected. Note: In order to control for chromatin input composition and quality (for protein, DNA, and RNA), a 1-mL aliquot was reserved before proceeding with the next step.
Affinity purification and LC-MS/MS
Protein A-IgG and biotin-streptavidin affinity purification steps followed by on-bead trypsin digestion, C18 column peptide purification, and LC-MS/MS were performed as described (Alekseyenko et al. 2014).
Phosphorylation identification
Mass spectrometric files were searched with the X!Tandem algorithm (Craig and Beavis 2004) for serine, threonine, and tyrosine phosphorylation. Fragment ion annotations for MS/MS mapped to unique phosphorylated residues were manually verified (Supplemental Data File 2).
DNA and RNA recovery for next-generation sequencing
DNA was recovered, and ChIP-seq libraries were prepared as described previously (Alekseyenko et al. 2014).
Direct RNA-seq by Helicos
RNA for direct RNA-seq (DRS) was recovered from beads as for DNA. After ethanol precipitation, the RNA sample was treated with TURBO DNase (Invitrogen, catalog no. AM1906) followed by phenol-chloroform and chloroform extraction and standard EtOH precipitation. Recovered RNA was purified with the RNeasy Plus minikit (Qiagen) using the “purification of total RNA containing miRNA” protocol. DRS was performed as described (Ozsolak et al. 2009).
Random-priming approach for cDNA synthesis and Illumina RNA-seq
RNA from pull-down and input samples was recovered as above (Helicos). An NEBNext ultradirectional RNA library kit (New England Biolabs, catalog no. E7420S) was used to make cDNA and RNA-seq libraries. Several minor modifications to the standard kit protocol were made based on (1) size (RNA fragmentation time was decreased from 15 to 5 min) and (2) the small amount of RNA (the number of PCR cycles were adjusted to 20 cycles in the USER excision and PCR library step).
Modified CRAC technique for RNA-seq
The CRAC technique (Granneman et al. 2009) was used for the on-bead ligation and library preparation for the RNA sample. In order to eliminate DNA contamination in RNA samples, the following changes to the protocol were made. Instead of DNA linkers, the following RNA oligos were used: for the 3′ end, /5rApp/rUrCrGrUrArUrGrCrCrGrUrCrUrUrCrUrGrCrUrUrGrUr/ddC/-3 (BiooScientific); and for the 5′ end, /5InvddT/GrGrUrUrCrArGrArGrUrUrCrUrArCrArGrUrCrCrGrArCrGrArUrC (Eisen et al. 2001). After both linkers were ligated, the RNA was recovered as described above for DNA recovery, and the sample was treated with TURBO DNase followed by phenol-chloroform and chloroform extraction and standard EtOH precipitation in the presence of 50 μg of glycogen before reverse transcription.
It is important to note that in each approach used for RNA preparation, no rRNA depletion kit or techniques were used in order to prevent additional potential biases.
Bamse
To ensure uniform and unambiguous assignment, the detected peptides were realigned to the D. melanogaster translation annotation (FlyBase 5.45) using NCBI blastp (with the following parameters: −M PAM30, −e 100, −W 1, −b 20, −v 20, −a 20). Only peptides whose top-scoring hits were unique among the annotated proteins were used in further analysis.
The number of peptide counts was modeled as a variable distributed according to a negative binomial distribution, with the rate determined by the log-linear model shown in Figure 2. A normal prior distribution [N(−2,0.1)] was assumed for the base-level abundance αt, and Γ prior [Γ(0.05,0.02)] was used for nonnegative enrichment factors β and γ, implemented using the R RJAGS package. The posterior distributions of parameters were determined using Gibbs sampling (3000 adaptations, 1500 sampling rounds, 10 independent chains). The empirical P-value for the presence of association was determined as a proportion of posterior samples with γ > 0.1. The statistical assessment of enrichment was performed separately on total and unique peptide counts.
Data accessibility
DNA, RNA, and peptide sequencing data in this study were submitted to the NCBI Gene Expression Omnibus public repository under the accession number GSE56101.
Additional methods
For transgenic constructs, Drosophila genetics, polytene chromosome staining, coimmunoprecipitation, Western blotting, histone peptide-binding assays, and ChIP-seq and RNA-seq data analysis, see the Supplemental Material.
Acknowledgments
We are grateful to Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School), Kristen Alexa, and Wolfram Goessling for help with photography of fly eyes, and Shigeki Iwase, Yang Shi, Sarah Elgin, and Peter Kaiser for reagents. We are grateful to Myles Brown, Sarah Elgin, Bob Kingston, and members of the Kuroda laboratory for critically reading an earlier version of the manuscript. This work was supported by Tufts University start-up funds to S.M.F., and grants from the National Institutes of Health (K25AG037596 to P.V.K., and GM045744 and GM101958 to M.I.K.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.241950.114.
