Abstract
Background
Genetic predisposition is the primary risk factor for familial breast cancer. For the majority of familial breast cancer, however, the genetic predispositions remain unknown. All newly identified predispositions occur rarely in disease population, and the unknown genetic predispositions are estimated to reach up to total thousands. Family unit is the basic structure of genetics. Because it is an autosomal dominant disease, individuals with a history of familial breast cancer must carry the same genetic predisposition across generations. Therefore, focusing on the cases in lineages of familial breast cancer, rather than pooled cases in disease population, is expected to provide high probability to identify the genetic predisposition for each family.
Methods
In this study, we tested genetic predispositions by analyzing the family-specific variants in familial breast cancer. Using exome sequencing, we analyzed three families and 22 probands with BRCAx (BRCA-negative) familial breast cancer.
Results
We observed the presence of family-specific, novel, deleterious germline variants in each family. Of the germline variants identified, many were shared between the disease-affected family members of the same family but not found in different families, which have their own specific variants. Certain variants are putative deleterious genetic predispositions damaging functionally important genes involved in DNA replication and damaging repair, tumor suppression, signal transduction, and phosphorylation.
Conclusions
Our study demonstrates that the predispositions for many BRCAx familial breast cancer families can lie in each disease family. The application of a family-focused approach has the potential to detect many new predispositions.
Background
Breast cancer is a leading cancer in women [1]. About 10-20% of breast cancer cases are family clustered, with multiple family members affected by the disease [2]. Genetic predispositions are the major risk factor for the disease. However, the genetic predispositions are currently known for only 30-40% of the familial breast cancer disease families. The remaining 60-70% of women with familial breast cancer have unknown predispositions and are diagnosed with BRCAx, for their unknown predisposition of familial breast cancer [3]. It is estimated the “missing” heredity trait for BRCAx families likely consists of thousands of rare variants, each presenting a minor disease risk [4]. Indeed, broadly screening the variants across disease populations has uncovered multiple new genetic predispositions for familial breast cancer. A consistent pattern among these newly classified predispositions is that they are always present at very-low frequencies in the given disease population [5-10]. Their extreme rarity implies that a greater sampling size of disease populations is required to identify the germline predispositions [10]. However, such an expansion is deemed to increase the complexity of data analysis, experimental costs, and time needed. As such, focusing only on the rare variants will not likely be able to determine the entire spectrum of genetic predispositions for BRCAx familial breast cancer families. New alternative hypotheses and approaches must be explored to improve the situation. For example, mosaic mutation has implications as potential predispositions for familial breast cancer [11].
Familial breast cancer is defined as an autosomal dominant genetic disease [12]. Although incidences of breast cancer often exhibit atypical Mendelian patterns due to the factors such as low penetrance of genetic predispositions, the predisposition in a disease-prone family is expected to transmit across generations and shared between family members. Focusing on each disease family with a history of the disease is expected to improve the chance to detect the predisposition in a family compared to screening the disease population of pooled cases without family relationships, which can dilute the predisposition highly prevalent in a disease family into insignificant level.
We hypothesize that the unknown predispositions for many BRCAx familial breast cancer are specific to each family with a history of the disease. Our previous exome study of a BRCAx familial breast cancer family shows the presence of rich genetic variants [13]. In the present study, we expand the exome sequencing study by analyzing three families with BRCAx familial breast cancer; 17 members had cancer, and five members were without cancer. Our study also includes 22 probands of BRCAx familial breast cancer. Our study reveals the presence of family-specific, novel, deleterious genetic variants as putative genetic predispositions in each family with BRCAx familial breast cancer.
Methods
Use of human subjects
The use of the patient samples for the study was approved by the Institutional Review Boards (IRB) of Creighton University School of Medicine (#00-12265 ) and University of Nebraska Medical Center (718-11-EP). All subjects signed the Consent to Participate Form for cancer genetic study.
