Abstract
In the nasal cavity, the nonmotile cilium of olfactory sensory neurons (OSNs) constitutes the chemosensory interface between the ambient environment and the brain. The unique sensory organelle facilitates odor detection for which it includes all necessary components of initial and downstream olfactory signal transduction. In addition to its function in olfaction, a more universal role in modulating different signaling pathways is implicated, for example, in neurogenesis, apoptosis, and neural regeneration. To further extend our knowledge about this multifunctional signaling organelle, it is of high importance to establish a most detailed proteome map of the ciliary membrane compartment down to the level of transmembrane receptors. We detached cilia from mouse olfactory epithelia via Ca2+/K+ shock followed by the enrichment of ciliary membrane proteins at alkaline pH, and we identified a total of 4,403 proteins by gel-based and gel-free methods in conjunction with high resolution LC/MS. This study is the first to report the detection of 62 native olfactory receptor proteins and to provide evidence for their heterogeneous expression at the protein level. Quantitative data evaluation revealed four ciliary membrane-associated candidate proteins (the annexins ANXA1, ANXA2, ANXA5, and S100A5) with a suggested function in the regulation of olfactory signal transduction, and their presence in ciliary structures was confirmed by immunohistochemistry. Moreover, we corroborated the ciliary localization of the potassium-dependent Na+/Ca2+ exchanger (NCKX) 4 and the plasma membrane Ca2+-ATPase 1 (PMCA1) involved in olfactory signal termination, and we detected for the first time NCKX2 in olfactory cilia. Through comparison with transcriptome data specific for mature, ciliated OSNs, we finally delineated the membrane ciliome of OSNs. The membrane proteome of olfactory cilia established here is the most complete today, thus allowing us to pave new avenues for the study of diverse molecular functions and signaling pathways in and out of olfactory cilia and thus to advance our understanding of the biology of sensory organelles in general.
Odorant perception in vertebrates is initiated by binding of volatile odor-conferring small molecules to olfactory receptors (ORs)1 located in the membrane of sensory cilia that extend from primary olfactory sensory neurons (OSNs) into the mucus covering the olfactory epithelium (OE) in the nasal cavity (1). ORs are G protein-coupled receptors (GPCRs) and represent the largest family of GPCRs with more than 1,000 distinct receptors in mice and rats and ∼400 in humans (2–4). Activation of ORs upon odorant-binding triggers a signal transduction cascade in olfactory cilia eventually leading to the generation of action potentials in the soma of OSNs (5) and transmission of the signal to the brain.
The canonical ciliary signal transduction pathway following OR activation is well established and includes activation of adenylyl cyclase (AC) III via the α subunit of the heterotrimeric olfactory G protein G(olf). This leads to increased levels of the second messenger cAMP, which in turn results in opening of cyclic nucleotide-gated (CNG) Na+/Ca2+ channels (6, 7) in the ciliary membrane. The resulting influx of Ca2+ ions causes depolarization of the membrane and induces opening of Ca2+-activated Cl− channels such as anoctamin 2 (ANO2) in the ciliary membrane (8–11). The following efflux of chloride ions from the cilia enhances depolarization of the OSN, thereby amplifying the signal and facilitating the generation of an action potential (12, 13).
In addition to the components involved in olfactory signal perception and transduction, cilia of OSNs are required to contain further distinct sets of proteins involved in various cellular functions such as regulation and termination of the signal response, odor adaptation, exchange of solutes with the mucus, intraciliary protein transport, and targeting of ciliary membrane proteins. Furthermore, OSNs have a life span of a few weeks only and undergo lifelong renewal (13, 14), which requires proteins mediating and regulating the constant processes of apoptosis, adult neurogenesis, and ciliogenesis. For a complete understanding of the molecular processes governing the diverse functions of OSN cilia, of potential cross-talk between different processes, and to reveal protein interaction networks formed as well as to gain insight into dysfunctions of the olfactory system causing, for example, hyposmia or anosmia, it is necessary to identify the individual molecular players involved.
Previous proteomics studies targeting the sensory cilia membrane of OSN from rat (15, 16) and mouse (11) or focusing on proteins of the ciliary Ca2+-signaling pathways affinity-purified from a membrane preparation of rat OE (17) already facilitated the identification of new ciliary proteins such as SLC4A1, a Cl−/HCO3− exchanger involved in intraciliary Cl− accumulation required for olfactory signal amplification (18), ANO2, the main Ca2+-activated Cl− channels mediating the efflux of chloride ions upon activation by Ca2+ (10, 11), and NCKX4, a Na+/Ca2+ exchanger mediating termination of the signal response and odorant adaptation (11, 19). However, these studies were still considerably limited in their ability to detect the central low abundant components of olfactory cilia such as GPCRs; consequently, OR proteins remain elusive so far.
The focus of our study was the comprehensive analysis of the mouse olfactory membrane ciliome with high sensitivity permitting the discovery of new resident proteins as well as the identification of low abundant central components of OSN cilia down to the level of ORs. Because of each OSN expressing only one type of the roughly 1,000 functional ORs present in mice and rat (3, 20, 21), individual species of these seven transmembrane (TM) domain receptors are of very low abundance and are thus difficult to detect. Moreover, the hydrophobic nature of integral membrane proteins still represents a major challenge in current proteomics research (22). Our strategy included the selective enrichment of olfactory ciliary membrane proteins based on the established Ca2+/K+-shock method and subsequent Na2CO3 treatment at alkaline pH followed by alternative protein and peptide separation methods employing SDS-PAGE and strong cation exchange (SCX) chromatography, respectively, in conjunction with high resolution LC/MS. As a result, we report here for the first time the robust detection of 62 ORs and provide important data about the relative abundance levels of these ORs showing a considerable heterogeneity in their expression. We further confirmed the localization of four new candidate proteins (ANXA1, ANXA2, ANXA5, and S100A5) with putative function in the regulation of olfactory signal transduction to OSN cilia and corroborated the ciliary localization of the Ca2+ transporters NCKX4 (19) and PMCA1 (23) involved in olfactory signal termination. Comparison of our large scale proteomics dataset with transcriptome data of ciliated mature OSN (24) finally led to the establishment of the so far most complete olfactory membrane ciliome consisting of 178 proteins. This ciliome includes all known components of the canonical signal transduction pathway and its regulation as well as a plethora of proteins with various functions in, for example, ion transport in and out of the cilia, response to nonolfactory stimuli, cytoskeletal organization, apoptosis, and cell survival. The extensive data reported here provide a wealth of information for subsequent functional studies of individual proteins with potentially important functions in olfactory cilia. It therefore serves as an important resource contributing to a better understanding of the biology of sensory organelles in general.
EXPERIMENTAL PROCEDURES
Enrichment of Ciliary Membrane Proteins
The isolation of cilia from murine OE was performed based on the Ca2+/K+-shock method described previously (16). Briefly, four OE per replicate were prepared from 10-week-old male CD1 mice (Charles River, Sulzfeld, Germany), washed in ice-cold solution A (20 mm HEPES, 140 mm NaCl, 3 mm KCl, 2 mm MgSO4, 7.5 mm d-glucose, 5 mm EGTA (pH 7.4)) containing Complete protease inhibitor mixture (Roche Diagnostics), and resuspended carefully in solution B (solution A containing 20 mm CaCl2 and 30 mm KCl). Cilia were detached from OE by gentle stirring for 20 min at 4 °C and separated from the remaining tissue by centrifugation for 5 min at 5,000 × g and 4 °C. The supernatant was collected, and deciliation was repeated twice. Supernatants were combined and centrifuged for 30 min at 26,000 × g and 4 °C. The resulting cilia-containing pellet was resuspended in 100 mm ice-cold Na2CO3 (pH 11.2). Following incubation on ice for 20 min, a membrane fraction enriched in ciliary membrane proteins was gained by centrifugation for 30 min at 100,000 × g and 4 °C and resuspended in 10 mm Tris (pH 7.4) containing 3 mm MgCl2 and 2 mm EGTA. Protein concentrations were determined using the Bradford assay with bovine serum albumin (BSA) as standard (25).
Gel Electrophoresis
Following enrichment of ciliary membrane proteins, 10 μg of protein were separated by one-dimensional SDS-PAGE on a 4–12% NuPAGE BisTris gradient gel (Invitrogen) according to the manufacturer's instructions and visualized by colloidal Coomassie Brilliant Blue G-250. Gel lanes were cut into 29 slices. Destaining, tryptic in-gel digestion, peptide extraction, and further sample preparation steps were performed as described previously (26).
Strong Cation Exchange Chromatography
10 μg of protein were precipitated using chloroform/methanol (27), resuspended in 40 μl of 60% (v/v) methanol, 20 mm NH4HCO3, and digested overnight at 37 °C using 0.5 μg of trypsin dissolved in 50 mm NH4HCO3 (pH 7.8). Peptide mixtures were then dried in vacuo and reconstituted in 60 μl of SCX chromatography solvent A (25% (v/v) acetonitrile, 10 mm KH2PO4). SCX chromatography was performed using an Ultimate classic system (Dionex LC Packings, Idstein, Germany (now Thermo Fisher Scientific)) equipped with a polysulfoethyl aspartamide column (1.0 mm inner diameter × 150 mm, PolyLC Inc., Columbia, MD). Peptides were loaded onto the column, washed for 20 min with SCX solvent A, and separated using the following KCl gradient: 0–30% SCX solvent B (25% (v/v) acetonitrile, 10 mm KH2PO4, 0.5 m KCl) in 40 min followed by 30–100% SCX solvent B in 10 min at a flow rate of 50 μl/min. A total of 25 fractions was collected: 6-min fractions from minute 1–30 and 2-min fractions from minute 31–70. Peptide fractions were dried in vacuo and reconstituted in 15 μl of 0.1% (v/v) trifluoroacetic acid (TFA), 5% (v/v) acetonitrile.
Mass Spectrometry
Peptide mixtures were analyzed by nano-HPLC-ESI-MS/MS using the UltiMate 3000 RSLCnano system (Dionex LC Packings/Thermo Fisher Scientific) on line-coupled to an LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen, Germany). Peptide mixtures were washed and preconcentrated for 25 min on a C18 precolumn (Acclaim PepMap 100, 75 μm inner diameter × 20 mm; 3-μm particle size (Thermo Fisher Scientific)) with 0.1% (v/v) TFA at a flow rate of 7 μl/min. Peptides were then separated on a C18 nano-LC column (Acclaim PepMap RSLC, 75 μm inner diameter × 250 mm, 2-μm particle size; Thermo Fisher Scientific) at a flow rate of 400 nl/min and with a column oven temperature of 40 °C. A binary solvent system consisting of 0.1% (v/v) formic acid (solvent A) and 0.1% (v/v) formic acid in 84% (v/v) acetonitrile (solvent B) was used with the following gradient: 5–30% solvent B in 70 min and 30–95% solvent B in 2 min.
