Abstract
We report the orthologous screening, engineering, and optimization of amino acid conversion enzymes for cell-specific proteomic labeling. Intracellular endoplasmic-reticulum-anchored Mycobacterium tuberculosis diaminopimelate decarboxylase (DDCM.tub-KDEL) confers cell-specific meso-2,6-diaminopimelate-dependent proliferation to multiple eukaryotic cell types. Optimized lysine racemase (LyrM37-KDEL) supports D-lysine specific proliferation and efficient cell-specific isotopic labeling. When ectopically expressed in discrete cell types, these enzymes confer 90% cell-specific isotopic labeling efficiency after 10 days of co-culture. Moreover, DDCM.tub-KDEL and LyrM37-KDEL facilitate equally high cell-specific labeling fidelity without daily media exchange. Consequently, the reported novel enzyme pairing can be used to study cell-specific signaling in uninterrupted, continuous co-cultures. Demonstrating the importance of increased labeling stability for addressing novel biological questions, we compare the cell-specific phosphoproteome of fibroblasts in direct co-culture with epithelial tumor cells in both interrupted (daily media exchange) and continuous (no media exchange) co-cultures. This analysis identified multiple cell-specific phosphorylation sites specifically regulated in the continuous co-culture. Given their applicability to multiple cell types, continuous co-culture labeling fidelity, and suitability for long-term cell–cell phospho-signaling experiments, we propose DDCM.tub-KDEL and LyrM37-KDEL as excellent enzymes for cell-specific labeling with amino acid precursors.
Cell–cell communication is an essential component of tissue homeostasis and is frequently deregulated in disease (1). Despite advances in the understanding of intracellular signaling networks in monoculture, our capacity to biochemically investigate communication between cells in direct co-culture remains limited. This is primarily due to the technical difficulty of discerning cell-specific protein signaling events from a co-culture. Stable isotope labeling with amino acids in cell culture (SILAC)1 (2) can be used to identify the discrete proteomes of different cell populations in co-culture (3). However, as L-lysine and L-arginine are equally metabolized by all cell types in a co-culture, SILAC technology is not suitable for investigating long-term cell-specific proteomic changes in proliferating co-cultures.
To address this limitation, Gauthier et al. recently reported an alternative cell-specific isotopic labeling technology (4). This approach, entitled cell-type-specific labeling with amino acid precursors (CTAP), relies on the ectopic expression of non-mammalian amino acid processing enzymes to convert lysine precursors into essential L-lysine. Two distinct enzymes specifically convert their respective lysine precursor substrates into L-lysine: diaminopimelate decarboxylase (DDC) converts meso-2,6-diaminopimelate (DAP) into L-lysine (5), and lysine racemase (Lyr) converts D-lysine into L-lysine (6). When these enzymes are combined with isotopically labeled D-lysine, their cell-specific expression confers perpetual and cell-specific labeling of two cell populations in co-culture (supplemental Figs. S1A and S1B).
As this technique uses amino acid processing enzymes to confer isotopic labeling, the ectopic performance of DDC and Lyr is directly responsible for cell-specific labeling fidelity. Thus, for biologically relevant CTAP experiments, optimal DDC and Lyr enzymes are essential. Optimal cell-specific labeling enzymes should (i) confer excellent precursor-specific proliferation, (ii) be strictly intracellular, (iii) be applicable to multiple cell types, (iv) avoid transcriptional cell stress, and (v) facilitate continuous (uninterrupted) co-culture experiments (supplemental Fig. S1C).
Conceptually, cell-specific labeling with amino acid precursors is an extremely powerful approach for proteomic analysis of long-term cell–cell communication. The approach has broad applications in cell biology and facilitates the investigation of previously inaccessible processes. However, the nascent methodology is restricted by suboptimal amino acid processing enzymes that hamper the labeling fidelity, biological significance, and widespread adoption of CTAP. For example, Gauthier et al. (4) note that extracellular amino acid converting enzymes substantially reduce long-term cell-specific labeling fidelity. As a result, daily media exchange is required in order to achieve cell-specific labeling efficiencies of ∼80%. The constant removal of conditioned media might undermine the biological significance of continuous cell–cell communication because of the “interrupted” co-culture environment. Moreover, although Gauthier et al. (4) employed DDC from Arabidopsis thaliana (DDCA.tha), the authors note that several cell lines ectopically expressing this enzyme fail to grow efficiently on DAP. As a result, DDCA.tha is suboptimal for the widespread adoption of cell-specific labeling with amino acid precursors.
Given the broad potential application of cell-specific labeling with amino acid precursors, we sought to develop an optimal enzyme pairing capable of conferring high-fidelity, cell-specific, isotopic labeling to multiple cell types. Here we report the screening, engineering, and characterization of optimized DDC and Lyr enzymes for use in cell-specific labeling with amino acid precursors.
EXPERIMENTAL PROCEDURES
Cell Culture
All cells were obtained from ATCC Manassas, VA (except KPC cells, a kind gift from Professor Owen Sansom, Glasgow) and were grown in DMEM (deficient for L-lysine and L-arginine) (DMP49, Caisson, Utah) supplemented with 10% (v/v) dialyzed FBS (Invitrogen), 0.3 mm L-arginine (A8094, Sigma) and/or L-lysine (L5501, Sigma), D-lysine (L5876, Sigma), and DAP (07036, Sigma). To increase reliance on cell-derived cell–cell communication factors, all co-cultures were performed in reduced (0.5%) dialyzed FBS. For SILAC isotopic labeling, cells were grown in 2.5 mm “medium” L-lysine (Isotec 616192, Sigma) (delta mass: 4.025107; delta average mass: 4.0246) or “heavy” L-lysine (Isotec 608041, Sigma) (delta mass: 8.014199; delta average mass: 7.9427). For CTAP isotopic labeling, LyrM37-KDEL cells were grown in either 2.5 mm medium D-lysine-4,4,5,5-d4 HCl (C/D/N d-7334) (delta mass: 4.025107; delta average mass: 4.0246) or 2.5 mm heavy D-lysine-3,3,4,4,5,5,6,6-d8 2HCl (C/D/N d-6367) (delta mass: 8.050214; delta average mass: 8.0493).
Cloning, Gene Synthesis, and Mutagenesis
DDC from A. thaliana (DDCA.tha) (Q949X7), Methanocaldococcus jannaschii (DDCM.jan) (Q58497), Helicobacter pylori (DDCH.pyl) (B4XMC6), Mycobacterium tuberculosis (DDCM.tub) (P0A5M4), Mycobacterium leprae (DDCM.lep) (Q50140), and Mycobacterium avium (DDCM.avi) (A0QCV9) were custom synthesized by GeneArt (Invitrogen) (codon optimized for mouse expression) and cloned into pCDNA3.1 Zeo(+) via EcoRI/NotI. A carboxyl-terminal HA tag was included to facilitate protein detection. A KDEL endoplasmic reticulum (ER) retention motif was added to the C terminus of DDCM.tub via PCR (custom primers (Sigma)). The DDCM.tub Cys93Ala mutation was inserted using the QuikChange II XL Site-Directed Mutagenesis Kit (200521, Agilent Technologies, Santa Clara, CA) and custom primers (Sigma).
