The ATR checkpoint transmits DNA damage signals through the ATR–Chk1 kinase cascade. However, whether post-translational modifications other than phosphorylation are important for this pathway remains largely unknown. Zou and colleagues show that protein SUMOylation plays a key role in the ATR pathway. The ATR regulatory partner ATRIP is modified by SUMO, and a mutant lacking SUMOylation sites fails to support efficient ATR activation. Interestingly, the data reveal that ATRIP SUMOylation promotes ATR activation by boosting multiple protein interactions that ensure efficient DNA damage signaling.
Keywords: ATR, ATRIP, checkpoint, SUMOylation
Abstract
The ATR (ATM [ataxia telangiectasia-mutated]- and Rad3-related) checkpoint is a crucial DNA damage signaling pathway. While the ATR pathway is known to transmit DNA damage signals through the ATR–Chk1 kinase cascade, whether post-translational modifications other than phosphorylation are important for this pathway remains largely unknown. Here, we show that protein SUMOylation plays a key role in the ATR pathway. ATRIP, the regulatory partner of ATR, is modified by SUMO2/3 at K234 and K289. An ATRIP mutant lacking the SUMOylation sites fails to localize to DNA damage and support ATR activation efficiently. Surprisingly, the ATRIP SUMOylation mutant is compromised in the interaction with a protein group, rather than a single protein, in the ATR pathway. Multiple ATRIP-interacting proteins, including ATR, RPA70, TopBP1, and the MRE11–RAD50–NBS1 complex, exhibit reduced binding to the ATRIP SUMOylation mutant in cells and display affinity for SUMO2 chains in vitro, suggesting that they bind not only ATRIP but also SUMO. Fusion of a SUMO2 chain to the ATRIP SUMOylation mutant enhances its interaction with the protein group and partially suppresses its localization and functional defects, revealing that ATRIP SUMOylation promotes ATR activation by providing a unique type of protein glue that boosts multiple protein interactions along the ATR pathway.
The maintenance of genomic stability requires not only DNA repair machineries but also signal transduction pathways that regulate and coordinate the DNA damage response (DDR) (Ciccia and Elledge 2010). In human cells, DNA damage signaling is primarily initiated by the ataxia telangiectasia-mutated (ATM) and the ATM- and Rad3-related (ATR) kinases. Whereas ATM is activated by DNA double-stranded breaks (DSBs), ATR is elicited by a much broader spectrum of DNA damage and replication stress (Cimprich and Cortez 2008; Marechal and Zou 2013; Shiloh and Ziv 2013). Once activated, ATM and ATR phosphorylate and activate their effector kinases, Chk2 and Chk1, respectively. Together, the ATM–Chk2 and ATR–Chk1 kinase cascades phosphorylate a number of substrates involved in DNA repair, DNA replication, and cell cycle transitions, coordinating these processes to suppress genomic instability. In addition to the phosphorylation events mediated by the ATM and ATR pathways, several other types of post-translational modifications (PTMs), such as ubiquitylation, SUMOylation, methylation, acetylation, and poly-ADP ribosylation, are also implicated in the DDR (Bekker-Jensen and Mailand 2010; Huen et al. 2010; Lukas et al. 2011; Jackson and Durocher 2013). We recently found that the efficient activation of ATR relies on a ubiquitylation circuitry mediated by RPA-ssDNA (RPA-coated ssDNA) and PRP19 (Marechal et al. 2014). This new finding raises a question as to whether other PTMs also participate in DNA damage signaling through the ATR pathway.
The activation of the ATR pathway by DNA damage and replication stress is a multistep process. This process is initiated by the induction of RPA-ssDNA by DNA damage and replication stress. ATR, through its functional partner, ATRIP, is recruited to RPA-ssDNA at sites of DNA damage and stressed replication forks (Costanzo et al. 2003; Zou and Elledge 2003; Ball et al. 2005; Namiki and Zou 2006). ATRIP not only binds RPA-ssDNA directly but also recognizes the ubiquitin chains (Marechal et al. 2014). Three regulators of ATR, the RAD17 complex, the RAD9–RAD1–HUS1 (9-1-1) complex, and RHINO, are recruited to junctions of ssDNA and dsDNA (Zou et al. 2002, 2003; Ellison and Stillman 2003; Cotta-Ramusino et al. 2011). A fourth ATR regulator, the MRE11–RAD50–NBS1 (MRN) complex, acts at ssDNA/dsDNA junctions in Xenopus extracts and binds RPA-ssDNA in human cells (Olson et al. 2007b; Oakley et al. 2009; Duursma et al. 2013; Shiotani et al. 2013). Both 9-1-1 and MRN interact with TopBP1, an activator of ATR–ATRIP, enabling it to stimulate the ATR kinase on DNA (Kumagai et al. 2006; Delacroix et al. 2007; Mordes et al. 2008; Yan and Michael 2009; Lee and Dunphy 2010, 2013; Liu et al. 2011; Duursma et al. 2013). Through a process that is still not fully elucidated, activated ATR recognizes Chk1 with the help of mediators (e.g., Claspin, Timeless, and Tipin), allowing activation of the ATR–Chk1 kinase cascade (Kumagai and Dunphy 2003; Kumagai et al. 2004; Unsal-Kaçmaz et al. 2005, 2007; Chou and Elledge 2006; Wang et al. 2006; Smith et al. 2009).
Protein SUMOylation has recently emerged as a PTM critical for the DDR (Morris 2010; Jackson and Durocher 2013). The accumulation of SUMO at sites of DNA damage is important for the efficient recruitment of DDR proteins such as BRCA1 and 53BP1 (Galanty et al. 2009; Guzzo et al. 2012; Hu et al. 2012). Furthermore, a group of proteins involved in homologous recombination (HR), including BRCA1, MDC1, RPA70, and BLM, is directly modified by SUMO (Morris et al. 2009; Ouyang et al. 2009b; Dou et al. 2010; Galanty et al. 2012; Luo et al. 2012; Yin et al. 2012). SUMOylation regulates DDR proteins in several different ways. The modification of RPA70 and BLM by SUMO promotes their interactions with RAD51 (Ouyang et al. 2009b; Dou et al. 2010). The SUMOylation of BRCA1 stimulates its ubiquitin ligase activity (Morris et al. 2009). SUMOylated MDC1 is recognized by the ubiquitin ligase RNF4, which promotes MDC1 degradation and regulates the association of RPA and RAD51 with DNA lesions (Galanty et al. 2012; Luo et al. 2012; Yin et al. 2012). Although it is evident that protein SUMOylation plays an important role in DNA repair, whether SUMOylation is directly involved in DNA damage signaling is still unclear. A recent genome-wide RNAi screen in Drosophila identified ATR, RPA, and TopBP1 as regulators of the G2/M checkpoint (Kondo and Perrimon 2011). Interestingly, the same screen also implicated the E2 SUMO-conjugating enzyme Ubc9 and the E3 SUMO ligase PIAS in the G2/M checkpoint, raising the possibility that SUMOylation participates in DNA damage signaling in addition to DNA repair.