The LTQ-Orbitrap Velos was equipped with a nano-electrospray ion source (Thermo Fisher Scientific) and distal coated SilicaTips (New Objective, Woburn, MA). The spray voltage applied was 1.4–1.5 kV, and the capillary temperature was 275 °C. External calibration of the instrument was performed with standard compounds, and for internal calibration, lock masses derived from a set of specific air contaminants were used. For data-dependent MS/MS analyses, the software Xcalibur 2.1 (Thermo Fisher Scientific) was used. Full scan MS spectra were acquired in the Orbitrap with the following parameters: mass range, m/z 300–2,000; resolution, 30,000 (at m/z 400); automatic gain control, 106 ions; maximum fill time, 500 ms. Fragmentation of the 20 most intense multiple charged ions by low energy collision-induced dissociation was carried out in the linear ion trap (automatic gain control, 3 × 104 ions; maximum fill time, 50 ms; normalized collision energy, 35%; activation q, 0.25; activation time, 10 ms). The dynamic exclusion time was set to 30 s.
Data Analysis
Mass spectrometric data of all replicates were processed together using the software MaxQuant (version 1.2.0.18) (28) and its integrated search engine Andromeda (29). For peptide and protein identification, peak lists of MS/MS spectra generated by MaxQuant using default settings were searched against the UniProt reference proteome set for Mus musculus. The species was restricted to M. musculus because exclusively proteins from mice were analyzed. In addition, MS/MS data were searched against a list of common contaminant proteins provided by MaxQuant.
Database searches were performed with tryptic specificity allowing two missed cleavages and 7 ppm initial mass tolerance for precursor ions and 0.5 Da for fragment ions. Oxidation of methionine was considered as variable modification; fixed modifications were not included. Peptides and proteins were identified with a false discovery rate of 0.01 calculated as described previously (28). At least one unique peptide with a minimum length of six amino acids per protein was required. Proteins identified by the same set of peptides were combined to a single protein group by MaxQuant. Only proteins identified with a minimum of two peptides, one of which was unique, in at least two out of three replicates for either the gel-based or the gel-free dataset are reported in this work (supplemental Table S1A). Gene ontology (GO) terms in the “biology process,” “molecular function,” and “cellular component” ontology were obtained using the Perseus software (version 1.4.1.3). Further information about protein and peptide identifications provided by MaxQuant are given in supplemental Tables S1, B and C.
IBAQ values defined as the sum of all peptide intensities of a protein divided by the number of theoretically observable tryptic peptides of the protein (i.e. full tryptic peptides with a length of 6–30 amino acids; (30)) were calculated by MaxQuant. The subcellular localization of proteins was determined based on information provided by UniProt and the GO database. For the prediction of the number of TM domains, the Phobius server (31) was used. Analysis of enriched GO terms was carried out using the Cytoscape (32) plugin BiNGO 2.44 (33). The significance level was set to p < 0.01 as determined by hypergeometric test and Benjamini and Hochberg false discovery rate correction.
Immunohistochemistry
Mice were sacrificed by cervical dislocation at P14, and the snouts were fixed in 4% paraformaldehyde in PBS (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4) overnight at 4 °C. Fixed tissue was cryoprotected in 30% sucrose in PBS for 16 h at 4 °C and embedded and frozen in Tissue-Tek OCT (Sakura Finetek Europe, Leiden, The Netherlands). 13-μm coronal cryosections on superfrost slides (Thermo Fisher Scientific) were obtained using a Leica CM3050S cryomicrotome. Tissue slices were permeabilized with 0.1% (v/v) Triton X-100 in PBS containing 1% (w/v) cold-water fish skin gelatin (Sigma-Aldrich). Cryosections were incubated with primary antibodies in PBS, 0.1% Triton X-100, 1% cold-water fish gelatin overnight at 4 °C, washed three times with PBS, incubated with fluorescently labeled secondary antibodies in PBS for 1 h at room temperature, and mounted in Roti Mount-FluorCare (Carl Roth, Karlsruhe, Germany). The following primary antibodies were used: affinity-purified polyclonal goat anti-AC III (N-14; Santa Cruz Biotechnology, Dallas, TX; catalogue number sc-32113; raised against a peptide mapping to the N-terminal cytoplasmic domain of human AC III (34)); affinity-purified polyclonal rabbit anti-ANXA1 (Abnova, Heidelberg, Germany; PAB7948; raised against a synthetic peptide corresponding to the amino acids at the C terminus of human ANXA1); affinity-purified polyclonal rabbit anti-ANXA2 (Abnova; PAB19088; raised against a synthetic peptide mapping to the internal region of human ANXA2); affinity-purified polyclonal rabbit anti-ANXA5 (Abcam, Cambridge, UK; ab14196; raised against the recombinant full-length protein (35)); polyclonal rabbit anti-PMCA1 antiserum (Novus Biologicals Inc., Littleton, CO; NBP1-46512; raised against a synthetic peptide from the C-terminal region of human PMCA1); affinity-purified polyclonal rabbit anti-SLC24A4/NCKX4 (Assay Biotechnology, Sunnyvale, CA; C18849; raised against a synthetic peptide derived from the internal region of human SLC24A4); and affinity-purified polyclonal rabbit anti-S100A5 (ProteinTech, Chicago; 17924-1-AP; raised against a human S100A5-GST fusion protein). Alexa Fluor-labeled secondary antibodies (goat anti-rabbit Alexa Fluor 594, A-11012 and donkey anti-goat Alexa Fluor 633, A-21082) were obtained from Invitrogen and used in a 1:1,000 dilution. All antibodies directed against the proteins of interest showed a cilia-specific staining pattern in cryosections. No staining was observed in cryosections incubated with secondary antibodies only (data not shown). Fluorescence images were obtained with a confocal microscope (LSM510 Meta; Carl ZEISS AG, Oberkochen, Germany) equipped with a ×40 objective lens (Zeiss) and further processed with CorelPHOTO-PAINT (Corel Corp.).
Immunoblot Analysis
Equal fractions of samples collected from distinct steps of a ciliary membrane preparation were subjected to SDS-PAGE on a 4–12% NuPAGE BisTris gradient gel (Invitrogen) following the manufacturer's instructions. Proteins were transferred to polyvinylidene difluoride (PVDF) membrane using semi-dry blotting at a constant current of 1.2 mA per cm2 for 2 h. The PVDF membrane was blocked with Roti-Block (Carl Roth) for 1 h and subsequently cut horizontally into defined sections to allow for the detection of several subcellular marker proteins of different molecular weight using the same PVDF membrane (i.e. AC III, 170–180 kDa in its glycosylated form; ezrin, 69 kDa; acetylated α-tubulin, 55 kDa; and COX IV, 16 kDa). The PVDF membrane sections were then incubated with the respective primary antibodies (diluted in 4% (w/v) BSA/PBS) for 1 h at room temperature. After extensive washing (four times with PBS, 0.1% (v/v) Tween 20), the membrane sections were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Following incubation with the ECL plus reagent (GE Healthcare), chemiluminescence signals were detected on Hyperfilm (GE Healthcare).
To assess the enrichment of olfactory cilia membranes, we employed primary antibodies that recognize well established marker proteins for ciliary membranes (AC III), microvilli (ezrin), the axoneme (acetylated α-tubulin), and mitochondrial membranes (COX IV); affinity-purified polyclonal rabbit anti-AC III (C-20, Santa Cruz Biotechnology; catalogue number sc-588; raised against a peptide mapping at the C terminus of mouse AC III (16, 36); dilution, 1:200); affinity-purified polyclonal rabbit anti-ezrin (Acris Antibodies, San Diego; AP21103PU-N; dilution, 1:200); mouse monoclonal anti-acetylated α-tubulin (Abcam; ab24610; raised against acetylated α-tubulin from the axoneme of sea urchin sperm flagella (37); dilution, 1:1,000); and monoclonal mouse anti-COX IV (Abcam, ab14744; raised against purified mitochondrial complex IV from cow (38); dilution, 1:200). HRP-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany; goat anti-mouse, 115-035-062; goat anti-rabbit, 111-035-144) and used at a dilution of 1:10,000. For all primary antibodies used for Western blot analyses, a single signal corresponding to the molecular weight of the target protein was detected in the respective section of the PVDF membrane. The intensities of the signals detected for each marker protein in distinct fractions collected during the preparation of ciliary membranes reflect the expected enrichment or depletion of each marker protein across these fractions (supplemental Fig. S1).
RESULTS AND DISCUSSION
Experimental Strategy
Our strategy for the in-depth analysis of the olfactory membrane ciliome was based on the selective enrichment of ciliary membranes from mouse OE followed by protein- and peptide-based separation in conjunction with high resolution LC/MS analysis (Fig. 1A). In the first step, cilia were detached from the OE (n = 4 per replicate) using the Ca2+/K+-shock method as previously used for the preparation of a sensory cilia-enriched fraction from rat OE (16). To confirm the detachment of olfactory cilia from mouse tissue through Ca2+/K+ shock, we used transgenic mice expressing the green fluorescent protein (GFP) under control of the olfactory marker protein (OMP) promoter (39). Analysis of OE cryosections from OMP-GFP mice by immunohistochemistry using an antibody against the sensory ciliary marker protein AC III and confocal microscopy revealed that cilia were effectively detached, leaving the epithelial surface intact and virtually free of cilia (Fig. 1B). In the second step, ciliary fractions from three independent replicates were treated with Na2CO3 at alkaline pH to further enrich for integral membrane proteins (40). The protein content of the resulting ciliary membrane fractions was in the range of 20–30 μg per replicate. The successful enrichment of olfactory cilia membranes was further substantiated by immunoblot analysis using antibodies against the cilia membrane protein AC III and marker proteins for microvilli (ezrin), the axoneme (acetylated tubulin), and mitochondrial membranes (COX IV) (supplemental Fig. S1).
Fig. 1.
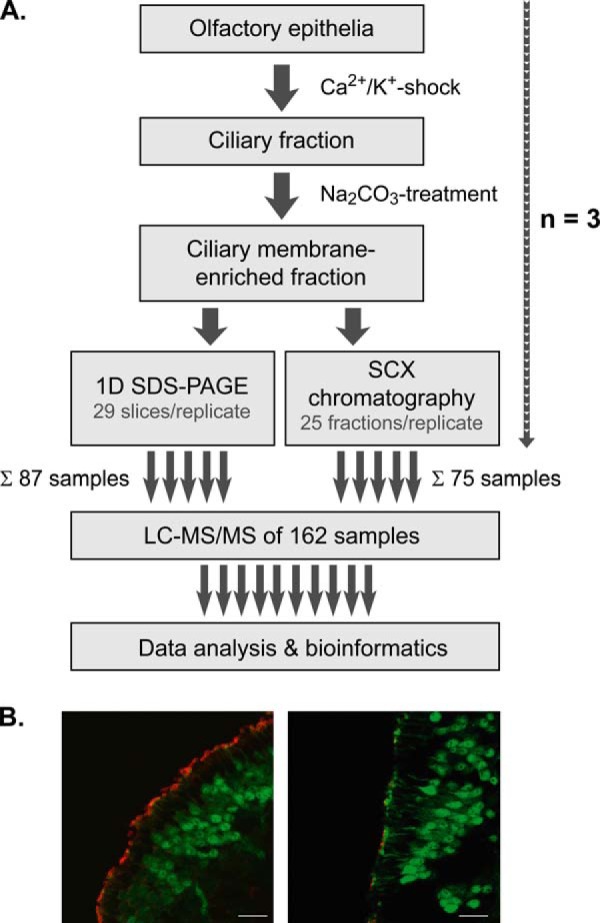
Experimental strategy. A, cilia were detached from OE of mice by Ca2+/K+ shock and further subjected to Na2CO3 washes at alkaline pH. Aliquots of ciliary membrane-enriched fractions were separated by SDS-PAGE or SCX chromatography followed by high resolution LC/MS and bioinformatics data analysis. In total, 162 samples from three independent replicates were analyzed. B, immunostaining of cryosections of OE from transgenic OMP-GFP mice. Mature olfactory sensory neurons as well as dendritic knobs at the ciliary layer are labeled green by GFP. Olfactory cilia were stained specifically using an antibody against adenylyl cyclase III. Confocal micrographs show OE sections before (left) and after (right) Ca2+/K+ shock. Scale bars indicate 20 μm. For further details, refer to main text.