Individuals from three families with BRCAx breast cancer were used to generate exome sequences as we have previously described [13]. Family I included six individuals with breast cancer and two individuals without breast cancer. Family II included five individuals with breast cancer, one obligate carrier and two individuals without breast cancer. Family III included five individuals with breast cancer and one individual without breast cancer. Additionally, 22 probands for BRCAx familial breast cancer were included in exome sequencing. All cases used in the study were BRCA1-negative, and BRCA2-negative, 41 were female and 3 were male, the average age is 42 years old (Figure 1, Table 1).
Figure 1.
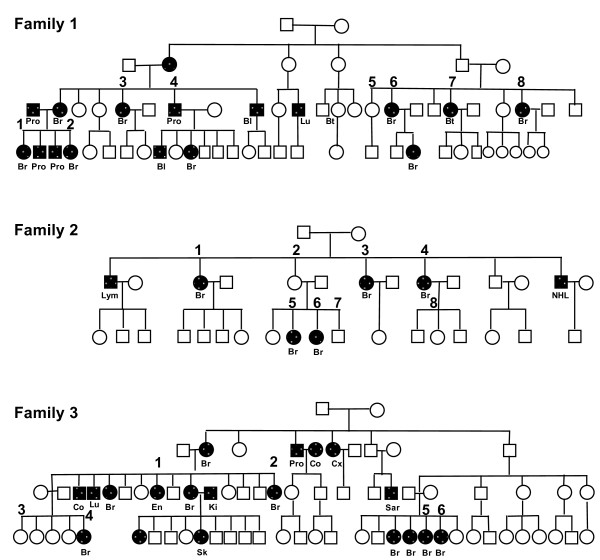
Pedigrees of the three families used in the study. BC (breast cancer), Bt (brain tumor), CRC (colorectal cancer), Lu (lung cancer), En (endometrium cancer), Ki (kidney cancer), Lym (lymphoma), NHL (non-Hodgkin lymphoma), OC (ovarian cancer), Pro (prostate cancer). Sar (sarcoma), Sk (skin cancer).
Table 1.
BRCAx familial breast cancer cases used in the study
|
Family |
Cancer type |
Pathology |
BRCA1/2 |
Exome |
||||
|---|---|---|---|---|---|---|---|---|
| Reads | Bases | Bases map rate (%) | Coverage | Variant called | ||||
| Family 1 |
|
|
|
|
|
|
|
|
| 1 |
Breast |
Infiltrating ductal |
- |
42,973,730 |
4,340,346,730 |
97.6 |
70 |
184,865 |
| 2 |
Breast |
Not available |
- |
40,158,059 |
4,055,963,959 |
98.3 |
65 |
152,692 |
| 3 |
Breast |
Infiltrating ductal |
- |
46,240,754 |
4,670,316,154 |
97.2 |
75 |
176,554 |
| 4 |
Prostate |
Adenocarcinoma |
- |
23,418,595 |
2,365,278,095 |
98.1 |
38 |
207,103 |
| 5 |
No Cancer |
|
- |
40,313,161 |
4,071,629,261 |
98.0 |
66 |
213,347 |
| 6 |
Breast, Colon |
Adenocarcinoma |
- |
17,496,012 |
1,767,097,212 |
97.9 |
28 |
183,741 |
| 7 |
Brain |
Not available |
- |
36,166,319 |
3,652,798,219 |
99.5 |
59 |
171,425 |
| 8 |
Breast |
Adenocarcinoma |
- |
27,830,687 |
2,810,899,387 |
96.3 |
45 |
104,343 |
| Family 2 |
|
|
|
|
|
|
|
|
| 1 |
Breast, Breast |
Medullary, infiltrating ductal |
- |
33,419,098 |
3,375,328,898 |
92.9 |
54 |
113,079 |
| 2 |
Obligated carrier |
|
- |
27,261,117 |
2,753,372,817 |
92.4 |
44 |
115,328 |
| 3 |
Breast |
Infiltrating ductal |
- |
40,973,473 |
4,138,320,773 |
99.6 |
67 |
127,272 |
| 4 |
Breast |
Ductal carcinoma in situ |
- |
29,561,523 |
2,985,713,823 |
91.5 |
48 |
108,655 |
| 5 |
Breast |
Infiltrating ductal |
- |
25,790,969 |
2,604,887,869 |
93.1 |
42 |
84,687 |
| 6 |
Breast |
Infiltrating ductal |
- |
37,657,589 |
3,803,416,489 |
91.6 |
61 |
139,891 |
| 7 |
No Cancer |
|
- |
17,433,912 |
1,760,825,112 |
91.6 |
28 |
131,786 |
| 8 |
No Cancer |
|
- |
35,977,512 |
3,633,728,712 |
97.3 |
59 |
128,680 |
| Family 3 |
|
|
|
|
|
|
|
|
| 1 |
Endometrial |
Adenocarcinoma |
- |
33,662,978 |
3,399,960,778 |
93.2 |
55 |
129,754 |
| 2 |
Breast, Skin |
Basal, infiltrating ductal |
- |
29,648,460 |
2,994,494,460 |
98.3 |
48 |
198,862 |
| 3 |
No Cancer |
|
- |
53,411,156 |
5,394,526,756 |
98.8 |
87 |
