Abstract
Hepatitis C virus (HCV) is a global health problem and one of the main reasons for chronic liver diseases such as cirrhosis and hepatocellular carcinoma. The HCV genome is translated into a polyprotein which is proteolytically processed into 10 viral proteins. The interactome of the HCV proteins with the host cell has been worked out; however, it remains unclear how viral proteins interact with each other. We aimed to generate the interaction network of these 10 HCV proteins using a flow-cytometry-based FRET assay established in our laboratory (Banning, C., Votteler, J., Hoffmann, D., Koppensteiner, H., Warmer, M., Reimer, R., Kirchhoff, F., Schubert, U., Hauber, J., and Schindler, M. (2010) A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5, e9344).
HCV proteins were constructed as fusions with the chromophores CFP and YFP. All HCV fusions were expressed and localized to specific subcellular compartments, indicating that they were functional. FACS-FRET measurements identified a total of 20 interactions; 13 of these were previously described and have now been confirmed in living cells via our method. Among the seven novel protein binding pairs, HCV p7 plays a pivotal role. It binds to the HCV capsid protein Core and the two glycoproteins E1 and E2. These interplays were further demonstrated in the relevant context of Huh7.5 liver cells expressing infectious HCV.
Our work demonstrates the feasibility of rapidly generating small interaction networks via FACS-FRET and defines the network of intra-HCV protein interactions. Furthermore, our data support an important role of p7 in HCV assembly.
Hepatitis C virus (HCV)1 belongs to the family of Flaviviridae and is the only member of the genus Hepacivirus. The ∼9.5-kB positive-strand RNA genome is directly translated via an internal ribosomal entry site into a polyprotein. This is proteolytically processed by cellular and viral proteases into structural (Core, E1, E2) and nonstructural (p7, NS2, NS3, NS4A/B, and NS5A/B) proteins (1). In recent decades, light was shed on the importance and biological relevance of most HCV proteins, which ultimately led to the development of the first specific antiviral therapy involving inhibition of the NS3 serine protease (2). However, because HCV is highly variable and because of the rapid emergence of drug resistance, additional therapeutic approaches are urgently needed (2). An impressive body of data was derived from protein interaction or siRNA screens investigating the interplay of HCV proteins with cellular factors (3–5). Although these screens are essential in order for researchers to understand how HCV manipulates the host cell, their potential benefit for novel therapeutic approaches could be limited. HCV is a chronic viral infection, and targeting host factors might result in drugs with severe adverse effects. Thus, a promising strategy would be to specifically inhibit interactions among viral proteins. Surprisingly, until now, a comprehensive analysis of the putative interactions and the interplay of HCV proteins with each other in living human cells has been lacking.
In the present work, we did an extensive and thorough analysis of intra-HCV protein interactions. We used our novel flow-cytometry-based FRET assay that allows rapid assessment of the interplay between proteins in thousands of living cells (6). Therefore, this experimental approach enables quantification and statistical evaluation of all results. From the total of 20 interactions established by FACS-FRET, we chose to further investigate three that were not yet described in the literature. The putative HCV viroporin p7 binds to the structural proteins, and this was verified via biochemical methods in cells expressing fully infectious HCV.
The established network of intra-HCV protein interactions in living mammalian cells provides new insights into the biology of this important human pathogen. Furthermore, we identified several HCV protein interactions that could be targeted for antiviral therapy.
EXPERIMENTAL PROCEDURES
Generation of HCV Expression Plasmids and HCV Viral Constructs
Each HCV protein was constructed either as ECFP-fusion, acting as an energy donor, or as EYFP-fusion, responding as an energy acceptor. All 10 HCV proteins were amplified from the HCV JFH1 sequence (7) (UniProt Q99IB8, molecular clone kindly provided by T. Wakita) and ligated into the pECFP-C1 amd pEYFP-C1 vectors (Clontech) via the EcoRI and XhoI restriction sites essentially as described before (6). Mutation RR33/35QQ in p7 was introduced via site-directed mutagenesis. HCV E1, NS3, and NS5A contain an internal XhoI site, and these were cloned into pECFP-N1 and pEYFP-N1 vectors via BsrGI and NotI. HCV-Jc1-E1(A4) and Jc1-E1(A4)-p7(HA) were generated through reconstitution of the H77 strain E1 protein sequence (A4) (8) SSGLYHVTNDC by means of splice-overlap-extension-PCR within the HCV-Jc1 and Jc1-p7(HA) variants (9), kindly provided by T. Pietschmann. The HCV-Jc1-NS5A(GFP) molecular clone was generously contributed by R. Bartenschlager (10). All PCR-derived inserts were sequenced to confirm nucleotide identity.
Cell Culture and Transfection
293T and Huh7.5 cells (11) (kindly contributed by C. Rice) were cultivated with Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum plus antibiotics (and 1% (v/v) non-essential amino acids for Huh7.5 cells) and grown at 37 °C in a humidified atmosphere containing 5% CO2. 293T single and cotransfections with ECFP and EYFP fusions were performed using the calcium phosphate method. FACS analyses were done 24 h post-transfection. For this, 150,000 cells per well were seeded in a 12-well plate. In total, 2.5 μg of DNA was transfected; for cotransfection of the fusions, an ECFP:EYFP ratio of 1.5:1 was used to compensate for the overall lower fluorescence intensity of ECFP and achieve optimal rates of double positive cells. Huh7.5 single and cotransfection with ECFP and EYFP fusions was performed using MetafectenePro (Biontex, Martinsried, Germany). FACS analyses were done 48 h post-transfection. For this, 350,000 cells per well were seeded in a 12-well plate and transfected with 2.0 μg of DNA according to the protocol provided by the manufacturer.
FACS-based FRET
FACS-FRET was done with a FACS CantoII Cytometer (BD Biosciences) equipped with 405 nm, 488 nm, and 633 nm lasers essentially as described before (6). For EYFP detection, we excited the cells with 488 nm and detected the resulting signal with a 529/24 filter (Semrock, New York, NY). The ECFP signals were detected via the 450/40 filter (Semrock) after excitation at 405 nm. FRET cells were excited with 405 nm, and signal detection was conducted with the 529/24 filter again. We used five controls for each cotransfection setup. Mock cells were transfected with water instead of DNA, and the vectors pECFP and pEYFP were singly transfected as well as cotransfected to exclude false positive FRET signals and background. An ECFP–EYFP fusion construct was used as a positive control. A minimum of 3000 ECFP and EYFP double positive cells was analyzed per measurement.
Electroporation of Huh7.5 Cells
After in vitro transcription (T7 RiboMAXTM Express Large Scale RNA Production System, Promega, Madison, WI), HCVJc1 RNA was electroporated (Gene Pulser Xcell System Electroporator, Bio-Rad) into Huh7.5 liver cells essentially as described before (7). In brief, 6.5 × 106 Huh7.5 cells were washed with PBS and suspended in 400 μl of Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, pH 7.6, 25 mM Hepes, 2 mM EGTA, 5 mM MgCl2; pH adjusted to 7.6 with KOH) with freshly added 2 mM ATP and 5 mM glutathione (end concentration; pH 7.6). After transfer into electroporation cuvettes, 5 μg of RNA was pulsed with 975 μF and 270 V. Cells were seeded into well plates or cell tissue flasks (125 cm2). Medium was changed 4 or 16 h after electroporation; cells were analyzed 72 h later.
Co-immunoprecipitation and Western Blot
After lysis of electroporated cells with 800 μl of CoIP-lysis buffer (0.05 M Tris, 0.15 M NaCl, 1 mM EDTA, pH 7.4, 1% TritonX-100) for 20 min on a stirring wheel, cell debris was removed by 10 min of centrifugation at 14,000 rpm. Supernatants of the lysates were incubated with rotation overnight, together with protease inhibitor (Complete Mini) and either α-HA(ms) (Sigma) or α-HA(rb) (Cell Signaling, Cambridge, UK) antibody (1:150). 30 μl of protein plus Protein G Sepharose was washed three times with CoIP-lysis buffer prior to 4 h of incubation with the antibody-lysate mixture. All steps were performed at 4 °C. After being washed three times with CoIP-lysis buffer, Sepharose was suspended in 20 μl of TBS and 15 μl of 5× Laemmli buffer and boiled at 95 °C for 10 min. Samples were analyzed via SDS-PAGE and Western blot. After transfer of the separated proteins from the SDS gel to a nitrocellulose membrane (0.4 μm; Whatman) and blocking, the membrane was incubated with primary monoclonal antibodies (α-Core (1:1000; C7–50, Abcam, Cambridge, UK), α-E2 (1:1000; AP33, Genentech, San Francisco), α-A4 (1:1000; kindly provided by H. Greenberg and J. Dubuisson), and α-HA(ms) (1:1000)) overnight. Membranes were washed, incubated with HRP-conjugated secondary antibody (α-mouse, 1:10,000, Sigma) for 3 h, and washed once again before protein detection.