In this study, we show that protein SUMOylation is important for activation of the ATR but not the ATM pathway. The contribution of protein SUMOylation to ATR activation is at least in part attributed to the SUMOylation of ATRIP. ATRIP is modified by SUMO2/3 chains at K234 and K289. Elimination of ATRIP SUMOylation compromised the localization of ATRIP to sites of DNA damage and phosphorylation of Chk1 and RPA32. Surprisingly, the ATRIP mutant lacking the SUMOylation sites is defective for the interaction with a group of proteins in the ATR pathway, including ATR, RPA70, TopBP1, and MRN. Furthermore, we found that ATR, RPA70, TopBP1, and MRN all have affinity for SUMO2 chains in cell extracts and thatTopBP1 and MRN are capable of binding SUMO2 chains directly. Fusion of a SUMO2 chain to the ATRIP mutant lacking the SUMOylation sites enhanced its binding to the protein group and partially restored its localization and function. These findings reveal that ATRIP SUMOylation potentiates the ATR pathway by enhancing the interaction of ATRIP with the protein group, rather than an individual protein, providing a unique type of pathway-boosting protein glue that ensures the efficient signaling of DNA damage.
Results
UBC9 is required for efficient recruitment of ATRIP to sites of DNA damage
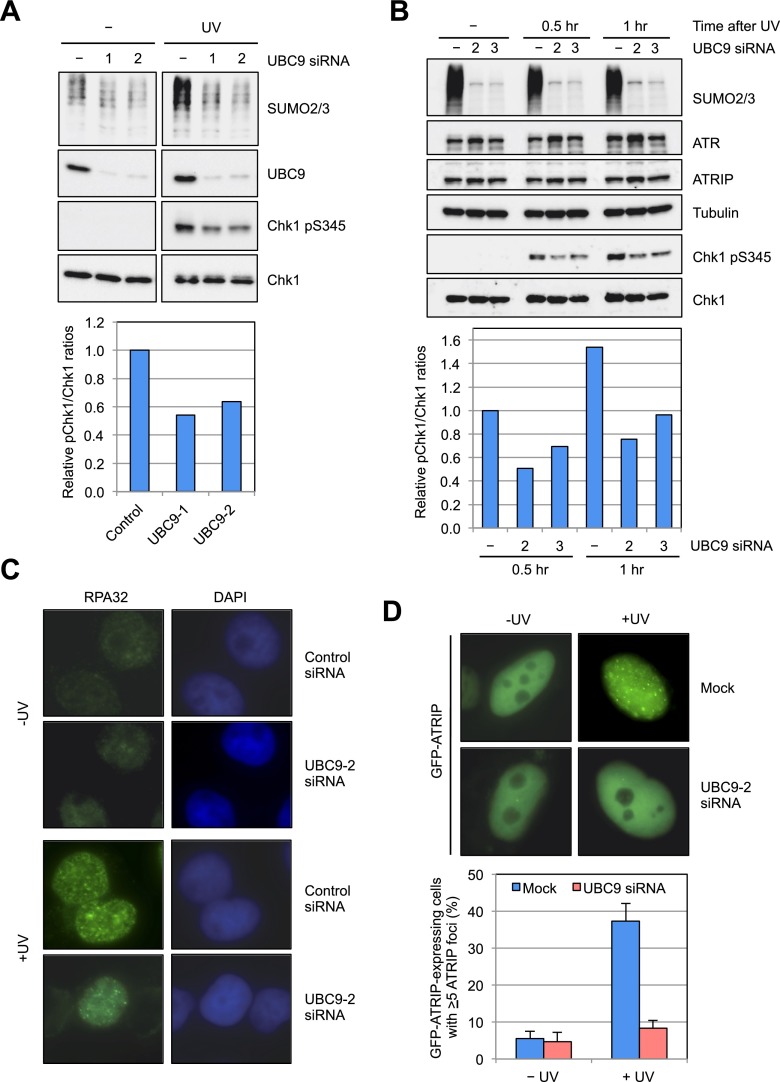
To investigate whether protein SUMOylation is important for DNA damage signaling, we first asked whether UBC9, the sole E2 SUMO-conjugating enzyme in human cells (Gareau and Lima 2010), is required for activation of the ATR and ATM pathways. We knocked down UBC9 in U2OS and HeLa cells with three independent siRNAs and analyzed the DNA damage-induced phosphorylation of Chk1 and Chk2, two specific substrates of ATR and ATM, respectively (Fig. 1A,B; Supplemental Fig. S1A–D). Cells were treated with ultraviolet (UV) light to activate the ATR pathway and with ionizing radiation (IR) to activate the ATM pathway. As expected, knockdown of UBC9 drastically reduced the overall levels of SUMO2/3 conjugates (Fig. 1A,B; Supplemental Fig. S1B,D). The UV-induced Chk1 phosphorylation was reduced by UBC9 knockdown (Fig. 1A,B; Supplemental Fig. S1B). The effects of UBC9 knockdown on Chk1 phosphorylation were observed 2 d after siRNA transfection when DNA synthesis was not significantly altered (Fig. 1A; Supplemental Fig. S1A–C). In contrast to Chk1 phosphorylation, the IR-induced Chk2 phosphorylation was unaffected in UBC9 knockdown cells (Supplemental Fig. S1D). These results suggest that UBC9 is required for efficient activation of the ATR but not the ATM pathway.
Figure 1.
UBC9 is required for efficient ATR activation. (A) U2OS cells were transfected with control, UBC9-1, or UBC9-2 siRNA. At 50 h after transfection, cells were treated with UV (20 J/m2) and analyzed 1 h later. The effects of UBC9 siRNAs on Chk1 phosphorylation were analyzed by Western. (B) HeLa cells were transfected with control, UBC9-2, or UBC9-3 siRNA. At 66 h after transfection, cells were treated with UV and analyzed as in A. (C) HeLa cells transfected with control or UBC9-2 siRNA were left undamaged or UV-irradiated (10 J/m2). The formation of RPA32 foci was analyzed by immunostaining 1 h after UV treatment. (D) HeLa cells were transfected with control or UBC9-2 siRNA and a plasmid expressing GFP-ATRIP. The formation of GFP-ATRIP foci 2 h after UV irradiation (10 J/m2) was analyzed in live cells. Quantifications of the GFP-ATRIP-expressing cells with ATRIP foci are shown in the bottom panel. Error bars represent SD from three independent experiments (n = 3).
To understand how UBC9 contributes to activation of the ATR pathway, we asked whether UBC9 knockdown affects any specific events during ATR activation. After UV treatment, both endogenous RPA32 and GFP-ATRIP were recruited to discrete nuclear foci in cells treated with control siRNA (Fig. 1C,D). The formation of RPA32 foci was not affected by UBC9 knockdown (Fig. 1C). However, GFP-ATRIP failed to form nuclear foci efficiently in UBC9 knockdown cells (Fig. 1D). Thus, although UBC9 is not required for generation of RPA-ssDNA in response to UV-induced DNA damage, it is required for the efficient recruitment of ATRIP. These results suggest a role for protein SUMOylation in ATRIP recruitment, an early event during ATR activation, prompting us to further investigate whether the proteins involved in this event are SUMOylated.