193,017 |
| 4 |
Breast |
Infiltrating ductal |
- |
31,736,845 |
3,205,421,345 |
98.3 |
52 |
130,941 |
| 5 |
Breast |
Ductal carcinoma in situ |
- |
35,014,538 |
3,536,468,338 |
98.4 |
57 |
129,754 |
| 6 |
Breast |
Not available |
- |
38,418,769 |
3,880,295,669 |
97.5 |
62 |
161,953 |
| Probands |
|
|
|
|
|
|
|
|
| 1 |
Breast |
Ductal carcinoma in situ |
- |
17,832,681 |
1,801,100,781 |
93.1 |
29 |
109,864 |
| 2 |
Breast |
Invasive ductal carcinoma |
- |
36,166,319 |
3,652,798,219 |
99.5 |
59 |
142,155 |
| 3 |
Breast |
Invasive ductal carcinoma |
- |
50,944,516 |
5,145,396,116 |
98.4 |
83 |
152,125 |
| 4 |
Breast |
Invasive ductal carcinoma |
- |
43,889,986 |
4,432,888,586 |
99.6 |
71 |
169,633 |
| 5 |
Breast |
Invasive ductal carcinoma |
- |
40,125,408 |
4,052,666,208 |
99.5 |
65 |
153,511 |
| 6 |
Breast |
Invasive lobular carcinoma |
- |
31,798,628 |
3,211,661,428 |
97.5 |
52 |
119,875 |
| 7 |
Breast |
Invasive ductal carcinoma |
- |
49,739,415 |
5,023,680,915 |
99.6 |
81 |
113,058 |
| 8 |
Breast |
Invasive ductal carcinoma |
- |
63,352,269 |
6,398,579,169 |
99.6 |
103 |
99,732 |
| 9 |
Breast |
Invasive ductal carcinoma |
- |
43,744,840 |
4,418,228,840 |
99.5 |
71 |
149,873 |
| 10 |
Breast |
Invasive ductal carcinoma |
- |
43,573,311 |
4,400,904,411 |
99.6 |
71 |
141,236 |
| 11 |
Breast |
Invasive ductal carcinoma |
- |
40,938,838 |
4,134,822,638 |
99.3 |
67 |
143,262 |
| 12 |
Breast |
Ductal carcinoma in situ |
- |
36,258,870 |
3,662,145,870 |
99.6 |
59 |
138,018 |
| 13 |
Breast |
Ductal carcinoma in situ |
- |
34,550,745 |
3,489,625,245 |
99.4 |
56 |
146,858 |
| 14 |
Breast |
Invasive ductal carcinoma |
- |
50,295,200 |
5,079,815,200 |
99.5 |
82 |
156,666 |
| 15 |
Breast |
Invasive ductal carcinoma |
- |
60,736,566 |
6,134,393,166 |
99.7 |
99 |
115,909 |
| 16 |
Breast |
Invasive ductal carcinoma |
- |
57,383,360 |
5,795,719,360 |
99.6 |
93 |
120,945 |
| 17 |
Breast |
Invasive ductal carcinoma |
- |
44,922,611 |
4,537,183,711 |
99.6 |
73 |
110,503 |
| 18 |
Breast |
Invasive ductal carcinoma |
- |
33,883,509 |
3,422,234,409 |
99.4 |
55 |
131,955 |
| 19 |
Breast |
Invasive ductal carcinoma |
- |
49,729,619 |
5,022,691,519 |
99.5 |
81 |
146,665 |
| 20 |
Breast |
Invasive ductal carcinoma |
- |
63,184,143 |
6,381,598,443 |
99.6 |
103 |
119,680 |
| 21 |
Breast |
Invasive ductal carcinoma |
- |
28,002,381 |
2,828,240,481 |
99.6 |
46 |
86,924 |
| 22 |
Breast |
Invasive ductal carcinoma |
- |
47,794,798 |
4,827,274,598 |
99.5 |
78 |
112,030 |
| Average | 38,941,211 | 3,933,062,277 | 97.7 | 63 | 140,187 | |||
Exome sequencing
For each sample, exome sequencing used DNA from blood cells. Exome libraries were constructed using the TruSeq Exome Enrichment Kit (62 Mb, Illumina, San Diego, CA) as per manufacturer’s procedures. Exome sequences were collected with a HiSeq™ 2000 sequencer (Illumina, San Diego, CA) with paired-end (2 × 100). All exome data were deposited in the Sequence Read Archive (SRA) database in the National Center for Biotechnology Information (NCBI) (Accession numbers SAMN02404413- SAMN02404456).