Comparison of a Gel-based and Gel-free Proteomics Approach for the Study of Olfactory Cilia Membranes
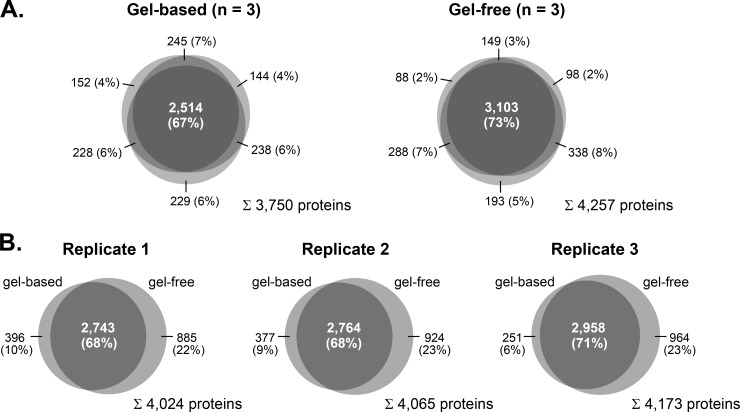
Because a major goal of this study was the establishment of a comprehensive protein inventory of olfactory ciliary membranes to the depth of low abundant ORs, we exploited the capacity of two alternative separation technologies. In the first approach, ciliary membrane fractions (10 μg of protein per replicate) were resolved by SDS-PAGE, and proteins in individual gel lanes were digested with trypsin prior to LC/MS analysis. In the second approach, tryptic peptides from cilia-enriched membranes (10 μg of protein per replicate) were separated by SCX, and the collected fractions were analyzed by LC/MS. Mass spectra were acquired on an LTQ Orbitrap Velos instrument followed by data analysis using the Andromeda/MaxQuant software suite (28, 29). Measurements of a total of 162 samples from three independent ciliary membrane preparations resulted in the identification of 4,403 unique protein groups with an false discovery rate of ≤1% and a minimum of two peptides, one of which was unique, detected in two or more replicates of either the gel-based or gel-free approach (supplemental Table S1A). Of these, 3,750 (85.2%) and 4,257 (96.7%) protein groups were covered by the gel-based and the gel-free approach, respectively (Fig. 2A). Both experimental tracks provided highly reproducible protein identification (ID) data. Using SDS-PAGE, 67% of the proteins were detected in all three replicates, and the overlap of proteins identified in at least two replicates was up to 86%. For the SCX approach, the overlap was 73% and up to 91% for proteins identified in all three and at least two replicates, respectively (Fig. 2A). Furthermore, the comparison of protein IDs from individual replicates showed an overlap of 68–71% between the gel-based and the gel-free approach (Fig. 2B). Of note, more than 20% of all protein IDs per replicate were exclusively obtained via SCX demonstrating the high performance of this approach for the study of the ciliary membrane proteome.
Fig. 2.
Venn diagrams showing the intersections of protein identifications from mouse olfactory cilia membranes. A, overlap of protein identifications (i.e. MaxQuant protein groups) obtained from three independent replicates by gel-based (left) and gel-free (right) proteomics analysis. B, comparison of protein identifications obtained by gel-based and gel-free analyses.
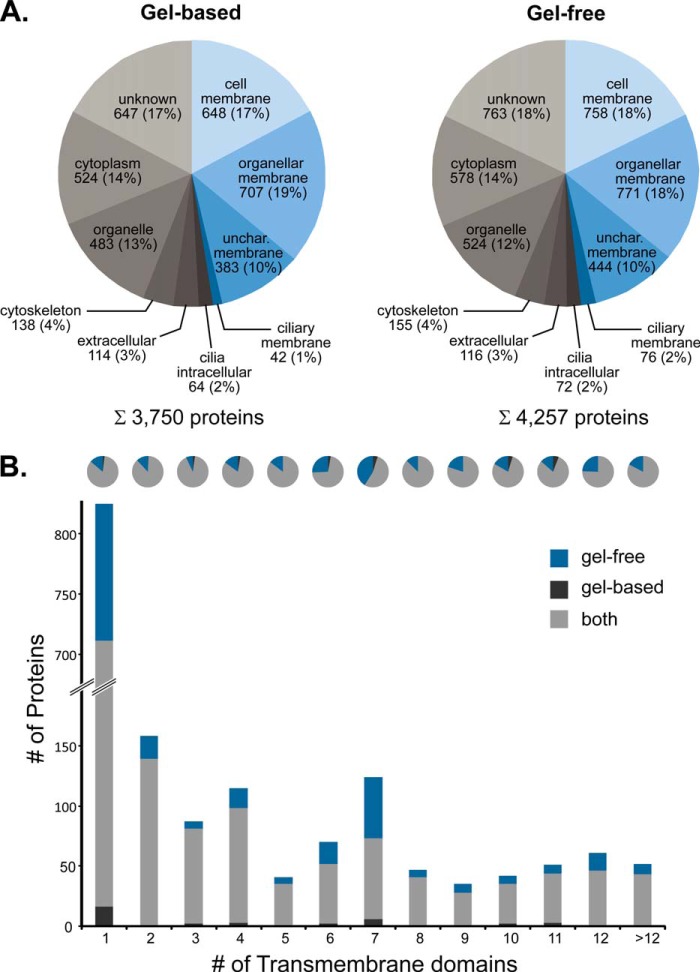
Analysis of the subcellular distribution of all protein IDs based on information derived from UniProt (41) and the Gene Ontology (42) database showed that the data are highly consistent between the two approaches. Approximately half of the proteins have been annotated as integral membrane proteins or membrane-associated (Fig. 3A and supplemental Table S1A). Of these, only 1–2% were annotated as ciliary membrane proteins, and the majority (36%) is located in the plasma membrane and membranes of various organelles. Furthermore, 10% were proteins of noncharacterized membranes, which may include potentially new ciliary proteins. The fraction of soluble proteins originated mainly from the cytoplasm, and various organelles or their localization was unknown, whereas only a small fraction (2%) was annotated as intracellular ciliary proteins (Fig. 3A). A comparison with proteomics data published previously revealed that our dataset covers a total of 373 unique proteins (79%; based on gene names) identified in ciliary membrane-enriched fractions of both mouse OSNs (325 proteins) (11) and rat OSNs (431 proteins) (16).
Fig. 3.
Distribution of proteins identified in olfactory cilia membrane fractions. A, subcellular localization of membrane proteins (blue) as well as soluble proteins (gray) identified by gel-based and gel-free LC/MS analyses. Information about the subcellular localization of identified proteins was derived from UniProt and the Gene Ontology database. Unchar., uncharacterized. B, distribution of membrane proteins according to the number of transmembrane domains predicted by Phobius (31) and according to the detection approach.
To predict the number of TM domains in proteins identified in this work, we used the Phobius server (31). A total of 1,708 proteins with putative TM domains were found with 1,426 proteins for the gel-based and 1,673 proteins for the gel-free approach (Fig. 3B and supplemental Table S1A). A large fraction (825 proteins, i.e. 48%) was predicted to contain a single TM span. Yet, the majority of proteins (883, i.e. 52%) was found to include multiple TM domains. Of particular interest is the class of proteins with seven TM domains characteristic for GPCRs, including low abundant ORs that are important functional components of olfactory sensory cilia. This group shows a local maximum with 124 proteins in our analysis (Fig. 3B). Interestingly, the SCX approach was found to be particularly well suited for the identification of 7-TM helix proteins often fulfilling important functions in signal transduction.
Detection of Olfactory Receptor Proteins in Sensory Cilia Membranes
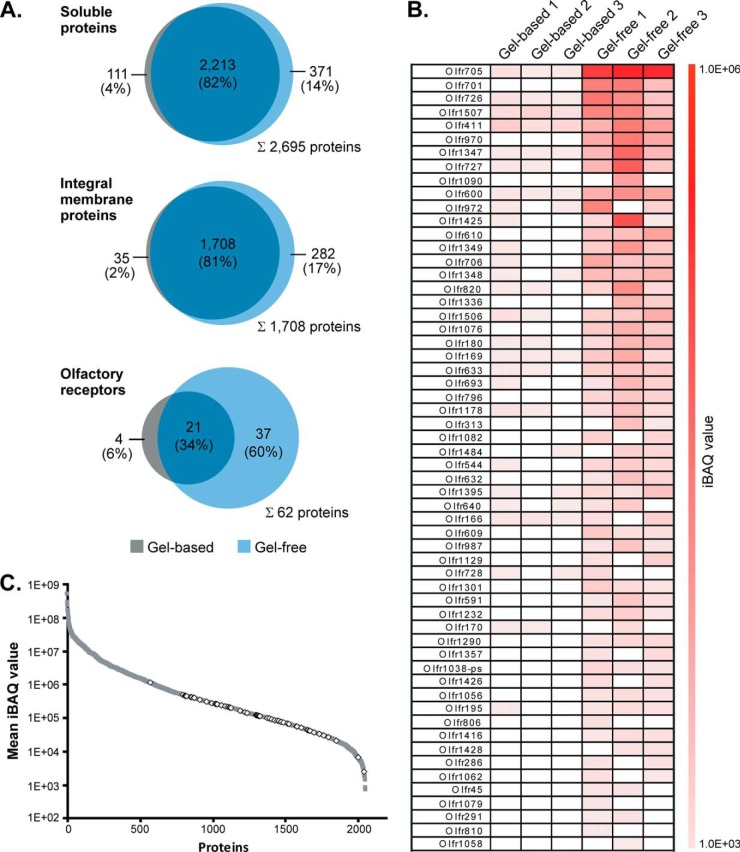
Further comparative analyses of the gel-based and gel-free datasets underscore the high potential of the SCX approach for the detection of low abundant ORs. Although the overlap of protein IDs for both soluble and the entirety of integral membrane proteins was higher than 80% between both approaches, we noticed that 37 (60%) of all the 62 identified ORs were exclusively detected by gel-free analysis (Fig. 4A). Only four ORs (6%) were uniquely identified in the gel-based approach, and a total of 21 ORs (34%) were found in both datasets. The limited ability to detect ORs via SDS-PAGE can presumably be ascribed to their tendency to oligomerize even under denaturing conditions (43–45). In addition, ORs have an N-terminal glycosylation site and may be further glycosylated at the extracellular domains, further impeding electrophoretic separation of ORs into distinct, focused bands (46). Accordingly, we failed to detect ORs in the gel region corresponding to their expected molecular mass of ∼32 kDa, an observation that is in line with previous data (11). The total peptide intensity profile showed that the majority of ORs was detected in gel bands of the upper molecular mass region (∼80 to >140 kDa), although only a minor peak was observed at ∼37 kDa (supplemental Fig. S2).