Confocal Microscopy, Co-localization Analyses, and Proximity Ligation Assay
293T cells or Huh7.5 cells were seeded on coverslips and transfected as described above. Subsequently cells were fixed for 30 min with 2% paraformaldehyde and mounted with Mowiol 4–88 (Carl Roth, Karlsruhe, Germany) on microscope slides. Confocal microscopy was done with a Zeiss LSM510 with Meta detector or with the Nikon Ti Eclipse equipped with the PerkinElmer UltraViewVox System (Yokogawa CSU-X1). If not otherwise indicated, we used HCS NuclearMask Deep Red Stain (Invitrogen) for identification of the nuclei. For co-localization studies and PLA, Huh7.5 cells were electroporated as described above and seeded on coverslips. 56 h post-electroporation, cells were fixed for 25 min with 2% paraformaldehyde, permeabilized for 15 min with 1% saponin, and blocked for 45 min with 5% BSA. Indicated primary antibodies (α-GFP (BioVision, San Francisco, CA), α-NS5A (clone 2F6/G11, IBT, Reutlingen, Germany), α-CD81 (Ancell, Bayport, MN), α-HA(rb), α-core, α-E2, and α-A4) were incubated 1:100 in 1% BSA for 2 h at room temperature. For co-localization studies, AlexaFluor 405, 488, or 555 anti-mouse or anti-rabbit was incubated for 1 h and mounted with Mowiol 4–88. For PLA secondary antibody probes, ligation reaction and amplification were assessed according to the manufacturer's protocol (Duolink, Sigma Aldrich). Spinning disc microscopy was done with the Nikon Ti Eclipse UltraViewVox System. Image analysis was done with the Volocity 6.2 software package. For co-localization, every cell was cropped and Pearson's R2 value was calculated according to Costes co-localization. For PLA software implemented automated spot counting was used.
Generation of the Interaction Network
The interaction map was constructed with the open source program Cytoscape. Intraviral HCV protein interactions found within the present study via FACS-FRET and published previously (compare Fig. 3) were summarized in one map (Fig. 9). Additionally, we incorporated the interaction of HCV proteins with the host cell. For this, interaction data from the VirusMint database and from de Chassey et al. (4), who did a proteome-wide interaction screen for HCV, were included. The VirusMint dataset was cleared of double and reverse tested interactions and those that were exclusively defined by co-localization studies.
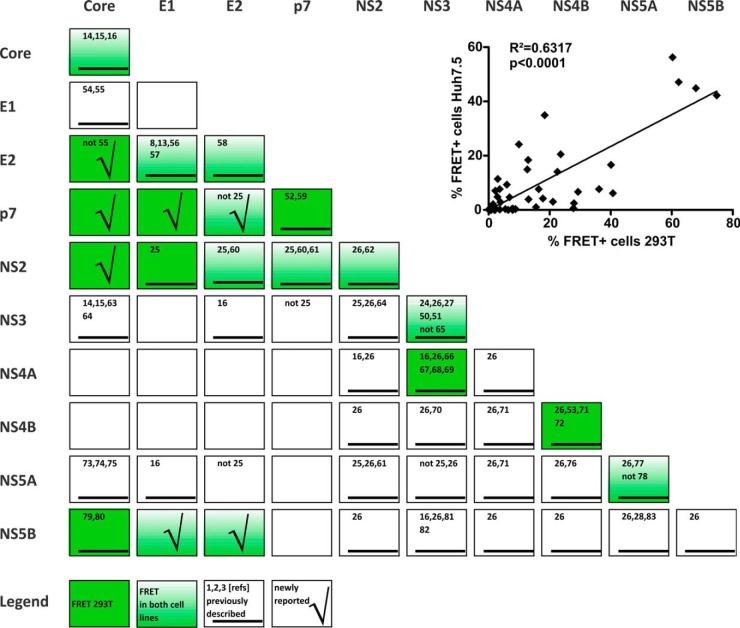
Fig. 3.
Overview of HCV protein interactions measured via FACS-FRET in both tested cell lines. Statistical significant interactions with FRET values ≥ 10% are presented (compare to supplemental Fig. S1). Interactions in 293T are highlighted by dark green boxes, and those that were also significant in Huh7.5 are colored in fading green. Furthermore, we marked interactions that were previously described by others (black underscored bar; numbers of the corresponding references are written in the boxes). Interactions that were newly reported here are highlighted by a check mark (5, 8, 13–16, 24–28, 50–83). The inlay depicts the correlation of FACS-FRET results generated for 293T and Huh7.5 liver cells. Mean values of the percentage of FRET+ cells derived from 293T transfection were plotted with the according values obtained from Huh7.5 transfections (n = 45; compare with supplemental Fig. S1). Pearson's correlation and the corresponding p value were calculated with Graph Pad Prism 5.0.
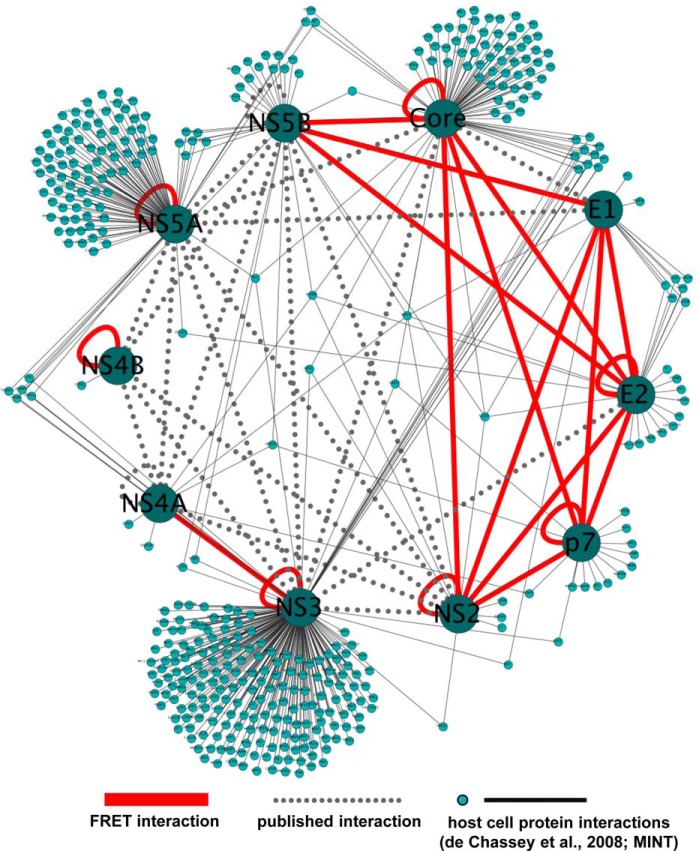
Fig. 9.
The HCV protein interaction network. HCV protein interactions measured in this study by FRET (red lines) or reported previously (dotted gray lines) were visualized as a network. Furthermore, HCV protein interactions with host cell factors were incorporated by using interaction data from the VirusMINT database and de Chassey et al. (4). This network was generated with Cytoscape (85).
Statistical Analyses
Statistical analyses were performed with Graph Pad Prism software, version 5.0. We used the two-tailed unpaired Student's t test to statistically assess differences between FRET and background signals (respective CFP-fusion protein cotransfected with YFP only). Pearson values were calculated for correlation analyses. For the co-localization and PLA results, we used one-way analysis of variance with Bonferroni post-test to assess significance levels.
RESULTS
Characterization of Fluorescently Tagged HCV Fusion Proteins
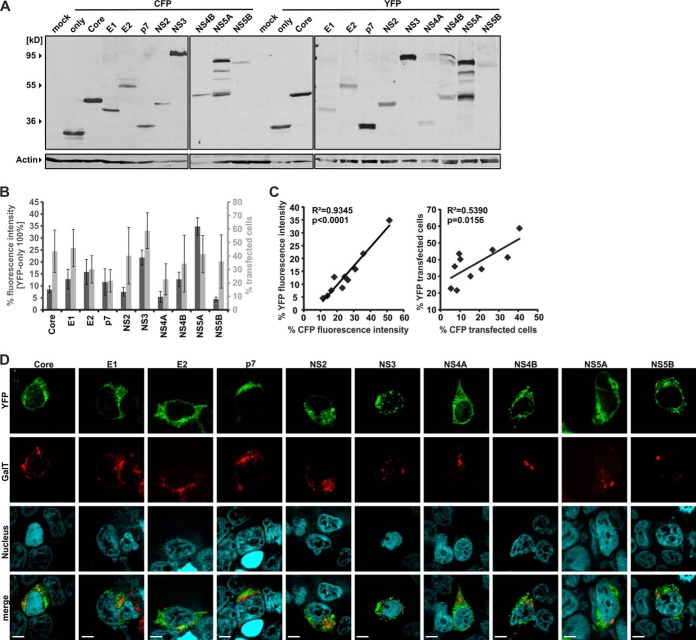
In order to define the network of intra-HCV protein interactions via FACS-FRET, we fused all 10 viral proteins of the JFH1 strain (7) with the chromophores CFP and YFP. Because a chromophore tag can alter the stability of a protein, we first checked proper expression by Western blotting. Linkage of CFP or YFP to the C terminus of the HCV Core, E1, and E2 completely abrogated protein expression, although we introduced a methionine start codon at the beginning of each ORF (data not shown). In contrast, all HCV proteins were expressed when we fused the chromophores at the N terminus, albeit with different efficiencies (Fig. 1A). Additionally, we measured the expression of the various HCV fusion proteins according to FACS parameters (i.e. the fluorescence intensity relative to YFP or CFP and the total percentage of transfected cells) (Fig. 1B). These parameters correlated significantly for the YFP and CFP HCV fusions (Fig. 1C) and allowed us to detect proteins that showed very weak expression in Western blots (e.g. YFP-NS4A; note that CFP-NS4A could not be detected via Western blot).
