Abstract
Ferritin heavy chain (FTH1) is a 21-kDa subunit of the ferritin complex, known for its role in iron metabolism, and which has recently been identified as a favorable prognostic protein for triple negative breast cancer (TNBC) patients. Currently, it is not well understood how FTH1 contributes to an anti-tumor response. Here, we explored whether expression and cellular compartmentalization of FTH1 correlates to an effective immune response in TNBC patients. Analysis of the tumor tissue transcriptome, complemented with in silico pathway analysis, revealed that FTH1 was an integral part of an immunomodulatory network of cytokine signaling, adaptive immunity, and cell death. These findings were confirmed using mass spectrometry (MS)-derived proteomic data, and immunohistochemical staining of tissue microarrays. We observed that FTH1 is localized in both the cytoplasm and/or nucleus of cancer cells. However, high cytoplasmic (c) FTH1 was associated with favorable prognosis (Log-rank p = 0.001), whereas nuclear (n) FTH1 staining was associated with adverse prognosis (Log-rank p = 0.019). cFTH1 staining significantly correlated with total FTH1 expression in TNBC tissue samples, as measured by MS analysis (Rs = 0.473, p = 0.0007), but nFTH1 staining did not (Rs = 0.197, p = 0.1801). Notably, IFN γ-producing CD8+ effector T cells, but not CD4+ T cells, were preferentially enriched in tumors with high expression of cFTH1 (p = 0.02). Collectively, our data provide evidence toward new immune regulatory properties of FTH1 in TNBC, which may facilitate development of novel therapeutic targets.
Triple negative breast cancer (TNBC)1 is a specific subtype of breast cancer, which lacks expression of estrogen receptor α (ESR1), progesterone receptor (PGR) and human epidermal growth factor receptor 2 (ERBB2). The incidence of TNBC accounts for ∼15% of all the breast cancer cases. It represents one of the most aggressive breast cancer subtypes, of which ∼30% of patients develop distant metastasis within 5 years after surgical removal of primary tumors (1). To date, no targeted therapy is available for TNBC patients. Therefore, the majority of TNBC patients is recommended to receive standard adjuvant chemotherapy, even when this is not beneficial to these patients. To reduce unnecessary administration of adjuvant chemotherapy, multiple prognostic signatures have been developed using gene expression (2–4) or proteomic techniques (5). These signatures are not only of clinical importance, but also implicate the underlying mechanisms of TNBC disease progression.
Modern mass spectrometry (MS) techniques facilitate clinical proteomic research as well as functional biochemical research. On one hand, hundreds of protein biomarkers can be identified for various diseases in a high throughput manner, which largely accelerates discovery of prognostic and predictive markers for clinical use (6–8). On the other hand, generated MS data sets can also be used to interpret molecular features of diseases and functions of protein markers (9, 10). Because proteins are the actual functional units, quantification of protein changes provides valuable functional evidence of disease phenotypes. However, routinely used MS based proteomic techniques enable quantification of limited numbers of proteins, covering only part of the human proteome (11). Therefore, interpretation of proteomic data needs to take into account analysis at the pathway level. A combination of genomic and proteomic approaches circumvents drawbacks of either technique and provides more confident biological insight into disease phenotypes. This approach uses genomic data to present global pathological events, and proteomic data to confirm changes with respect to potentially causative proteins.
Various molecular signatures for TNBC implicate positive immune regulation as a favorable prognostic feature in patients (2–4). This observation could be important to understand disease progression of TNBC, and also suggest that some of the immune regulatory proteins may serve as potential therapeutic targets for TNBC patients. Using nano-scale liquid chromatography hyphenated with high resolution tandem MS (nLC-MS/MS) technique, we previously identified an 11-protein signature that predicts prognosis of TNBC patients (5). Ferritin heavy chain (FTH1), one of the discriminatory proteins in this signature, has been suggested to display immunomodulatory functions. In order to better understand the role of immune-modulation in TNBC disease progression, potentially enhancing an effective anti-tumor immune response, we further investigated FTH1 function.
FTH1 is a 21 kDa subunit of the ferritin complex. The ferritin complex captures intracellular ferrous iron (Fe2+) and converts it into ferric iron (Fe3+) by the ferroxidase activity of FTH1, which potentially reduces DNA damage caused by Fe2+ induced reactive oxygen species (ROS) and protects cancer cells from cell death (12, 13). FTH1 interacts with some important pathways related to TNBC, such as the NK-κB pathway (14, 15) and apoptosis pathways (16), which indicates the importance of this protein in TNBC progression. Also, FTH1 has been suggested as an immunomodulatory protein in various other cancer types including melanoma (17) and non-TNBC (18). Interestingly, FTH1 expression is up-regulated in TNBC cell lines, especially in the chromatin-bound nuclear fraction of MDA-MB-231 (TNBC) in contrast to MCF-7 (non-TNBC) cell lines (13). Subcellular localization of this protein may be potentially important for TNBC disease progression. In the present study, we used a combined transcriptomic and proteomic approach together with immunohistochemistry (IHC) to investigate the function and subcellular localization of FTH1 in TNBC. Our data reveals a clear relationship between cytoplasmic localization of FTH1 and enhanced numbers of CD8+ but not CD4+ tumor-infiltrating T cells, which in turn is associated with less aggressive TNBC.
EXPERIMENTAL PROCEDURES
Transcriptomic and Proteomic Data Sets
Transcriptomic and proteomic data sets used in this study were previously published and can be accessed from public databases. The gene expression profiling data of 63 TNBC samples were extracted from publically available data set (Accession number: GSE2034, GSE5327) (19, 20). The global nLC-MS/MS data of 126 TNBC samples are available in ProteomeXchange database (Accession number: PXD000260) (5). A total of 47 samples were measured by both gene expression and nLC-MS/MS profiling.
Functional Analysis
Gene set enrichment analysis (GSEA) (21) was performed on gene expression data of all the 63 samples searched against TNBC subtype database (supplemental Table S1) constructed based on previously reported findings (22) and the Molecular Signatures Database (MSigDB) (version 3.1) (23). False discovery rates (FDRs) of enriched pathways were estimated based on 1,000 time permutation on defined phenotypes with fixed 149 seeds. Multiple probes assigned to the same gene were collapsed into single gene expression using the probes with the highest expression value for every tested sample. Genes were ranked using Student's t test when gene sets were compared between good and poor prognostic samples (dichotomized phenotype), and using Pearson correlation when gene sets were associated with expression of FTH1 (continuous phenotype). In accordance with previous findings (22), a less stringent cutoff of FDR<0.60 was used to enrich for biological pathways because of the batch effects of the two data set and intrinsic heterogeneity of TNBC.
We visualized the results of Gene Set Enrichment Analysis (21) from the gene expression data analyzed using EnrichmentMap software (24). Permissive p value cutoff of 0.05 and FDR cutoff of 0.25 were applied with MSigDB (version 3.1) (23) for the visualization, according to the online user guide (http://baderlab.org/Software/EnrichmentMap/UserManual).
To visualize gene and protein interaction map, we selected genes and proteins of which the expression significantly correlate with expression of FTH1 with a p value cutoff of 0.01 (Spearman's rank correlation) in transcriptomic and proteomic data sets, respectively. Subsequently, the gene list with corresponding correlation coefficients were submitted to IPA Knowledge Base using Ingenuity System (version 18030641), whereas protein interaction maps were visualized via STRING (version 9.1) (25). Correlation heat maps were created with an unsupervised hierarchical method with Euclidean distance and complete linkage using Cluster (version 3.0) (26).
Clustering Analysis of Proteomic Data Set
nLC-MS/MS data set normalized by “Combat” algorithm was used for clustering analysis. Proteins of “IFN γ pathway” (interferon gamma), “L1 pathway” (recycling of the cell adhesion molecular L1), “antigen presentation pathway,” and “apoptosis pathway” were clustered using an unsupervised hierarchical method with Euclidean distance and complete linkage. Based on heat map patterns, three clusters were defined for each pathway with high, medium or low expression values. These clusters were stratified with use of survival analyses in order to evaluate the association between pathways and patient prognosis.
Sample Selection
A tissue microarray (TMA) was constructed from a cohort of 412 primary TNBC tissues from local tissue bank. Triple negativity of formalin-fixed paraffin-embedded tissues was confirmed by IHC staining of ESR1, PGR and ERBB2. When IHC staining of ERBB2 protein was scored as 2+, fluorescence in situ hybridization (Dako Denmark A/S, Glostrup, Denmark) was used to assess amplification of the corresponding gene. For inclusion into the final analysis, we followed the same criteria as described in our previous study (5): All tissues were obtained from patients with negative lymph-node status, who did not receive systemic adjuvant therapy (n = 191). Twenty-six patients who had no distant metastasis were excluded because of insufficient time of clinical follow-up (<60 months). Of those patients who developed metastatic disease within 5 years after surgical removal of primary tumors, some patients (n = 10) had local relapse prior to distant metastasis. These patients were excluded from this study to limit the potential influence of local relapse on distant metastasis. An additional 8 tissues were excluded because of poor tissue quality. Therefore, a total of 147 samples were eligible for this study, including 39 poor prognostic patients who developed distant metastasis within 5 years after surgical removal of primary tumor. A subset of 48 samples were used for nLC-MS/MS profiling in our previous study (5). Therefore, this study can be defined as verification and pre-validation of TNBC biomarker FTH1. A detailed characterization of these 147 samples is available in Table I.