Fig. 4.
Identification and abundance of olfactory receptors. A, overlap of soluble proteins (top), integral membrane proteins (middle), and olfactory receptors (bottom) identified in olfactory cilia membrane fractions following the gel-based and gel-free approach. B, IBAQ values of all ORs identified in individual replicates of gel-based and gel-free LC/MS analyses. ORs are listed according to the mean of their iBAQ values derived from gel-free experiments. C, relative abundance of ORs. ORs (open diamonds) and all other proteins identified (filled squares) exhibiting a molecular mass of ≤50 kDa are plotted versus their mean iBAQ value determined across all gel-free replicates.
To further assess the capability of our proteomics approach for the detection of ORs from mouse ciliary membranes, we calculated iBAQ values defined as the sum of all peptide intensities divided by the number of theoretically observable tryptic peptides of a protein to estimate relative protein abundance levels (supplemental Table S1A) (30). A total of 62 ORs were ranked according to their mean iBAQ values determined in gel-free experiments and visualized in a heat map (Fig. 4B). In general, iBAQ values of ORs were considerably higher in gel-free compared with gel-based experiments with comparable values between individual replicates. Quantitative data thus confirm the higher capacity of the peptide-centric approach for the detection of low abundant OR proteins. Furthermore, an iBAQ intensity plot (representing proteins detected in gel-free experiments with a mass ≤50 kDa to preclude bias against low molecular mass components) revealed that abundance levels of ORs span more than 2 orders of magnitude (Fig. 4C). Our data therefore clearly point to a considerable heterogeneity in the expression of ORs, a finding that is consistent with real time PCR data revealing differences in mRNA levels for individual ORs of up to 300-fold (47). To conclude, we provide here for the first time detailed information about the relative abundance levels of native ORs in mouse tissue.
Quantitative Analysis of Proteins Involved in Olfactory Signal Transduction Based on iBAQ
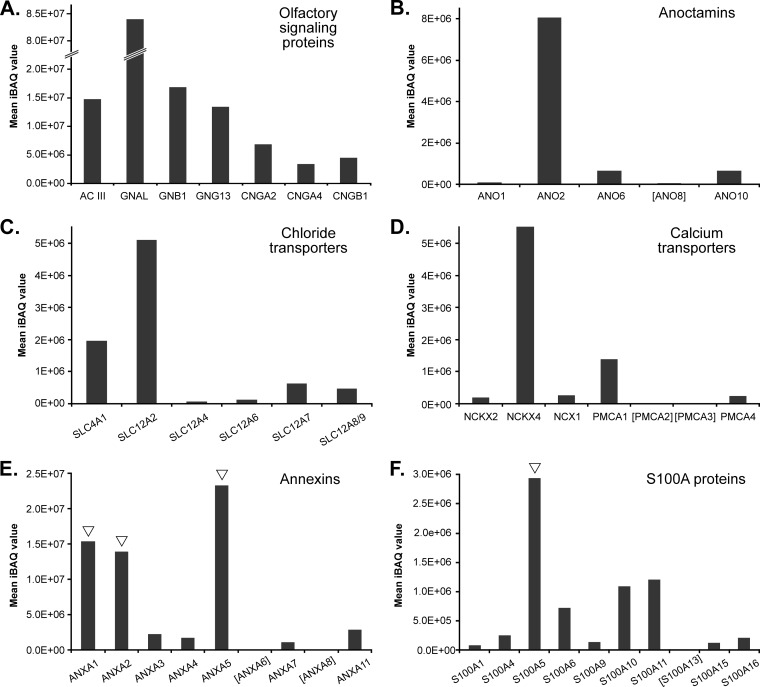
Activation of ORs in ciliary membranes initiates a signaling cascade that eventually leads to the conversion of a chemical cue to an electric current. In addition to the first successful detection of numerous ORs in sensory cilia, we further identified all the known constituents of the canonical pathway of olfactory signal transduction, namely AC III, the three CNG channel subunits CNGA2, CNGA4, and CNGB1, as well as the heterotrimeric G protein subunits GNAL, GNB1, and GNG13 (Fig. 5A and supplemental Table S1A). They all exhibited high iBAQ values (≥3.4 × 106) indicative for their high expression and important functions in olfactory cilia. Interestingly, GNAL (known as G(olf), the α subunit of the heterotrimeric G protein mediating signal transduction by activating AC III) was found to be ∼5-fold more abundant than the two other subunits GNB1 and GNG13. Moreover, AC III was ∼2-fold higher in abundance than the CNG channel subunit CNGA2. High concentrations of G(olf) and AC III guarantee effective signal amplification by the generation of cAMP. Consistent with published data (48), iBAQ-based quantification of the three CNG channel subunits indicated a stoichiometry of ∼2:1:1 (CNGA2:CNGA4:CNGB1).
Fig. 5.
Relative abundance of proteins with known and putative function in olfactory signaling. Olfactory signaling proteins (A), anoctamins (B), chloride and calcium transporters (C and D), annexins (E), and S100A proteins (F) detected in olfactory cilia membrane fractions are plotted against their mean iBAQ value determined across all replicates. ▿, new components of olfactory cilia validated in this work. Mean iBAQ values of proteins in square brackets were in the range of 2.82E + 03 to 9.03E + 04.
We further examined abundance levels of members of the ANO family of which we consistently identified five isoforms (ANO1, ANO2, ANO6, ANO8, and ANO10) (Fig. 5B). ANO1 and ANO2 are known to function as CaCCs (49–53). However, ANO2 was previously shown to be the only family member specifically expressed in the cilia of OSNs (10, 18, 54, 55). In agreement with these data, ANO2 was found to be highly enriched in ciliary membrane fractions. It was ∼12-fold more abundant than ANO6 and ANO10, although ANO1 and ANO8 were only detected in minor amounts. Similarly, the proteins SLC4A1 and SLC12A2 (also known as NKCC1), two solute transporters required for the accumulation of chloride ions in olfactory cilia (18), displayed high iBAQ values that clearly distinguished them from the other detected chloride transporters (i.e. SLC12A4, SLC12A6, SLC12A7, and SLC12A8/9) presumably with no function in olfactory signal transduction (Fig. 5C). Our data further indicate that SLC12A2 is ∼2.6-fold more abundant than SLC4A1, underscoring its important role in chloride-based signal amplification upon OR activation (18).
We next extended our quantitative analysis to membrane transport proteins with potential or known function in intraciliary Ca2+ homeostasis, including K+-dependent (NCKX) or -independent (NCX) Na+/Ca2+ exchangers (9, 56) as well as plasma membrane calcium ATPases (PMCAs) (23, 57, 58). Of the three Na+/Ca2+ exchangers identified, NCKX4 (also known as SLC24A4) exhibited highest abundance, although NCKX2 and NCX1 were detected only in minor amounts in sensory cilia membrane fractions (Fig. 5D). The pivotal function of the K+-dependent Na+/Ca2+ exchanger NCKX4 in olfactory signal termination has recently been demonstrated in NCKX4-deficient mice (19). However, previous work also showed the presence of NCX1 in isolated cilia (57), which was verified by immunohistochemistry revealing also its expression in cell bodies, dendrites, and dendritic knobs of OSNs (59). It was then shown that OMP, a hallmark of mature OSNs, regulates NCX activity by protein-protein interactions allowing it to modulate Ca2+ homeostasis and thus olfactory signal transduction (59). We further report here for the first time the presence of NCKX2 in ciliary membrane fractions of OSNs, yet at comparatively low levels. NCKX2 is a bidirectional transporter typically extruding intracellular Ca2+ and known to be expressed in retinal cone photoreceptors and in the brain where it locally controls neuronal Ca2+ levels allowing for the development of synaptic plasticity (60–62). Therefore, a function of NCKX2 in the modulation of the Ca2+ concentration in olfactory cilia appears to be likely.
In addition to the NCKX and NCX proteins, we detected all four PMCA (also referred to as ATP2B) isoforms in olfactory cilia membranes with PMCA1 being ∼6-fold more abundant than PMCA4, although PMCA2 and PMCA3 were only present in minute amounts (Fig. 5D). All four isoforms have been detected in mouse OSNs with a ciliary expression pattern for the PMCA isoforms 1, 2, and 4 (23). PMCA inhibition was shown to prolong the cAMP-elicited current (57) and to reduce the rate of Ca2+ clearance in the dendritic knobs of stimulated OSNs (58). Interestingly, in sperm, the most abundant form is PMCA4 with a role in hyperactivated motility and male fertility, although for PMCA1 a constitutive function has been proposed (63).
Further proteins with a suggested role in olfactory sensory signal transduction are annexins (ANXA) that bind phospholipids and cellular membranes depending on the intracellular Ca2+ concentration and interact with cytoskeletal proteins to modulate membrane formation and fusion processes (64, 65). Several members of the ANXA protein family have been detected in olfactory cilia membrane preparations by proteomic approaches (11, 15–17). In this work, nine different ANXA isoforms were identified, and of these, ANXA1, ANXA2, and ANXA5 exhibited the highest abundance levels (Fig. 5E). ANXA5 has previously been found to be up-regulated in mouse OE pulse-exposed to the odorant octanal (66). Interestingly, ANXA2 together with S100A10 protein interacts with Na+, K+, and Ca2+ channels to regulate their translocation and function (67–69), and the uniquely cAMP/protein kinase A (PKA)-dependent complex has been shown to specifically interact with the cystic fibrosis conductance regulator protein involved in cystic fibrosis (70) as well as the transient receptor potential vanilloid type 6 channel (TRPV6) in airway and gut epithelia (71). In frog olfactory epithelia, ANXA isoforms 1, 2, and 5 bind to dicalcin in a Ca2+-dependent manner (72). The annexin-dicalcin complexes lead to phospholipid aggregation regulated by Ca2+/calmodulin suggested to protect sensory cilia exposed to environmental and mechanical stress. Dicalcin includes two regions, each displaying more than 50% sequence identity with chicken S100A11 protein (72). In line with this, we identified here 10 different S100A proteins. S100A5 exhibited the highest abundance, and the two members A11 and A10 were ∼3-fold lower in abundance (Fig. 5F). Congruently, S100A5 is specifically expressed in OSNs at the transcript level (73), and its expression pattern correlates with the activity of odorant-stimulated OSNs (74). Moreover, the protein binds Ca2+ with particularly high affinity (75), making it an interesting candidate with potential function in Ca2+ buffering in active OSNs in mice.
Validation of Ciliary Candidate Proteins by Immunohistochemistry
Based on comparative, quantitative analysis of members of distinct protein families involved in olfactory signaling processes, we selected four new candidate proteins (i.e. ANXA1, ANXA2, ANXA5, and S100A5) of sensory cilia for further validation (see Fig. 5). To this end, we studied the subcellular location of candidate proteins in the mouse OE by immunohistochemistry. In addition, we sought to confirm the ciliary localization of the two Ca2+ transporters NCKX4 and PMCA1 exhibiting highest abundance levels among the various Ca2+ transporters detected here. It has previously been shown that NCKX4 mRNA is specifically expressed in the layer of mature OSNs (19) and that endogenous PMCA1 is present in the cilia as well as dendritic knobs, dendrite, and cell body of mouse OSNs (23).