Fig. 1.
Characterization of HCV-CFP and -YFP fusion proteins. All fusion proteins used in this study and characterized here carry an N-terminal chromophore tag. A, 300,000 293T cells were transfected with 5 μg of DNA of each HCV CFP/YFP fusion protein in a six-well format. Cellular lysates were generated for Western blotting, and expression of the fusion proteins was detected by an antibody specific for various chromophores including CFP and YFP. Actin was blotted as a loading control. B, 150,000 293T cells were transfected with 2.5 μg of DNA in a 12-well format. The mean fluorescence intensity and the percentage of cells transfected with the HCV-YFP fusion proteins were assessed via FACS analysis. Mean values and standard deviations were calculated from at least four independent transfections. C, transfections and measurements were done similarly to those described for B but with the HCV-CFP fusions. Then the mean values for fluorescence intensity and the percentage of transfected cells were correlated. Squared Pearson's correlation (R2) and corresponding p values were calculated with Graph Pad Prism 5.0. D, 293T cells transfected with the indicated HCV-YFP fusion proteins and GalT-CFP were grown on coverslips and embedded for confocal microscopy. The scale bar indicates a distance of 7 μm.
Chromophore tags are generally well tolerated (12). Nevertheless, they can affect the localization of a protein and thus its functionality. We therefore investigated the subcellular distribution of the YFP/CFP fused HCV proteins via confocal microscopy. HCV proteins are associated with intracellular membranes (e.g. the endoplasmic reticulum) and lipids. In line with this, all YFP/CFP HCV fusion proteins showed an endoplasmic-reticulum-like or punctuated subcellular distribution (Fig. 1D). In contrast, no fusions were diffusely expressed within the cell, nor did they localize to the nucleus.
We conclude that the generated YFP/CFP HCV fusion proteins are useful tools for assessing the network of intra-HCV protein interactions via FACS-FRET.
Analysis of Specific HCV Protein Interactions via FACS-FRET
As proof of principle, we next aimed to validate a set of previously described interactions with our method. In Fig. 2 we present confocal images of the subcellular localization of the fusion proteins (panel 1) and representative FACS plots (panel 2), as well as a summary generated from multiple independent biological replicates including statistics (panel 3). For FACS-FRET, we first gated on double positive cells expressing both YFP and CFP and then plotted the CFP intensity versus FRET (see Fig. 2, panel 2). Gates were generated according to the negative (cotransfection of YFP and CFP) and the positive control (transfection of a YFP–CFP fusion protein), which generally resulted in less than 0.5% of FRET+ cells for the negative control and more than 95% of FRET+ cells for the positive control (6). We furthermore analyzed at least 3000 double positive cells per transfection.
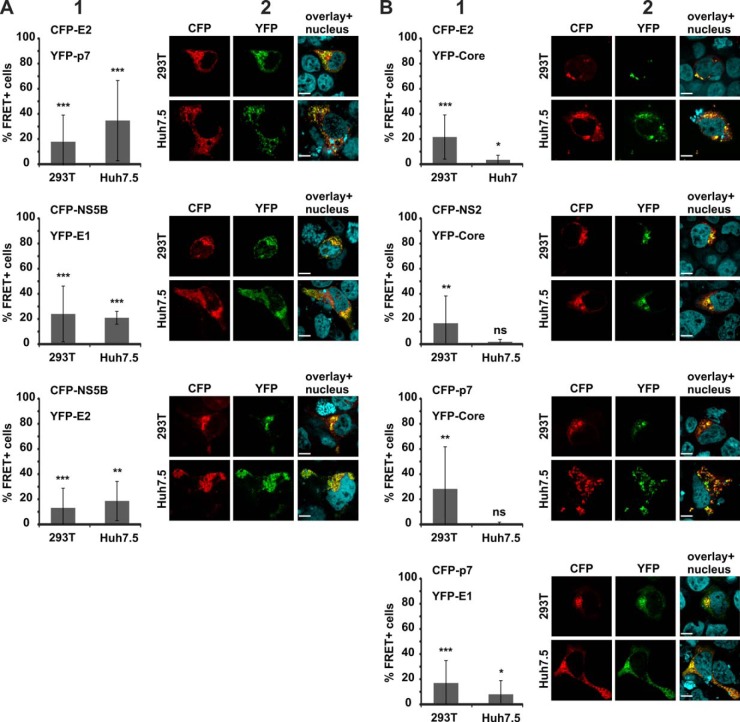
Fig. 2.
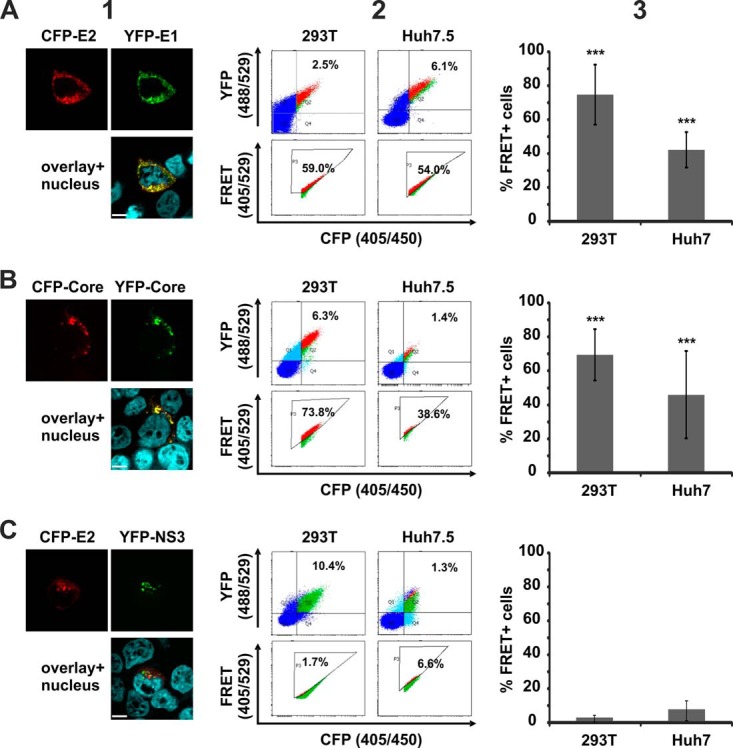
FRET analysis of specific HCV protein interactions. 293T and Huh7.5 cells were transfected with (A) CFP-E2 and YFP-E1, (B) CFP-Core and YFP-Core, or (C) CFP-E2 and YFP-NS3. Panel 1 shows the subcellular distribution of the respective YFP (green) and CFP (red) fusions, as well as areas of co-localization (yellow) in 293T cells. The scale bar represents a length of 7 μm. Panel 2 depicts examples of primary FACS plots acquired for FRET analysis. Panel 3 shows the mean value of the percentage of cells scoring FRET+ (MF) and the according standard deviation across multiple independent transfections (n = 3 to 17).
When we assessed heterodimer formation of the HCV glycoproteins E1 and E2 (8, 13) (Fig. 2A), we measured 74.8% FRET+ cells (S.D. ± 17.68%; n = 17) in 293T cells and confirmed this result in the liver cell line Huh7.5 (MF = 42.2%; S.D. ± 10.45; n = 7). Furthermore, we confirmed multimerization of the HCV Core protein (14, 15), which is essential for nucleocapsid formation (Fig. 2B; MF = 67.9%, S.D. ± 14.88, n = 9 in 293T cells; MF = 44.9%, S.D. ± 25.25, n = 8 in Huh7.5 cells). In contrast, we failed to verify the interaction of E2 with the NS3 protease (16) in our system (Fig. 2C; MF < 10% in both cell lines). Of note, in contrast to E1 and E2 heterodimer formation and Core multimerization, we also failed to detect areas of pronounced co-localization when we assessed the subcellular distributions of E2 and NS3 (Fig. 2). This is in line with the absence of robust FRET measured by flow cytometry.
Thus, our measurements revealed some differences from previous observations. Nevertheless, we were able to verify important and well-established interactions in living 293T and Huh7.5 liver cells via our FACS-FRET approach.