Exome sequence mapping and variant calling
Exome sequences were mapped to the human genome reference sequence hg19 by Bowtie2 with default parameters in paired mode [14]. The subsequent SAM files were converted to BAM files. Duplicates were removed using Picard (http://picard.sourceforge.net). The mapped reads were locally realigned using the genome mapping tool RealignerTargetCreator from the Genome Atlas Tool Kit (GATK) [15]. The base quality scores were recalibrated using BaseRecalibrator (GATK), with NCBI dbSNP build 137, in the GATK resource bundles for reference sequence hg19. VarScan 2 was used for variant calling, [16]. VarScan 2 was run on pileup data generated from BAM files using SAMtools utilities [17]. The mpileup command, with –B parameter to disable base alignment quality (BAQ) computation, and the default parameters were used, with the minimum read depth at 10 and the minimum base quality at 30. The called variants were annotated with ANNOVAR using the software-provided databases of the Reference Sequence (RefSeq; NCBI), dbSNP 137, the 1000 Genomes Project, and the NIH Heart, Lung and Blood Institute (NHLBI) Exome Sequencing Project (ESP) 6500 (http://evs.gs.washington.edu).
Those that matched in the databases were classified as known variants and removed. Family-specific normal variants were eliminated by removing the variants shared between the affected and the unaffected family members in each family. The remaining novel variants were classified into synonymous, non-synonymous, splicing site change, stop gain- or loss groups. The variants causing synonymous changes were then removed. For the remaining variants, PolyPhen-2 was used to identify variants causing deleterious effects in the affected genes [probably damaging score: 0.909-1; possibly damaging score: 0.447 - 0.908; Benign score: 0 - 0.446; HumVar score: [18]. The variants defined as benign were removed. These processes generated a list of novel, deleterious variants only present in the cancer-affected family members and probands, Note that the variants in probands were filtered by population databases only.
Power calculation
Using a two-sided paired t-test and assuming a genetic relative risk (GRR) equal to 5.8, disease prevalence equal to 0.03, a disease locus frequency equal to 0.01, and a sib recurrence ratio of 2, a sample size of 20 achieves 81% power to detect a mutation difference with a (standardized) effect size of 0.67 between the affected member and the unaffected member. The significance level (alpha) is, in turn, 0.05 [19,20].
Validation
Sanger sequencing was used to validate deleterious variants. Sense and antisense PCR primers for each selected variant were designed using the Primer3 program. The original DNA samples that were used in exome sequencing were served as PCR templates. PCR amplicons were subjected to BigDye sequencing. The resulting sequences were evaluated using CLC Genomics Workbench Program (Cambridge, MA) to confirm the variants called from exome sequences.