Table I. Clinical and pathological characteristics of patients and their tumor.
| Characteristics | TNBC cohort on TMA (n = 147)a |
|---|---|
| Age (years) | |
| Mean (S.D.) | 52.1 (13.3) |
| ≤40 | 34 (23.1%) |
| 41–55 | 50 (34.0%) |
| 56–70 | 48 (32.7%) |
| >70 | 15 (10.2%) |
| Menopausal status | |
| Premenopausal | 75 (51.0%) |
| Postmenopausal | 72 (49.0%) |
| Tumor size (cm) | |
| Mean (S.D.) | 2.5 (1.2) |
| pT1, ≤ 2 cm | 62 (42.2%) |
| pT2+pT3, >2 cm | 77 (52.4%) |
| Unknown | 8 (5.4%) |
| Grade | |
| Grade 1 | 1 (0.7%) |
| Grade 2 | 22 (15.0%) |
| Grade 3 | 121 (82.3%) |
| Unknown | 3 (2.0%) |
| Metastases within 5 years | |
| Yes | 39 (26.5%) |
| No | 108 (73.5%) |
a These 147 samples consist of samples (n = 48) overlapping with our LC-MS data set (5).
This study was approved by the Medical Ethics Committee of the Erasmus Medical Center Rotterdam, The Netherlands (MEC 02.953) and was performed in accordance to the Code of Conduct of the Federation of Medical Scientific Societies in The Netherlands, and wherever possible we adhered to the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) (27).
IHC Staining
IHC staining was performed with the above-described TMA of TNBC tissues. All tumor tissues on this TMA were evaluated by a dedicated pathologist (C.H.M.v.D.) to assess histologic tumor subtype and grade according to Bloom and Richardson (28). Each tissue in the TMA was prepared in biological triplicates from three invasive tumor regions based on hematoxylin and eosin staining of whole tissue sections. Tissue sections of 4 μm were incubated for 1 h at room temperature with anti-FTH1 antibody at a dilution of 1:100 against residues near the C terminus of the protein (rabbit-anti-human monoclonal antibody, clone name: EPR3005Y, GeneTex Inc., Irvine, CA, USA). Antigen retrieval was performed prior to antibody incubation by heating the slides for 40 min at 95 °C and washing with Dako retrieval solution (pH = 6) (DakoCytomation, Carpinteria, CA, USA) when the slides were cooled down to room temperature. Staining was visualized by anti-mouse EnVision+® System-HRP (DAB) (DakoCytomation). Cytoplasmic (c) and nuclear (n) FTH1 stainings were separately scored as percentages of positive invasive tumor cells (6 categories: ≤1%, 2–10%, 11–25%, 26–50%, 51–75%, >75%) by two independent observers. Average scores of biological triplicates were recorded for statistical analysis.
Next, tissue sections were prepared from 30 samples that represent different staining patterns of FTH1 on our TMA. Each of these sections was stained with anti-FTH1, -CD3, -CD4 and -CD8 antibodies. Anti-CD3 (rabbit-anti-human polyclonal antibody) and anti-CD8 (mouse-anti-human monoclonal antibody, clone name: C8/C144B) were purchased from Dako Denmark A/S (Glostrup, Denmark). Ready-to-use anti-CD4 antibody was purchased from Ventana (rabbit-anti-human monoclonal antibody, clone name: SP35, Ventana medical systems Inc., Tucson, AZ, USA). IHC staining of CD8 was performed with anti-CD8 antibody at a dilution of 1:100 and washed in a retrieval solution at pH = 9 for 40 min at room temperature. The rest of the protocol was the same as described for FTH1 staining. Slides were incubated with anti-CD3 antibody at a dilution of 1:150 for 32 min, and with ready-to-use anti-CD4 antibody for 16 min at room temperature. Antigen retrieval prior to CD3 and CD4 stainings was performed by heating slides for 64 min at 97 °C and washing with Tris-EDTA solution (pH = 8.4) when the slides were cooled down to room temperature. Staining was visualized by discoveryTM universal secondary antibody (Ventana, Ventana medical systems Inc., Tucson, AZ, USA). FTH1 staining on whole tissue sections was scored following the same method as used for TMA. CD3 was scored in density of positive staining cells, and CD4 and CD8 were scored as CD4/CD8 ratio. The density of CD4 and CD8 markers was calculated using the formula: density of CD4+ and CD8+ T-lymphocytes = density of CD3+ T-lymphocytes × percentage of CD4+ and CD8+ T-lymphocytes. All slides were scored by two independent observers, of whom one was a pathologist (C.H.M.v.D.).
Statistical Analysis
Univariate and multivariate Cox regression analyses were performed using Stata software (version 12.0). Hazard ratios (HR) and the corresponding 95% confidence interval (CI) were calculated based on stratified groups of patients. Kaplan-Meier analyses were performed using Stata software (version 12.0) and GraphPad Prism 5 (version 5.01). Based on visual observation of Kaplan-Meier curves, optimal cutoffs of cFTH1 and nFTH1 staining were determined for reporting. Nonparametric Spearman correlation and Wilcoxon rank-sum test were used for analyzing IHC staining data.
RESULTS
Expression of FTH1 Protein in TNBC Tissue is Related to Prognosis of Patients
Ferritin complex consists of 21 kDa heavy chain (FTH1) and 19 kDa light chain (FTL). The ratio of FTH1 and FTL can be altered under inflammatory and/or infectious conditions (12). Because the same conditions lead to recruitment of immune cells and their contribution to angiogenesis, cell proliferation and tumor invasiveness (29), we questioned how FTH1 expression exactly relates to immune parameters. In the MS data set of 126 TNBC tumors, we observed that protein expression of FTH1 was significantly higher in good prognostic patients (n = 82) than those with poor prognosis (n = 44) (p = 0.001), whereas neither expression of FTL (p = 0.054) nor FTH1/FTL ratio (p = 0.566) showed significant difference between patients with different clinical outcome (Fig. 1A). In Kaplan-Meier survival analysis, the 5-year metastasis-free survival rate of the TNBC patients was significantly associated with FTH1 expression [the first (43.75%, 14/32 patients), second (64.52%, 20/31 patients), third: (80.65%, 25/31 patients) and fourth (71.88%, 23/32 patients) quartile] [Log-rank trend p = 0.002, HR (95% CI) = 0.66 (0.50–0.87)] (Fig. 1B, upper panel). The patients with the lowest FTH1 expression (the first quartile) have significantly lower 5-year metastasis-free survival rate than the rest of the patients (the second-fourth quartile) [Log-rank p = 0.001, HR (95% CI) = 0.38 (0.21–0.70)] (Fig. 1B, lower panel). Therefore, aggressiveness of TNBC tumors appeared to be related to decreased FTH1 expression in our cohort of samples.
Fig. 1.