Proteus mirabilis lysine racemase (LyrWT) (M4GGR9) was custom synthesized by GeneArt (Invitrogen) (codon optimized for mouse expression) and cloned into pCDNA3.1 Zeo(+) (V86020, Invitrogen) via EcoRI/NotI. A carboxyl-terminal HA tag was included to facilitate protein detection. The putative leader sequence was removed (amino acids 1–36) (LyrM37) and a KDEL ER retention motif was added at the C terminus via PCR (custom primers (Sigma)) (LyrM37-KDEL). Full annotated sequences can be found in the supplemental material. CTAP constructs have been submitted to AddGene (DDCM.tub-KDEL: 51529 and LyrM37-KDEL: 51530).
Transfection and Proliferation Studies
Cells were transfected in a six-well plate with Lipofectamine 2000 (11668, Invitrogen) (1:4 DNA:lipofectamine ratio). After 24 h, cells were re-plated at 3 × 103 cells per well in a white 96-well plate (734–2002, Nunc, Roskilde, Denmark) and grown in the presence of titrated concentrations of L-lysine, D-lysine, or DAP. After 72 h, cell viability was measured using CellTiter-Glo® (G7570, Promega, Madison, WI). All results are expressed as a percentage of cells grown in 10 mm L-lysine. For stable selection, cells were transfected as above and grown for five passages in the presence of either 2.5 mm D-lysine (LyrM37-KDEL) or 5 mm DAP (DDCM.tub-KDEL). MDA-MB-231 cells were infected with pGIPZ lentivirus (Thermo Scientific) for GFP expression, and C3H10T1/2 cells were infected with pMSCV-pBabeMCS-IRES-RFP retrovirus (33337, Addgene, Cambridge, MA) for RFP expression. MDA-MB-231 GFP+ and C3H10T1/2 RFP+ expressing cells were then isolated using a FACSAria (BD Biosciences).
Western Blotting
Cells were lysed in PLC buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 1.5 mm MgCl2, 1 mm EGTA), and 50 μg of lysate was resolved via SDS-PAGE. All Western blotting was performed on the LiCor Odyssey platform (intensity = 5, gamma = 1). Primary antibodies were as follows: rabbit anti-HA (1:5000) (ab9110, Abcam, Cambridge, UK), mouse anti-β-actin (1:5000) (ab6276, Abcam), rabbit p-eIF2α (Ser51) (1:1000) (CST 3398, Cell Signaling Technology, Danvers, MA), and mouse eIF2α (1:1000) (CST 2103, Cell Signaling Technology). Secondary antibodies were anti-rabbit DyLight 800 (1:15,000) (CST 5151, Cell Signaling Technology) and anti-mouse DyLight 680 (1:15,000) (CST 5470, Cell Signaling Technology).
Microscopy
C3H10T1/2 cells were transfected as described above and plated on Ibidi 35-mm μ-Dishes (80136, Thistle Scientific, Glasgow, UK). After 24 h, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 2% BSA, 2% FBS. Cells were then immunostained for α-tubulin (MCA78G, AbD Serotec, Oxford, UK) (1:1000), protein disulfide isomerase (ab2792, Abcam) (1:1000), and HA tag (ab9110, Abcam) (1:5000). Alexa Fluor® 546 goat anti-rabbit (A-11035, Invitrogen), Alexa Fluor® 647 goat anti-mouse (A-21236, Invitrogen) and Alexa Fluor® 488 donkey anti-rat (A-21208, Invitrogen) were all used at 1:1000. Hoechst (33258, Sigma) nuclei staining was performed at 1 μg/ml. Cells were imaged on a Zeiss 710 confocal microscope, and images were analyzed using Zeiss Zen software Oberkochen, Germany.
Labeling Efficiency
For the triple-labeled comparison between L-lysine and D-lysine, C3H10T1/2 +LyrM37-KDEL cells were grown for five passages on 2.5 mm light, medium (+4 Da), or heavy (+8 Da) L-lysine or D-lysine. For MDA-MB-231 +DDCM.tub-KDEL/C3H10T1/2 +LyrM37-KDEL co-cultures, cells were seeded at 5 × 105 cells per cell type in a 10-cm dish, grown in the presence of 2.5 mm heavy (+8 Da) D-Lysine and 5 mm DAP for 10 days, and then separated via FACS (see below). Cells were washed in PBS, lysed in PLC buffer, and resolved via SDS-PAGE, and gel bands were excised. Proteins were reduced with 10 mm dithiothreitol, alkylated with 50 mm iodoacetamide, and digested with 50 ng of trypsin (V5111, Promega). Peptides were eluted in 5% trifluoroacetic acid (TFA), 50% acetonitrile, lyophilized, and resuspended in 0.1% TFA. Following LC-MS/MS (Orbitrap Velos) (see details below), each sample was searched for all three isotopomeric versions of lysine (Mascot, Matrix Sciences, London, UK) in Protein Discoverer 1.4 (Thermo Scientific). Raw intensity values from each label were summed, and the percentage contribution of each label was calculated per peptide.
Flow Cytometry
MDA-MB-231/C3H10T1/2 co-cultures were harvested using trypsin, and live cells were discerned using 0.2 μg/ml DAPI (Invitrogen). Cells were then sorted on a FACSAria (BD Biosciences) according to their GFP and RFP expression. The mCherry channel was used for better separation of the RFP-expressing C3H10T1/2 cells. Analyses were performed using the software FlowJo (Tree Star Inc., Ashland, OR).
Selected Reaction Monitoring
To empirically identify proteotypic Lyr/DDC peptides suitable for relative isotopic quantification, we transfected C3H10T1/2 cells with LyrM37-KDEL, DDCM.tub-KDEL, or an empty vector control; lysed them in PLC buffer; and resolved them via SDS-PAGE. Bands were excised between 35 and 50 kDa and digested as described above. Peptides were eluted in 5% TFA, 50% acetonitrile, lyophilized, and resuspended in 0.1% TFA. Samples were analyzed on a TSQ Vantage triple quadruple mass spectrometer (Thermo Scientific) coupled to a NanoLC-Ultra 1D (Eksigent, Dublin, CA). Reversed-phase chromatographic separation was performed on an Acclaim PepMap100 C18 Nano-Trap Column (100 μm inner diameter by 2 cm packed with C18 (5-μm bead size, 100 Å)) (Thermo Scientific) and an NTCC-360 packed tip column (75 μm inner diameter, 15 cm, 3-μm particle size) (Nikkyo Technos, Tokyo, Japan) with a 30-min linear gradient of 5% to 50% solvent B (100% acetonitrile + 0.1% formic acid). The TSQ Vantage was operated with a Q1 unit resolution of 0.7 full width at half-maximum, a Q3 of 0.7 full width at half-maximum, an ion spray voltage of 2200 V, and a capillary inlet temperature of 270 °C. Peptide fragmentation was carried out in Q2 at 1.5 millitorr, and collision energies for each peptide were predicted (7). Each selected reaction monitoring (SRM) transition had a minimum dwell time of 20 ms, with cycle times of 1.2 s. The raw data files were produced in Xcalibur 2.1 (Thermo Scientific), and all data were processed using Skyline 2.1 (8). The three most intense peptides from each enzyme were selected for use in relative isotopic protein quantification (supplemental Table S1). Representative product ion chromatograms for these peptides are shown in supplemental Fig. S5.