ATRIP is SUMOylated at K234 and K289
We noticed that ATRIP was among the proteins that were captured by TAP-tagged SUMO2 in a proteomic study (Golebiowski et al. 2009). Using the SUMOsp 2.0 program that predicts sites of SUMOylation (Ren et al. 2009), we identified three putative SUMOylation sites in human ATRIP (K125, K234, and K289) (Fig. 2A; Supplemental Fig. S2A). To verify whether ATRIP is SUMOylated in vivo, we transiently expressed SFB (S/Flag/Streptavidin-binding peptide)-tagged ATRIP in cells and immunoprecipitated it under a denaturing condition. Under this condition, the interactions of SFB-ATRIP with ATR and RPA were disrupted (Fig. 2B). Analysis of the precipitated SFB-ATRIP with anti-SUMO antibodies revealed that it was conjugated with SUMO2/3 but not SUMO1 (Fig. 2B; Supplemental Fig. S2B,C). As expected, SUMO2/3 conjugates of SFB-ATRIP were drastically reduced by UBC9 knockdown (Fig. 2B). In addition, immunoprecipitation of endogenous ATRIP or endogenous SUMO2/3 captured SUMOylated ATRIP (Fig. 2C; Supplemental Fig. S2D). Consistent with the SUMOylation of ATRIP in cells, SUMO2 was conjugated to SFB-ATRIP in vitro in the presence of E1, E2, and ATP (Fig. 2D). Together, these results establish ATRIP as a target of SUMOylation in the ATR pathway.
Figure 2.
ATRIP is SUMOylated at K234 and K289. (A) A schematic representation of the domain structure of ATRIP and the three putative SUMOylation sites. K234 and K289 are predicted to be canonical SUMOylation sites, whereas K125 is a noncanonical site. (B) HeLa cells were transfected with control or UBC9-2 siRNA and a plasmid expressing SFB-ATRIP. SFB-ATRIP was immunoprecipitated using Flag antibody under a denaturing condition, and the precipitated ATRIP was analyzed using SUMO2/3 antibody. (C) HeLa cells were transfected with control or UBC9-2 siRNA and irradiated with UV (50 J/m2). Thirty minutes after irradiation, endogenous ATRIP was immunoprecipitated with ATRIP antibody under a denaturing condition and analyzed by Western using SUMO2/3 antibody. (D) SFB-ATRIP was expressed in 293T cells and purified using Flag antibody. Purified SFB-ATRIP was incubated with E1, E2, SUMO2, and ATP. SUMOylated SFB-ATRIP was detected using SUMO2/3 antibody. (E) SFB-tagged ATRIPWT, ATRIP2KR, and ATRIP3KR were expressed in HeLa cells and immunoprecipitated using Flag antibody under a denaturing condition. SUMOylated SFB-ATRIP was analyzed using SUMO2/3 antibody.
We next asked whether ATRIP SUMOylation is regulated by DNA damage and replication stress. The SUMO2/3 conjugates of endogenous ATRIP were detectable even in the absence of DNA damage (Fig. 2C; Supplemental Fig. S2D). In response to UV irradiation, the levels of SUMOylated ATRIP were clearly elevated (Fig. 2C). However, the levels of SUMOylated ATRIP were not significantly affected by the DNA replication inhibitor hydroxyurea (HU) (Supplemental Fig. S2D). Thus, ATRIP is SUMOylated in the absence of DNA damage, and this modification is stimulated by UV but not HU.
To determine whether ATRIP is SUMOylated at the three predicted SUMOylation sites, we generated a set of ATRIP mutants in which the lysines of the predicted sites are replaced with arginines. The ATRIPK234R and ATRIPK289R mutants were SUMOylated at much lower levels than the ATRIPK125R mutant (Supplemental Fig. S2E). K234 contributes to ATRIP SUMOylation most significantly and is conserved in higher vertebrates (Supplemental Fig. S2A). Importantly, ATRIP2KR, the ATRIP mutant carrying both K234R and K289R mutations, was SUMOylated at a much lower level than wild-type ATRIP (ATRIPWT) (Fig. 2E; Supplemental Fig. S2B,E). Compared with ATRIP2KR, the ATRIP3KR mutant lacking all three predicted sites did not display a further reduction in SUMOylation (Fig. 2E), suggesting that K125 does not contribute to ATRIP SUMOylation appreciably. Consistent with the SUMOylation of ATRIP at K234 and K289 in cells, a recombinant ATRIP fragment encompassing these two sites was SUMOylated in vitro (Supplemental Fig. S2F). Introduction of the K234R and K289R mutations to this ATRIP fragment substantially reduced the SUMOylation in vitro. These results suggest that K234 and K289 are the primary SUMOylation sites of ATRIP.
ATRIP SUMOylation is critical for ATR activation
To assess the functional significance of ATRIP SUMOylation, we generated a set of HeLa derivative cell lines that stably express SFB-ATRIPWT or SFB-ATRIP2KR or only carry the corresponding vector. Upon knockdown of endogenous ATRIP using an siRNA targeting the 3′ untranslated region (UTR) of ATRIP mRNA, the levels of SFB-ATRIPWT and SFB-ATRIP2KR were similar to that of endogenous ATRIP prior to the knockdown (Fig. 3A,B). Importantly, cells expressing ATRIPWT or ATRIP2KR did not show significant differences in the cell cycle and DNA synthesis (Supplemental Fig. S3A). Even in the presence of endogenous ATRIP, ATRIP2KR exerted dominant-negative effects on the UV-induced Chk1 phosphorylation (Fig. 3A). In the absence of endogenous ATRIP, the UV-induced Chk1 phosphorylation was significantly reduced in vector-containing cells (Fig. 3A). The defect in Chk1 phosphorylation was suppressed by ATRIPWT but not ATRIP2KR (Fig. 3A,B). Similar results were obtained from three independently derived cell lines expressing SFB-ATRIP2KR, ruling out possible clonal variations (Fig. 3A,B; Supplemental Fig. S3B). The HU-induced Chk1 phosphorylation was also compromised in cells expressing ATRIP2KR (Fig. 3C), suggesting that ATRIP SUMOylation is a general requirement for ATR activation in response to DNA damage and replication stress. RPA32, another substrate of ATR, was also phosphorylated less efficiently in cells expressing ATRIP2KR than in cells expressing ATRIPWT (Fig. 6D, below). Consistent with its defect in ATR activation, ATRIP2KR failed to suppress the UV sensitivity of cells lacking endogenous ATRIP and to inhibit DNA synthesis upon DNA damage (Fig. 3D; Supplemental Fig. S3C). To determine whether the functional defects of ATRIP2KR are due to misfolding of the protein, we compared the cleavage of SFB-ATRIPWT and SFB-ATRIP2KR by partial protease digestion (Supplemental Fig. S3D). No difference in protease cleavage was detected between SFB-ATRIPWT and SFB-ATRIP2KR. Thus, while the SUMOylation of ATRIP is not strictly dependent on DNA damage and replication stress, it plays an important role in ATR activation.
Figure 3.