HCV Protein Interactions Determined via FACS-FRET
To establish the complete network of intra-HCV protein interactions, we tested all fusion proteins in two combinations: YFP–protein A with CFP–protein B, and YFP–protein B with CFP–protein A. Extensive FACS-FRET measurements and co-localization analyses were performed after cotransfection of both constructs in 293T cells (see supplemental data). The FRET signal of the negative control (i.e. YFP only cotransfected with CFP only) generally resulted in a signal of less than 2% FRET+ cells (average value, 0.49%; S.D. ± 0.93; n = 190).
The quantitative results of all measurements are summarized in supplemental Fig. S1, and a qualitative overview is presented in Fig. 3. For each of the 100 possible combinations, we indicated the following parameters: n, number of biological replicates; MF, mean percentage of FRET+ cells; S.D., standard deviation; and significance value. We used the two-tailed unpaired Student's t test to statistically challenge observed differences between FRET and background signals. Background was defined by an additional negative control (i.e. the respective CFP-fusion protein cotransfected with YFP only for each experiment). We furthermore arbitrarily introduced an additional stringency threshold of 10% FRET+ cells for interactions (supplemental Fig. S1; values in red). Therefore, some FRET signals below the threshold were significantly higher than the background but were not considered relevant (supplemental Fig. S1; values in green).
293T cells are kidney derived and were used for FACS-FRET here because they are an established and easy-to-transfect mammalian cell system that allows overexpression of proteins (6). However, HCV has strong liver cell tropism (12). Thus, interactions of viral proteins could be different in the presence of liver cell specific factors. We therefore used FACS-FRET to assess a total of 45 interactions in Huh7.5 liver cells (11). These analyses included all those that were over the 10% threshold in 293T cells. The background signal of the negative control for Huh7.5 transfections was 0.29% FRET+ cells (±0.97; n = 82). The results were analyzed similarly to the data obtained from 293T cells and incorporated in supplemental Fig. S1. Although the absolute percentage of FRET+ cells varied between the two cell types, interactions found in 293T cells could generally be confirmed in Huh7.5 cells. Thus the results obtained from both cell lines correlated significantly (see inset in Fig. 3; R2 = 0.6317; p < 0.0001; n = 45).
In all, as depicted in the consolidated summary in Fig. 3, we report a total of 20 interactions in living mammalian cells determined via FACS-FRET (p < 0.05 and MF ≥ 10%). 11 could be detected in both cell lines (Core/Core, E1/E2, E1/NS5B, E2/E2, E2/p7, E2/NS2, E2/NS5B, p7/NS2, NS2/NS2, NS3/NS3, NS5A/NS5A), whereas the other 9 interactions met our criteria only in 293T cells (Core/E2, Core/p7, Core/NS2, Core/NS5B, E1/p7, E1/NS2, p7/p7, NS3/NS4A, NS4B/NS4B). Most important, 7 of the 20 protein interactions determined via FACS-FRET had not been described in the literature before (Fig. 3 and Fig. 4). Binding of E2/p7, NS5B/E1, and NS5B/E2 was observed in both cell lines (Fig. 4A). In contrast, FRET signals between E2/Core, NS2/Core, p7/Core, and p7/E1 were only significant and above the 10% threshold in 293T cells (Fig. 4B).
Fig. 4.
Novel HCV protein interactions detected via FACS-FRET. Panel 1 gives mean values of the percentage of FRET+ cells and standard deviations of multiple independent experiments (for the number of replicates (n), mean FRET values (MF), and standard deviations (S.D.), as well as statistical calculations, please refer to supplemental Fig. S1). We further present representative confocal images of 293T and Huh7.5 cells that were transfected with the indicated CFP (red) and YFP (green) fusion proteins in panel 2. Co-localization in the overlay appears yellow. The scale bar is 7 μm. A, CFP-E2 and YFP-p7, CFP-NS5B and YFP-E1, and CFP-NS5B and YFP-E2 showed significant FRET in both cell lines, whereas (B) CFP-E2 and YFP-Core, CFP-NS2 and YFP-Core, CFP-p7 and YFP-Core, and CFP-p7 and YFP-E1 only exerted FRET in 293T cells.
In sum, we established the intra-HCV interactome with FACS-FRET. Thereby we succeeded in confirming a variety of interactions in the context of living mammalian cells. Moreover, our novel approach revealed a set of previously unrecognized HCV protein interactions that might be important during viral replication.
Residues RR33 and RR35 in p7 Mediate Interaction with E2
It has been reported that p7 is important for HCV production and that mutation of a dibasic motif RR33/35QQ in HCV-JFH1 p7 disrupts virus production without a defect at the level of E2-p7-NS2 processing (17). However, the molecular determinants for the defect of the p7-RR/QQ mutant are still elusive, and we hypothesized that the mutation might affect intraviral p7 protein interactions. We thus changed the arginines at positions 33 and 35 in JFH1 p7-CFP/YFP to glutamine. Next, we investigated the multimerization of the p7-RR/QQ mutant and wild-type JFH-1 p7, as well as their interaction with Core, E1, and E2, via FACS-FRET (Fig. 5). Changing the dibasic R motif to Q slightly—albeit significantly—affected the interaction of p7 with Core and E1, as well as oligomerization (see the primary FACS plots in Fig. 5A and the quantitative analyses of multiple replicates in Fig. 5B). In contrast, mutation of p7 RR33/35QQ strongly disrupted p7 interaction with E2 (Fig. 5).
Fig. 5.
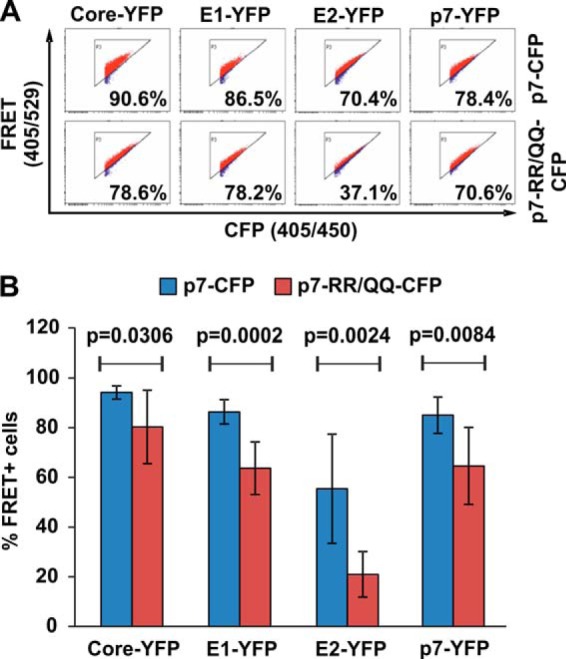
JFH1 p7-RR33/35QQ differentially interacts with Core, E1, and E2. A, primary FACS-FRET plots of 293T cells transfected with the indicated CFP and YFP fusion proteins. Please note that for p7 self-interaction we cotransfected either the p7-CFP and p7-YFP plasmid or the p7-RR/QQ-CFP together with the p7-RR/QQ-YFP. B, mean values of the percentage of FRET+ cells and standard deviations of seven independent transfections performed as indicated in A. For calculation of the statistical significance, mean FRET signals were compared with the two-tailed unpaired Student's t test (Graph Pad Prism 5.0).
We concluded that the dibasic arginine motif at positions 33 and 35 in JFH1 p7 might be involved in interaction with E2. Moreover, these data demonstrate that our system can contribute to the elucidation of molecular determinants of virological phenotypes.
Detection of p7 Binding to Core, E1, and E2 via CoIP
For FACS-FRET, viral proteins were fused with chromophores and overexpressed in 293T or Huh7.5 liver cells. Previously, we were able to confirm in a variety of studies that positive FACS-FRET results can be verified by biochemical methods and with untagged proteins (6, 18–20). Nevertheless, in the context of a dynamic viral infection, expression levels might vary substantially, and other viral proteins might affect specific interactions.
We aimed to verify interactions discovered via FACS-FRET in HCV expressing Huh7.5 cells. As far as we know, no specific antibodies are available for detection of either E1 or p7 from JFH1 or Jc1. However, it is possible to stain the E1-A4 epitope derived from HCV-H77 via immunoblot (8). Thus, we reconstituted the E1-A4 sequence in the HCV-Jc1 backbone and the HCV-Jc1 HA-p7 construct (9) in order to be able to analyze E1 and p7 with biochemical techniques.