To discern cell-specific labeling fidelity, C3H10T1/2 cells stably transfected with LyrM37-KDEL and MDA-MB-231 cells stably transfected with DDCM.tub-KDEL were seeded at 5 × 105 cells per cell type in a 10-cm dish and directly co-cultured for 7 days. Each day a population of cells were lysed and tryptic peptides were prepared as described above. Unscheduled SRM for both light and heavy (+8 Da) LyrM37-KDEL and DDCM.tub-KDEL lysine-containing peptides was performed. The top two transitions from each peptide were selected for relative isotopic quantification of each enzyme, light and heavy transitions were summed, and the heavy percentage was calculated per peptide across technical duplicates.
SRM transitions have been uploaded to PeptideAtlas (www.peptideatlas.org/PASS/PASS00410).
Growth Factor Array
MDA-MB-231/C3H10T1/2 co-cultures were seeded at 5 × 105 cells per cell type in a 10-cm dish and grown with (interrupted) or without (continuous) daily media exchange (+0.5% dialyzed FBS) (n = 4). After 5 days, conditioned media was collected from each co-culture culture, and relative changes in growth factors were determined using a reverse-phase glass slide antibody capture array (AAH-CYT-G2000–8, RayBiotech, Norcross, GA) (174 proteins quantified in technical duplicate per sample).
Phosphopeptide Enrichment
MDA-MB-231/C3H10T1/2 co-cultures were seeded at 5 × 105 cells per cell type in a 10-cm dish, grown for 5 days (with or without daily media exchange (+0.5% dialyzed FBS)), lysed in 6 m urea, pooled, sonicated, centrifuged to clear cell debris, and digested with Lys-C/trypsin using the filter-aided sample preparation (FASP) method (9). Phosphopeptides were enriched by adding 1 mg of magnetic TiO2 (MR-TID010, ReSyn Bio, South Africa) or Ti-IMAC (MR-TIM010, ReSyn Bio) beads to 200 μg of tryptic peptides (following the manufacturer's protocol) and desalted using OLIGO™-R3 resin (Invitrogen). Each biological replicate contained 3× TiO2 and 3× Ti-IMAC enrichments (6× LC-MS/MS technical replicates per biological replicate). LC-MS/MS phosphopeptide identification was performed as described below.
Discovery LC-MS/MS (Data-dependent Acquisition)
All samples were run on an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) coupled to a NanoLC-Ultra 2D (Eksigent). Reverse-phase chromatographic separation was performed on a 100 μm inner diameter × 20 mm trap column packed in-house with C18 (5-μm bead size, Reprosil-Gold, Dr Maisch, Ammerbuch-Entringen, Germany) and a 75 μm inner diameter × 30 cm column packed in-house with C18 (5-μm bead size, Reprosil-Gold, Dr Maisch) using a 120-min linear gradient of 0% to 50% solvent B (100% acetonitrile + 0.1% formic acid) against solvent A (100% H2O + 0.1% formic acid) with a flow rate of 300 nL/min. The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap MS and MS/MS acquisition. Survey full-scan MS spectra (m/z 375–2000) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 and a Fourier transform target value of 1 × 106 ions. The 20 most abundant ions were selected for fragmentation using collision-induced dissociation and dynamically excluded for 8 s. For phosphopeptide samples, the 10 most abundant ions were selected for fragmentation using higher-energy collisional dissociation and scanned in the Orbitrap at a resolution of 7500 at m/z 400. Selected ions were dynamically excluded for 8 s. For accurate mass measurement, the lock mass option was enabled using the polydimethylcyclosiloxane ion (m/z 445.120025) as an internal calibrant. For peptide identification, raw data files produced in Xcalibur 2.1 (Thermo Scientific) were processed in Proteome Discoverer 1.4 (Thermo Scientific) and searched against Swiss-Prot human (20012 02 release, 20,662,136 entries) and mouse (2011 03 release, 15,082,690 entries) databases using Mascot (v2.2). Searches were performed with a precursor mass tolerance of 10 ppm, a fragment mass tolerance of 0.5 Da, and a maximum of two missed cleavages. Static modifications were limited to carbamidomethylation of cysteine, and variable modifications used were oxidation of methionine; deamidation of asparagine/glutamine; isotopomeric labeled lysine (+4 and +8); and phosphorylation of serine, threonine, and tyrosine residues. Peptides were further filtered using a Mascot significance threshold of <0.05, a peptide ion score of >20, and a false discovery rate of <0.01 (evaluated by Percolator (10)). Phospho-site localization probabilities were calculated with phosphoRS 3.1 (11). For relative phosphopeptide quantification, heavy/medium ratios were calculated by Proteome Discoverer 1.4 (normalized to protein median). Only lysine-containing peptides were included in the quantitative analysis.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (12) with the dataset identifiers PXD000870 (labeling efficiency) and PXD000871 (cell-specific phosphoproteomics).
RESULTS
Improved DDC for Cell-specific Labeling with Amino Acid Precursors
DDC converts DAP into L-lysine (5). Currently, only A. thaliana (thale cress) DDC (DDCA.tha) has been successfully reported to confer DAP-dependent growth in co-culture (4). However, only one cell line has been successfully established with DAP-dependent growth, and the authors note that several other cell lines do not proliferate efficiently with DDCA.tha. Moreover, there is currently no empirical evidence that DDCA.tha is retained within eukaryotic cells, which could lead to compromised labeling fidelity in co-culture. Given the broad potential applications of cell-specific labeling with amino acid precursors, a DDC enzyme that confers efficient, intracellular, DAP-dependent growth in a variety of cell lines is essential.
Given the poor activity of DDCA.tha, we hypothesized that DDC enzymes sourced from alternative species may confer improved DAP-dependent growth to eukaryotic cells. To investigate this, we compared DDCA.tha with orthologous DDC from M. jannaschii (a thermophilic archaea) (DDCM.jan), H, pylori (a microaerophilic stomach bacterium) (DDCH.pyl), and M. tuberculosis (a pathological mycobacterium) (DDCM.tub) (Fig. 1A). These enzymes were screened because of their distinct dimerization profiles and active site loop structures (13) (supplemental Fig. S2). The orthologous enzymes all expressed to similar levels (Fig. 1B), and despite the absence of a predicted signal peptide (supplemental Fig. S3A) (SignalP 4.1 (14)), small traces of each DDC were detected in conditioned media. Notably, DDCM.tub conferred dramatically improved DAP-dependent growth relative to all other enzymes. As a result, DDCM.tub was advanced for further optimization and characterization.
Fig. 1.