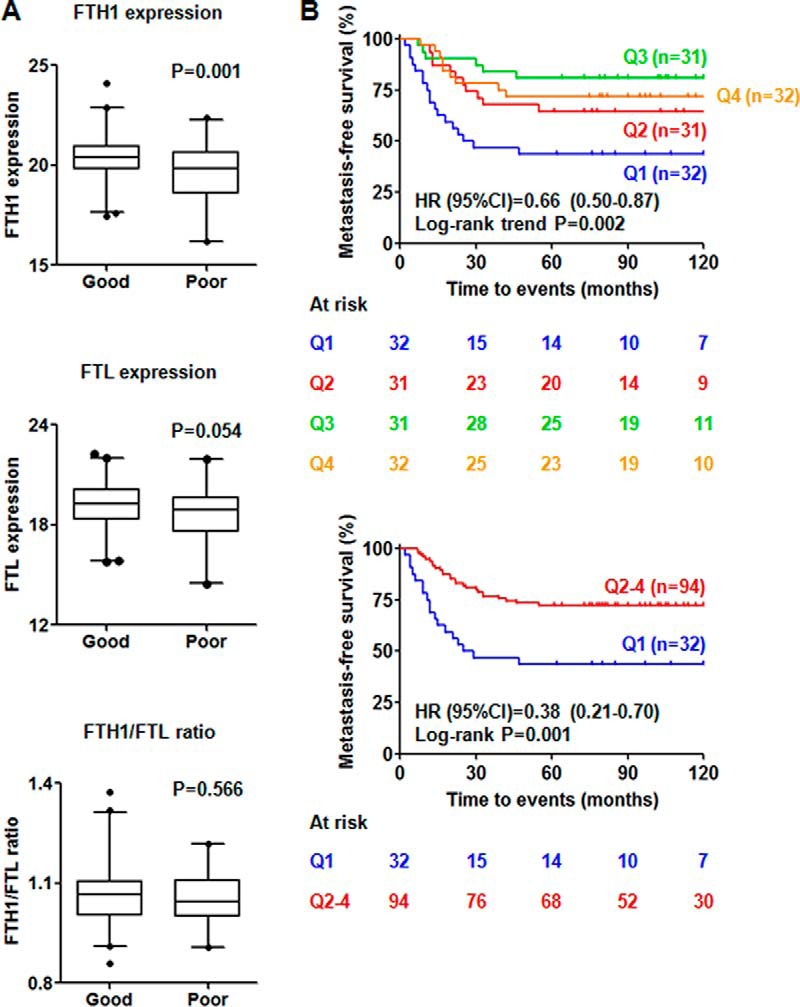
Prognostic values of FTH1, FTL, and FTH1/FTL ratio in the MS data set of 126 TNBC tumors. A, Expression of FTH1 is significantly higher in good (n = 82) prognostic TNBC patients, but there is no significant difference in expression of FTL and FTH1/FTL ratio between the two groups of patients; B, Expression of FTH1 significantly correlates to metastasis-free survival in Kaplan-Meier survival analyses (Q1- Q4 = first, second, third and fourth quartile. HR = hazard ratio, CI = confidence interval).
Transcriptomic and Proteomic Approach Identifies FTH1 as an Immunomodulatory Protein
Proteins are the actual functional units, therefore protein level information is important for functional understanding of underlying mechanisms of disease phenotype. However, proteomic techniques still suffer from low identification rate of the human proteome (11). To provide a first analysis of molecular mechanisms that drive TNBC progression, we compared the gene expression patterns of 63 TNBC samples to those genes that were differentially expressed between different TNBC subtypes (22). Two molecular subtypes were enriched under an FDR of 0.25. We observed that genes that showed down-regulated expression in the mesenchymal (M) subtype were enriched in good prognostic samples (Fig. 2A upper panel, supplemental Table S2), and genes that showed up-regulated expression in the M subtype were enriched in poor prognostic samples (Fig. 2A lower panel, supplemental Table S2). Genes that showed up-regulated expression in the immunomodulatory (IM) subtype were enriched in good prognostic samples (Fig. 2B, supplemental Table S2). This observation indicates that the M subtype was associated with poor prognosis and the IM subtype was related to good prognosis, in line with previous findings (22).
Fig. 2.
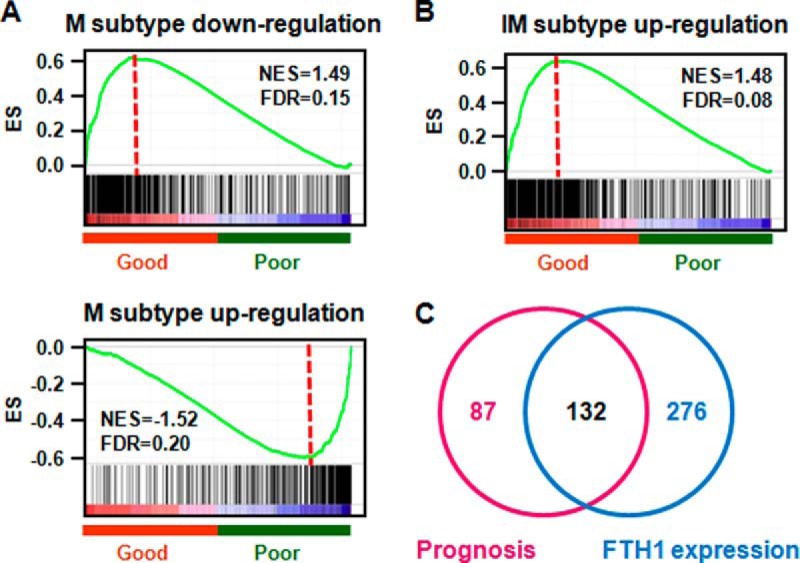
GSEA analyses reveal TNBC prognosis and FTH1-associated gene sets. A, Regulation of mesenchymal (M) subtype specific genes in TNBC patients; B, Regulation of immunomodulatory subtype (IM) specific genes in TNBC patients; C, Overlap between TNBC prognosis-associated pathways and FTH1 expression related pathways.
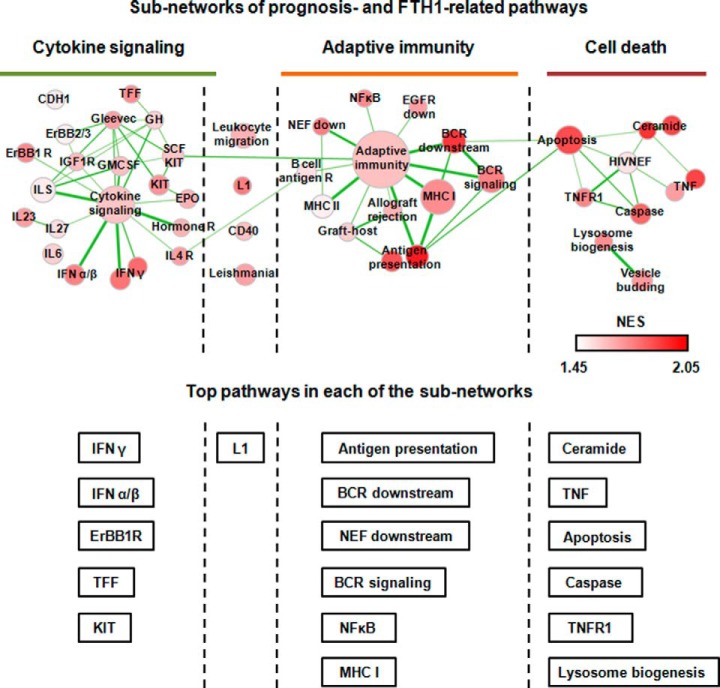
Next, we examined overlap biological pathways that are significantly associated with prognosis of TNBC patients and expression of the FTH1 gene. Because of the complexity of TNBC molecular features, a less stringent cutoff of FDR<0.60 was used to enrich for biological pathways. With this criterion, 219 and 408 biological pathways were identified to be related to TNBC aggressiveness and expression of FTH1 gene, respectively, of which 132 pathways were shared between these two parameters (Fig. 2C, supplemental Table S3). These 132 pathways were then filtered with permissive p value cutoff of 0.05 and FDR cutoff of 0.25 and visualized in EnrichmentMap Software (24). We identified 4 subnetworks that are associated with expression of FTH1: (1) cytokine signaling, (1–2) link between cytokine signaling and adaptive immunity, (2) adaptive immunity, and (3) cell death (Fig. 3). In the cytokine signaling subnetwork, interferon and interleukin signaling was activated as well as some of the cell adhesion pathways, which induce downstream activation of adaptive immunity (e.g. antigen processing and presentation, B-cell receptor signaling) and subsequently immunomodulated cell death (e.g. ceramide pathway, apoptosis pathway) (Fig. 3, supplemental Fig. 1).
Fig. 3.
Visualization of the network between TNBC prognosis and FTH1 related pathways in EnrichmentMap software. All the enriched pathways were divided into 4 major subnetworks: cytokine signaling, link between innate and adaptive immunity, adaptive immunity, and cell death. Node size represents the gene-set size. Density of node color is related to enrichment of pathways. Thickness of connective lines indicates the degree of overlap between two gene-sets. Top-ranking pathways based on the normalized enrichment score (NES) were listed under each of the subnetworks.