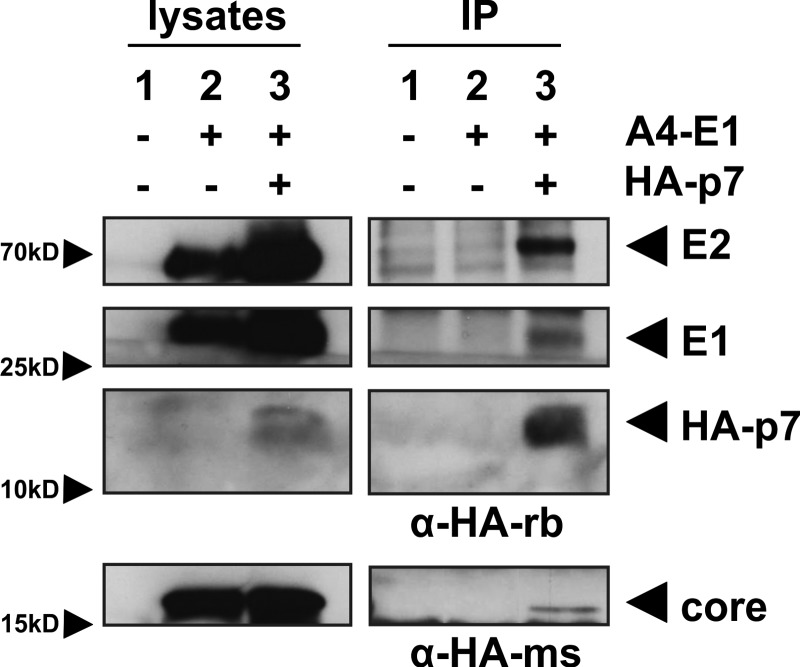
To achieve high expression levels, Huh7.5 cells were electroporated with in vitro transcribed RNA of the respective construct. Quantification of electroporation efficiency via intracellular FACS staining confirmed that generally more than 40% of cells were HCV positive (not shown). CoIP with an anti-HA antibody from rabbit revealed specific interaction of p7 with E2 and E1 (Fig. 6, upper bands). Unfortunately, the migration pattern of the light chain of this antibody did not allow detection of Core at ∼21 kDa. Thus, we repeated the immunoprecipitation with a mouse-derived anti-HA antibody and were then able to detect specific interaction of HA-p7 with Core (Fig. 6, lower band). These results demonstrate that p7 interacts with the HCV structural proteins in liver cells expressing fully infectious HCV.
Fig. 6.
Viral structural proteins co-immunoprecipitate with p7 in lysates of HCV expressing Huh7.5 cells. Huh7.5 cells were RNA electroporated with (1) no RNA, (2) HCV Jc1-E1(A4), or (3) HCV Jc1-E1(A4)-p7(HA), allowing detection of E1 by the A4 antibody (8) and p7 with an anti-HA antibody. Then p7(HA) was immunoprecipitated with a rabbit-derived anti-HA antibody (upper bands) or a mouse-derived anti-HA antibody (lower bands). Core, E1, E2, and p7 were detected in lysates and post-precipitation via immunoblotting.
The HCV Structural Proteins Interact with p7 in Virus Expressing Intact Huh7.5 Liver Cells
FRET allows the detection of transient dynamic interactions in the physiological environment of the cell (6, 12). Because two putative interaction partners need to be in close proximity for robust FRET—usually less than 10 nm apart—there has to be a substantial degree of co-localization between two binding partners. We thus investigated the co-localization of HCV proteins in virus expressing Huh7.5 cells.
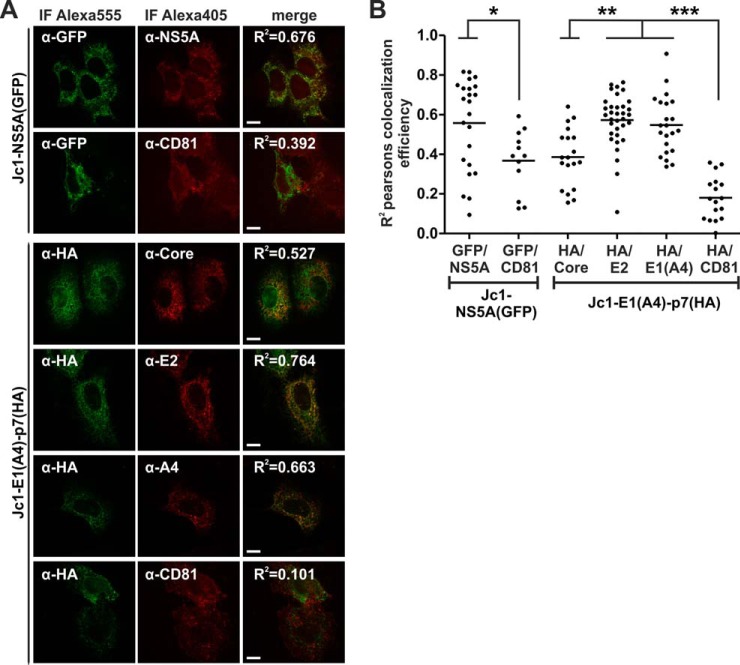
As a positive control, we electroporated Huh7.5 cells with the HCV Jc1-NS5A(GFP) construct expressing an NS5A-GFP fusion protein (10). Then we did immunofluorescence staining with NS5A and GFP specific antibodies, necessarily reflecting a high degree of co-localization quantified by the squared Pearson's co-localization coefficient (R2 = 0.676, Fig. 7A, and mean R2 = 0.5581 ± 0.0479 S.E., Fig. 7B). In contrast, we were surprised to see some co-localization between NS5A-GFP and CD81, although R2 values were significantly lower (see Figs. 7A and 7B).
Fig. 7.
Viral structural proteins co-localize with p7 in HCV expressing Huh7.5 cells. Huh7.5 cells were RNA electroporated with the indicated HCV constructs. Then immunofluorescence staining with specific antibodies was performed to detect areas of protein co-localization by means of confocal microscopy. We quantified co-localization using Costes Pearson's correlation (84). A, examples of confocal images. The squared Pearson's correlation coefficient (R2) is indicated in the merged image. The scale bar has a length of 5 μm. B, each dot represents the R2 correlation for one analyzed cell. Mean values of at least 12 measured cells per co-localization analysis were plotted and assessed for significant differences with one-way analysis of variance (Graph Pad Prism 5.0): p < 0.01 (*), p < 0.001 (**), and p < 0.0001 (***).
Next we used the Jc1-E1(A4)-p7(HA) variant to quantify co-localization between p7 via HA staining and the HCV structural proteins. There was a complete absence of co-localization between p7(HA) and CD81 (mean R2 = 0.1804 ± 0.0264 S.E.). In contrast, Core, E2, and E1(A4) co-localized with p7(HA), and the mean R2 values were significantly greater than those measured for p7(HA) and CD81 (see Figs. 7A and 7B; mean R2 values were 0.3859 for p7(HA)/Core, 0.5731 for p7(HA)/E2, and 0.5476 for p7(HA)/E1(A4), respectively). Thus, the viral structural proteins co-localized with p7 in HCV expressing Huh7.5 cells.
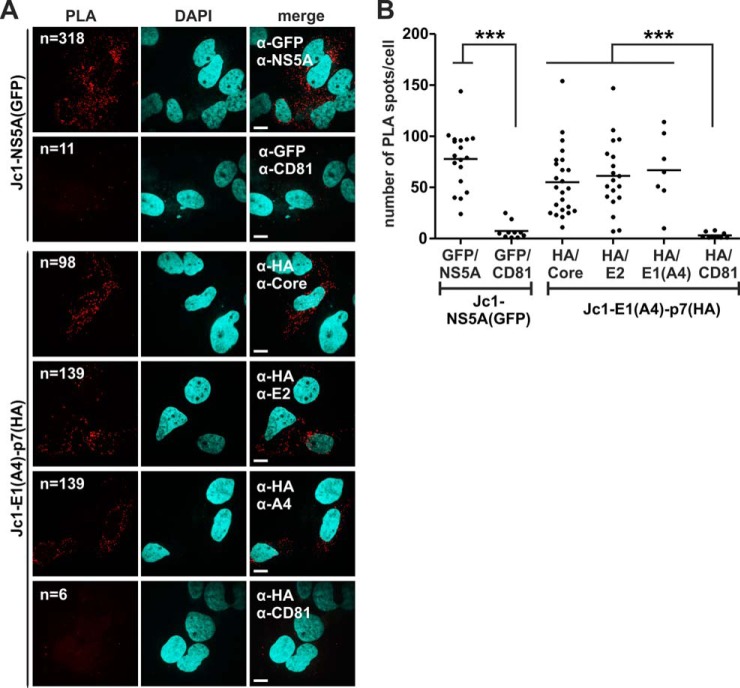
Co-localization indicates the presence of two putative binding partners in the same subcellular region and is a hint, but not proof, of direct interaction. We therefore decided to exploit PLA (21) to demonstrate direct interactions of the viral proteins in intact cells. PLA can be done with the same primary antibodies that were used for immunofluorescence. When these are in close proximity, the secondary PLA antibodies trigger an enzymatic reaction followed by a rolling circle amplification of fluorescent oligonucleotide probes. Thus, when two proteins interact within a cell, a bright fluorescent spot visualizes this event. We first did control stainings in Huh7.5 cells with the single primary antibodies and the PLA probes to exclude unspecific binding, which did not result in detectable PLA spots (data not shown). We then repeated the setting of the co-localization experiment presented in Fig. 7 but used PLA probes as secondary antibodies. As a readout, we counted the amount of PLA spots per cell and analyzed a minimum of 10 cells per staining (Fig. 8). The positive control, which was detection of the NS5A-GFP fusion by specific NS5A and GFP antibodies, resulted in an average number of 78 spots per cell, whereas NS5A-GFP showed no PLA signal with CD81 (Figs. 8A and 8B). Thus, despite a certain degree of co-localization between NS5A-GFP and CD81 (compare Fig. 7), the negative PLA result argued against the direct interaction of these two proteins. In contrast, all structural HCV proteins gave high PLA signals with p7(HA) that were in the range of the positive control (see Figs. 8A and 8B; the mean number of PLA spots per cell was 55 for p7(HA)/Core, 61 for p7(HA)/E2, and 67 for p7(HA)/E1(A4)). Importantly, the negative control, which was PLA measured between p7(HA) and CD81, gave only background signals (three spots per cell), and calculated differences were highly significant (Fig. 8B).