Optimized cell-specific diaminopimelate decarboxylase. A, C3H10T1/2 proliferation on titrated concentrations of meso-2,6-diaminopimelate (DAP) when transfected with HA-tagged diaminopimelate decarboxylase (DDC) from Arabidopsis thaliana (DDCA.tha), Methanocaldococcus jannaschii (DDCM.jan), Helicobacter pylori (DDCH.pyl), or Mycobacterium tuberculosis (DDCM.tub). Results are expressed as a percentage of cells grown in 10 mm L-lysine. DDCM.tub substantially outperformed all other enzymes. B, all DDC enzymes were primarily found in the cell lysate, although small traces could be detected in conditioned media. C, DDCM.tub and DDCM.tub-KDEL expression in C3H10T1/2 fibroblasts. D, DDCM.tub and DDCM.tub-KDEL DAP-dependent proliferation in C3H10T1/2 fibroblasts. E, immunofluorescent DDCM.tub localization in C3H10T1/2 fibroblasts. “α-Tubulin” denotes total cell shape, and “PDI” (protein disulfide isomerase) discerns the endoplasmic reticulum (ER). DDCM.tub was found throughout the cell, whereas DDCM.tub-KDEL located to the ER. F, stable C3H10T1/2+DDCM.tub-KDEL cells were grown on titrated concentrations of L-lysine, D-lysine, or DAP for 72 h. DDCM.tub-KDEL conferred growth comparable to L-lysine when the DAP concentration was >2.5 mm. DDCM.tub-KDEL cells did not grow on D-lysine. G, C3H10T1/2+DDCM.tub-KDEL cells were grown for 24 h in 10 mm L-lysine, the absence of lysine, or titrated concentrations of DAP. The transcriptional arrest marker pS51 eIF2α was used as a marker of amino acid starvation and measured by Western blotting. Concentrations of DAP greater than 1 mm rescued the amino acid starvation response. H, proliferation of multiple cell lines on 5 mm DAP following transient transfection with an empty vector, DDCA.tha, or DDCM.tub-KDEL. I, C3H10T1/2 proliferation on titrated concentrations of DAP when transfected with an empty vector or DDC enzymes from DDCM.tub, Mycobacterium leprae (DDCM.lep), Mycobacterium avium (DDCM.avi), or DDCA.tha. Cell viability was measured after 72 h. J, C3H10T1/2 proliferation on titrated concentrations of DAP when transfected with wild-type DDCM.tub, DDCM.tub Cys93Ala mutant, DDCA.tha, or DDCH.ply. The Cys93Ala mutation dramatically reduced DAP-dependent cell proliferation, suggesting that disulfide homodimerization is critical to the improved survival conferred by DDCM.tub. K, expression of DDC enzymes from J.
As intracellular DDC is required to ensure true cell-specific labeling, we anchored DDCM.tub in the ER by adding a C-terminal KDEL retention sequence (15) (DDCM.tub-KDEL). DDCM.tub-KDEL exhibited reduced extracellular levels relative to wild-type DDCM.tub (Fig. 1C) and conferred excellent DAP-dependent growth (Fig. 1D). DDCM.tub was found throughout the cell, whereas DDCM.tub-KDEL was retained within the ER (Fig. 1E). These results confirm that DDC can function efficiently within the eukaryotic ER. Stably transfected DDCM.tub-KDEL cells conferred proliferation comparable to that with L-lysine when grown on concentrations of DAP greater than 2.5 mm and did not grow on D-lysine at any concentration (Fig. 1F).
To ensure that DDCM.tub-KDEL did not induce an amino acid stress response, stable DDCM.tub-KDEL cells were grown on titrated concentrations of DAP, and the phosphorylation of eIF2α on serine 51 (pS51 eIF2α) was used as an early translational arrest stress marker (16). In accordance with our earlier proliferation studies, DDCM.tub-KDEL conferred non-stressed cell growth at DAP concentrations between 1 and 10 mm (Fig. 1G).
To test their applicability across multiple cell types, we transfected DDCA.tha and DDCM.tub-KDEL into eight eukaryotic cell lines and measured cell viability following growth on 5 mm DAP (Fig. 1H). Relative to cells grown on 10 mm L-lysine, DDCA.tha transfected cells typically achieved between 15% and 40% proliferation when grown on 5 mm DAP. In contrast, all cell lines transfected with DDCM.tub-KDEL achieved between 80% and 100% proliferation. Moreover, cells transfected with DDCA.tha failed to grow beyond passage 2, whereas all cells transfected with DDCM.tub-KDEL proliferated on DAP continuously. These observations suggest that DDCM.tub-KDEL is a broadly applicable DAP-processing enzyme for cell-specific labeling with amino acid precursors.
Although DDC from M. tuberculosis conferred superior DAP-dependent proliferation to multiple eukaryotic cells, the mechanism underlying this improvement is currently unclear. To investigate whether this phenotype was unique to DDCM.tub or represented a general feature of Mycobacterium DDC enzymes, DDC from the closely related M. leprae (DDCM.lep) and M. avium (DDCM.avi) were also expressed in eukaryotic cells. Interestingly, all Mycobacterium DDC enzymes substantially outperformed DDCA.tha (Fig. 1I). Mycobacterium DDC enzymes contain a unique cysteine residue at position 93 that forms a disulfide bond with a cysteine common to all other DDCs at DDCM.tub position 375 (supplemental Fig. S2). As DDC is thought to function as a homodimer (17), we hypothesized that the collectively improved performance of Mycobacterium DDC enzymes could be due to their common intermonomer disulfide bridges. To test this hypothesis, we compared wild-type DDCM.tub to a Cys93Ala mutant DDCM.tub. The DDCM.tub Cys93Ala point mutation severely impaired DDC activity (Fig. 1J) independent of the expression level (Fig. 1K). This observation implies that the intermonomer disulfide bridges found in Mycobacterium DDC enzymes are responsible for their improved activity in eukaryotic cells.
Given its dramatically improved catalytic activity, intracellular localization, and applicability to multiple eukaryotic cells, we propose DDCM.tub-KDEL as an optimal disulfide-homodimer enzyme for cell-specific labeling with amino acid precursors.
Improved Lyr for Cell-specific Labeling with Amino Acid Precursors
Lyr catalyzes the conversion of D-lysine into L-lysine (6). P. mirabilis wild-type Lyr (LyrWT) contains a putative signal peptide (amino acids 1–30) that has been predicted to facilitate Lyr secretion from eukaryotic cells (supplemental Fig. S3B) (SignalP 4.1 (14)). Gauthier et al. (4) note that wild-type P. mirabilis Lyr is prolifically secreted from eukaryotic cells. Extracellular Lyr converts labeled D-lysine to labeled L-lysine in conditioned media and severely compromises co-culture labeling efficiency. Consequently, growth medium has to be changed daily in order to maintain cell-specific labeling efficiency, which undermines continuous co-culture cell–cell communication. As with DDC, a truly intracellular Lyr enzyme is required for high-fidelity cell-specific labeling of continuous co-cultures.