ATRIP SUMOylation is critical for ATR activation. (A,B) HeLa cells stably expressing SFB-ATRIPWT or SFB-ATRIP2KR or carrying the vector were transfected with control or ATRIP 3′ UTR siRNA and irradiated with UV (8 J/m2). The levels of the indicated proteins and the phosphorylation of Chk1 were analyzed by Western 1 h after UV treatment. Two independently derived SFB-ATRIP2KR-expressing cell lines (2KR and 2KR-20) were tested in A and B. (C) Cells were treated with 1 mM HU and analyzed as in A 1 h after the treatment. (D) Cells stably expressing SFB-ATRIPWT or SFB-ATRIP2KR or carrying the vector were transfected with ATRIP 3′ UTR siRNA and irradiated with different doses of UV. Cell viability was measured using the clonogenic assay.
Figure 6.
Fusion of a SUMO2 chain to ATRIP2KR partially bypasses ATRIP SUMOylation. (A) HeLa cells transfected with plasmids expressing HA-ATRIP2KR, HA-ATRIP3S-2KR, or HA-ATRIP3S2KA-2KR were irradiated with UV (10 J/m2). Cells extracts were generated 1 h after UV treatment and subjected to immunoprecipitation using HA antibody. The proteins precipitated were analyzed by Western and quantified. SD was obtained from two independent experiments. (B) HeLa cells transfected with plasmids expressing SFB-tagged ATRIPWT, ATRIP2KR, and ATRIP3S-2KR were microirradiated with laser. The localization of SFB-ATRIP was analyzed using anti-Flag antibody 30 min after microirradiation. RPA32 served as a marker of DNA damage. (C) Quantification of the fractions of RPA32 stripe-positive cells with ATRIP stripes. Error bars represent SD from three independent experiments (n = 3). (D,E) HeLa cells expressing SFB-tagged ATRIPWT, ATRIP2KR-20 or ATRIP3S-2KR or carrying the vector were transfected with ATRIP 3′ UTR siRNA and irradiated with UV (10 J/m2). The levels of the indicated proteins as well as the phosphorylation of Chk1 and RPA32 were analyzed by Western 1 h after UV treatment. The bands of pChk1, Chk1, pRPA32, and RPA32 were quantified from the Western blot, and the relative ratios of pChk1/Chk1 and pRPA32/RPA32 are shown in E.
ATRIP SUMOylation promotes localization of ATRIP to sites of DNA damage
Given that both UBC9 and ATRIP SUMOylation are required for efficient ATR activation and that knockdown of UBC9 compromises the recruitment of ATRIP to sites of DNA damage, we asked whether the SUMOylation of ATRIP is important for the localization of ATRIP after DNA damage. Using cells that express GFP-ATRIPWT or GFP-ATRIP2KR at similar levels, we compared the localizations of these two proteins (Supplemental Fig. S4). Nuclear foci of GFP-ATRIPWT were readily detected in cells treated with UV (Fig. 4A,B). In contrast, GFP-ATRIP2KR did not form foci efficiently (Fig. 4A,B), suggesting that the SUMOylation of ATRIP is required for the proper localization of ATRIP to sites of DNA damage. To measure the localization defect of ATRIP2KR more quantitatively, we irradiated cells expressing GFP-ATRIPWT or GFP-ATRIP2KR with a focal 355-nm laser and followed the localization of GFP-ATRIP in a time lapse (Fig. 4C). Accumulation of GFP-ATRIPWT was detected in the irradiated areas as early as 10 min after irradiation, whereas GFP-ATRIP2KR was not detected until 20 min after irradiation (Fig. 4C). Comparison of signals of GFP-ATRIPWT and GFP-ATRIP2KR in the irradiated areas showed that the recruitment of ATRIP2KR was reduced and delayed (Fig. 4D). However, we noted that the localization defect of ATRIP2KR was partial, raising the possibility that this specific defect of ATRIP2KR may not be fully accountable for its defect in Chk1 phosphorylation. Following this idea, we further investigated whether ATRIP2KR is defective for additional events that are required for efficient ATR activation.
Figure 4.
ATRIP SUMOylation promotes localization of ATRIP to sites of DNA damage. (A) HeLa cells transfected with plasmids expressing GFP-ATRIPWT or GFP-ATRIP2KR were irradiated with UV (50 J/m2), and the formation of GFP-ATRIP foci was analyzed 1 h after UV treatment in live cells. (B) Quantification of the fractions of GFP-ATRIP-expressing cells with ATRIP foci. Error bars represent SD from three independent experiments (n = 3). (C) Cells transiently expressing GFP-ATRIPWT or GFP-ATRIP2KR were microirradiated with a focal laser. The localization of GFP-ATRIP to the irradiated areas was analyzed in a time lapse as described in the Materials and Methods. (D) The increase of GFP-ATRIP signals in the irradiated areas were quantified. Each data point is the average of 10 independent measurements, and error bars represent SEM (n = 10).
ATRIP SUMOylation is involved in multiple protein interactions in the ATR pathway
The efficient activation of ATR by DNA damage relies on a series of protein interactions. For instance, the interaction between ATR and ATRIP is critical for the assembly of the ATR–ATRIP kinase complex (Cortez et al. 2001). The interaction between ATRIP and RPA plays a key role in recruiting ATR–ATRIP to RPA-ssDNA (Zou and Elledge 2003), and the interaction between TopBP1 and ATR–ATRIP allows TopBP1 to stimulate the kinase activity of ATR (Kumagai et al. 2006; Mordes et al. 2008; Liu et al. 2011). Furthermore, the MRN complex, which interacts with ATR–ATRIP, RPA, and TopBP1, also contributes to ATR activation (Stiff et al. 2005; Jazayeri et al. 2006; Myers and Cortez 2006; Olson et al. 2007a,b; Yoo et al. 2009; Duursma et al. 2013; Shiotani et al. 2013). To investigate whether ATRIP SUMOylation is involved in these protein interactions, we first tested the effects of UBC9 knockdown. Cells expressing HA-ATRIPWT were treated with UBC9 or control siRNA and irradiated with UV, and the proteins associated with HA-ATRIPWT were analyzed (Fig. 5A). Unexpectedly, the amounts of ATR, RPA70, TopBP1, MRE11, and RAD50 associated with HA-ATRIPWT were all significantly reduced by UBC9 knockdown (Fig. 5A). Consistently, the interactions of endogenous ATRIP with ATR, RPA70, TopBP1, MRE11, and NBS1 were also compromised by UBC9 knockdown (Fig. 5B). In marked contrast, UBC9 knockdown did not affect the interaction of ATM with NBS1 (Supplemental Fig. S5A). These surprising results suggest that protein SUMOylation promotes multiple protein interactions specifically in the ATR pathway.
Figure 5.