To allow for the visualization of mature OSNs by fluorescence microscopy, we used cryosections of transgenic OMP-GFP mice (39). Thus, OSNs and dendritic knobs at the ciliary layer were labeled green in confocal micrographs (Fig. 6, merged). In addition, we used specific antisera directed against the six proteins of interest (Fig. 6, red) and the ciliary marker protein AC III (blue). In the OE, the dendrites of OSNs extend to the apical surface of the olfactory tissue and multiple cilia project from the dendritic knobs to provide a large chemosensory surface area for the detection of chemical cues entering the nasal cavity. For the ciliary marker AC III and all the endogenous candidate proteins studied here, we consistently observed discrete immunosignals present at the apical surface of the mouse OE and on top of the dendritic knobs of OSNs. The staining pattern of candidate proteins in the respective cryosections corresponded well to the pattern of AC III in the overlay with mature OSNs. For PMCA1 and S100A5, we also observed immunosignals across OSNs, an observation that is consistent with previous reports (23, 76). Taken together, our immunohistochemistry data confirm the presence of ANXA1, ANXA2, ANXA5, S100A5, PMCA1, and NCKX4 in the cilia of mouse OSNs. Based on our quantitative proteomic analysis, we can further conclude that these proteins either binding or transporting Ca2+ ions are specifically associated with membranes of olfactory cilia.
Fig. 6.
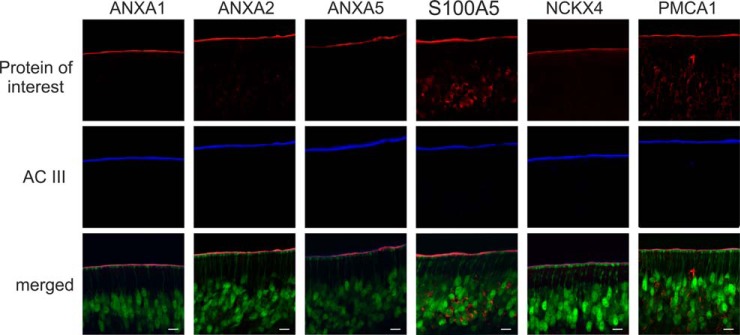
Localization of the candidate proteins ANXA1, ANXA2, ANXA5, and S100A5 to the olfactory cilia membrane. ANXA1, ANXA2, ANXA5, and S100A5 as well as the known ciliary proteins NCKX4 and PMCA1 (all depicted in red) show co-localization with the olfactory cilia marker protein AC III (blue) in cryosections of olfactory epithelia from transgenic OMP-GFP mice. In the overlay, mature olfactory sensory neurons and dendritic knobs are labeled green. Scale bars indicate 10 μm.
Toward the Delineation of the Ciliary Membrane Proteome of OSNs
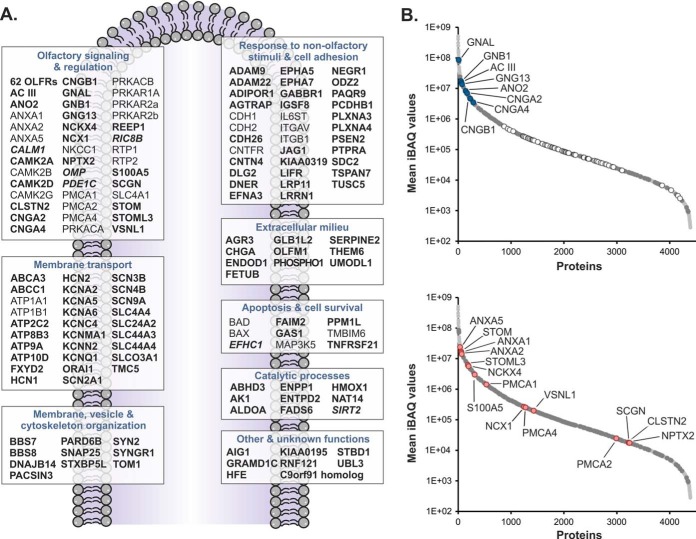
In this work, we sought to specifically delineate the ciliary membrane proteome of OSNs. However, olfactory cilia and membrane fractions thereof cannot be purified to homogeneity, and therefore, the respective protein inventory still contains numerous co-enriched contaminants such as proteins of various intracellular organelles, different forms of tubulin, ribosomal proteins, and various xenobiotic-metabolizing enzymes. These contaminants are known to be commonly present in ciliary membrane preparations of OSNs (15, 16). To address this issue and eventually identify those proteins with specific functions associated with olfactory cilia membranes, we compared our dataset with a catalogue of 691 genes reported to be specifically expressed in mature, ciliated OSNs by transcriptome analysis (24). This resulted in a cilium-specific set of 246 proteins to which we added known olfactory signaling components (59 ORs, CNGB1, GNB1, and ANO2) whose mRNA levels were not represented in the transcriptome data (supplemental Table S1A). GO enrichment analysis confirmed significant over-representation of functional terms related to ciliogenesis, olfactory perception, signal transduction, ion transport, as well as cAMP, ATP, and calmodulin binding, providing a valid representation of known structural and functional characteristics of cilia of OSNs (supplemental Fig. S3). Of these 308 proteins, 178 (58%) were annotated as integral or peripheral components of cell membranes, cilia membranes, and unspecified membranes, or they represent extracellular proteins according to GO and UniProt (supplemental Table S1A). For comprehensiveness, we finally included an additional 35 membrane or regulatory proteins identified in this work with ciliary localization or associated with ciliary functions but not defined mature OSN-specific by transcriptome analysis (24). The molecular blueprint of the ciliary membrane of OSNs established here reflects the different functions of the sensory organelle located at the interface between the brain and the ambient environment (Fig. 7A). It includes essentially all proteins known to be involved in olfactory signal transduction and its regulation as well as numerous proteins with a role in adult neurogenesis. In addition to 62 ORs, we also identified factors enhancing OR expression at the surface (REEP1, RPT1, and RTP2) (77, 78) and different subunits of PKA attenuating OR activity by phosphorylation (79).
Fig. 7.
Extended olfactory membrane ciliome. A, depicted are all 178 proteins defined as olfactory cilium-specific membrane and membrane-associated proteins as well as 35 further membrane or regulatory proteins with ciliary localization or associated with ciliary functions identified in this work but not defined mature OSN-specific by transcriptome analysis (24). For more details about the proteins, please refer to the text and supplemental Table S1A. Bold, cilium-specific membrane proteins; bold and italic, cilium-specific soluble proteins; regular type, non-cilium-specific proteins. B, components of the olfactory membrane ciliome (dark gray circles) and all other proteins identified in olfactory cilia membrane-enriched fractions (light gray circles) are plotted versus their mean iBAQ value. Relative abundance levels of known proteins of the canonical pathway of olfactory signal transduction (blue circles) including 62 ORs (white circles) and components of the Ca2+ signaling network (red circles) are highlighted.
Constituents of the canonical ciliary cAMP-dependent signal transduction pathway (i.e. GNAL, GNB1, GNG13, AC III, CNGB1, CNGA2, CNGB1, and ANO2) were found to be highly abundant with iBAQ values placing them among the top 10% of all proteins (Fig. 7B, top, and supplemental Table S1A). These signaling components are indispensable for the cellular answer to the activation of ORs by binding of an odorant and thus are present ubiquitously among all functional OSNs. As highlighted by these central factors and the 62 ORs, the abundant distribution of the olfactory cilium-specific membrane proteome established in this work covers ∼5 orders of magnitude (Fig. 7B, top). For individual ORs, we found considerable differences in abundance. This finding is consistent with previous data showing that mRNA expression levels for ORs differ significantly and therefore strongly depend on the receptor type (47, 80–84). Differences in OR abundance can generally be accounted for by different mRNA expression levels for a given OR per OSN (47, 85) as well as variations in the number of OSNs expressing an individual OR gene (47, 80–82, 84). Interestingly, for the majority of ORs identified in this work, we observed no clear correlation between protein abundance and mRNA expression levels (data not shown) (83). Among the few exceptions were Olfr705 and Olfr1507, both strongly expressed at the transcript and protein levels. More studies are certainly needed to better understand how OR expression is modulated in OSNs.
The ciliary membrane proteome of OSNs established in this work also includes numerous proteins that constitute a regulatory Ca2+ signaling network (Fig. 7, A and B, bottom panels). Ca2+ signaling is a central theme in different functional aspects of OSNs including the following: (i) olfactory signaling (86–88); (ii) neuroprotection and adaptation (89); and (iii) apoptosis and neural regeneration (90, 91). Accordingly, we found many of these components to be highly abundant (Fig. 7B). The ciliary Ca2+ signaling network includes proteins modulating intracellular Ca2+ concentration such as NCKX4 (19), NCX1 (57), and PMCAs (23, 57, 58) as well as several Ca2+-binding proteins (ANXA1, ANXA2, ANXA5, CLSTN2, VSNL1, STOM, STOML3, NPTX2and SCGN) whose function in the context of sensory cilia have not been well established as yet. For example, visinin-like protein 1 (VSNL1) has been described as an EF-hand-containing Ca2+ sensor that induces neurite outgrowth and is suggested to have neurotoxic and neuroprotective roles in neurons (92). Although a function of different annexins for Ca2+-dependent regulation of ion channels and transporters has been discussed before (67–71), stomatin (STOM) and stomatin-like protein 3 (STOML3) may represent additional new factors. Of note, both proteins have previously been reported to be part of the Ca2+ signaling network of OSNs (17). Furthermore, although STOM is more widely expressed in OSNs (73), STOML3 specifically localizes to sensory cilia where it is proposed to function in olfactory signal transduction (93, 94) and regulates mechanoreceptor sensitivity (95–97). Further low abundant Ca2+-binding proteins of interest are secretagogin (SCGN), calsyntenin 2 (CLSTN2), and neuropentraxin II (NPTX2) (Fig. 7B, bottom). SCGN is related to calbindin and calretinin and modulates neuronal differentiation in the olfactory system (98). Both CLSTN2 and NPTX2 have a potential role in synaptic plasticity (99, 100), and a homologue of NPTX2 is involved in intracellular protein sorting to the acrosome in testis (101).
In the following, the discussion will be focused on further selected components and distinct functional aspects of the olfactory cilia membrane proteome reported in this work (Fig. 7).
Because OSNs are functionally renewed in less than a month (13, 14), a tight balance between apoptosis and survival has to be ensured to maintain sensory function. In this context, our ciliary proteome of OSNs provides several molecular links. For example, we identified the tumor necrosis factor receptor superfamily member 21 (TNFRSF21), also known as death receptor 6, which has been reported to induce apoptosis through a new BAX-dependent pathway (102). A further factor is the growth arrest-specific 1 (GAS1) protein that induces apoptosis by dephosphorylation of BAD (103). Of note, we also detected BAX, BAD, and additional apoptosis regulators as well as the BAX inhibitor protein TMBIM6 at high abundance (supplemental Table S1A). Further identified membrane-associated proteins with an anti-apoptotic function are Fas apoptotic inhibitory molecule 2 (FAIM2) (104) and the Ser/Thr protein phosphatase PPM1L (105). We further detected the EF-hand-containing Ca2+-binding protein EFHC1 executing a pro-apoptotic effect through interaction with a voltage-dependent Ca2+ channel in cultured primary neurons; mutations in EFHC1 cause juvenile myoclonic epilepsy (106). It is of interest to note here that the mouse orthologue of EFHC1 is a conserved axonemal protein mainly expressed in sperm flagella and tracheal cilia, suggesting a potential role of cilia in the development of juvenile myoclonic epilepsy (107). The protein was reported to be absent from immotile primary cilia (107). Yet, we provide here clear evidence for its presence in olfactory cilia that is consistent with transcriptome data (24).