Structural studies suggest P. mirabilis Lyr contains a globular catalytic core (amino acids 37–407) distinct from the putative signal peptide (18). In an attempt to limit extracellular Lyr (while retaining the catalytic activity), we removed amino acids 1–36 from the enzyme (LyrM37) and added a C-terminal KDEL ER retention motif (LyrM37-KDEL) (Fig. 2A).
Fig. 2.
Optimized lysine racemase for cell-specific labeling with D-lysine. A, Proteus mirabilis lysine racemase (Lyr) constructs. Wild-type Lyr (LyrWT) contains a putative signal peptide (amino acids 1–36) (green) and a globular catalytic domain (amino acids 37–407) (light blue). Catalytic residues K74 and Y299 are highlighted (blue). To limit secretion of Lyr, constructs were prepared without the signal peptide (LyrM37) and with a KDEL endoplasmic retention motif (red) (LyrM37-KDEL). B, C3H10T1/2 proliferation on titrated concentrations of D-lysine when transfected with an empty vector, LyrWT, LyrM37, or LyrM37-KDEL. Results are expressed as a percentage of cells grown in 10 mm L-lysine. C, full-length LyrWT was readily secreted from cells (solid arrow), whereas cleaved LyrWT remained intracellular (hollow arrow). LyrM37 limited extracellular Lyr, and the addition of a KDEL motif ablated Lyr secretion. D, LyrWT located to the ER (for secretion), LyrM37 was spread throughout the cell, and LyrM37-KDEL resided within the ER (for retention). E, stable C3H10T1/2+LyrM37-KDEL cells were grown on titrated concentrations of L-lysine, D-lysine, or meso-2,6-diaminopimelate (DAP). LyrM37-KDEL conferred cell proliferation comparable to L-lysine when the D-lysine concentration was >1 mm. LyrM37-KDEL cells did not grow on DAP. F, C3H10T1/2+LyrM37-KDEL cells were grown for 24 h in 10 mm L-lysine, the absence of lysine, or titrated concentrations of D-lysine. The transcriptional arrest marker pS51 eIF2α was measured by Western blotting. Concentrations of D-lysine greater than 1 mm rescued the amino acid starvation response. G, isotopic labeling efficiency of C3H10T1/2+LyrM37-KDEL cells grown on 2.5 mm light, medium (+4 Da), or heavy (+8 Da) L-lysine (i.e. SILAC) and D-lysine (five passages). Each data point represents an individual peptide, and mean labeling percentages are indicated by a red line. Error bars = standard deviation.
When transfected into C3H10T1/2 (C3) eukaryotic fibroblasts, LyrM37 and LyrM37-KDEL enzymes conferred improved proliferation on lower concentrations of D-lysine relative to LyrWT (Fig. 2B). Only full-length LyrWT protein could be detected in conditioned media, whereas a smaller, cleaved version of LyrWT was found in cell lysates. LyrM37 protein was largely located in cell lysates, although small traces of LyrM37 could still be detected in conditioned media (Fig. 2C). Thus, although the signal peptide appears to enhance Lyr secretion, overexpressed Lyr can still escape the cell of origin independently of the signal peptide. In contrast, LyrM37-KDEL was exclusively located in cell lysates and could not be detected in conditioned media. LyrWT located to the ER (for secretion), LyrM37 was indiscriminately dispersed through the cell, and LyrM37-KDEL resided within the ER (for retention) (Fig. 2D). Collectively, these observations confirm that P. mirabilis Lyr activity is independent of its putative signal peptide and can function in the eukaryotic ER. Thus, unlike LyrWT (or “modified” Lyr (4)), LyrM37-KDEL is a truly intracellular D-lysine conversion enzyme.
Cells stably transfected with LyrM37-KDEL achieved proliferation comparable to that with L-lysine when grown on concentrations of D-lysine greater than 1 mm (Fig. 2E). To ensure that LyrM37-KDEL did not induce an amino acid stress response, LyrM37-KDEL cells were grown on titrated concentrations of D-lysine, and the early translational arrest stress marker pS51 eIF2α (16) was quantified (Fig. 2F). In accordance with earlier proliferation studies, LyrM37-KDEL conferred non-stressed cell growth at D-lysine concentrations between 1 and 10 mm. Stable LyrM37-KDEL cells proliferated on multiple D-lysine heavy isotopes (+4 Da and +8 Da) and achieved labeling efficiencies comparable to those of SILAC when grown in monoculture (Fig. 2G). Given its catalytic proficiency and improved intracellular localization, we propose LyrM37-KDEL as an excellent D-lysine processing enzyme for cell-specific labeling with amino acid precursors.
LyrM37-KDEL and DDCM.tub-KDEL Isotopic Labeling Stability in Interrupted and Continuous Co-cultures
The primary application of cell-specific labeling with amino acid precursors is the study of continuous cell–cell communication in proliferating co-cultures. Previous CTAP methodology requires daily media exchange to maintain labeling stability and thus does not permit the study of continuous cell–cell communication. To examine the ability of our improved cell-specific labeling enzymes to support studies of continuous cell–cell communication, we first tested the influence of continuous versus interrupted co-culture on cell-specific labeling fidelity.