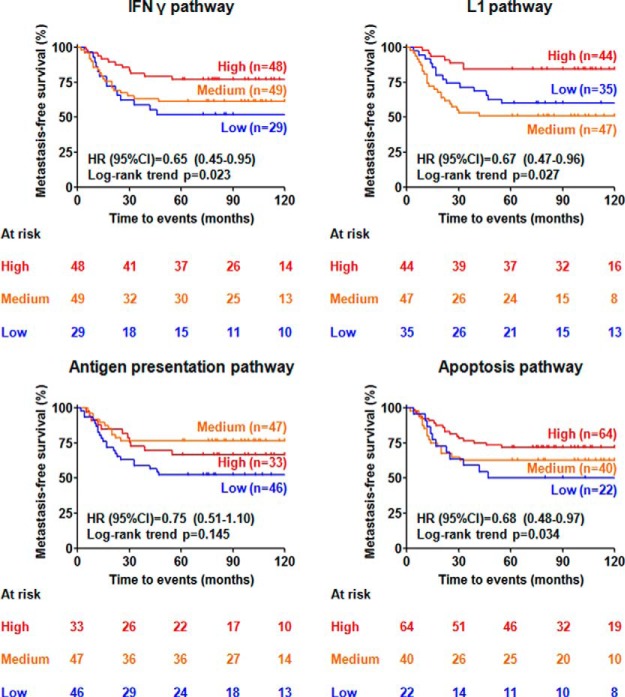
To verify pathway analysis findings at protein level, we applied unsupervised clustering analyses on the top pathway from each of the four subnetworks using the level of protein abundance quantified by nLC-MS/MS, and searched for correlations between different clusters and metastasis-free survival of patients. The four top pathways, IFN γ, L1, antigen processing and presentation, and apoptosis, were clearly clustered into high, medium and low groups based on expression of proteins involved in these pathways (Fig. 4). We observed that IFN γ (Log-rank trend p = 0.023), L1 (Log-rank p = 0.027), antigen processing and presentation (Log-rank p = 0.145), and apoptosis (Log-rank trend p = 0.034) pathways significantly correlated or showed a trend to correlate with prognosis of TNBC patients, where higher expression of these pathways went hand-in-hand with a favorable 5-year metastasis-free survival (Fig. 5).
Fig. 4.
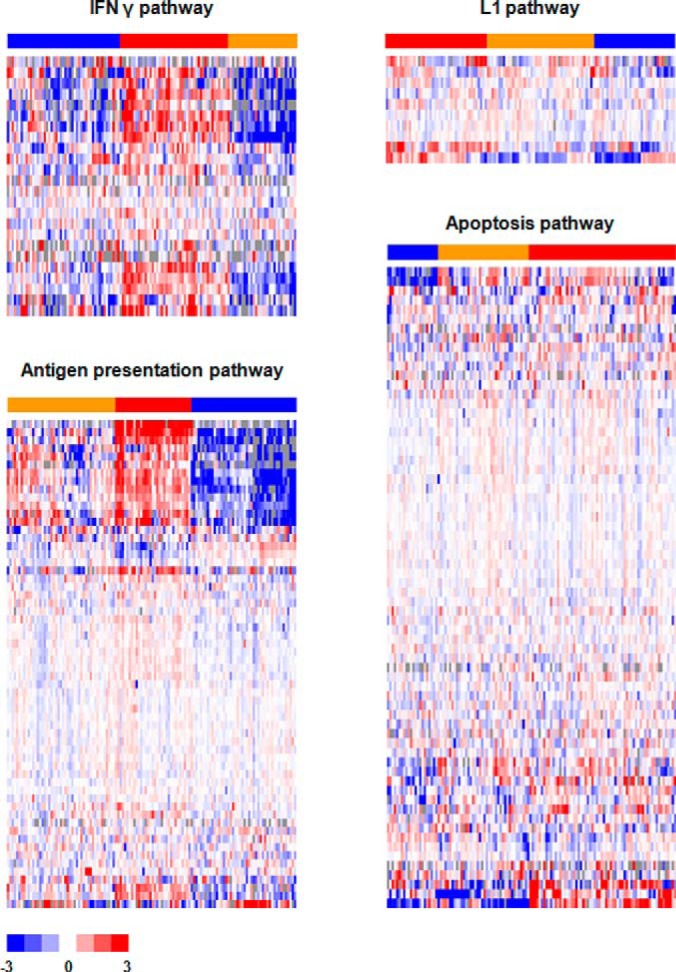
Unsupervised hierarchical clustering of the proteins involved in IFN γ, L1, antigen presentation, and apoptosis pathways. Bars of sample clusters: high (red), medium (orange), and low (blue) expression of the pathways.
Fig. 5.
Expression level of proteins involved in IFN γ, L1, antigen presentation, and apoptosis pathways and their association with metastasis-free survival of the TNBC patients.
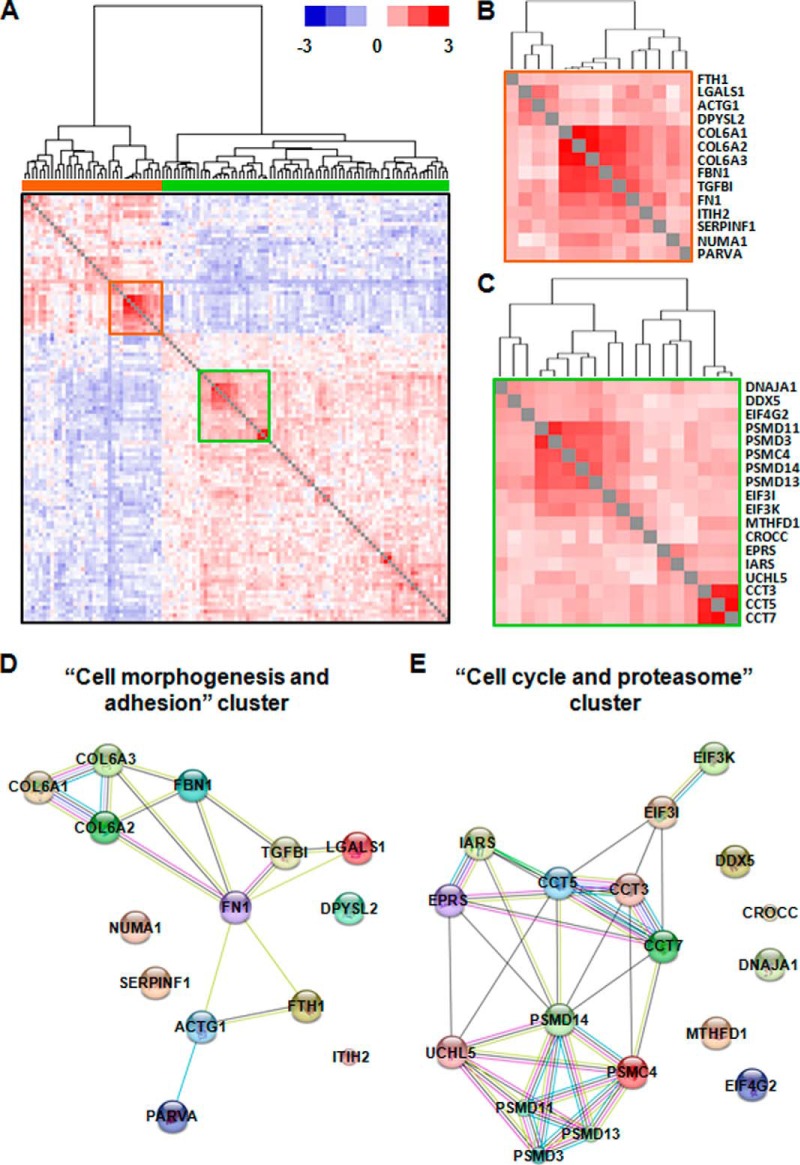
To study the interactome of FTH1, we performed interaction analyses using the transcriptomic and proteomic data sets. Over-representation analysis using all the genes which significantly correlate with FTH1 expression (Spearman's rank correlation, p < 0.01) showed that two immunomodulatory networks (“dermatological/inflammatory disease” and “antigen presentation, infectious disease”) are over-represented behind these genes (supplemental Fig. 2A, orange bars). We further focused on the “antigen processing, infectious disease” network because this pathway was significantly associated with TNBC prognosis and FTH1 expression in GSEA analysis. Interestingly, multiple key genes, which regulate “antigen process, infectious disease” network, such as proteosome genes, TAP1, HLA molecules, and STAT1, positively correlated with FTH1 expression (supplemental Fig. 2B). Furthermore, we examined those proteins of which the expression is significantly associated with FTH1 expression. As expected, antigen processing and presentation pathways, especially major histocompatibility complex class I (MHC I), were over-represented behind these proteins (supplemental Fig. 3, orange bars). Clustering analysis showed that these proteins could roughly be divided into two major clusters: those that positively (Fig. 6A, orange bar) and negatively (Fig. 6A, green bar) correlate with FTH1 expression. The positive cluster mainly over-represented the biological process of “cell morphogenesis and adhesion,” whereas the negative one mainly over-represented “cell cycle and proteasome” (data not shown). Furthermore, two highly correlated regions were identified from these two clusters (Fig. 6A, red and green squares). The first region consisted of cell adhesion proteins such as collagens (COL6A1, COL6A2, COL6A3), fibrillin-1 (FBN1), transforming growth factor-beta-induced protein ig-h3 (TGFBI), fibronectin (FN1), and Alpha-parvin (PARVA), cytoskeleton protein Gamma-actin (ACTG1) and immunoregulatory protein galectin-1 (LGALS1) (Fig. 6B and 6D), indicating that FTH1 induced adhesion contact between tumor cells and immune cells via creating a cell adhesion environment. From the second region (Fig. 6C and 6E), we observed the enrichment of cell cycle regulatory proteins such as eukaryotic translation initiation factor 3 subunit I/K (EIF3I and EIF3K), chaperonins such as T-complex protein 1 subunit gamma/epsilon/eta (CCT3, CCT5, and CCT7), proteasome subunits and interaction enzyme such as 26S protease regulatory subunit 6B (PSMC4), 26S proteasome non-ATPase regulatory subunit 3/11/13/14 (PSMD3, PSMD11, PSMD13 and PSMD14) and ubiquitin carboxyl-terminal hydrolase isozyme L5 (UCHL5), and tRNA multisynthetase complex such as cytoplasmic isoleucine-tRNA ligase (IARS) and bifunctional glutamate/proline-tRNA ligase (EPRS). Among these proteins, proteasome complex is known to regulate the cell cycle by degradation of ubiquitylated key proteins involved in cell cycle progression or arrest (30). Also, EIF3 and tRNA multisynthetase complex are known to be involved in an inflammatory mRNA translation process initiated by IFN γ (31). However, this process is highly dependent on phosphorylation of EPRS (31), and therefore may not be fully explained by our proteomic data. Interestingly, proteasome complex has also shown to be involved in MHC I antigen processing via epitope production (32). However, this immunoregulatory function is limited to a special type of proteasome called “immunoproteasome,” which is induced by IFN γ stimulation (33). The immunoproteasome may represent a small proportion of proteasomes and be dynamically regulated. Therefore, it has little impact on the total proteasome level in cancer cells.