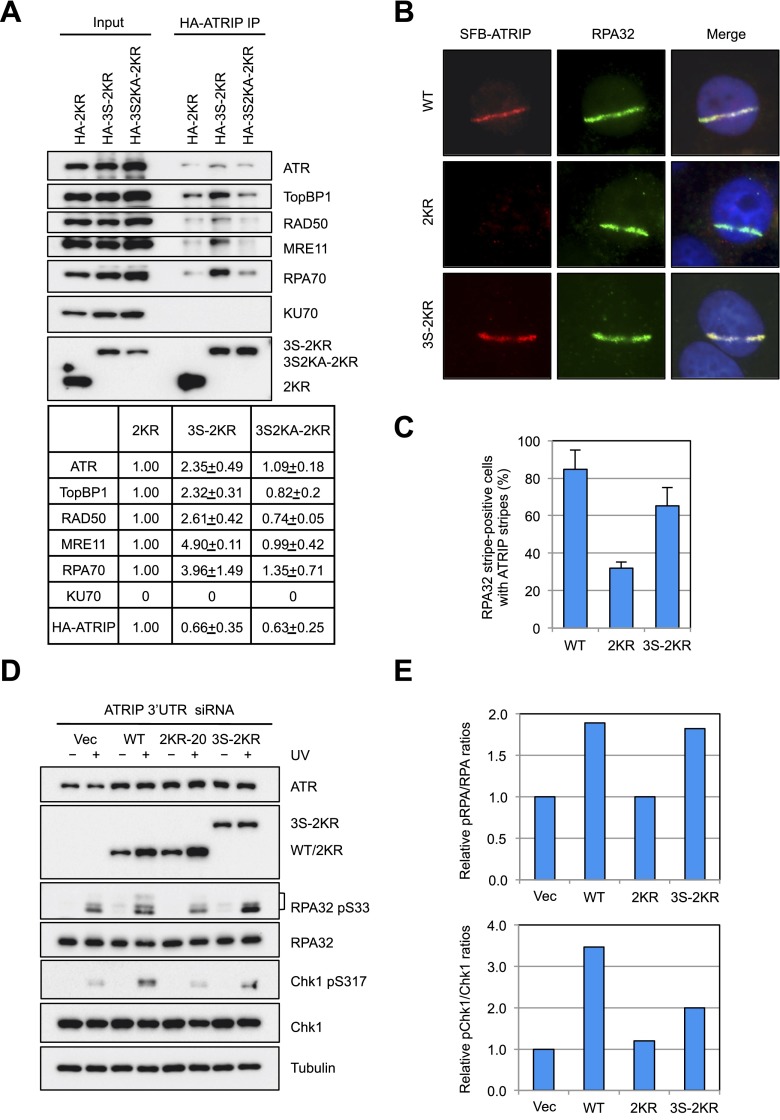
ATRIP SUMOylation enhances multiple protein interactions in the ATR pathway. (A) HeLa cells were transfected with control or two different UBC9 siRNAs and a plasmid expressing HA-ATRIPWT and irradiated with UV (10 J/m2). Cell extracts were generated 1 h after UV treatment, HA-ATRIPWT was immunoprecipitated using HA antibody, and the proteins associated with HA-ATRIPWT were analyzed by Western. (B) HeLa cells were transfected with control or UBC9-2 siRNA and treated with UV (10 J/m2). Endogenous ATRIP was immunoprecipitated 1 h after UV treatment, and the indicated proteins in the precipitates were analyzed by Western. (C) HeLa cells transfected with plasmids expressing HA-ATRIPWT or HA-ATRIP2KR or with the corresponding vector were irradiated with UV (10 J/m2). Cell extracts were generated 1 h after UV treatment and subjected to immunoprecipitation using HA antibody. The proteins precipitated were analyzed by Western and quantified. SD was obtained from two independent experiments. (D) HeLa nuclear extracts were incubated with purified GST-1xSUMO2 or GST-4xSUMO2. The input extracts, GST-1xSUMO2, and GST-4xSUMO2 were visualized with Ponceau S staining in the bottom panel. The indicated proteins in the input and pull-downs of GST-1xSUMO2 or GST-4xSUMO2 were analyzed by Western. (E) Purified TopBP1 or MRN complexes were incubated with purified His-1xSUMO2 or His-4xSUMO2. The TopBP1 and MRN components pulled down by His-tagged SUMO2 were detected by Western.
To determine whether ATRIP SUMOylation is important for the protein interactions in the ATR pathway, we immunoprecipitated HA-ATRIPWT and HA-ATRIP2KR from cells depleted of endogenous ATRIP and irradiated with UV (Fig. 5C). Compared with HA-ATRIPWT, HA-ATRIP2KR captured less ATR, RPA70, TopBP1, and MRN (Fig. 5C), showing that the SUMOylation of ATRIP indeed contributes to these interactions. In addition, Timeless, a mediator of the ATR pathway, also associated with HA-ATRIPWT more efficiently than with HA-ATRIP2KR (Fig. 5C). Thus, in contrast to the PTMs that regulate interactions between specific protein pairs, the SUMOylation of ATRIP enhances the interaction of ATRIP with multiple functional partners in the ATR pathway. While this result provides a possible explanation for the severe defect of ATRIP2KR in Chk1 phosphorylation, it raises a new question as to how ATRIP SUMOylation enhances the interaction of ATRIP with multiple partners.
To understand how ATRIP SUMOylation enhances multiple protein interactions, we asked whether some of the ATRIP-interacting proteins have affinity for SUMO. First, we tested whether these proteins can be captured by recombinant SUMO2 chains (GST-4xSUMO2) from nuclear extracts. Because many SUMO2/3-binding proteins have higher affinity for SUMO2/3 chains than mono-SUMO2/3, we used mono-SUMO2 (GST-1xSUMO2) as a control. Unexpectedly, ATR, RPA70, TopBP1, MRN, and Timeless all bound to 4xSUMO2 preferentially (Fig. 5D). In contrast, ATM, Chk2, and MDC1 failed to bind 4xSUMO2 in extracts (Supplemental Fig. S5B). Furthermore, KU70, an abundant protein that binds DNA breaks, was not captured by 4xSUMO2 (Fig. 5D). These results reveal a surprising feature of ATRIP-interacting proteins: They all share the ability to bind SUMO2/3 chains. Next, we tested whether some of the ATRIP-interacting proteins can bind SUMO2 chains directly. Both purified TopBP1 and MRN bound to purified His-4xSUMO2 (Fig. 5E), showing that multiple ATRIP-interacting proteins are capable of recognizing SUMO2/3 chains directly. The common ability of ATRIP-interacting proteins to bind SUMO2/3 chains provides a possible mechanism to enhance their interaction with SUMOylated ATRIP.
Fusion of a SUMO2 chain to ATRIP partially bypasses ATRIP SUMOylation
If ATRIP SUMOylation increases the avidity of ATRIP to other proteins in the ATR pathway, one would expect that fusing a SUMO2/3 chain to ATRIP2KR might alleviate the defects of ATRIP2KR. To test this possibility, we generated ATRIP3S-2KR by fusing 3xSUMO2 to the N terminus of ATRIP2KR. Using this fusion protein, we first compared the abilities of ATRIP2KR and ATRIP3S-2KR to associate with other proteins in the ATR pathway (Fig. 6A). Compared with HA-ATRIP2KR, HA-ATRIP3S-2KR coprecipitated greater amounts of ATR, RPA70, TopBP1, and MRN, showing that the presence of a SUMO2 chain on ATRIP indeed increases the association of ATRIP with these proteins. To ascertain that the increased binding of ATRIP3S-2KR to proteins in the ATR pathway is attributed to the SUMO2 chain, we introduced the K33A and K35A mutations to each of the three SUMO2s in HA-ATRIP3S-2KR, resulting in HA-ATRIP3S2KA-2KR. The K33A and K35A mutations of SUMO2 are known to disrupt the interaction of SUMO2 with its binding proteins (Chupreta et al. 2005; Ouyang et al. 2009a). The ability of HA-ATRIP3S2KA-2KR to bind ATR, RPA70, TopBP1, and MRN is lower than HA-ATRIP3S-2KR and similar to HA-ATRIP2KR (Fig. 6A), confirming that the SUMO2 chain of ATRIP3S-2KR is responsible for the increased binding to the protein group in the ATR pathway.