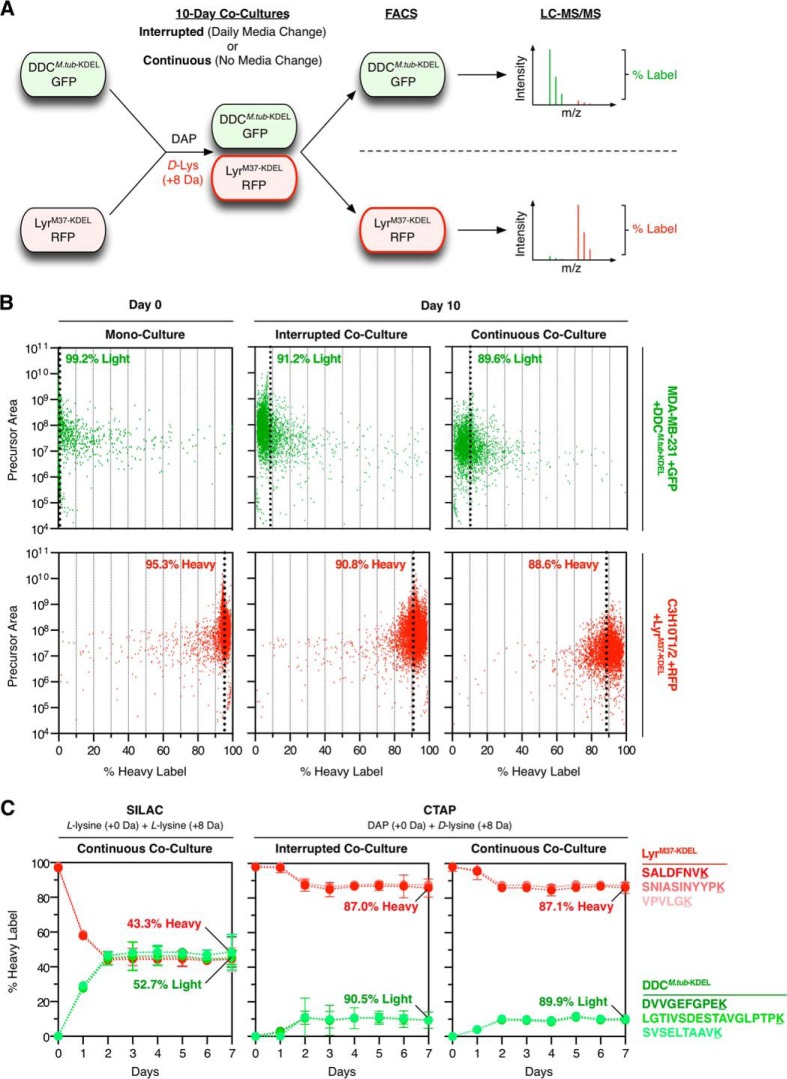
To test the long-term labeling stability of the optimized enzymes in co-culture, we transfected LyrM37-KDEL into RFP+ C3 fibroblasts and DDCM.tub-KDEL into GFP+ MDA-MB-231 epithelial tumor cells. C3 +LyrM37-KDEL cells were then labeled as heavy (+8 Da) and directly co-cultured with light MDA-MB-231 +DDCM.tub-KDEL cells in the presence of heavy D-lysine and light DAP. To investigate the influence of media exchange on labeling efficiency, we maintained cells as either “interrupted” (with daily media exchange) or “continuous” (without media exchange) co-culture. After 10 days, cells were separated into monoculture via FACS (using RFP and GFP) (supplemental Fig. S4), lysed, and resolved via SDS-PAGE, and tryptic peptides were analyzed via LC-MS/MS (Fig. 3A). Light and heavy precursor ions were quantified for each peptide, and a mean labeling efficiency percentage was calculated for each population. Critically, DDCM.tub-KDEL and LyrM37-KDEL conferred ∼90% cell-specific labeling fidelity in both interrupted (91.2% light/90.8% heavy) and continuous (89.6% light/88.6% heavy) 10-day co-cultures (Fig. 3B).
Fig. 3.
Cell-specific co-culture isotopic labeling stability. A, light MDA-MB-231 +GFP+ DDCM.tub-KDEL cells were co-cultured with heavy C3H10T1/2 +RFP +LyrM37-KDEL cells in the presence of DAP (+0 Da) + D-lysine (+8 Da) for 10 days. Cells were maintained as either “interrupted” (with daily media exchange) or “continuous” co-culture (without media exchange). Cell populations were separated via FACS, lysed, and resolved via SDS-PAGE, and gel bands were excised and digested. Tryptic peptides were analyzed via LC-MS/MS, and labeling efficiency was calculated for each cell population. B, interrupted and continuous co-culture labeling fidelity. Each data point represents an individual peptide, and mean labeling percentages are indicated by a dashed line. C, cell-specific co-culture labeling stability time course. Heavy C3H10T1/2+LyrM37-KDEL fibroblasts were combined with light MDA-MB-231+DDCM.tub-KDEL tumor cells and co-cultured for 7 days in L-lysine (+0 Da) + L-lysine (+8 Da) (SILAC amino acids) or DAP (+0 Da) + D-lysine (+8 Da) (CTAP). Each day, a cell population was lysed and light and heavy versions of three proteotypic peptides from each enzyme were analyzed via selected reaction monitoring. As each enzyme is only ectopically expressed in one cell type, the light:heavy ratio for each peptide provides a discrete proxy for cell-specific isotopic labeling efficiency. SILAC co-cultures rapidly lost cell-specific labeling fidelity (<2 days), as L-lysine can be metabolized by both cell types. In contrast, both interrupted and continuous CTAP environments maintained cell-specific labeling fidelity over a 7-day co-culture. Error bars = S.E. of technical replicates (n = 3).
As DDCM.tub-KDEL is specifically expressed in light MDA-MB-231 cells and LyrM37-KDEL is specifically expressed in heavy C3 cells, the isotopic lysine composition of each enzyme provides a discrete proxy for general cell-specific labeling efficiency. To compare the temporal labeling stability of this CTAP method with that of traditional SILAC labeling, we combined heavy C3 +LyrM37-KDEL cells with light MDA-MB-231 +DDCM.tub-KDEL cells and co-cultured for 7 days in either L-lysine (+0 Da) + L-lysine (+8 Da) (SILAC amino acids) or DAP (+0 Da) + D-lysine (+8 Da) (CTAP amino acid precursors). Temporal labeling efficiencies for the ectopic LyrM37-KDEL and DDCM.tub-KDEL enzymes were subsequently determined for each co-culture using SRM (Fig. 3C) (supplemental Fig. S5). As expected, light and heavy L-lysine was metabolized by both cell types in the SILAC co-culture and resulted in a rapid loss of cell-specific labeling fidelity (1 to 2 days) (7 days = 47.3% light DDCM.tub-KDEL/44.3% heavy LyrM37-KDEL). These observations confirm that SILAC labeling is unsuitable for long-term analysis of cell-specific proteomes in co-culture. In contrast, both interrupted (7 days = 9.5% light DDCM.tub-KDEL/87.0% heavy LyrM37-KDEL) and continuous (7 days = 10.1% light DDCM.tub-KDEL/87.1% heavy LyrM37-KDEL) CTAP conditions conferred stable cell-specific labeling fidelity throughout their 7-day co-cultures. These results further confirm that optimized DDCM.tub-KDEL and LyrM37-KDEL enzymes are suitable for the long-term cell-specific quantitative proteomic analysis of uninterrupted, continuous co-cultures.
Phosphoproteomic Analysis of Cell-specific Signaling in Continuous Co-culture
As DDCM.tub-KDEL and LyrM37-KDEL support the long-term cell-specific labeling stability of both interrupted and continuous co-cultures, we next sought to investigate the cell-signaling implications of daily media exchange. First, we monitored the relative abundance of cell-signaling factors present in conditioned media from both interrupted and continuous 5-day co-cultures. In contrast to interrupted co-cultures, multiple growth factors and cytokines accumulated in continuous co-cultures (Fig. 4A). This soluble signaling potential is not established in interrupted co-cultures and highlights the need for new technologies to study cell–cell communication in co-culture. To investigate the cell-specific signaling consequences of the two approaches, we compared the phosphoproteomic profile of an interrupted MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL medium co-culture with a that of a continuous MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL heavy co-culture (Fig. 4B). An interrupted versus interrupted experiment was also performed to control for stochastic phospho-signaling differences that might have arisen during the 5-day co-culture. Cell-specific phosphoproteomic analysis of C3 cells revealed substantial signaling differences between the interrupted and continuous co-cultures (Fig. 4C) (supplemental Table S2).
Fig. 4.