Fig. 6.
Over-represented network from all the proteins correlated with FTH1 expression. A, A hierarchical cluster of proteins significantly correlating with FTH1 expression forms two distinct clusters, representing to those which show positive (orange bar) and negative (green bar) correlation. The red and green squares highlight two highly correlated subclusters; B, The red subcluster positively correlates with FTH1 expression; C, The green subcluster negatively correlates with FTH1 expression; D, The protein interaction map from the red subcluster is associated with biological process of “cell morphogenesis and adhesion”; E, The protein interaction map from the green subcluster is associated with the biological process of “cell cycle and proteasome.”
Subcellular Localization of FTH1 Predicts Clinical Outcome in TNBC Patients
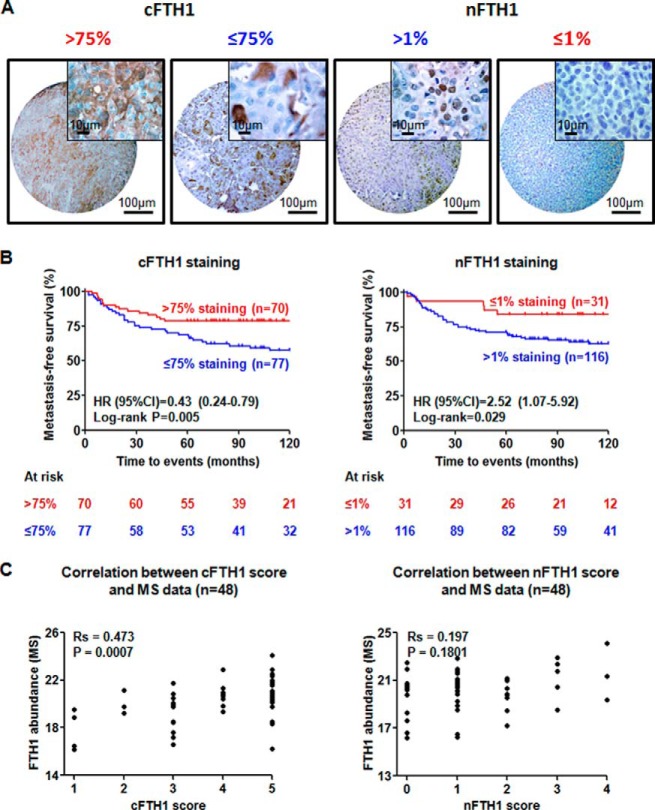
Some previous studies indicated that subcellular localization of FTH1 might be potentially important for cancer aggressiveness (13). To investigate the prognostic value of subcellular localization of FTH1, IHC staining of FTH1 was performed on dedicated TNBC TMAs. To investigate whether and how FTH1 relates to the ability of TNBC tumors to metastasize, we selected 147 tumor samples from an initial cohort of 412 primary TNBC tumors in order to eliminate the following confounding variables: lymph node positivity and adjuvant treatment, which may affect distant metastatic patterns of the TNBC patients. As previously reported in breast cancer cell lines (13), besides cytoplasmic (c) staining of FTH1 in the majority of TNBC tissues, nuclear (n) staining was also observed in some TNBC tissues (Fig. 7A). Kaplan-Meier survival analyses showed that expression of cFTH1 in >75% of tumor cells clearly related to favorable prognosis (Log-rank p = 0.005), whereas expression of nFTH1 in >1% of tumor cells clearly related to adverse prognosis (Log-rank p = 0.029) (Fig. 7B). In addition, patients with high expression of cFTH1 showed significantly lower risk of developing distant metastasis than those with low expression of cFTH1 according to univariate [HR (95% CI): 0.43 (0.24–0.79), p = 0.006] and multivariate [HR (95% CI): 0.42 (0.23–0.78), p = 0.006] Cox regression analyses. In contrast, patients with higher nFTH1 expression had higher risk of developing distant metastases than those with lower nFTH1 expression, as shown in univariate [HR (95% CI): 2.52 (1.07–5.92), p = 0.035] and multivariate [HR (95% CI): 2.27 (0.95–5.44), p = 0.065] Cox regression analyses (Table II). From the 147 samples studied on TMA, we had MS data available for 48 samples. We examined the correlation between cFTH1 or nFTH1 and FTH1 abundance measured by MS in the 48 overlapping samples. Interestingly, cFTH was significantly correlated with total FTH1 abundance as measured previously by MS (Spearman correlation: Rs = 0.47, p < 0.001), whereas nFTH1 did not (Rs = 0.20, p = 0.18) (Fig. 7C). This observation verifies that nFTH1 is relatively low abundant and has little influence on total FTH1 abundance in cancer cells. Therefore, the immunomodulatory role of FTH1 as observed by MS data analysis may be predominantly determined by cFTH1.
Fig. 7.
Subcellular localization of FTH1 and its prognostic significance. A, Cytoplasmic and nuclear staining of FTH1 on TMA and their corresponding cutoff points for Kaplan-Meier analyses; B, Kaplan-Meier analyses show that >75% of tumor cells positive for cFTH1 was associated with favorable prognosis (left panel), whereas >1% positive nFTH1 was related to unfavorable prognosis of TNBC patients (right panel); C, A significant correlation (Spearman correlation) was observed between total FTH1 as measured by LC-MS (5) and cFTH1 staining (left panel), but not nFTH1 staining.
Table II. Univariate and multivariate analyses of cFTH1 and nFTH1 related to metastasis-free survival (n = 147).
| TNBC cohort on TMA |
Univariate |
Multivariate |
||
|---|---|---|---|---|
| Variables | Hazard ratio (95% CI) | p value | Hazard ratio (95% CI) | p value |
| (A) Conventional prognostic factors | ||||
| Age (Continuous variable) | 1.00 (0.98–1.02) | 0.913 | 1.02 (0.98–1.05) | 0.356 |
| Menopausal status (Post- versus Pre-menopausal) | 0.85 (0.49–1.48) | 0.571 | 0.56 (0.21–1.49) | 0.244 |
| Tumor size (>2 cm and unknown versus ≤2 cm) | 1.69 (0.94–3.02) | 0.078 | 1.89 (1.04–3.43) | 0.035 |
| Tumor grade (Grade 3 and unknown versus Grade 1 and 2) | 0.57 (0.30–1.10) | 0.093 | 0.56 (0.29–1.10) | 0.092 |
| (B) FTH1 expression | ||||
| cFTH1 expression (>75% versus ≤75% staining) | 0.43 (0.24–0.79) | 0.006 | 0.42 (0.23–0.78) | 0.006a |
| nFTH1 expression (>1% versus ≤1% staining) | 2.52 (1.07–5.92) | 0.035 | 2.27 (0.95–5.44) | 0.065a |
a cFTH1 and nFTH1 staining were separately added into conventional prognostic factors for multivariate assessment.