Next, we compared the localizations of SFB-tagged ATRIPWT, ATRIP2KR, and ATRIP3S-2KR in cells microirradiated with a UV laser (Fig. 6B). Compared with ATRIPWT, the localization of ATRIP2KR to the RPA-positive DNA damage stripes was compromised at 30 min after microirradiation (Fig. 6B,C). In contrast to ATRIP2KR, ATRIP3S-2KR was more readily detected in DNA damage stripes at 30 min, suggesting that the localization defect of ATRIP2KR is suppressed by the fusion of 3xSUMO2 to ATRIP2KR. The localization of ATRIP3S-2KR to DNA damage stripes was slightly less efficient than ATRIPWT (Fig. 6C), indicating that the 3xSUMO2 fusion of ATRIP2KR did not fully recapitulate the natural SUMOylation of ATRIPWT. Nonetheless, UBC9 knockdown did not reduce the localization of ATRIP3S-2KR to DNA damage stripes at 30 min after microirradiation (Supplemental Fig. S6A), suggesting that the recruitment of ATRIP3S-2KR is largely independent of UBC9. These results provide direct evidence that the presence of a SUMO2 chain on ATRIP partially bypasses the requirement of SUMOylation for ATRIP localization.
Last, we tested whether the defect of ATRIP2KR in ATR activation is suppressed by the 3xSUMO2 fusion. Even in cells treated with control siRNA, ATRIP2KR exerted dominant-negative effects on UV-induced Chk1 and RPA32 phosphorylation, possibly through its interaction with endogenous ATRIP (Supplemental Fig. S6B,C). Unlike ATRIP2KR, ATRIP3S-2KR did not interfere with Chk1 and RPA32 phosphorylation (Supplemental Fig. S6C). In cells depleted of endogenous ATRIP, ATRIP3S-2KR restored the phosphorylation of RPA32 and modestly rescued the phosphorylation of Chk1 (Fig. 6D,E). Thus, the defect of ATRIP2KR in ATR activation is partially suppressed by the fusion of 3xSUMO2 to ATRIP2KR. The inability of ATRIP3S-2KR to fully rescue Chk1 phosphorylation may be due to the suboptimal position or length of 3xSUMO2, altered conformation of ATRIP3S-2KR, or lack of dynamic ATRIP SUMOylation. Nevertheless, the fusion of a SUMO2 chain to ATRIP2KR indeed enhances its binding to proteins in the ATR pathway and partially bypasses the role for ATRIP SUMOylation in ATRIP localization and ATR function. These results strongly suggest that ATRIP SUMOylation promotes ATR activation by providing a unique type of protein glue that boosts multiple protein interactions along the ATR pathway.
Discussion
The ATR pathway is not simply a kinase cascade
The ATR pathway has been long viewed as a kinase cascade (Cimprich and Cortez 2008; Ciccia and Elledge 2010; Flynn and Zou 2011; Marechal and Zou 2013). We recently showed that the full activation of ATR pathway is dependent on a ubiquitylation circuitry mediated by RPA-ssDNA and the ubiquitin ligase PRP19 (Marechal et al. 2014). In this study, we found that UBC9 is required for efficient ATR activation, suggesting that the ATR pathway is also intertwined with the SUMOylation circuitry. It should be noted that knockdown of UBC9 reduces the SUMOylation of many proteins, some of which may affect ATR activation indirectly. Furthermore, although a partial defect of Chk1 phosphorylation was detected in cells with reduced UBC9 before the cell cycle was altered, prolonged UBC9 knockdown eventually altered the cell cycle, making it difficult to reveal the full impact of UBC9 depletion on ATR activation. Importantly, we found that the SUMOylation of ATRIP, the regulatory partner of ATR, is critical for activation of the ATR pathway, demonstrating the role for SUMOylation in ATR activation without the confounding effects of UBC9 depletion.
Interestingly, multiple ATRIP-interacting proteins in the ATR pathway have affinity for SUMO2 chains in vitro. In contrast, several proteins in the ATM pathway do not share this property. These findings suggest that SUMO binding is a unique property of the protein group that functions in the ATR pathway. In yeast, resection of DNA ends, the process that drives Mec1 (the yeast counterpart of ATR) activation at DNA breaks, promotes SUMOylation of numerous DDR proteins (Cremona et al. 2012; Psakhye and Jentsch 2012). Inactivation of Ubc9 or SUMO ligases in yeast reduced the DNA damage-induced phosphorylation of Rad53, the effector kinase of Mec1, suggesting that the interplay between protein SUMOylation and the ATR/Mec1 pathway is evolutionarily conserved (Cremona et al. 2012). However, it should be noted that the role for UBC9 in DNA damage signaling is more prominent in humans than in yeast. Importantly, we identified ATRIP as one of the critical targets of SUMOylation in the ATR pathway, revealing the first specific link between the SUMOylation and the DNA damage signaling circuitries.
ATRIP SUMOylation acts as a pathway-boosting protein glue
ATRIP is an obligated functional partner of ATR. Even in the absence of DNA damage, ATRIP forms a complex with ATR (Cortez et al. 2001). During the DDR, ATRIP plays multiple roles in the ATR pathway. ATRIP binds RPA-ssDNA and targets the ATR–ATRIP complex to sites of DNA damage (Zou and Elledge 2003). ATRIP also interacts with TopBP1, which stimulates the kinase activity of ATR–ATRIP (Kumagai et al. 2006; Mordes et al. 2008). Furthermore, ATRIP interacts with the MRN complex, which is important for the efficient phosphorylation of ATR substrates (Olson et al. 2007a). Together, these findings suggest that ATRIP is a critical protein-interacting module of the ATR–ATRIP complex that acts at multiple steps of DNA damage signaling. In the absence of ATRIP SUMOylation, the interactions of ATRIP with ATR, RPA, TopBP1, and MRN are all compromised, suggesting that ATRIP SUMOylation is not a mechanism that facilitates the binding of ATRIP to a specific protein, but a mechanism that increases the interaction of ATRIP with multiple partners. As exemplified by the partial defect of ATRIP2KR to localize to sites of DNA damage, loss of ATRIP SUMOylation impairs but does not abolish the individual events during ATR activation. Nonetheless, the partial defects of ATRIP2KR at multiple steps of ATR activation collectively lead to a severe defect in Chk1 phosphorylation, revealing a previously unappreciated mechanism that ensures the overall efficiency of DNA damage signaling.