Fig. 8.
Viral structural proteins interact with p7 in HCV expressing intact Huh7.5 cells. Huh7.5 cells were RNA electroporated with the indicated HCV constructs. Then staining for proximity ligation assay (PLA) was done with the same antibodies that were used for co-localization analyses (compare with Fig. 7). We quantified positive PLA events through software-based counting of red dots and normalized the resulting value to the number of cells (nuclear DAPI staining). A, representative examples of PLA images. Indicated are the antibodies used for the primary PLA stain and the number of PLA dots identified by counting software. The scale bar has a length of 5 μm. B, each dot represents the number of PLA spots counted for one cell per image. Mean values of at least eight measurements per antibody pair were plotted and assessed for significant differences with one-way analysis of variance (Graph Pad Prism 5.0): p < 0.01 (*), p < 0.001 (**), and p < 0.0001 (***).
Together, the cumulative data from the co-localization and PLA experiments strongly suggest that HCV p7 interacts with the viral structural proteins Core, E1, and E2 in intact and HCV expressing Huh7.5 liver cells.
DISCUSSION
We performed a comprehensive assessment of intra-HCV protein interactions with a FACS-based FRET assay in intact cells. This analysis revealed a set of 20 protein–protein interactions that met our stringency threshold and thus exerted robust FRET. Through thorough literature mining, we found that 13 interactions were previously described with alternative techniques (compare Fig. 3). Therefore, we have confirmed for the first time their association in living cells. In addition, the seven protein interaction pairs newly discovered by us comprise mainly structural proteins. HCV p7 binds to Core, E1, and E2, and interaction of p7 with E2 seems important for virus production. Furthermore, Core was found to exert FRET with E2 and NS2, and this whole complex might have a pivotal role in HCV assembly and egress. In addition, we report significant FRET for NS5B together with E1 and E2. From a biological point of view, the possible importance of the latter interactions remains elusive.
FACS-FRET has a variety of striking advantages that render this technique superior to other methods for the detection of protein interactions (6, 22). Nevertheless, one serious constraint is the necessity of using fusion proteins with chromophores. These tags can impair the functionality and expression of the native proteins. We extensively tested expression levels via FACS and Western blot and checked the subcellular localization of the transfected fusions. HCV Core, E1, and E2 with a C-terminal chromophore tag were not expressed at all, and as expected, expression levels of the N-terminal fusions varied (see Fig. 1 for details). However, the specific localization to distinct subcellular compartments indicated that the HCV fusion proteins were properly expressed, although we cannot exclude impairments in functionality. At least for the nonstructural proteins, a large body of published data on tagged HCV fusion proteins from work employing HIS, FLAG, HA, GST, and GFP tags supports no major functional impairments as a consequence of the tag (9, 10, 16, 23–29).
In contrast to the various in situ and precipitation techniques, FRET works in living and intact cells, supporting interactions in the physiological conditions of the cellular environment. Furthermore, proteins bind each other in their natural subcellular compartment, which is a clear advantage in comparison to yeast two-hybrid systems (6). Nevertheless, cellular proteins are important determinants for HCV replication, and we have to consider the possibility that liver cell specific factors also influence intraviral protein interactions (3, 5). We thus conducted experiments in 293T and Huh7.5 cells and revealed in general a significant correlation between the results for the two cell types. In line with this, it was recently demonstrated that the exogenous expression of HCV entry receptors, the microRNA miR122, and apolipoprotein E is sufficient to achieve completion of the whole HCV life cycle in 293T cells (30). We therefore postulate that major differences in FRET, which we observed occasionally between 293T and Huh7.5 cells (compare Fig. 4B), are most likely attributable to the lower levels of protein expression in the liver cell line.
Another problem we encountered was the major differences in the percentages of FRET+ cells depending on the usage of CFP and YFP as either donor or acceptor. One extreme example is the well-established interaction between the glycoproteins E1 and E2 (8, 13, 31). Assessing FRET between CFP-E2 and YFP-E1 resulted in 74.8% FRET+ cells, whereas transfection of CFP-E1 together with YFP-E2 did not give any FRET at all (Fig. 2, supplemental Fig. S1). This phenomenon has been described before and can be explained by the donor:acceptor stoichiometry (32, 33). FRET is generally more efficient when there is an excess of acceptor molecules. Therefore, a ratio of 1:2 gives higher FRET than 2:1 (32). In addition, Koushik and colleagues performed elegant experiments demonstrating continuously higher FRET correlating with increasing amounts of acceptor (33). Thus, we generally tested both possible donor–acceptor combinations to eliminate a potential loss of FRET as a reason for different donor:acceptor quantities.
Apart from the restrictions addressed and discussed above, we postulate that FACS-based FRET is currently one of the gold-standard techniques for investigating protein interactions. We established and used this technique in the past to demonstrate and map, for instance, the interaction of HIV-1 Vpu and Ebola GP2 with the antiviral factor Tetherin/Bst-2 (6, 18, 19) and to show the interplay between HIV-1 Gag and tetraspanins (34). Also, other groups used our FACS-FRET approach to show direct protein interactions, confirming independently the robustness of the assay (35–38). Of note, Kim and coworkers systematically compared FACS-FRET, bioluminescence resonance energy transfer, and fluorescence lifetime imaging microscopy (22). In this study, the same potential interaction partners of the amyloid precursor protein were studied using all three different methods. They concluded that FACS-FRET is the most sensitive and reliable approach for investigating protein interactions with a superior Z-score as an indicator for high-throughput screening compatibility.
Nevertheless, we analyzed the results from our FACS-FRET approach with high stringency. Experiments were performed in two cell lines, and an extensive number of biological replicates were conducted by different individuals. Subsequently, statistics were used to assess the significance of identified interactions versus the negative control. Furthermore, an additional stringency threshold of 10% was introduced. Only percentages of FRET+ cells higher than 10% were considered as “true.” Thus, the interactions discussed and presented have high statistical confidence. The reliability of the whole screening approach is also reflected in the fact that we confirmed three of the seven newly found interactions with alternative techniques. Strikingly, interaction of p7 with Core, E1, and E2 could be demonstrated by CoIP, immunofluorescence, and PLA in Huh7.5 liver cells expressing replicating and fully infectious HCV. Apart from that, E1 and E2 showed significant FRET with NS5B in 293T and Huh7.5 cells (Fig. 4A). Indeed, interaction of NS5B with the two glycoproteins and Core, which we also found via FACS-FRET, could participate in correct coordination of assembly and RNA genome incorporation. However, this hypothesis has to be experimentally followed up in further experiments, although a recent study suggests a role of NS5B in HCV assembly (39).
It is remarkable that most of the interactions reported herein refer to structural proteins and NS2 and p7 (Figs. 3 and 9). In contrast, we detected nearly no FRET between the nonstructural proteins as well as the nonstructural and structural proteins. The discrepancy with regard to published nonstructural interactions could be explained by the techniques used. In contrast to the previously exploited CoIP, FRET cannot detect higher molecular complexes because of the increasing Foerster's radius (12, 40). Thus it is tempting to speculate that the HCV nonstructural proteins form higher molecular complexes among themselves and with the structural proteins, whereas the structural proteins directly interact with each other. This further emphasizes that the interactions newly described herein might be critically involved in HCV assembly. In this context, direct binding of p7 and Core was postulated before (41), and a variety of hints regarding interaction of these proteins are published in the literature (42–44). For instance, Popescu and colleagues reported that Core, the envelope proteins, and p7 influence subcellular NS2 localization (23). Using FACS-FRET, CoIP, and PLA, we were able to demonstrate for the first time that there is a direct interaction between p7 and Core, as well as between NS2 and Core. These findings could give a functional rationale for previously published data. Furthermore, it was suggested that E2 and p7 might interact (9, 44, 45), and Gentzsch and colleagues postulated that the p7-RR/QQ mutant has a defect at the step of capsid envelopment. Strikingly, our data suggest that mutation of the dibasic motif in p7 selectively impairs the interaction with E2 (44). Thus, our FACS-FRET analyses provide a mechanistic explanation for the virological phenotype of this mutant.