Further components of ciliary membranes identified in this work are factors involved in the following: (i) vesicle trafficking and intraflagellar transport, e.g. BBS7 and BBS8, two membrane-associated components of the BBSome essential for the sorting of proteins to the cilia (108); (ii) membrane transport processes to maintain ion concentration gradients across the ciliary membrane (numerous K+ and Na+ channels or Na+/K+ ATPase subunits such as FXYD2 (alias ATP1G1), ATP1A1, and ATP1B1); and (iii) catalytic processes, e.g. AK1 and ALDOA, both known to localize to sperm flagella where they contribute to meet the high energy demand of the motile organelle (109, 110).
Our data also indicate an extensive communication between olfactory cilia and the extracellular milieu in response to nonolfactory stimuli. Previous studies of primary cilia provided functional connections to the Hedgehog (111), Wnt (112), and Notch (113–115) signaling pathways as well as evidence for the presence of both adhesion receptors of the integrin family and Ca2+-dependent adhesion proteins, the cadherins (116). Moreover, odorant-induced Notch signaling in Drosophila brain has been reported (117). Consistently, we detected the Notch ligands JAG1 and DNER (118) and the Notch signaling component γ-secretase PSEN2 involved in the migration of OSNs (119). In addition to several integrins (ITGB1, ITGAV, and PCDHB1) and cadherins (CDH1, CDH2, and CDH26), we further report teneurin-2 (ODZ2) functioning in neurogenesis and cell adhesion processes (120, 121) and ephrin-A5 (EPHA5) involved in neuronal guidance by regulating ITGB1 activity (122). Other members detected are the EPHA7 receptor and the membrane-anchored ephrin EFNA3.
The ciliary membrane dataset also includes plexin A3 (PLXNA3) and A4 (PLXNA4), two semaphorin receptors involved in neuronal development and axon path finding (123, 124), as well as the leukemia inhibitory factor receptor. Evidence for a putative function of PLXNA3 in the development of sensory receptor neurons in the inner ear (125) and the olfactory system has recently been provided (126). Furthermore, leukemia inhibitory factor receptor-interleukin 6 receptor (IL6ST) heterodimers interacting with the ciliary neurotrophic factor receptor (127) mediate LIF-induced signaling that is involved in neurogenesis and regeneration of OSNs under conditions leading to degenerative cell death (128, 129).
Finally, we would like to point to the new ciliary candidate tetraspanin 7 (TSPAN7), a cell surface glycoprotein with four TM domains, that is reported to form signaling complexes at the plasma membrane where it localizes to distinct microdomains to possibly modulate neurite growth and neuronal migration in interplay with integrins (130). Tetraspanins have also been suggested to regulate the activity of matrix metalloproteinases (131) with a role in cell adhesion and neurogenesis (132). Consistently, we report here for the first time the presence of two disintegrin and metalloproteinase domain-containing proteins, ADAM9 and ADAM22, in olfactory cilia membranes. The latter protein has recently been found in brain as a component of voltage-gated Ca2+ channel environments (133).
CONCLUSIONS
A detailed understanding of the functions of olfactory cilia requires in-depth knowledge about the molecular components constituting this unique sensory organelle. In this study, we concentrated on the comprehensive analysis of the membrane compartment of olfactory cilia isolated from mice to learn more about the proteins central for forming this important chemical interface to convey information from the exterior environment to the inside of OSNs. Isolation of cilia diminutive in size from mouse olfactory epithelia is a highly demanding task, and no protocols exist that facilitate the purification of primary cilia and membranes thereof to homogeneity. The strategy applied here is universally applicable to the study of primary cilia. Classical techniques based on chemical stresses to gently detach cilia from tissues and prepare ciliary membrane sheets are combined with powerful protein/peptide separation methods followed by high resolution LC/MS for deep proteome sequencing analysis. The approach showed excellent reproducibility and yielded the largest, so far, cilia-derived dataset consisting of 4,403 protein entries. Gel-free analyses enabled us for the first time to reliably detect a total of 62 ORs revealing a substantial heterogeneity in their abundance levels. We identified and further verified the four Ca2+-binding proteins ANXA1, ANXA2, ANXA5, and S100A5 as true resident proteins of olfactory cilia providing new functional targets to investigate how olfactory signal transduction is modulated. With the methods at hand, we will now be able to decipher stimulus-induced changes in the olfactory cilia membrane proteome, including low abundant signaling components like ORs, and thus to decipher processes governing long term adaptation and/or sensitization of the peripheral olfactory system toward chemical cues. Because pure ciliary membranes cannot be obtained to date, we exploited an existing transcriptome dataset specific for mature, ciliated OSNs to verify and establish the currently most complete membrane ciliome of OSNs revealing a remarkable compositional complexity and high functional diversity of this unique sensory organelle. We expect that our findings will be consequential in promoting signaling research on olfactory cilia as well as primary cilia in general.
The mass spectrometry proteomics data as well as MaxQuant result files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (134) with the dataset identifier PXD000514.
Supplementary Material
Acknowledgments
We thank Eva M. Neuhaus (Charité Berlin) and Günter Gisselmann (Ruhr-Universität Bochum) for scientific discussion.
Footnotes
Author contributions: B.W. designed research; K.K., A.T., and H.W. performed the research; H.E.M., H.H.H., and B.W. contributed new reagents or analytic tools; K.K., A.T., H.W., M.E., S.O., and B.W. analyzed data; and K.K., S.O., and B.W. wrote the paper.
* This work was supported by Deutsche Forschungsgemeinschaft Grant SFB642, Excellence Initiative of the German Federal and State Governments Grant EXC 294 BIOSS Centre for Biological Signaling Studies (to B.W.), and P.U.R.E (Protein Unit for Research in Europe), a project funded by the federal state of Nordrhein-Westfalen, Germany (to M.E. and H.E.M.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- OR
- olfactory receptor
- AC III
- adenylyl cyclase type III
- CNG
- cyclic nucleotide-gated cation channel
- GFP
- green fluorescent protein
- GO
- Gene Ontology
- OE
- olfactory epithelium
- OSN
- olfactory sensory neuron
- SCX
- strong cation exchange
- TM
- transmembrane
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GPCR
- G protein-coupled receptor
- ID
- identification
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. McEwen D. P., Jenkins P. M., Martens J. R. (2008) Olfactory cilia: our direct neuronal connection to the external world. Curr. Top. Dev. Biol. 85, 333–370 [DOI] [PubMed] [Google Scholar]
- 2. Buck L., Axel R. (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 [DOI] [PubMed] [Google Scholar]
- 3. Godfrey P. A., Malnic B., Buck L. B. (2004) The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. U.S.A. 101, 2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niimura Y., Nei M. (2003) Evolution of olfactory receptor genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 100, 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breer H. (2003) Sense of smell: recognition and transduction of olfactory signals. Biochem. Soc. Trans 31, 113–116 [DOI] [PubMed] [Google Scholar]
- 6. Su C.-Y., Menuz K., Carlson J. R. (2009) Olfactory perception: receptors, cells, and circuits. Cell 139, 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeMaria S., Ngai J. (2010) The cell biology of smell. J. Cell Biol. 191, 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frings S., Reuter D., Kleene S. J. (2000) Neuronal Ca2+-activated Cl− channels–homing in on an elusive channel species. Prog. Neurobiol. 60, 247–289 [DOI] [PubMed] [Google Scholar]
- 9. Reisert J., Bauer P. J., Yau K.-W., Frings S. (2003) The Ca-activated Cl channel and its control in rat olfactory receptor neurons. J. Gen. Physiol. 122, 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rasche S., Toetter B., Adler J., Tschapek A., Doerner J. F., Kurtenbach S., Hatt H., Meyer H., Warscheid B., Neuhaus E. M. (2010) Tmem16b is specifically expressed in the cilia of olfactory sensory neurons. Chem. Senses 35, 239–245 [DOI] [PubMed] [Google Scholar]
- 11. Stephan A. B., Shum E. Y., Hirsh S., Cygnar K. D., Reisert J., Zhao H. (2009) ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc. Natl. Acad. Sci. U.S.A. 106, 11776–11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Firestein S. (2001) How the olfactory system makes sense of scents. Nature 413, 211–218 [DOI] [PubMed] [Google Scholar]
- 13. Hatt H. (2004) Molecular and cellular basis of human olfaction. Chem. Biodivers 1, 1857–1869 [DOI] [PubMed] [Google Scholar]
- 14. Murray R. C., Calof A. L. (1999) Neuronal regeneration: lessons from the olfactory system. Semin. Cell Dev. Biol. 10, 421–431 [DOI] [PubMed] [Google Scholar]
- 15. Mayer U., Ungerer N., Klimmeck D., Warnken U., Schnölzer M., Frings S., Möhrlen F. (2008) Proteomic analysis of a membrane preparation from rat olfactory sensory cilia. Chem. Senses 33, 145–162 [DOI] [PubMed] [Google Scholar]
- 16. Mayer U., Küller A., Daiber P. C., Neudorf I., Warnken U., Schnölzer M., Frings S., Möhrlen F. (2009) The proteome of rat olfactory sensory cilia. Proteomics 9, 322–334 [DOI] [PubMed] [Google Scholar]
- 17. Klimmeck D., Mayer U., Ungerer N., Warnken U., Schnölzer M., Frings S., Möhrlen F. (2008) Calcium-signaling networks in olfactory receptor neurons. Neuroscience 151, 901–912 [DOI] [PubMed] [Google Scholar]
- 18. Hengl T., Kaneko H., Dauner K., Vocke K., Frings S., Möhrlen F. (2010) Molecular components of signal amplification in olfactory sensory cilia. Proc. Natl. Acad. Sci. U.S.A. 107, 6052–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stephan A. B., Tobochnik S., Dibattista M., Wall C. M., Reisert J., Zhao H. (2012) The Na+/Ca2+ exchanger NCKX4 governs termination and adaptation of the mammalian olfactory response. Nat. Neurosci. 15, 131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niimura Y., Nei M. (2007) Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fleischer J., Breer H., Strotmann J. (2009) Mammalian olfactory receptors. Front. Cell. Neurosci. 3, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whitelegge J. P. (2013) Integral membrane proteins and bilayer proteomics. Anal. Chem. 85, 2558–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weeraratne S. D., Valentine M., Cusick M., Delay R., Van Houten J. L. (2006) Plasma membrane calcium pumps in mouse olfactory sensory neurons. Chem. Senses 31, 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nickell M. D., Breheny P., Stromberg A. J., McClintock T. S. (2012) Genomics of mature and immature olfactory sensory neurons. J. Comp. Neurol. 520, 2608–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Wiese S., Reidegeld K. A., Meyer H. E., Warscheid B. (2007) Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 7, 340–350 [DOI] [PubMed] [Google Scholar]