Continuous co-culture cell–cell communication. A, conditioned media from 5-day interrupted and continuous co-cultures (n = 4) was analyzed for cell signaling factors. Multiple growth factors, cytokines, and extracellular matrix ligands accumulated in continuous co-cultures in contrast to interrupted co-cultures. B, an interrupted MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL medium 5-day co-culture was compared with a continuous MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL heavy 5-day co-culture. Co-cultures were lysed, combined, trypsin digested, enriched for phosphopeptides, and subjected to LC-MS/MS analysis. C, quantitative cell-specific phosphoproteomic analysis of 5-day co-cultures. Each point represents the mean (± S.E., n = 3) of one cell-specific phosphopeptide. Multiple cell-specific phosphosites were differentially regulated in continuous co-cultures. Aligned phosphopeptides from control interrupted MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL medium and interrupted MDA-MB-231 +DDCM.tub-KDEL light/C3 +LyrM37-KDEL heavy 5-day co-cultures are shown for comparison. Regulated phosphopeptides can be seen in supplemental Table S2.
As these phosphorylation events would not have been detected using existing interrupted co-culture methodology, this observation suggests that novel biological questions now can be addressed through the enhanced CTAP technology. Given their improved intracellular localization, applicability to multiple cell types, and robust cell-specific labeling stability, we propose DDCM.tub-KDEL and LyrM37-KDEL as excellent enzymes for the phosphoproteomic analysis of long-term continuous cell–cell communication.
DISCUSSION
Cell-specific labeling with amino acid precursors permits discrete proteomic labeling of proliferating cell types in co-culture. In order for this technology to function successfully, two discrete enzymes must confer distinct cell-specific L-lysine precursor proliferation to eukaryotic cells. In this study, we proposed novel lysine-precursor converting enzymes for efficient, stable, and continuous cell-specific labeling with amino acid precursors.
A primary finding of this study is that DDC enzymes from the Mycobacterium genus confer excellent DAP-dependent proliferation to eukaryotic cells. Although this study identified DDCM.tub as the optimal enzyme, DDCM.lep and DDCM.avi also conferred improved DAP-dependent growth to eukaryotic cells relative to alternative species. This superior performance appears to be facilitated by an intermonomer disulfide bond that stabilizes the active DDC homodimer in Mycobacterium DDC enzymes.
Although DDCM.tub-KDEL provided excellent DAP conversion for cell-specific labeling with amino acid precursors, we note that growing stable DDC cells on DAP (EC50 = 0.6 mm) is 15-fold less efficient than growing cells on L-lysine (EC50 = 0.04 mm). It is currently unclear why higher concentrations of DAP are required. Previous work has suggested that the rate of DAP uptake could be rate-limiting (19). However, as we observed identical performance between DDCM.tub (un-localized) and DDCM.tub-KDEL (ER-localized), DAP localization from the eukaryotic cytoplasm to the ER does not appear to be a rate-limiting step. Future efforts to improve DDC+DAP efficiency could focus on improving DAP uptake into the eukaryotic cytoplasm (e.g. by modulating amino acid transceptors (20)). Another explanation could be poor DDC enzyme performance. Although DDCM.tub substantially outperformed all other DDC enzymes tested, future efforts to improve DAP-dependent proliferation could also expand beyond the multiple DDC orthologs described here. However, a C-terminal KDEL motif should be added to ensure intracellular enzyme retention.
Our data confirm that P. mirabilis Lyr effectively converted D-lysine to L-lysine in eukaryotic cells. Moreover, we show that by removing the putative signal peptide (M37) and anchoring the enzyme in the ER (via KDEL motif), LyrM37-KDEL can act as a suitable intracellular enzyme for cell-specific labeling with amino acid precursors. LyrM37-KDEL confers efficient cell proliferation on concentrations of D-lysine greater than 1 mm and can convert isotopically labeled medium (+4 Da) and heavy (+8 Da) D-lysine into labeled L-lysine. As a result, when combined with DDCM.tub-KDEL, LyrM37-KDEL can be used for triple-labeled comparisons between co-cultures.
Although LyrM37-KDEL provided sufficient D-lysine conversion for cell-specific labeling, we note that growing Lyr cells on D-lysine (EC50 = 0.38 mm) is around 10-fold less efficient than growing cells directly on L-lysine (EC50 = 0.03 mm). Again, it is currently unclear why higher concentrations of D-lysine are required. As with DDC, one explanation could be enzyme performance. Although P. mirabilis Lyr is clearly suitable for cell-specific labeling (as shown in Figs. 3B and 3C), it is possible that orthologous Lyr enzymes could confer more efficient D-lysine-dependent growth. Our success with orthologous DDC screening suggests that future studies might benefit from testing enzymes from alternative species. However, if undertaking such a screen, one should consider removing any putative signal sequences and adding a C-terminal KDEL motif for intracellular retention. Another explanation for the discrepancy might be poor intracellular import of D-lysine. However, as we observed substantially improved activity of intracellular LyrM37 and LyrM37-KDEL (Fig. 2B) relative to extracellular LyrWT, our data suggest that intracellular D-lysine-to-L-lysine conversion is actually more effective than extracellular conversion. Crucially, when the DAP concentration was >5 mm and that of D-lysine was >2.5 mm, neither DDCM.tub-KDEL nor LyrM37-KDEL experienced growth-limiting proliferation relative to traditional growth conditions. Thus, as long as sufficient DAP and D-lysine are supplied in the growth media, the performance of DDCM.tub-KDEL or LyrM37-KDEL should remain suitable for cell-specific labeling with amino acid precursors.
Given the broad applicability of the revised enzymes, the reported methodology now extends the study of cell–cell communication to a diverse selection of different cell types. To this end, we have deposited DDCM.tub-KDEL and LyrM37-KDEL expression constructs in Addgene (DDCM.tub-KDEL: 51529 and LyrM37-KDEL: 51530) for widespread application by the proteomic community.
The optimized enzyme pairing described here provides stable proteomic labeling of specific cell types in continuous co-culture. However, we did observe a modest reduction in labeling efficiency for co-cultures relative to pre-labeled monocultures. Temporal analysis of labeling fidelity indicated that this decrease occurred during the first 2 days of co-culture. The small reduction in labeling efficiency could have been due to multiple factors. For example, secreted proteins can be taken up by cells of a different label during normal paracrine signaling (21). Moreover, although we experienced decreased extracellular enzyme secretion from live cells, it is possible that dead cells could release active enzymes into co-culture media. Furthermore, gap-junctions between adjacent cells can transport small molecules (<1 kDa) (22) and could theoretically leak labeled amino acids between confluent co-cultures. Such events could represent a biological limit to long-term cell-specific labeling in co-culture, and interfering with these processes (i.e. blocking gap-junctions) might undermine the biological significance of co-culture experiments. However, as illustrated by our triple-labeled phosphoproteomic co-culture comparison (Figs. 4B and 4C), the superior labeling stability of LyrM37-KDEL and DDCM.tub-KDEL capably enables high-fidelity cell-specific experiments from continuous co-cultures.