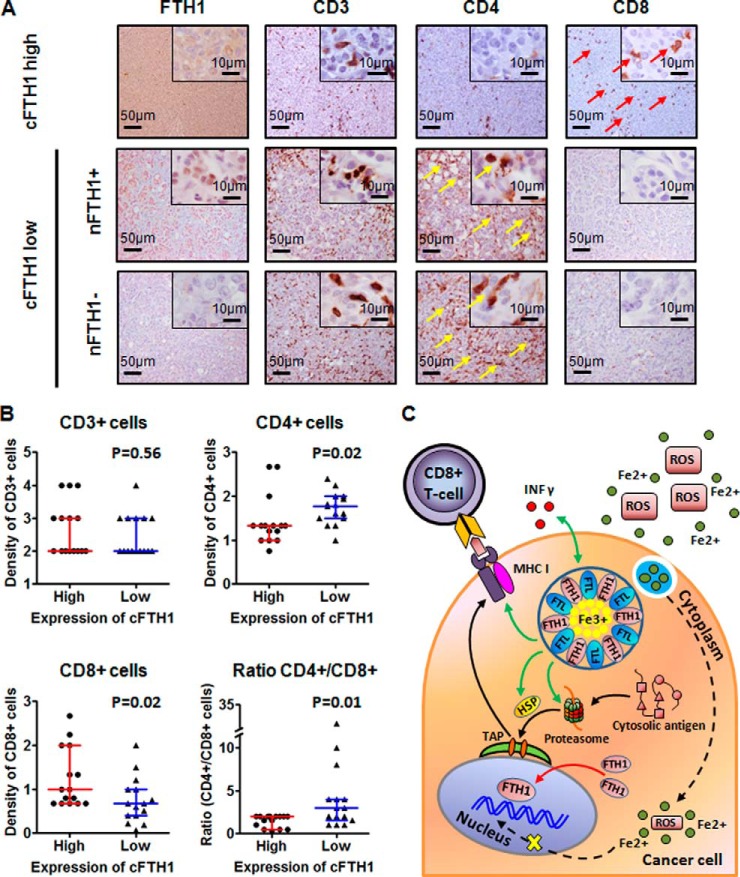
TNBC Tissue with cFTH1 Expression Show Accumulation of CD8+ T Cells
The antigen processing and presentation pathway was identified as a top pathway in the FTH1-regulatory network. This pathway consists of MHC I and MHC II subpathways (34), which are directly related to recognition of tumor antigens by T cells (CD4+ and/or CD8+ cells) and T-cell mediated tumor clearance. Previously, the number of infiltrating CD4+ helper T cells, CD8+ cytotoxic T cells, and CD4+/CD8+ ratio has been used to evaluate the immune status of cancer patients (35). Here, we examined whether cFTH1 levels take part in immune responses in TNBC tumors by staining whole tissue sections (n = 30) with known expression levels and subcellular localization of FTH1 for various T-cell markers (Fig. 8A). Although the density of T-lymphocytes (CD3+) did not differ between tumors with different expression levels of cFTH1 (p = 0.56), we observed that the density of CD4+ T cells was found to be significantly decreased (p = 0.02), whereas that of CD8+ T cells was significantly increased (p = 0.02) in the tumors with cFTH1 in >75% tumor cells. Consequently, TNBC with high cFTH1 levels presented with clearly decreased ratios between CD4+ and CD8+ T cells (p = 0.01) (Fig. 8B). The similar pattern of CD8+ T-cell enrichment was also observed in close conjunction to the tumor region with high cFTH1 expression (data not shown). These findings favor the following model of cFTH1 in TNBC tumors (Fig. 8C): Ferritin complex captures intracellular Fe2+ ions (green dots) and converts them into Fe3+ ions (yellow dots). Accumulation of Fe3+ iron in tumor cells increases in response to inflammation, and IFN γ (red dots) can enhance the inflammatory response of tumor cells. Together, inflammatory signals can enhance processing of cytosolic antigens through the proteasome, heat shock proteins (HSP), and antigen peptide transporters (TAP), resulting in antigen presentation by MHC I. This in turn activates CD8+ T-lymphocytes that recognize tumor cells and eventually induce the apoptotic process of the tumor cells.
Fig. 8.
Accumulation of CD8+ T-lymphocytes in tumor regions with expression of cFTH1. A, IHC staining of FTH1, CD3, CD4 and CD8 in TNBC whole tissue sections (yellow arrows: CD4+ T-lymphocytes; red arrows: CD8+ T-lymphocytes); B, Box plots summarize the observation in (A). CD4+/CD8+ ratio was significantly increased in TNBC samples with high expression of cFTH1. Data are presented as the median ± interquartile range (Mann-Whitney U test, n = 15 per group); C, A proposed functional model of cFTH1 in the context of the antigen processing and presentation pathway: Ferritin complex captures intracellular Fe2+ ions (green dots) and converts them into Fe3+ ions (yellow dots). Accumulation of Fe3+ iron in tumor cells increases in response to inflammation, and IFN γ (red dots) can enhance the inflammatory response of tumor cells. Together, inflammatory signals can enhance processing of cytosolic antigens through the proteasome, heat shock proteins (HSP), and antigen peptide transporters (TAP), resulting in antigen presentation by major histocompatibility complex class I (MHC I). This in turn attracts CD8+ T-lymphocytes that recognize tumor cells and induce the apoptotic process. On the other hand, cFTH1 can be transported from the cytoplasm to the nucleus. nFTH1 protects from DNA damage caused by reactive oxygen species (ROS) (dashed arrow). Solid lines: observed interactions; Dashed line: hypothetical events; Green arrows: molecular functions of cFTH1; Red arrow: molecular function of nFTH1; Black arrows: pathways driven by FTH1.
DISCUSSION
In the current study, we followed up a previously reported 11-protein prognostic signature (5) and investigated the potential mechanism behind the prognostic value of FTH1 in TNBC patients. To this end, we used a combined transcriptomic and proteomic approach by performing integrative analyses on previously generated transcriptomic and proteomic data of the TNBC tissues, and identified an immunomodulatory network regulated by FTH1 in the TNBC tumor tissues. The immunomodulatory function of FTH1 appeared to be regulated by cFTH1 and not nFTH1, with the former being related to good prognosis and the latter to poor prognosis of TNBC patients. Immune stainings of TNBC tissues for T lymphocyte subsets further substantiated the link between cFTH1 and effective immune responses in TNBC.
Transcriptomic and proteomic approaches are both powerful techniques to investigate cancer biology. However, these two techniques also have significant drawbacks to answer disease phenotypes. Microarray-based transcriptomics covers a major part of the coding region of the human genome, and therefore provides a complete picture of gene expression of measured samples. However, this technique does not take into account protein stability and turn-over, and multiple posttranscriptional events which may result in alteration of the translational process, such as micro-RNA interference (36) and posttranslational modification (37, 38). Therefore, actively transcribed genes may not lead to translation of functional protein units. MS-based proteomics measure protein abundance to provide more direct information related to disease phenotype. However, current MS-based proteomics, even by applying the most sophisticated chromatographic and MS techniques, does not cover the complete human proteome (11, 39). However, a combination of transcriptomic and proteomic data may more accurately allow interpretation of actual pathological process during disease progression.
In this study, we pinpointed that FTH1 is prognostic, whereas FTL and FTH1/FTL ratio has no prognostic value in our TNBC cohort. Previously, FTL has been identified as a prognostic marker in node-negative breast cancer cases (40). In addition, FTH1/FTL ratio has been shown to be elevated in the blood of melanoma patients compared with normal subjects (17). This evidence implies that both FTL and the FTH1/FTL ratio are potentially important to (breast) cancer. However, we were not able to recapitulate these findings in our cohort of samples. On one hand, negative correlations between FTL or FTH1/FTL ratio and prognosis of the TNBC patients may be caused by sampling error between our TNBC cohort and the actual TNBC population. On the other hand, negative correlations may also indicate that only abnormal expression of FTH1 is essential for disease progression of TNBC patients, which needs to be confirmed in a large independent cohort of samples.
The prognostic value of FTH1 was verified using an antibody-based TMA technique as an independent technical confirmation. Intriguingly, we revealed the importance of subcellular localization of FTH1 in disease progression of TNBC. Patients with high expression of cFTH1 (>75% positive staining of invasive cells) showed significantly better 5-year metastasis-free survival, whereas the patients with high expression of nFTH1 (>1% positive staining of invasive cells) showed significantly worse 5-year metastasis-free survival. In the majority of TNBC tissues, FTH1 dominantly localized in the cytoplasm of invasive cancer cells, and therefore expression of cFTH1 correlated well with total FTH1 levels measured by MS. This indicates that we can use cFTH1 as a surrogate marker for total FTH1 when studying pathways and their relation to FTH1 in the TNBC tissue.