- 27. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 30. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 31. Käll L., Krogh A., Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 32. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess over-representation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 34. Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., Hass P. E., Sandoval W., Peterson A. S. (2012) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14, 61–72 [DOI] [PubMed] [Google Scholar]
- 35. Li W., Li H., Sanders P. N., Mohler P. J., Backs J., Olson E. N., Anderson M. E., Grumbach I. M. (2011) The multifunctional Ca2+/calmodulin-dependent kinase II δ (CaMKIIδ) controls neointima formation after carotid ligation and vascular smooth muscle cell proliferation through cell cycle regulation by p21. J. Biol. Chem. 286, 7990–7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He J., Tian H., Lee A. C., Ma M. (2012) Postnatal experience modulates functional properties of mouse olfactory sensory neurons. Eur. J. Neurosci. 36, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toth C., Shim S. Y., Wang J., Jiang Y., Neumayer G., Belzil C., Liu W.-Q., Martinez J., Zochodne D., Nguyen M. D. (2008) Ndel1 promotes axon regeneration via intermediate filaments. PLoS ONE 3, e2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yatsuga S., Suomalainen A. (2012) Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum. Mol. Genet. 21, 526–535 [DOI] [PubMed] [Google Scholar]
- 39. Potter S. M., Zheng C., Koos D. S., Feinstein P., Fraser S. E., Mombaerts P. (2001) Structure and emergence of specific olfactory glomeruli in the mouse. J. Neurosci. 21, 9713–9723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Magrane M., Consortium U. (2011) UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cook B. L., Ernberg K. E., Chung H., Zhang S. (2008) Study of a synthetic human olfactory receptor 17–4: expression and purification from an inducible mammalian cell line. PLoS ONE 3, e2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cook B. L., Steuerwald D., Kaiser L., Graveland-Bikker J., Vanberghem M., Berke A. P., Herlihy K., Pick H., Vogel H., Zhang S. (2009) Large-scale production and study of a synthetic G protein-coupled receptor: human olfactory receptor 17-4. Proc. Natl. Acad. Sci. U.S.A. 106, 11925–11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leck K.-J., Zhang S., Hauser C. A. (2010) Study of bioengineered zebra fish olfactory receptor 131-2: receptor purification and secondary structure analysis. PLoS ONE 5, e15027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lanctot P. M., Leclerc P. C., Clément M., Auger-Messier M., Escher E., Leduc R., Guillemette G. (2005) Importance of N-glycosylation positioning for cell-surface expression, targeting, affinity, and quality control of the human AT1 receptor. Biochem. J 390, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young J. M., Shykind B. M., Lane R. P., Tonnes-Priddy L., Ross J. A., Walker M., Williams E. M., Trask B. J. (2003) Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 4, R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng J., Zagotta W. N. (2004) Stoichiometry and assembly of olfactory cyclic nucleotide-gated channels. Neuron 42, 411–421 [DOI] [PubMed] [Google Scholar]
- 49. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. J. (2008) TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 [DOI] [PubMed] [Google Scholar]
- 50. Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y. (2008) Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y. D., Cho H., Koo J. Y., Tak M. H., Cho Y., Shim W.-S., Park S. P., Lee J., Lee B., Kim B.-M., Raouf R., Shin Y. K., Oh U. (2008) TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 [DOI] [PubMed] [Google Scholar]
- 52. Pifferi S., Dibattista M., Menini A. (2009) TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch. 458, 1023–1038 [DOI] [PubMed] [Google Scholar]
- 53. Stöhr H., Heisig J. B., Benz P. M., Schöberl S., Milenkovic V. M., Strauss O., Aartsen W. M., Wijnholds J., Weber B. H., Schulz H. L. (2009) TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J. Neurosci. 29, 6809–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sagheddu C., Boccaccio A., Dibattista M., Montani G., Tirindelli R., Menini A. (2010) Calcium concentration jumps reveal dynamic ion selectivity of calcium-activated chloride currents in mouse olfactory sensory neurons and TMEM16b-transfected HEK 293T cells. J. Physiol. 588, 4189–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Billig G. M., Pál B., Fidzinski P., Jentsch T. J. (2011) Ca2+-activated Cl− currents are dispensable for olfaction. Nat. Neurosci. 14, 763–769 [DOI] [PubMed] [Google Scholar]
- 56. Pyrski M., Koo J. H., Polumuri S. K., Ruknudin A. M., Margolis J. W., Schulze D. H., Margolis F. L. (2007) Sodium/calcium exchanger expression in the mouse and rat olfactory systems. J. Comp. Neurol. 501, 944–958 [DOI] [PubMed] [Google Scholar]
- 57. Castillo K., Delgado R., Bacigalupo J. (2007) Plasma membrane Ca2+-ATPase in the cilia of olfactory receptor neurons: possible role in Ca2+ clearance. Eur. J. Neurosci. 26, 2524–2531 [DOI] [PubMed] [Google Scholar]
- 58. Saidu S. P., Weeraratne S. D., Valentine M., Delay R., Van Houten J. L. (2009) Role of plasma membrane calcium ATPases in calcium clearance from olfactory sensory neurons. Chem. Senses 34, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kwon H. J., Koo J. H., Zufall F., Leinders-Zufall T., Margolis F. L. (2009) Ca extrusion by NCX is compromised in olfactory sensory neurons of OMP mice. PLoS ONE 4, e4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dong H., Light P. E., French R. J., Lytton J. (2001) Electrophysiological characterization and ionic stoichiometry of the rat brain K+-dependent Na+/Ca2+ exchanger, NCKX2. J. Biol. Chem. 276, 25919–25928 [DOI] [PubMed] [Google Scholar]
- 61. Schnetkamp P. P. (2004) The SLC24 Na+/Ca2+-K+ exchanger family: vision and beyond. Pflugers Arch. 447, 683–688 [DOI] [PubMed] [Google Scholar]
- 62. Li X.-F., Kiedrowski L., Tremblay F., Fernandez F. R., Perizzolo M., Winkfein R. J., Turner R. W., Bains J. S., Rancourt D. E., Lytton J. (2006) Importance of K+-dependent Na+/Ca2+-exchanger 2, NCKX2, in motor learning and memory. J. Biol. Chem. 281, 6273–6282 [DOI] [PubMed] [Google Scholar]
- 63. Okunade G. W., Miller M. L., Pyne G. J., Sutliff R. L., O'Connor K. T., Neumann J. C., Andringa A., Miller D. A., Prasad V., Doetschman T., Paul R. J., Shull G. E. (2004) Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J. Biol. Chem. 279, 33742–33750 [DOI] [PubMed] [Google Scholar]
- 64. Gerke V., Moss S. E. (2002) Annexins: from structure to function. Physiol. Rev. 82, 331–371 [DOI] [PubMed] [Google Scholar]
- 65. Santamaria-Kisiel L., Rintala-Dempsey A. C., Shaw G. S. (2006) Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 396, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barbour J., Neuhaus E. M., Piechura H., Stoepel N., Mashukova A., Brunert D., Sitek B., Stühler K., Meyer H. E., Hatt H., Warscheid B. (2008) New insight into stimulus-induced plasticity of the olfactory epithelium in Mus musculus by quantitative proteomics. J. Proteome Res. 7, 1594–1605 [DOI] [PubMed] [Google Scholar]
- 67. Girard C., Tinel N., Terrenoire C., Romey G., Lazdunski M., Borsotto M. (2002) p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 21, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Okuse K., Malik-Hall M., Baker M. D., Poon W.-Y., Kong H., Chao M. V., Wood J. N. (2002) Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature 417, 653–656 [DOI] [PubMed] [Google Scholar]
- 69. van de Graaf S. F., Hoenderop J. G., Gkika D., Lamers D., Prenen J., Rescher U., Gerke V., Staub O., Nilius B., Bindels R. J. (2003) Functional expression of the epithelial Ca2+ channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. EMBO J. 22, 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Borthwick L. A., McGaw J., Conner G., Taylor C. J., Gerke V., Mehta A., Robson L., Muimo R. (2007) The formation of the cAMP/protein kinase A-dependent annexin 2-S100A10 complex with cystic fibrosis conductance regulator protein (CFTR) regulates CFTR channel function. Mol. Biol. Cell 18, 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Borthwick L. A., Neal A., Hobson L., Gerke V., Robson L., Muimo R. (2008) The annexin 2-S100A10 complex and its association with TRPV6 is regulated by cAMP/PKA/CnA in airway and gut epithelia. Cell Calcium 44, 147–157 [DOI] [PubMed] [Google Scholar]
- 72. Uebi T., Miwa N., Kawamura S. (2007) Comprehensive interaction of dicalcin with annexins in frog olfactory and respiratory cilia. FEBS J. 274, 4863–4876 [DOI] [PubMed] [Google Scholar]
- 73. Yu T.-T., McIntyre J. C., Bose S. C., Hardin D., Owen M. C., McClintock T. S. (2005) Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J. Comp. Neurol. 483, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bennett M. K., Kulaga H. M., Reed R. R. (2010) Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol. Cell. Neurosci. 43, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schäfer B. W., Fritschy J. M., Murmann P., Troxler H., Durussel I., Heizmann C. W., Cox J. A. (2000) Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J. Biol. Chem. 275, 30623–30630 [DOI] [PubMed] [Google Scholar]
- 76. McIntyre J. C., Davis E. E., Joiner A., Williams C. L., Tsai I.-C., Jenkins P. M., McEwen D. P., Zhang L., Escobado J., Thomas S., Szymanska K., Johnson C. A., Beales P. L., Green E. D., Mullikin J. C., NISC Comparative Sequencing Program, Sabo A., Muzny D. M., Gibbs R. A., Attié-Bitach T., Yoder B. K., Reed R. R., Katsanis N., Martens J. R. (2012) Gene therapy rescues cilia defects and restores olfactory function in a mammalian ciliopathy model. Nat. Med. 18, 1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saito H., Kubota M., Roberts R. W., Chi Q., Matsunami H. (2004) RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691 [DOI] [PubMed] [Google Scholar]
- 78. Matsunami H., Mainland J. D., Dey S. (2009) Trafficking of mammalian chemosensory receptors by receptor-transporting proteins. Ann. N.Y. Acad. Sci. 1170, 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mashukova A., Spehr M., Hatt H., Neuhaus E. M. (2006) β-Arrestin2-mediated internalization of mammalian odorant receptors. J. Neurosci. 26, 9902–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ressler K. J., Sullivan S. L., Buck L. B. (1993) A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73, 597–609 [DOI] [PubMed] [Google Scholar]