Incorporating interaction data from the VirusMINT database and the work by de Chassey and colleagues (4) into our intra-HCV protein interaction network leads to suggestions of the existence of three major nodes HCV uses to manipulate the host cell (Fig. 9). As already pointed out, most proteins might indirectly interact via higher molecular complexes. However, the direct interaction network of Core, E1, E2, p7, NS2, and NS5B is connected to the host cell mainly by the multiple virus–host interactions exerted by Core. Two other clusters comprise the NS3:NS4 protease with NS3 as the main communicator with host factors and the HCV NS5A, which is an important cofactor for HCV replication (46, 47). Thus, the single viral proteins Core, NS3, and NS5A within the direct interaction clusters could serve as interfaces for HCV-mediated manipulation of the host cell.
In sum, we postulate that the newly described interactions herein contribute to the formation of an HCV assembly network. In addition, our data underline and support the pivotal role of p7 in HCV morphogenesis (9, 17, 41–44, 48, 49). In the future, it will be of high interest to use FACS-FRET in order to map interaction domains important for the diverse intra-HCV protein interactions and to correlate mutations in viral proteins with virological parameters, as demonstrated here for the p7-RR/QQ mutant. Similar experiments could be done for patient-derived and thus evolutionarily shaped viral proteins. Such approaches will shed light on the biological relevance of the diverse intra-HCV protein interactions and are highly warranted.
Supplementary Material
Acknowledgments
We thank Ulrike Protzer for constant support and encouragement, Ruth Brack-Werner for helpful discussions, and Eike Steinmann and Eva Herker for critical reading of the manuscript. In addition, we are very grateful to Thomas Pietschmann, Ralf Bartenschlager, Charles Rice, Takaji Wakita, Harry Greenberg, and Jean Dubuisson for the generous contribution of reagents.
Footnotes
Author contributions: N.H., K.B., and M.S. designed research; N.H., K.B., and K.R. performed research; N.H., K.B., and M.S. contributed new reagents or analytic tools; N.H., K.B., and M.S. analyzed data; M.S. wrote the paper; M.S. planned and conceived the overall study.
* Nicole Hagen was supported by a scholarship from the Structure and Dynamics in Infection (SDI) Graduate School Hamburg and a stipend from Pro Exzellenzia (Hamburger Hochschulen für Frauen). This work was supported by institutional funding received from the Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg and the Helmholtz Zentrum Munich, German Research Center for Environmental Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- HCV
- hepatitis C virus
- CoIP
- co-immunoprecipitation
- PLA
- proximity ligation assay
- MF
- mean value of the percentage of cells scoring FRET+.
REFERENCES
- 1. Moradpour D., Penin F., Rice C. M. (2007) Replication of hepatitis C virus. Nat. Rev. Microbiol. 5, 453–463 [DOI] [PubMed] [Google Scholar]
- 2. Kronenberger B., Zeuzem S. (2012) New developments in HCV therapy. J. Viral Hepatitis 19 Suppl 1, 48–51 [DOI] [PubMed] [Google Scholar]
- 3. Tai A. W., Benita Y., Peng L. F., Kim S. S., Sakamoto N., Xavier R. J., Chung R. T. (2009) A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Chassey B., Navratil V., Tafforeau L., Hiet M. S., Aublin-Gex A., Agaugue S., Meiffren G., Pradezynski F., Faria B. F., Chantier T., Le Breton M., Pellet J., Davoust N., Mangeot P. E., Chaboud A., Penin F., Jacob Y., Vidalain P. O., Vidal M., Andre P., Rabourdin-Combe C., Lotteau V. (2008) Hepatitis C virus infection protein network. Mol. Syst. Biol. 4, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Q., Brass A. L., Ng A., Hu Z., Xavier R. J., Liang T. J., Elledge S. J. (2009) A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U.S.A. 106, 16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banning C., Votteler J., Hoffmann D., Koppensteiner H., Warmer M., Reimer R., Kirchhoff F., Schubert U., Hauber J., Schindler M. (2010) A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5, e9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Krausslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubuisson J., Hsu H. H., Cheung R. C., Greenberg H. B., Russell D. G., Rice C. M. (1994) Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68, 6147–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vieyres G., Brohm C., Friesland M., Gentzsch J., Wolk B., Roingeard P., Steinmann E., Pietschmann T. (2013) Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 87, 1664–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schaller T., Appel N., Koutsoudakis G., Kallis S., Lohmann V., Pietschmann T., Bartenschlager R. (2007) Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81, 4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blight K. J., McKeating J. A., Rice C. M. (2002) Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76, 13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siegel R. M., Chan F. K., Zacharias D. A., Swofford R., Holmes K. L., Tsien R. Y., Lenardo M. J. (2000) Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci. STKE 2000, pl1. [DOI] [PubMed] [Google Scholar]
- 13. Ralston R., Thudium K., Berger K., Kuo C., Gervase B., Hall J., Selby M., Kuo G., Houghton M., Choo Q. L. (1993) Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J. Virol. 67, 6753–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsumoto M., Hwang S. B., Jeng K. S., Zhu N., Lai M. M. (1996) Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218, 43–51 [DOI] [PubMed] [Google Scholar]
- 15. Mousseau G., Kota S., Takahashi V., Frick D. N., Strosberg A. D. (2011) Dimerization-driven interaction of hepatitis C virus core protein with NS3 helicase. J. Gen. Virol. 92, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspe G., Tiollais P., Transy C., Legrain P. (2000) A genomic approach of the hepatitis C virus generates a protein interaction map. Gene 242, 369–379 [DOI] [PubMed] [Google Scholar]
- 17. Steinmann E., Penin F., Kallis S., Patel A. H., Bartenschlager R., Pietschmann T. (2007) Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bolduan S., Votteler J., Lodermeyer V., Greiner T., Koppensteiner H., Schindler M., Thiel G., Schubert U. (2011) Ion channel activity of HIV-1 Vpu is dispensable for counteraction of CD317. Virology 416, 75–85 [DOI] [PubMed] [Google Scholar]
- 19. Kuhl A., Banning C., Marzi A., Votteler J., Steffen I., Bertram S., Glowacka I., Konrad A., Sturzl M., Guo J. T., Schubert U., Feldmann H., Behrens G., Schindler M., Pohlmann S. (2011) The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J. Infect. Dis. 204 Suppl 3, S850–S860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoffmann D., Schwarck D., Banning C., Brenner M., Mariyanna L., Krepstakies M., Schindler M., Millar D. P., Hauber J. (2012) Formation of trans-activation competent HIV-1 Rev:RRE complexes requires the recruitment of multiple protein activation domains. PLoS One 7, e38305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S. M., Ostman A., Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 [DOI] [PubMed] [Google Scholar]
- 22. Kim J., Lee J., Kwon D., Lee H., Grailhe R. (2011) A comparative analysis of resonance energy transfer methods for Alzheimer related protein-protein interactions in living cells. Mol. Biosyst. 7, 2991–2996 [DOI] [PubMed] [Google Scholar]
- 23. Popescu C. I., Callens N., Trinel D., Roingeard P., Moradpour D., Descamps V., Duverlie G., Penin F., Heliot L., Rouille Y., Dubuisson J. (2011) NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7, e1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frick D. N., Rypma R. S., Lam A. M., Gu B. (2004) The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J. Biol. Chem. 279, 1269–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma Y., Anantpadma M., Timpe J. M., Shanmugam S., Singh S. M., Lemon S. M., Yi M. (2011) Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dimitrova M., Imbert I., Kieny M. P., Schuster C. (2003) Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77, 5401–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cho H. S., Ha N. C., Kang L. W., Chung K. M., Back S. H., Jang S. K., Oh B. H. (1998) Crystal structure of RNA helicase from genotype 1b hepatitis C virus. A feasible mechanism of unwinding duplex RNA. J. Biol. Chem. 273, 15045–15052 [DOI] [PubMed] [Google Scholar]
- 28. Shimakami T., Hijikata M., Luo H., Ma Y. Y., Kaneko S., Shimotohno K., Murakami S. (2004) Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78, 2738–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolk B., Buchele B., Moradpour D., Rice C. M. (2008) A dynamic view of hepatitis C virus replication complexes. J. Virol. 82, 10519–10531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Da Costa D., Turek M., Felmlee D. J., Girardi E., Pfeffer S., Long G., Bartenschlager R., Zeisel M. B., Baumert T. F. (2012) Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. J. Virol. 86, 11919–11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yi M., Nakamoto Y., Kaneko S., Yamashita T., Murakami S. (1997) Delineation of regions important for heteromeric association of hepatitis C virus E1 and E2. Virology 231, 119–129 [DOI] [PubMed] [Google Scholar]
- 32. Chen H., Puhl H. L., 3rd, Koushik S. V., Vogel S. S., Ikeda S. R. (2006) Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys. J. 91, L39–L41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koushik S. V., Blank P. S., Vogel S. S. (2009) Anomalous surplus energy transfer observed with multiple FRET acceptors. PLoS One 4, e8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koppensteiner H., Banning C., Schneider C., Hohenberg H., Schindler M. (2012) Macrophage internal HIV-1 is protected from neutralizing antibodies. J. Virol. 86, 2826–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thyrock A., Stehling M., Waschbusch D., Barnekow A. (2010) Characterizing the interaction between the Rab6 GTPase and Mint3 via flow cytometry based FRET analysis. Biochem. Biophys. Res. Commun. 396, 679–683 [DOI] [PubMed] [Google Scholar]