Previously, it has been demonstrated that the IM subtype of TNBC associates with favorable prognosis (22). Intriguingly, FTH1 appears to be an immunomodulatory protein, which was shown here to be linked to immune response and downstream cell death pathways. Gene and protein network analyses also revealed that immunoregulation networks were over-represented in our transcriptomic and proteomic data. From our data, a potential molecular functional model could be derived: Accumulation of FTH1, especially cFTH1, may enhance the inflammatory microenvironment of cancer cells, which can result in activation of CD8+ cytotoxic T cells, activation of NK cells, and inhibition of angiogenesis. As direct evidence, we first observed upstream activation of cell adhesion pathways and different cytokine signaling such as IFN γ, interleukins and tumor necrosis factors (Fig. 3). More importantly, antigen processing and presentation pathway was identified as one of the most enriched pathways in the TNBC patients with good prognosis. It is known that this pathway is composed of MHC I and MHC II parts, which directly regulates the activation of CD8+ and CD4+ T cells (34). As confirmation, the correlation between cFTH1 expression and enrichment of CD8+ T cells in the tumor area was observed, which indicates an immunoregulatory function of this protein. Related to our findings, a recent publication has shown that enrichment of CD8+ T cells is a marker for favorable prognosis of basal breast cancer (41). Moreover, CD8+ T-cell infiltration with respect to favorable prognosis in patients have also been observed in other types of cancer such as colorectal cancer (42, 43) and ovarian cancer (44). In addition, lower density of CD4+ T cells in the tumors with high cFTH1 expression may be related to lower density of immune suppressive CD4+/CD25+ regulatory T-cell (Treg) population (45), and the latter is expected to suppress CD4+CD25- and CD8+ T-cell responses against tumors. Interestingly, the ratio between CD8+ effector T cells and Treg cells may not only have prognostic value, but also predicts the impact of chemotherapy and immune therapy (46). In addition, although ferritin complex stores Fe3+ iron, these irons can be reduced back to Fe2+ in the presence of strong reducing agents such as xanthine oxidareductase. As a consequence, ROS will be regenerated and therefore tumor cells will be exposed to activated immune cells such as neutrophils (47). On the other hand, we also found nFTH1 to be related to an adverse clinical outcome of TNBC patients. Because FTH1 prevents DNA damage from ROS through conversion of Fe2+ into Fe3+ irons (12, 13), nuclear localization of FTH1 may protect from DNA damage (e.g. induced by chemotherapeutics) and is thus favorable for survival of cancer cells (Fig. 8C). In addition, nFTH1 has been shown to interact with death domain-associated protein (DAXX) and inhibit DAXX-mediated apoptosis (16). Interestingly, a time-dependent cytoplasm-to-nucleus switching of FTH1 has been observed in HEK-293 cell lines through activation of the CXCL12-CXCR4 signaling pathway (48). Because CXCL12-CXCR4 signaling promotes migration and invasion of malignant cells and leads to metastasis of breast cancer (49, 50), we argue that this signaling stimulates malignancy through nuclear translocation of FTH1.
However, actual immunoregulatory role of FTH1 may be much more complex than simply induction of CD8+ T-cell related immune response. Our data cannot exclude the probability that FTH1, based on its cellular compartmentalization, may serve as a positive regulator or negative inhibitor of immune response. Balance between the positive and negative feedback determines whether tumor cells are in response or resistant to the activated CD8+ T cells, and therefore results in different aggressiveness of TNBC between individual patients. This balance of positive or negative feedback has been previously reported in other types of cancer, such as B7-H1 in melanoma cells (51). In breast cancer, a controversial finding may indicate a similar mechanism: A variant of FTH1, placental immunoregulatory ferritin, serves as an immune suppressor by inhibition of CD8+ T-cell infiltration in ESR1+ MCF-7 breast cancer cells (18). Alternatively, FTH1 may also regulate the immune system in two other ways. As a secretory protein (12), the level of secreted FTH1 may positively correlate with expression of cellular FTH1. Therefore, FTH1 can possibly regulate immune response through the tumor microenvironment. Furthermore, it is known that FTH1 interacts with different transcription factors which modulate the immune system, such as NF-κB (52) and AP-1 (53). Hence, FTH1 can modulate the immune system through downstream signaling of these transcription factors. Therefore, further studies (e.g. immunoprecipitation and functional silencing) are required to comprehensively understand immunomodulatory function of FTH1, and may lead to targeted immune therapy to treat TNBC patients.
In conclusion, this study confirms the prognostic value of FTH1 and pinpoints the importance of its subcellular localization. Our data also suggest that cFTH1 regulates the MHC I part of the antigen processing and presentation pathway and subsequently attracts CD8+ T cells to target tumor cells, whereas nFTH1 is beneficial to survival of cancer cells. We speculate that blockade of cytoplasm-to-nucleus switching of FTH1 may suppress tumor metastasis and therefore serve as a potential therapeutic target for TNBC, which should be further investigated.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Elizabeth A. McClellan and Dr. Andrew Stubbs for technical support on bioinformatics analysis, and Ms. Mandy van Brakel for helpful discussion.
Footnotes
Author contributions: N.L., R.D., J.W.M., J.A.F., and A.U. designed research; N.L., T.D., A.M. Timmermans, R.B., A.M. Trapman-Jansen, R.F., and C.H.v. performed research; P.N.S., F.C.S., J.B.B., and V.T. contributed new reagents or analytic tools; N.L. and M.P.L. analyzed data; N.L., R.D., J.W.M., J.A.F., and A.U. wrote the paper; N.L., T.D., A.M. Timmermans, R.B., A.M. Trapman-Jansen, R.F., M.P.L., C.H.v., P.N.S., F.C.S., J.B.B., V.T., R.D., J.W.M., J.A.F., and A.U. reviewed and revised the manuscript.
* This study was financed through the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific research (NWO), the Dutch Cancer Society, EMCR 2009-4319, and the Center for Translational Molecular Medicine, Breast CARE project (030-104).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S3.
1 The abbreviations used are:
- TNBC
- triple negative breast cancer
- nLC-MS/MS
- nano-scale liquid chromatography hyphenated with high resolution tandem mass spectrometry
- FTH1
- ferritin heavy chain
- ROS
- reactive oxygen species
- IHC
- immunohistochemistry/immunohistochemical
- MSigDB
- molecular signatures database
- FDR
- false discovery rate
- IFN γ
- interferon gamma
- L1
- recycling of the cell adhesion molecular L1
- TMA
- tissue microarray
- cFTH1
- cytoplasmic FTH1
- nFTH1
- nuclear FTH1
- HR
- hazard ratios
- CI
- confidence interval
- FTL
- ferritin light chain
- M
- mesenchymal subtype
- IM
- immunomodulatory subtype
- MHC
- I/II major histocompatibility complex class I and class II
- HSP
- heat shock proteins
- TAP
- antigen peptide transporters
- Treg
- regulatory T cell.