- 81. Iwema C. L., Fang H., Kurtz D. B., Youngentob S. L., Schwob J. E. (2004) Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J. Neurosci. 24, 356–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rimbault M., Robin S., Vaysse A., Galibert F. (2009) RNA profiles of rat olfactory epithelia: individual and age related variations. BMC Genomics 10, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shiao M.-S., Chang A. Y., Liao B.-Y., Ching Y.-H., Lu M.-Y., Chen S. M., Li W.-H. (2012) Transcriptomes of mouse olfactory epithelium reveal sexual differences in odorant detection. Genome Biol. Evol. 4, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Khan M., Vaes E., Mombaerts P. (2013) Temporal patterns of odorant receptor gene expression in adult and aged mice. Mol. Cell. Neurosci. 57, 120–129 [DOI] [PubMed] [Google Scholar]
- 85. Zhao S., Tian H., Ma L., Yuan Y., Yu C. R., Ma M. (2013) Activity-dependent modulation of odorant receptor gene expression in the mouse olfactory epithelium. PLoS ONE 8, e69862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Menini A. (1999) Calcium signalling and regulation in olfactory neurons. Curr. Opin. Neurobiol. 9, 419–426 [DOI] [PubMed] [Google Scholar]
- 87. Matthews H. R., Reisert J. (2003) Calcium, the two-faced messenger of olfactory transduction and adaptation. Curr. Opin. Neurobiol. 13, 469–475 [DOI] [PubMed] [Google Scholar]
- 88. Kleene S. J. (2008) The electrochemical basis of odor transduction in vertebrate olfactory cilia. Chem. Senses 33, 839–859 [DOI] [PubMed] [Google Scholar]
- 89. Friedman L. K. (2006) Calcium: a role for neuroprotection and sustained adaptation. Mol. Interv. 6, 315–329 [DOI] [PubMed] [Google Scholar]
- 90. Schwob J. E. (2002) Neural regeneration and the peripheral olfactory system. Anat. Rec. 269, 33–49 [DOI] [PubMed] [Google Scholar]
- 91. Beites C. L., Kawauchi S., Crocker C. E., Calof A. L. (2005) Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp. Cell Res. 306, 309–316 [DOI] [PubMed] [Google Scholar]
- 92. Braunewell K.-H., Klein-Szanto A. J., Szanto A. J. (2009) Visinin-like proteins (VSNLs): interaction partners and emerging functions in signal transduction of a subfamily of neuronal Ca2+-sensor proteins. Cell Tissue Res. 335, 301–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kobayakawa K., Hayashi R., Morita K., Miyamichi K., Oka Y., Tsuboi A., Sakano H. (2002) Stomatin-related olfactory protein, SRO, specifically expressed in the murine olfactory sensory neurons. J. Neurosci. 22, 5931–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Goldstein B. J., Kulaga H. M., Reed R. R. (2003) Cloning and characterization of SLP3: a novel member of the stomatin family expressed by olfactory receptor neurons. J. Assoc. Res. Otolaryngol. 4, 74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wetzel C., Hu J., Riethmacher D., Benckendorff A., Harder L., Eilers A., Moshourab R., Kozlenkov A., Labuz D., Caspani O., Erdmann B., Machelska H., Heppenstall P. A., Lewin G. R. (2007) A stomatin-domain protein essential for touch sensation in the mouse. Nature 445, 206–209 [DOI] [PubMed] [Google Scholar]
- 96. Martinez-Salgado C., Benckendorff A. G., Chiang L.-Y., Wang R., Milenkovic N., Wetzel C., Hu J., Stucky C. L., Parra M. G., Mohandas N., Lewin G. R. (2007) Stomatin and sensory neuron mechanotransduction. J. Neurophysiol. 98, 3802–3808 [DOI] [PubMed] [Google Scholar]
- 97. Moshourab R. A., Wetzel C., Martinez-Salgado C., Lewin G. (2013) Stomatin-domain protein interactions with acid sensing ion channels modulate nociceptor mechanosensitivity. J. Physiol. 591, 5555–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Attems J., Alpar A., Spence L., McParland S., Heikenwalder M., Uhlén M., Tanila H., Hökfelt T. G., Harkany T. (2012) Clusters of secretagogin-expressing neurons in the aged human olfactory tract lack terminal differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 6259–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Omeis I. A., Hsu Y. C., Perin M. S. (1996) Mouse and human neuronal pentraxin 1 (NPTX1): conservation, genomic structure, and chromosomal localization. Genomics 36, 543–545 [DOI] [PubMed] [Google Scholar]
- 100. Preuschhof C., Heekeren H. R., Li S.-C., Sander T., Lindenberger U., Bäckman L. (2010) KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia 48, 402–408 [DOI] [PubMed] [Google Scholar]
- 101. Reid M. S., Blobel C. P. (1994) Apexin, an acrosomal pentaxin. J. Biol. Chem. 269, 32615–32620 [PubMed] [Google Scholar]
- 102. Zeng L., Li T., Xu D. C., Liu J., Mao G., Cui M.-Z., Fu X., Xu X. (2012) Death receptor 6 induces apoptosis not through type I or type II pathways, but via a unique mitochondria-dependent pathway by interacting with Bax protein. J. Biol. Chem. 287, 29125–29133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zarco N., González-Ramírez R., González R. O., Segovia J. (2012) GAS1 induces cell death through an intrinsic apoptotic pathway. Apoptosis 17, 627–635 [DOI] [PubMed] [Google Scholar]
- 104. Fernández M., Segura M. F., Solé C., Colino A., Comella J. X., Ceña V. (2007) Lifeguard/neuronal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. J. Neurochem. 103, 190–203 [DOI] [PubMed] [Google Scholar]
- 105. Saito J., Toriumi S., Awano K., Ichijo H., Sasaki K., Kobayashi T., Tamura S. (2007) Regulation of apoptosis signal-regulating kinase 1 by protein phosphatase 2Cε. Biochem. J. 405, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Suzuki T., Delgado-Escueta A. V., Aguan K., Alonso M. E., Shi J., Hara Y., Nishida M., Numata T., Medina M. T., Takeuchi T., Morita R., Bai D., Ganesh S., Sugimoto Y., Inazawa J., Bailey J. N., Ochoa A., Jara-Prado A., Rasmussen A., Ramos-Peek J., Cordova S., Rubio-Donnadieu F., Inoue Y., Osawa M., Kaneko S., Oguni H., Mori Y., Yamakawa K. (2004) Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat. Genet. 36, 842–849 [DOI] [PubMed] [Google Scholar]
- 107. Ikeda T., Ikeda K., Enomoto M., Park M. K., Hirono M., Kamiya R. (2005) The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 579, 819–822 [DOI] [PubMed] [Google Scholar]
- 108. Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., Nachury M. V. (2010) The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cao W., Haig-Ladewig L., Gerton G. L., Moss S. B. (2006) Adenylate kinases 1 and 2 are part of the accessory structures in the mouse sperm flagellum. Biol. Reprod. 75, 492–500 [DOI] [PubMed] [Google Scholar]
- 110. Krisfalusi M., Miki K., Magyar P. L., O'Brien D. A. (2006) Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 75, 270–278 [DOI] [PubMed] [Google Scholar]
- 111. Huangfu D., Anderson K. V. (2005) Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. U.S.A. 102, 11325–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bisgrove B. W., Yost H. J. (2006) The roles of cilia in developmental disorders and disease. Development 133, 4131–4143 [DOI] [PubMed] [Google Scholar]
- 113. Stubbs J. L., Davidson L., Keller R., Kintner C. (2006) Radial intercalation of ciliated cells during Xenopus skin development. Development 133, 2507–2515 [DOI] [PubMed] [Google Scholar]
- 114. Krebs L. T., Iwai N., Nonaka S., Welsh I. C., Lan Y., Jiang R., Saijoh Y., O'Brien T. P., Hamada H., Gridley T. (2003) Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 17, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ezratty E. J., Stokes N., Chai S., Shah A. S., Williams S. E., Fuchs E. (2011) A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 145, 1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Seeger-Nukpezah T., Golemis E. A. (2012) The extracellular matrix and ciliary signaling. Curr. Opin. Cell Biol. 24, 652–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lieber T., Kidd S., Struhl G. (2011) DSL-Notch signaling in the Drosophila brain in response to olfactory stimulation. Neuron 69, 468–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Eiraku M., Tohgo A., Ono K., Kaneko M., Fujishima K., Hirano T., Kengaku M. (2005) DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat. Neurosci. 8, 873–880 [DOI] [PubMed] [Google Scholar]
- 119. Maier E., Nord H., von Hofsten J., Gunhaga L. (2011) A balance of BMP and notch activity regulates neurogenesis and olfactory nerve formation. PLoS ONE 6, e17379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Rubin B. P., Tucker R. P., Martin D., Chiquet-Ehrismann R. (1999) Teneurins: a novel family of neuronal cell surface proteins in vertebrates, homologous to the Drosophila pair-rule gene product Ten-m. Dev. Biol. 216, 195–209 [DOI] [PubMed] [Google Scholar]
- 121. Rubin B. P., Tucker R. P., Brown-Luedi M., Martin D., Chiquet-Ehrismann R. (2002) Teneurin 2 is expressed by the neurons of the thalamofugal visual system in situ and promotes homophilic cell-cell adhesion in vitro. Development 129, 4697–4705 [DOI] [PubMed] [Google Scholar]
- 122. Davy A., Robbins S. M. (2000) Ephrin-A5 modulates cell adhesion and morphology in an integrin-dependent manner. EMBO J. 19, 5396–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cheng H. J., Bagri A., Yaron A., Stein E., Pleasure S. J., Tessier-Lavigne M. (2001) Plexin-A3 mediates semaphorin signaling and regulates the development of hippocampal axonal projections. Neuron 32, 249–263 [DOI] [PubMed] [Google Scholar]
- 124. Schwarz Q., Waimey K. E., Golding M., Takamatsu H., Kumanogoh A., Fujisawa H., Cheng H.-J., Ruhrberg C. (2008) Plexin A3 and plexin A4 convey semaphorin signals during facial nerve development. Dev. Biol. 324, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Katayama K., Imai F., Suto F., Yoshida Y. (2013) Deletion of Sema3a or plexinA1/plexinA3 causes defects in sensory afferent projections of statoacoustic ganglion neurons. PLoS ONE 8, e72512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Young J., Metay C., Bouligand J., Tou B., Francou B., Maione L., Tosca L., Sarfati J., Brioude F., Esteva B., Briand-Suleau A., Brisset S., Goossens M., Tachdjian G., Guiochon-Mantel A. (2012) SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Hum. Reprod. 27, 1460–1465 [DOI] [PubMed] [Google Scholar]
- 127. Skiniotis G., Lupardus P. J., Martick M., Walz T., Garcia K. C. (2008) Structural organization of a full-length gp130/LIF-R cytokine receptor transmembrane complex. Mol. Cell 31, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Getchell T. V., Shah D. S., Partin J. V., Subhedar N. K., Getchell M. L. (2002) Leukemia inhibitory factor mRNA expression is upregulated in macrophages and olfactory receptor neurons after target ablation. J. Neurosci. Res. 67, 246–254 [DOI] [PubMed] [Google Scholar]
- 129. Nan B., Getchell M. L., Partin J. V., Getchell T. V. (2001) Leukemia inhibitory factor, interleukin-6, and their receptors are expressed transiently in the olfactory mucosa after target ablation. J. Comp. Neurol. 435, 60–77 [DOI] [PubMed] [Google Scholar]
- 130. Berditchevski F. (2001) Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114, 4143–4151 [DOI] [PubMed] [Google Scholar]
- 131. Lafleur M. A., Xu D., Hemler M. E. (2009) Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol. Biol. Cell 20, 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zucker S., Pei D., Cao J., Lopez-Otin C. (2003) Membrane type-matrix metalloproteinases (MT-MMP). Curr. Top. Dev. Biol. 54, 1–74 [DOI] [PubMed] [Google Scholar]
- 133. Müller C. S., Haupt A., Bildl W., Schindler J., Knaus H.-G., Meissner M., Rammner B., Striessnig J., Flockerzi V., Fakler B., Schulte U. (2010) Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 107, 14950–14957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.