- 36. Somvanshi R. K., Chaudhari N., Qiu X., Kumar U. (2011) Heterodimerization of beta2 adrenergic receptor and somatostatin receptor 5: implications in modulation of signaling pathway. J. Mol. Signal. 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asbach B., Ludwig C., Saksela K., Wagner R. (2012) Comprehensive analysis of interactions between the Src-associated protein in mitosis of 68 kDa and the human Src-homology 3 proteome. PLoS One 7, e38540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hassinen A., Pujol F. M., Kokkonen N., Pieters C., Kihlstrom M., Korhonen K., Kellokumpu S. (2011) Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J. Biol. Chem. 286, 38329–38340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gouklani H., Bull R. A., Beyer C., Coulibaly F., Gowans E. J., Drummer H. E., Netter H. J., White P. A., Haqshenas G. (2012) Hepatitis C virus nonstructural protein 5B is involved in virus morphogenesis. J. Virol. 86, 5080–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Piston D. W., Kremers G. J. (2007) Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem. Sci. 32, 407–414 [DOI] [PubMed] [Google Scholar]
- 41. Boson B., Granio O., Bartenschlager R., Cosset F. L. (2011) A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 7, e1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gouklani H., Beyer C., Drummer H., Gowans E. J., Netter H. J., Haqshenas G. (2013) Identification of specific regions in hepatitis C virus core, NS2 and NS5A that genetically interact with p7 and coordinate infectious virus production. J. Viral Hepatitis 20, e66–e71 [DOI] [PubMed] [Google Scholar]
- 43. Bentham M. J., Foster T. L., McCormick C., Griffin S. (2013) Mutations in hepatitis C virus p7 reduce both the egress and infectivity of assembled particles via impaired proton channel function. J. Gen. Virol. 94, 2236–2248 [DOI] [PubMed] [Google Scholar]
- 44. Gentzsch J., Brohm C., Steinmann E., Friesland M., Menzel N., Vieyres G., Perin P. M., Frentzen A., Kaderali L., Pietschmann T. (2013) Hepatitis C virus p7 is critical for capsid assembly and envelopment. PLoS Pathog. 9, e1003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atoom A. M., Jones D. M., Russell R. S. (2013) Evidence suggesting that HCV p7 protects E2 glycoprotein from premature degradation during virus production. Virus Res. 176, 199–210 [DOI] [PubMed] [Google Scholar]
- 46. Tellinghuisen T. L., Foss K. L., Treadaway J. (2008) Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4, e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang L., Hwang J., Sharma S. D., Hargittai M. R., Chen Y., Arnold J. J., Raney K. D., Cameron C. E. (2005) Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280, 36417–36428 [DOI] [PubMed] [Google Scholar]
- 48. Wozniak A. L., Griffin S., Rowlands D., Harris M., Yi M., Lemon S. M., Weinman S. A. (2010) Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6, e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brohm C., Steinmann E., Friesland M., Lorenz I. C., Patel A., Penin F., Bartenschlager R., Pietschmann T. (2009) Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 83, 11682–11693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Serebrov V., Pyle A. M. (2004) Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430, 476–480 [DOI] [PubMed] [Google Scholar]
- 51. Locatelli G. A., Spadari S., Maga G. (2002) Hepatitis C virus NS3 ATPase/helicase: an ATP switch regulates the cooperativity among the different substrate binding sites. Biochemistry 41, 10332–10342 [DOI] [PubMed] [Google Scholar]
- 52. Luik P., Chew C., Aittoniemi J., Chang J., Wentworth P., Jr., Dwek R. A., Biggin P. C., Venien-Bryan C., Zitzmann N. (2009) The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 12712–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Welker M. W., Welsch C., Meyer A., Antes I., Albrecht M., Forestier N., Kronenberger B., Lengauer T., Piiper A., Zeuzem S., Sarrazin C. (2010) Dimerization of the hepatitis C virus nonstructural protein 4B depends on the integrity of an aminoterminal basic leucine zipper. Protein Sci. 19, 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Merola M., Brazzoli M., Cocchiarella F., Heile J. M., Helenius A., Weiner A. J., Houghton M., Abrignani S. (2001) Folding of hepatitis C virus E1 glycoprotein in a cell-free system. J. Virol. 75, 11205–11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lo S. Y., Selby M. J., Ou J. H. (1996) Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70, 5177–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. (1993) Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lanford R. E., Notvall L., Chavez D., White R., Frenzel G., Simonsen C., Kim J. (1993) Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology 197, 225–235 [DOI] [PubMed] [Google Scholar]
- 58. Whidby J., Mateu G., Scarborough H., Demeler B., Grakoui A., Marcotrigiano J. (2009) Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J. Virol. 83, 11078–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clarke D., Griffin S., Beales L., Gelais C. S., Burgess S., Harris M., Rowlands D. (2006) Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 281, 37057–37068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stapleford K. A., Lindenbach B. D. (2011) Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85, 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tedbury P., Welbourn S., Pause A., King B., Griffin S., Harris M. (2011) The subcellular localization of the hepatitis C virus non-structural protein NS2 is regulated by an ion channel-independent function of the p7 protein. J. Gen. Virol. 92, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lorenz I. C., Marcotrigiano J., Dentzer T. G., Rice C. M. (2006) Structure of the catalytic domain of the hepatitis C virus NS2–3 protease. Nature 442, 831–835 [DOI] [PubMed] [Google Scholar]
- 63. Jones D. M., Atoom A. M., Zhang X., Kottilil S., Russell R. S. (2011) A genetic interaction between the core and NS3 proteins of hepatitis C virus is essential for production of infectious virus. J. Virol. 85, 12351–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schregel V., Jacobi S., Penin F., Tautz N. (2009) Hepatitis C virus NS2 is a protease stimulated by cofactor domains in NS3. Proc. Natl. Acad. Sci. U.S.A. 106, 5342–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gallinari P., Brennan D., Nardi C., Brunetti M., Tomei L., Steinkuhler C., De Francesco R. (1998) Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J. Virol. 72, 6758–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bartenschlager R., Lohmann V., Wilkinson T., Koch J. O. (1995) Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Failla C., Tomei L., De Francesco R. (1994) Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68, 3753–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin C., Pragai B. M., Grakoui A., Xu J., Rice C. M. (1994) Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68, 8147–8157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tanji Y., Hijikata M., Satoh S., Kaneko T., Shimotohno K. (1995) Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69, 1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Paredes A. M., Blight K. J. (2008) A genetic interaction between hepatitis C virus NS4B and NS3 is important for RNA replication. J. Virol. 82, 10671–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lin C., Wu J. W., Hsiao K., Su M. S. (1997) The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 71, 6465–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu G. Y., Lee K. J., Gao L., Lai M. M. (2006) Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80, 6013–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Masaki T., Suzuki R., Murakami K., Aizaki H., Ishii K., Murayama A., Date T., Matsuura Y., Miyamura T., Wakita T., Suzuki T. (2008) Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82, 7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 75. Goh P. Y., Tan Y. J., Lim S. P., Lim S. G., Tan Y. H., Hong W. J. (2001) The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290, 224–236 [DOI] [PubMed] [Google Scholar]
- 76. Lundin M., Lindstrom H., Gronwall C., Persson M. A. (2006) Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J. Gen. Virol. 87, 3263–3272 [DOI] [PubMed] [Google Scholar]
- 77. Love R. A., Brodsky O., Hickey M. J., Wells P. A., Cronin C. N. (2009) Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83, 4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tellinghuisen T. L., Marcotrigiano J., Rice C. M. (2005) Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435, 374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Uchida M., Hino N., Yamanaka T., Fukushima H., Imanishi T., Uchiyama Y., Kodama T., Doi T. (2002) Hepatitis C virus core protein binds to a C-terminal region of NS5B RNA polymerase. Hepatol. Res. 22, 297–306 [DOI] [PubMed] [Google Scholar]
- 80. Kang S. M., Choi J. K., Kim S. J., Kim J. H., Ahn D. G., Oh J. W. (2009) Regulation of hepatitis C virus replication by the core protein through its interaction with viral RNA polymerase. Biochem. Biophys. Res. Commun. 386, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang C., Cai Z., Kim Y. C., Kumar R., Yuan F., Shi P. Y., Kao C., Luo G. (2005) Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J. Virol. 79, 8687–8697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ishido S., Fujita T., Hotta H. (1998) Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244, 35–40 [DOI] [PubMed] [Google Scholar]
- 83. Shirota Y., Luo H., Qin W., Kaneko S., Yamashita T., Kobayashi K., Murakami S. (2002) Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277, 11149–11155 [DOI] [PubMed] [Google Scholar]
- 84. Costes S. V., Daelemans D., Cho E. H., Dobbin Z., Pavlakis G., Lockett S. (2004) Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86, 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Lotia S., Pico A. R., Bader G. D., Ideker T. (2012) A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.