REFERENCES
- 1. Dent R., Trudeau M., Pritchard K. I., Hanna W. M., Kahn H. K., Sawka C. A., Lickley L. A., Rawlinson E., Sun P., Narod S. A. (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 13, 4429–4434 [DOI] [PubMed] [Google Scholar]
- 2. Rody A., Karn T., Liedtke C., Pusztai L., Ruckhaeberle E., Hanker L., Gaetje R., Solbach C., Ahr A., Metzler D., Schmidt M., Müller V., Holtrich U., Kaufmann M. (2011) A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 13, R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hallett R. M., Dvorkin-Gheva A., Bane A., Hassell J. A. (2012) A Gene Signature for Predicting Outcome in Patients with Basal-like Breast Cancer. Sci. Rep. 2, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yau C., Esserman L., Moore D. H., Waldman F., Sninsky J., Benz C. C. (2010) A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 12, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu N. Q., Stingl C., Look M. P., Smid M., Braakman R. B. H., De Marchi T., Sieuwerts A. M., Span P. N., Sweep F. C. G. J., Linderholm B. K., Mangia A., Paradiso A., Dirix L. Y., Van Laere S. J., Luider T. M., Martens J. W. M., Foekens J. A., Umar A. (2014) Comparative proteome analysis revealing an 11-protein signature for aggressive triple negative breast cancer. J. Natl. Cancer Inst. 106, djt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Warmoes M., Jaspers J. E., Pham T. V., Piersma S. R., Oudgenoeg G., Massink M. P. G., Waisfisz Q., Rottenberg S., Boven E., Jonkers J., Jimenez C. R. (2012) Proteomics of mouse BRCA1-deficient mammary tumors identifies DNA repair proteins with potential diagnostic and prognostic value in human breast cancer. Mol. Cell. Proteomics 11, M111.013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geiger T., Madden S. F., Gallagher W. M., Cox J., Mann M. (2012) Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 72, 2428–2439 [DOI] [PubMed] [Google Scholar]
- 8. Umar A., Kang H., Timmermans A. M., Look M. P., Meijer-van Gelder M. E., Den Bakker M. A., Jaitly N., Martens J. W. M., Luider T. M., Foekens J. A., Pasa-Tolić L. (2009) Identification of a putative protein profile associated with tamoxifen therapy resistance in breast cancer. Mol Cell Proteomics 8, 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ambrosino C., Tarallo R., Bamundo A., Cuomo D., Franci G., Nassa G., Paris O., Ravo M., Giovane A., Zambrano N., Lepikhova T., Jänne O. A., Baumann M., Nyman T. A., Cicatiello L., Weisz A. (2010) Identification of a hormone-regulated dynamic nuclear actin network associated with estrogen receptor alpha in human breast cancer cell nuclei. Mol. Cell. Proteomics 9, 1352–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cha S., Imielinski M. B., Rejtar T., Richardson E. A., Thakur D., Sgroi D. C., Karger B. L. (2010) In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol. Cell. Proteomics 9, 2529–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michalski A., Cox J., Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 12. Knovich M. A., Storey J. A., Coffman L. G., Torti S. V., Torti F. M. (2009) Ferritin for the clinician. Blood Rev. 23, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shpyleva S. I., Tryndyak V. P., Kovalchuk O., Starlard-Davenport A., Chekhun V. F., Beland F. a, Pogribny I. P. (2011) Role of ferritin alterations in human breast cancer cells. Breast Cancer Res. Treat. 126, 63–71 [DOI] [PubMed] [Google Scholar]
- 14. Pham C. G., Bubici C., Zazzeroni F., Papa S., Jones J., Alvarez K., Jayawardena S., De Smaele E., Cong R., Beaumont C., Torti F. M., Torti S. V., Franzoso G. (2004) Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell 119, 529–542 [DOI] [PubMed] [Google Scholar]
- 15. Kiessling M. K., Klemke C. D., Kaminski M. M., Galani I. E., Krammer P. H., Gülow K. (2009) Inhibition of constitutively activated nuclear factor-kappaB induces reactive oxygen species- and iron-dependent cell death in cutaneous T-cell lymphoma. Cancer Res. 69, 2365–2374 [DOI] [PubMed] [Google Scholar]
- 16. Liu F., Du Z.-Y., He J.-L., Liu X.-Q., Yu Q.-B., Wang Y.-X. (2012) FTH1 binds to Daxx and inhibits Daxx-mediated cell apoptosis. Mol. Biol. Rep. 39, 873–879 [DOI] [PubMed] [Google Scholar]
- 17. Gray C. P., Arosio P., Hersey P. (2003) Association of Increased Levels of Heavy-Chain Ferritin with Increased CD4 + CD25 + Regulatory T-Cell Levels in Patients with Melanoma Association of Increased Levels of Heavy-Chain Ferritin with Patients with Melanoma 1. Clin. Cancer Res. 9, 2551–2559 [PubMed] [Google Scholar]
- 18. Halpern M., Zahalka M. A., Traub L., Moroz C. (2007) Antibodies to Placental Immunoregulatory Ferritin with Transfer of Polyclonal Lymphocytes Arrest MCF-7 Human Breast Cancer Growth in a Nude Mouse Model. Neoplasia 9, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Klijn J. G. M., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M. J. J., Atkins D., Foekens J. A. (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 20. Minn A. J., Gupta G. P., Padua D., Bos P., Nguyen D. X., Nuyten D., Kreike B., Zhang Y., Wang Y., Ishwaran H., Foekens J. A., Van de Vijver M., Massagué J. (2007) Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl. Acad. Sci. U.S.A. 104, 6740–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lehmann B. D., Bauer J. A., Chen X., Sanders M. E., Chakravarthy A. B., Shyr Y., Pietenpol J. A. (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P. (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merico D., Isserlin R., Stueker O., Emili A., Bader G. D. (2010) Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS One 5, e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., Von Mering C., Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McShane L. M., Altman D. G., Sauerbrei W., Taube S. E., Gion M., Clark G. M. (2005) Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst. 97, 1180–1184 [DOI] [PubMed] [Google Scholar]
- 28. Elston C. W., Ellis I. O. (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410 [DOI] [PubMed] [Google Scholar]
- 29. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 30. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 31. Mukhopadhyay R., Jia J., Arif A., Ray P. S., Fox P. L. (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamano T., Mizukami S., Murata S., Chiba T., Tanaka K., Udono H. (2008) Hsp90-mediated assembly of the 26 S proteasome is involved in major histocompatibility complex class I antigen processing. J. Biol. Chem. 283, 28060–28065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloetzel P.-M. (2004) The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta 1695, 225–233 [DOI] [PubMed] [Google Scholar]
- 34. Vyas J. M., Van Der Veen A. G., Ploegh H. L. (2008) The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 8, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brivio F., Fumagalli L., Parolini D., Messina G., Rovelli F., Rescaldani R., Vigore L., Vezzo R., Vaghi M., Di Bella S., Lissoni P. (2008) T-helper/T-regulator lymphocyte ratio as a new immunobiological index to quantify the anticancer immune status in cancer patients. In vivo 22, 647–650 [PubMed] [Google Scholar]
- 36. Lowery A. J., Miller N., McNeill R. E., Kerin M. J. (2008) MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin. Cancer Res. 14, 360–365 [DOI] [PubMed] [Google Scholar]
- 37. Naro C., Sette C. Phosphorylation-Mediated Regulation of Alternative Splicing in Cancer. Int. J. Cell Biol. 2013, 151839, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vucic D., Dixit V. M., Wertz I. E. (2011) Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 12, 439–452 [DOI] [PubMed] [Google Scholar]
- 39. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jézéquel P., Campion L., Spyratos F., Loussouarn D., Campone M., Guérin-Charbonnel C., Joalland M.-P., André J., Descotes F., Grenot C., Roy P., Carlioz A., Martin P.-M., Chassevent A., Jourdan M.-L., Ricolleau G. (2012) Validation of tumor-associated macrophage ferritin light chain as a prognostic biomarker in node-negative breast cancer tumors: A multicentric 2004 national PHRC study. Int. J. Cancer 131, 426–437 [DOI] [PubMed] [Google Scholar]
- 41. Liu S., Lachapelle J., Leung S., Gao D., Foulkes W. D., Nielsen T. O. (2012) CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast Cancer Res. 14, R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diederichsen A. C. P., Hjelmborg J. v B., Christensen P. B., Zeuthen J., Fenger C. (2003) Prognostic value of the CD4+/CD8+ ratio of tumour infiltrating lymphocytes in colorectal cancer and HLA-DR expression on tumour cells. Cancer Immunol. Immunother. 52, 423–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P.-H., Trajanoski Z., Fridman W.-H., Pagès F. (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 [DOI] [PubMed] [Google Scholar]
- 44. Zhang L., Conejo-Garcia J. R., Katsaros D., Gimotty P. A., Massobrio M., Regnani G., Makrigiannakis A., Gray H., Schlienger K., Liebman M. N., Rubin S. C., Coukos G. (2003) Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 348, 203–213 [DOI] [PubMed] [Google Scholar]
- 45. Gray C. P. (2002) Heavy chain ferritin activates regulatory T cells by induction of changes in dendritic cells. Blood 99, 3326–3334 [DOI] [PubMed] [Google Scholar]
- 46. Quezada S. A., Peggs K. S., Simpson T. R., Allison J. P. (2011) Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol. Rev. 241, 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ward P. A., Varani J. (1993) Mechanisms of neutrophil-mediated injury. Clin. Exp. Immunol. 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li R., Luo C., Mines M., Zhang J., Fan G.-H. (2006) Chemokine CXCL12 induces binding of ferritin heavy chain to the chemokine receptor CXCR4, alters CXCR4 signaling, and induces phosphorylation and nuclear translocation of ferritin heavy chain. J. Biol. Chem. 281, 37616–37627 [DOI] [PubMed] [Google Scholar]
- 49. Luker K. E., Lewin S. A., Mihalko L. A., Schmidt B. T., Winkler J. S., Coggins N. L., Thomas D. G., Luker G. D. (2012) Scavenging of CXCL12 by CXCR7 promotes tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene 31, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang X. H.-F., Jin X., Malladi S., Zou Y., Wen Y. H., Brogi E., Smid M., Foekens J. A., Massagué J. (2013) Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Taube J. M., Anders R. A., Young G. D., Xu H., Sharma R., McMiller T. L., Chen S., Klein A. P., Pardoll D. M., Topalian S. L., Chen L. (2012) Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 4, 127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lin M., Rippe R. A., Niemelä O., Brittenham G., Tsukamoto H. (1997) Role of iron in NF-kappa B activation and cytokine gene expression by rat hepatic macrophages. Am. J. Physiol. 272, G1355–G1364 [DOI] [PubMed] [Google Scholar]
- 53. Kaomongkolgit R., Cheepsunthorn P., Pavasant P., Sanchavanakit N. (2008) Iron increases MMP-9 expression through activation of AP-1 via ERK/Akt pathway in human head and neck squamous carcinoma cells. Oral Oncol. 44, 587–594 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.