References
- Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC 1998. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol 143: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI 2014. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci 111: 2488–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Axel R 1997. Genes expressed in neurons of adult male Drosophila. Cell 88: 459–469 [DOI] [PubMed] [Google Scholar]
- Ashburner M (1989). Drosophila, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Badugu R, Shareef MM, Kellum R 2003. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem 278: 34491–34498 [DOI] [PubMed] [Google Scholar]
- Bassett AR, Cooper SE, Ragab A, Travers AA 2008. The chromatin remodelling factor dATRX is involved in heterochromatin formation. PLoS ONE 3: e2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead RB, Ner SS, Hales KG, Grigliatti TA, Baker BS, Bellen HJ 2005. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics 169: 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Barbash DA 2011. Functional conservation of the Drosophila hybrid incompatibility gene Lhr. BMC Evol Biol 11: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC 2009. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181: 1303–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, Khochbin S 2003. Cdyl: a new transcriptional co-repressor. EMBO Rep 4: 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Cuartero S, Fresan U, Reina O, Planet E, Espinas ML 2014. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J 33: 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B 2007. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, Ragozin S, Jeltsch A 2011. The ATRX–ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet 20: 2195–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R, Szidonya J, Korge G, Sehnert M, Taubert H, Archoukieh E, Tschiersch B, Morawietz H, Wustmann G, Hoffmann G, et al. 1993. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics 133: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem 276: 3484–3491 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC 2000. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10: 204–210 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet 30: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Ge YW, Hartnett T 1994. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J Biol Chem 269: 21315–21321 [PubMed] [Google Scholar]
- Elgin SC, Reuter G 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV 2010. Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric β-heterochromatin in vivo. J Biol Chem 285: 15027–15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18: 777–782 [DOI] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S 2008. HP1: a functionally multifaceted protein. Curr Opin Genet Dev 18: 169–174 [DOI] [PubMed] [Google Scholar]
- Franz H, Mosch K, Soeroes S, Urlaub H, Fischle W 2009. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J Biol Chem 284: 35049–35059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev PG, Gerasimova TI 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet 220: 121–126 [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, de Wit E, Bussemaker HJ, van Steensel B 2007. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. EMBO J 26: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K, Cook KR, Karpen GH 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev 15: 1334–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yu Z, Zhang S, Liang X, Chen J, Li C, Ma J, Jiao R 2010. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin. J Cell Sci 123: 2853–2861 [DOI] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- James TC, Elgin SC 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL 2010. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev 24: 2133–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC 2002. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci 99: 8707–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ 3rd 2005. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 331: 929–937 [DOI] [PubMed] [Google Scholar]
- Le Douarin B, You J, Nielsen AL, Chambon P, Losson R 1998. TIF1α: a possible link between KRAB zinc finger proteins and nuclear receptors. J Steroid Biochem Mol Biol 65: 43–50 [DOI] [PubMed] [Google Scholar]
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL 2008. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 32: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. 2011. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet 43: 220–227 [DOI] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL 1997. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457 [DOI] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4: 529–540 [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18: 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107 [DOI] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, Kimura H, Obuse C 2010. Human POGZ modulates dissociation of HP1α from mitotic chromosome arms through Aurora B activation. Nat Cell Biol 12: 719–727 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM 2009. Direct RNA sequencing. Nature 461: 814–818 [DOI] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol 161: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindyurin AV, Boldyreva LV, Shloma VV, Kolesnikova TD, Pokholkova GV, Andreyeva EN, Kozhevnikova EN, Ivanoschuk IG, Zarutskaya EA, Demakov SA, et al. 2008. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J Cell Sci 121: 1693–1703 [DOI] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G 2004. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J 23: 3516–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Wolff I 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet 182: 516–519 [DOI] [PubMed] [Google Scholar]
- Reuter G, Dorn R, Wustmann G, Friede B, Rauh G 1986. Third chromosome suppressor of position-effect variegation loci in Drosophila melanogaster. Mol Gen Genet 202: 481–487 [Google Scholar]
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 26: 103–115 [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ 3rd 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 19: 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HW, Lee DH, Florens L, Swanson SK, Washburn MP, Kwon SH 2014. Analysis of the heterochromatin protein 1 (HP1) interactome in Drosophila. J Proteomics 102: 137–147 [DOI] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet 10: e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman JI, Sakai A, Goldstein S, Ahmad K 2009. The XNP remodeler targets dynamic chromatin in Drosophila. Proc Natl Acad Sci 106: 14472–14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman JI, Goldstein S, Ahmad K 2010. Perturbation analysis of heterochromatin-mediated gene silencing and somatic inheritance. PLoS Genet 6: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res 22: 2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ 3rd, Spierer P, Bontron S 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet 3: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer CD, Stephens GE, Thompson BA, Funches L, Bernat JA, Craig CA, Elgin SCR 2002. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc Natl Acad Sci 99: 14332–14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol 20: 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G 2012. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734 [DOI] [PubMed] [Google Scholar]
- Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED 2004. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J 23: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadez-Graham V, Yoshioka Y, Velazquez O, Kawamori A, Vazquez M, Neumann A, Yamaguchi M, Zurita M 2012. XNP/dATRX interacts with DREF in the chromatin to regulate gene expression. Nucleic Acids Res 40: 1460–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton LM, Elledge SJ, Kharchenko PV, Garcia BA, Kuroda MI 2013. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol 20: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS 2007. E(var)3-9 of Drosophila melanogaster encodes a zinc finger protein. Genetics 177: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal T, Reuter G 2002. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 160: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Res 7: 1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA, RNA, and peptide sequencing data in this study were submitted to the NCBI Gene Expression Omnibus public repository under the accession number GSE56101.