How does ATRIP SUMOylation influence the interaction of ATRIP with multiple partners? One possibility is that ATRIP SUMOylation contributes to these interactions by stabilizing the ATR–ATRIP complex and facilitating its localization to DNA damage sites. Although the formation of the ATR–ATRIP complex is not dependent on DNA damage, it is regulated by other factors. For example, the TEL2–TTI1–TTI2 complex and the NEK1 kinase have been implicated in the assembly and/or stabilization of the ATR–ATRIP complex (Takai et al. 2007, 2010; Hurov et al. 2010; Liu et al. 2013). ATRIP SUMOylation enhances the interaction between ATR and ATRIP, and ATR has affinity for SUMO2 chains in cell extracts. These results suggest that ATR may recognize the SUMO2 chains on ATRIP, facilitating the assembly and/or stabilization of the ATR–ATRIP complex and its localization to DNA damage sites.
While the SUMOylation of ATRIP may enhance the interaction of ATRIP with other proteins by stabilizing the ATR–ATRIP complex, our data also suggest that ATRIP SUMOylation likely has a more direct role in these interactions. In addition to ATR, RPA70, TopBP1, and MRN displayed affinity for SUMO2 chains in cell extracts, and purified TopBP1 and MRN interacted with SUMO2 chains directly. Furthermore, even in undamaged cells where ATRIP is not localized to sites of DNA damage, fusion of a functional SUMO2 chain to ATRIP2KR enhanced its interaction with proteins in the ATR pathway (data not shown), suggesting that the SUMOylation of ATRIP increases its avidity for RPA, TopBP1, and MRN independently of ATRIP localization. The SUMO-mediated interactions of ATRIP with RPA, TopBP1, and MRN may facilitate the recruitment of ATR–ATRIP to RPA-ssDNA, the stimulation of ATR–ATRIP by TopBP1, and the functions of MRN in ATR activation. Based on our findings, we propose that the SUMOylation of ATRIP potentiates ATR activation by creating versatile protein glue on ATRIP, enhancing the function of ATRIP at multiple steps of ATR activation (Fig. 7). In this model, while ATRIP SUMOylation is not absolutely required for any individual interaction of ATRIP in the ATR pathway, it makes the protein interactions of ATRIP more efficient as a whole by providing a unique type of pathway-boosting protein glue (Fig. 7). ATRIP SUMOylation may contribute to not only the initial activation of ATR but also the amplification and maintenance of ATR-mediated signaling. ATRIP SUMOylation may occur transiently and be spatially or temporally regulated. Although only a small fraction of ATRIP is SUMOylated, this fraction of ATRIP may play a key role in driving specific events during ATR activation.
Figure 7.
A model of the role for ATRIP SUMOylation in the ATR pathway. ATRIP SUMOylation potentiates DNA damage signaling by enhancing multiple protein interactions in the ATR pathway. The SUMO-mediated protein interactions of ATRIP stabilize the ATR–ATRIP complex, promote the localization of ATR–ATRIP to RPA-ssDNA, facilitate the stimulation of ATR–ATRIP by TopBP1, and enable the function of MRN in ATR activation. In the absence of ATRIP SUMOylation, multiple steps of ATR activation are compromised, leading to severely defective Chk1 activation.
The individual and group properties of ATRIP SUMOylation
Recent studies in yeast have shown that a large number of DDR proteins, such as the HR proteins, are SUMOylated in response to DNA damage (Cremona et al. 2012; Psakhye and Jentsch 2012). Furthermore, many yeast HR proteins possess possible SUMO-binding motifs. Interestingly, only “wholesale” elimination of SUMOylation of several HR proteins impaired HR significantly, leading to a model in which SUMOylation promotes HR by regulating the HR proteins as a group, rather than individual proteins (Psakhye and Jentsch 2012). Consistent with this model, we found that multiple proteins in the ATR pathway have affinity for SUMO2 chains. Furthermore, the SUMOylation of ATRIP enhances its interaction with a protein group, rather than a specific protein, in the ATR pathway. These findings suggest that, like in yeast, in human cells, SUMOylation is also a mechanism to regulate the protein groups that function in the DDR.
Our results also revealed some interesting differences between the roles for SUMOylation in the yeast HR pathway and the human ATR pathway. Unlike the SUMOylation of yeast HR proteins, the SUMOylation of human ATRIP is not strictly dependent on DNA damage. UV stimulates ATRIP SUMOylation, providing a possible mechanism to enhance the ATR response. However, ATRIP SUMOylation is not obviously increased by HU yet is critical for HU-induced Chk1 phosphorylation, suggesting that ATRIP SUMOylation is a priming event for ATR activation in this context. In yeast, although SUMOylation of the HR protein group is critical for repair, elimination of SUMOylation of individual HR proteins did not compromise repair significantly. In human cells, knockdown of UBC9 severely reduced the interaction of HA-ATRIP with other proteins in the ATR pathway, whereas elimination of ATRIP SUMOylation alone only modestly affected these interactions. These results suggest that ATRIP is only part of the protein group in the ATR pathway that is regulated by SUMOylation. Nonetheless, elimination of ATRIP SUMOylation severely compromised Chk1 phosphorylation, showing that the SUMOylation of a key protein in a pathway can have a profound impact on the function of the pathway. The SUMOylation of ATRIP provides an interesting example of how SUMOylation of individual proteins can regulate the protein groups in which they function, linking the individual and group properties of this modification (Fig. 7). It is tempting to speculate that SUMOylation of many proteins other than ATRIP may act similarly in different pathways, boosting the efficiency of various cellular processes.
Materials and methods
Cell culture, plasmids, and transfection
HeLa and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and L-Glutamine. HeLa derivative cells stably expressing SFB-ATRIPWT or SFB-ATRIP2KR or carrying the vector were cultured in the presence of 1 μg/mL puromycin.
Plasmids expressing ATRIP with different tags (SFB, 3xHA, and GFP) were generated using the Gateway Technology (Invitrogen). Various point mutants of ATRIP and SUMO2 were generated by site-directed mutagenesis. To tag ATRIP with a 3xSUMO2 chain, SUMO2 (amino acids 12–91) was amplified by PCR and cloned into the destination vector. The insertion of three copies of SUMO2 to the vector was confirmed by DNA sequencing. For plasmid transfection, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer’s protocol. All siRNAs (control, ATRIP, and UBC9) used in this study were purchased from Invitrogen and transfected using Lipofectamine RNAiMAX (Invitrogen). The sequences of the siRNAs are shown in the Supplemental Material.
Immunoprecipitation and antibodies
For testing the SUMOylation of ATRIP, cell extracts were prepared in lysis buffer (1% SDS, 20 mM Tris-HCl at pH 8.0, 10% glycerol, 1 mM DTT, 15 mM N-ethylmaleimide [NEM] [Sigma-Aldrich], protease inhibitor cocktail [Sigma-Aldrich]). The extracts were denatured by boiling for 10 min and then sonicated. After centrifugation at 13,800 rpm for 10 min, the supernatants were collected and diluted in 9 vol of modified RIPA buffer (20 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 0.5% Igepal CA-630, 10% glycerol) supplemented with 10 mM NEM and protease inhibitors. SFB-ATRIP was immunoprecipitated using anti-Flag M2 agarose beads (Sigma-Aldrich). For immunoprecipitation of HA-ATRIP (wild-type, 2KR, 3S-2KR, and 3S2KA-2KR) proteins and endogenous ATRIP, ATM, and NBS1, cell extracts were prepared in NETN buffer (20 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 0.5% Igepal CA-630, 10% glycerol, 1 mM DTT, 15 mM NEM, 5 mM NaF, protease inhibitor cocktail). After sonication and centrifugation, the supernatants (2 mg) were subjected to immunoprecipitation using HA, ATRIP, ATM, and NBS1 antibodies and Dynabead Protein G (Life Technologies).