Unlike previous methodology, the revised DDCM.tub-KDEL and LyrM37-KDEL enzymes now permit long-term cell-specific labeling of continuous co-cultures. When comparing the cell-specific phosphoproteomes of both continuous and interrupted co-cultures, we observed phosphorylation events unique to co-cultures with no media exchange (Fig. 4C) (supplemental Table S2). Several of the regulated cell-specific phosphosites are involved in transducing extracellular signals. For example, phosphorylated RPS6 (pS235/pS236) is a downstream target of both RAS-MAPK-p70S6K and PI3K-mTOR-p70S6K mitogen-activated signaling pathways (23, 24). Moreover, phosphorylated eIF4B is also regulated by both MAPK-p70S6K and mTOR-p70S6K mitogen-activated signaling pathways (25). CDK1 phosphorylation of SQSTM (pT269/pT272) is downstream of MAPK signaling (26) and controls mitotic progression (27). AB1IP transduces activated RAS-signaling to the cytoskeleton (28), and NGF reduces DPYL2 phosphorylation (pS522) (29). Although our experiments did not allow us to discern the precise source of these regulations, the new DDCM.tub-KDEL and LyrM37-KDEL CTAP enzymes now permit the quantitative study of continuous cell-specific co-culture phosphorylation. We envisage this technology as enabling future researchers to study the discrete mechanisms of continuous cell–cell signaling.
Given their applicability to multiple cell types, continuous co-culture labeling fidelity, and suitability for long-term cell–cell phospho-signaling experiments, we propose DDCM.tub-KDEL and LyrM37-KDEL as excellent enzymes for cell-specific labeling with amino acid precursors.
Supplementary Material
Acknowledgments
We acknowledge colleagues at the ICR and the Cell Communication Team for valuable input and useful discussion. We also thank the PRIDE Team for facilitating MS/MS data distribution.
Footnotes
Author contributions: C.J.T., I.C.N., and C.J. designed research; C.J.T. and I.C.N. performed research; J.D.W. and L.L. contributed new reagents or analytic tools; C.J.T. and I.C.N. analyzed data; C.J.T. and C.J. wrote the paper; D.A.L. and C.J. oversaw the project.
* C.T. is funded by a Sir Henry Wellcome Postdoctoral Fellowship (098847/Z/12/Z). C.J. holds a CR-UK Career establishment award (C37293/A12905).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SILAC
- stable isotope labeling by amino acids in cell culture
- CTAP
- cell-type-specific labeling with amino acid precursors
- DAP
- meso-2,6-diaminopimelate
- DDC
- diaminopimelate decarboxylase
- ER
- endoplasmic reticulum
- H. pyl
- Helicobacter pylori
- Lyr
- lysine racemase
- LyrWT
- Proteus mirabilis lysine racemase
- RFP
- red fluorescent protein
- M. avi
- Mycobacterium avium
- M. jan
- Methanocaldococcus jannaschii
- M. lep
- Mycobacterium leprae
- M. tub
- Mycobacterium tuberculosis
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Garden G. A., La Spada A. R. (2012) Intercellular (mis)communication in neurodegenerative disease. Neuron 73, 886–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 3. Jorgensen C., Sherman A., Chen G. I., Pasculescu A., Poliakov A., Hsiung M., Larsen B., Wilkinson D. G., Linding R., Pawson T. (2009) Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326, 1502–1509 [DOI] [PubMed] [Google Scholar]
- 4. Gauthier N. P., Soufi B., Walkowicz W. E., Pedicord V. A., Mavrakis K. J., Macek B., Gin D. Y., Sander C., Miller M. L. (2013) Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat. Methods 10(8), 768–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dewey D. L., Work E. (1952) Diaminopimelic acid decarboxylase. Nature 169, 533–534 [DOI] [PubMed] [Google Scholar]
- 6. Huang H. T., Kita D. A., Davisson J. W. (1952) Racemization of lysine by Proteus vulgaris. J. Am. Chem. Soc. 80, 1006–1007 [Google Scholar]
- 7. Maclean B., Tomazela D. M., Abbatiello S. E., Zhang S., Whiteaker J. R., Paulovich A. G., Carr S. A., Maccoss M. J. (2010) Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 82, 10116–10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 10. Kall L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 11. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 12. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu T., Wu D., Chen J., Ding J., Jiang H., Shen X. (2008) The catalytic intermediate stabilized by a “down” active site loop for diaminopimelate decarboxylase from Helicobacter pylori. Enzymatic characterization with crystal structure analysis. J. Biol. Chem. 283, 21284–21293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 15. Munro S., Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 [DOI] [PubMed] [Google Scholar]
- 16. Samuel C. E. (1993) The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268, 7603–7606 [PubMed] [Google Scholar]
- 17. Gokulan K., Rupp B., Pavelka M. S., Jr., Jacobs W. R., Jr., Sacchettini J. C. (2003) Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J. Biol. Chem. 278, 18588–18596 [DOI] [PubMed] [Google Scholar]
- 18. Wu H. M., Kuan Y. C., Chu C. H., Hsu W. H., Wang W. C. (2012) Crystal structures of lysine-preferred racemases, the non-antibiotic selectable markers for transgenic plants. PLoS One 7, e48301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saqib K. M., Hay S. M., Rees W. D. (1994) The expression of Escherichia coli diaminopimelate decarboxylase in mouse 3T3 cells. Biochim. Biophys. Acta 1219, 398–404 [DOI] [PubMed] [Google Scholar]
- 20. Hundal H. S., Taylor P. M. (2009) Amino acid transceptors: gate keepers of nutrient exchange and regulators of nutrient signaling. Am. J. Physiol. Endocrinol. Metab. 296, E603–E613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rechavi O., Kalman M., Fang Y., Vernitsky H., Jacob-Hirsch J., Foster L. J., Kloog Y., Goldstein I. (2010) Trans-SILAC: sorting out the non-cell-autonomous proteome. Nat. Methods 7, 923–927 [DOI] [PubMed] [Google Scholar]
- 22. Herve J. C., Derangeon M. (2013) Gap-junction-mediated cell-to-cell communication. Cell Tissue Res. 352, 21–31 [DOI] [PubMed] [Google Scholar]
- 23. Bandi H. R., Ferrari S., Krieg J., Meyer H. E., Thomas G. (1993) Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J. Biol. Chem. 268, 4530–4533 [PubMed] [Google Scholar]
- 24. Roux P. P., Shahbazian D., Vu H., Holz M. K., Cohen M. S., Taunton J., Sonenberg N., Blenis J. (2007) RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shahbazian D., Roux P. P., Mieulet V., Cohen M. S., Raught B., Taunton J., Hershey J. W., Blenis J., Pende M., Sonenberg N. (2006) The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 25, 2781–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee S. J., Pfluger P. T., Kim J. Y., Nogueiras R., Duran A., Pages G., Pouyssegur J., Tschop M. H., Diaz-Meco M. T., Moscat J. (2010) A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 11, 226–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Linares J. F., Amanchy R., Greis K., Diaz-Meco M. T., Moscat J. (2011) Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol. Cell. Biol. 31, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jenzora A., Behrendt B., Small J. V., Wehland J., Stradal T. E. (2005) PREL1 provides a link from Ras signalling to the actin cytoskeleton via Ena/VASP proteins. FEBS Lett. 579, 455–463 [DOI] [PubMed] [Google Scholar]
- 29. Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.