The antibodies to ATR, ATM, 53BP1, MDC1, NBS1, MRE11, RPA70, TopBP1, Timeless, and phospho-RPA32 (Ser33) were from Bethyl Laboratories; the antibodies to phospho-Chk1 (Ser317 and Ser345), phospho-Chk2 (Thr68), RAD50, UBC9, and Tubulin were from Cell Signaling; the antibodies to Chk1, HA, GST, and SUMO1 were from Santa Cruz Biotechnology; the SUMO2/3 antibody was from Abcam; the HA antibody used for immunoprecipitation was from Covance; the RPA32 antibody was from ThermoScientific; the Chk2 antibody was from Millipore; the KU70 was from GenTex; and the Flag antibody was from Sigma-Aldrich. ATRIP antibody was previously described (Cortez et al. 2001).
Laser microirradiation and immunofluorescence analysis
For laser microirradiation, cells were cultured in BrdU-containing medium for 24 h and then in phenol red-free DMEM (Invitrogen) before being irradiated by a 355-nm UV laser. For detection of GFP-ATRIP or RPA foci, cells were directly irradiated with 256-nm UV using a calibrated UV lamp (UVP). At the indicated times after irradiation, cells were extracted with 1× PBS containing 0.5% Triton X-100 for 5 min on ice, fixed in 3% paraformaldehyde/2% sucrose for 15 min, and extracted again with 1× PBS containing 0.5% Triton X-100 for 3 min on ice. Cells were subsequently incubated with RPA32 antibody in 1× PBS containing 3% BSA/0.05% Tween 20 for 1 h at 37°C and with Flag antibody in the same buffer overnight at 4°C. After the incubation with primary antibodies, cells were washed three times with 1× PBS containing 0.05% Tween 20 and incubated with secondary antibodies for 1 h at 37°C. Cells were then washed three times with 1× PBS containing 0.05% Tween 20 and stained by DAPI. Imaging was performed using a Nikon Eclipse 90i epifluorescence microscope. Additional details on focal laser microirradiation and live-cell imaging are provided in the Supplemental Material.
Acknowledgments
We thank Dr. J. Chen and Dr. G. Gill for reagents, R. Buisson for assistance with FACS data analysis, and members of the Zou laboratory for helpful discussions. L.Z. is a Jim and Ann Orr Massachusetts General Hospital Research Scholar. This work was supported by grants from National Institutes of Health (NIH; GM076388) and the Federal Share of Massachusetts General Hospital Proton Program to L.Z., and NIH grants (CA092584 and CA162804) to D.J.C.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.238535.114.
References
- Ball HL, Myers JS, Cortez D 2005. ATRIP binding to replication protein A–single-stranded DNA promotes ATR–ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 9: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci 103: 18143–18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA 2005. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol 25: 4272–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D 2008. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol Cell 11: 203–213 [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER 3rd, Hurov K, Sowa ME, Harper JW, Elledge SJ 2011. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332: 1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona CA, Sarangi P, Yang Y, Hang LE, Rahman S, Zhao X 2012. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell 45: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM 2007. The Rad9–Hus1–Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Huang C, Singh M, Carpenter PB, Yeh ET 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol Cell 39: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duursma AM, Driscoll R, Elias JE, Cimprich KA 2013. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 50: 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V, Stillman B 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 1: E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Zou L 2011. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 36: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26: 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT 2009. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2: ra24. [DOI] [PubMed] [Google Scholar]
- Guzzo CM, Berndsen CE, Zhu J, Gupta V, Datta A, Greenberg RA, Wolberger C, Matunis MJ 2012. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal 5: ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Paul A, Wang B 2012. Rap80 protein recruitment to DNA double-strand breaks requires binding to both small ubiquitin-like modifier (SUMO) and ubiquitin conjugates. J Biol Chem 287: 25510–25519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J 2010. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov KE, Cotta-Ramusino C, Elledge SJ 2010. A genetic screen identifies the triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev 24: 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49: 795–807 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Kondo S, Perrimon N 2011. A genome-wide RNAi screen identifies core components of the G(2)–M DNA damage checkpoint. Sci Signal 4: rs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG 2003. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol 5: 161–165 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Kim SM, Dunphy WG 2004. Claspin and the activated form of ATR–ATRIP collaborate in the activation of Chk1. J Biol Chem 279: 49599–49608 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG 2006. TopBP1 activates the ATR–ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Lee J, Dunphy WG 2010. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9–Hus1–Rad1 complex at stalled replication forks. Mol Biol Cell 21: 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dunphy WG 2013. The Mre11–Rad50–Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell 24: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L 2011. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell 43: 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ho CK, Ouyang J, Zou L 2013. Nek1 kinase associates with ATR–ATRIP and primes ATR for efficient DNA damage signaling. Proc Natl Acad Sci 110: 2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Luo K, Zhang H, Wang L, Yuan J, Lou Z 2012. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 31: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Zou L 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5: a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J, Zou L 2014. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell 53: 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D 2008. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR 2010. SUMO in the mammalian response to DNA damage. Biochem Soc Trans 38: 92–97 [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Myers JS, Cortez D 2006. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281: 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki Y, Zou L 2006. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci 103: 580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley GG, Tillison K, Opiyo SA, Glanzer JG, Horn JM, Patrick SM 2009. Physical interaction between replication protein A (RPA) and MRN: involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry 48: 7473–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Lee AY, Chen L, Wu X 2007a. The Mre11–Rad50–Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem 282: 22939–22952 [DOI] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X 2007b. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol 27: 6053–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Shi Y, Valin A, Xuan Y, Gill G 2009a. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 34: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA 2009b. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol 7: e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820 [DOI] [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y 2009. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics 9: 3409–3412 [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14: 197–210 [PubMed] [Google Scholar]
- Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H, Zou L 2013. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep 3: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Fu MA, Brown EJ 2009. Tim–Tipin dysfunction creates an indispensible reliance on the ATR–Chk1 pathway for continued DNA synthesis. J Cell Biol 187: 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O’Driscoll M, Jeggo PA 2005. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J 24: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T 2007. Tel2 regulates the stability of PI3K-related protein kinases. Cell 131: 1248–1259 [DOI] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, Pavletich NP 2010. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev 24: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A 2005. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol 25: 3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK 2007. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27: 3131–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zou L, Lu T, Bao S, Hurov KE, Hittelman WN, Elledge SJ, Li L 2006. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell 23: 331–341 [DOI] [PubMed] [Google Scholar]
- Yan S, Michael WM 2009. TopBP1 and DNA polymerase-α directly recruit the 9-1-1 complex to stalled DNA replication forks. J Cell Biol 184: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 26: 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG 2009. The Mre11–Rad50–Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell 20: 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou L, Cortez D, Elledge SJ 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev 16: 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci 100: 13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]