Results
Mapping exome data and calling variants
Exome sequences were collected via a blood sample from each study participant and mapped to the human genome reference sequence hg19. Variants were called from the mapping data. We focused on single-base, non-synonymous variants that affect protein coding, splicing, and stop gain- or loss mutations, which are reliably detectable by exome analysis [21]. The average exome coverage was 63x, and the average number of variants called was 140,187 per case (Table 1).
To increase the likelihood that the variants identified in the breast cancer-affected family members are breast cancer-associated, variants in each data set were filtered by: 1) removal of common variants present in human populations. All variants matching to population-derived variant databases (i.e., dbSNP137, ESP6500, and 1000 genomes) were removed; 2) Removal of family-specific normal variants. For the three families in the study, the variants shared between the affected and the unaffected members in the same family were removed. To identify those causing deleterious effects in the affected genes, the remaining variants were analyzed using the Polyphen-2 Program [18]. A total of 337 novel, deleterious variants present only in the affected members of Families I, II, and III were identified at an average of 112 variants per family (Table 2, Additional files 1: Table S1A, B, C); 689 novel, deleterious variants were identified in the 22 probands at an average of 30 variants per proband (Table 2, Additional files 2: Table S2A, B). Sanger sequencing validated the mapped variants at a validation rate of 83% (53/64), highlighting the reliability of the variants identified by exome mapping analysis (Additional file 1: Table S1D).
Table 2.
Novel, deleterious variants detected in breast cancer-affected cases*
| Family | Total (%) | Individual (%) | Shared**(%) |
|---|---|---|---|
| Family 1 |
|
|
|
| 1 |
37 |
35 |
2 |
| 2 |
26 |
26 |
0 |
| 3 |
25 |
15 |
10 |
| 4 |
48 |
39 |
9 |
| 6 |
29 |
17 |
12 |
| 7 |
12 |
6 |
6 |
| 8 |
14 |
6 |
8 |
| Subtotal |
143 (199) |
123 (86) |
20 (14) |
| Family 2 |
|
|
|
| 1 |
22 |
13 |
9 |
| 2 |
15 |
5 |
10 |
| 3 |
21 |
9 |
12 |
| 4 |
21 |
12 |
9 |
| 5 |
16 |
8 |
8 |
| 6 |
8 |
2 |
6 |
| Subtotal |
66 (100) |
47 (71) |
19 (29) |
| Family 3 |
|
|
|
| 1 |
39 |
13 |
26 |
| 2 |
48 |
27 |
21 |
| 4 |
21 |
12 |
9 |
| 5 |
32 |
12 |
20 |
| 6 |
41 |
19 |
22 |
| Subtotal |
128 (100) |
83 (65) |
45 (35) |
| Total |
337 (100) |
253 (75) |
84 (25) |
|
Probands |
|
|
|
| 1 |
35 |
10 |
25 |
| 2 |
58 |
22 |
36 |
| 3 |
74 |
28 |
46 |
| 4 |
77 |
49 |
28 |
| 5 |
70 |
28 |
42 |
| 6 |
41 |
16 |
25 |
| 7 |
31 |
24 |
7 |
| 8 |
43 |
27 |
16 |
| 9 |
51 |
19 |
32 |
| 10 |
61 |
30 |
31 |
| 11 |
70 |
35 |
35 |
| 12 |
51 |
12 |
39 |
| 13 |
55 |
15 |
40 |
| 14 |
60 |
30 |
30 |
| 15 |
51 |
31 |
20 |
| 16 |
41 |
31 |
10 |
| 17 |
32 |
18 |
14 |
| 18 |
57 |
25 |
32 |
| 19 |
58 |
18 |
40 |
| 20 |
47 |
23 |
24 |
| 21 |
33 |
25 |
8 |
| 22 |
34 |
22 |
12 |
| Total |
689 (100) |
568 (82) |
121 (18) |
| Per proband | 30 | 26 | 6 |
*The counts in subtotal and total are the unique number of variants.
**Shared with family members in the families, or shared with other probands.
Novel deleterious variants are mostly family-specific
We compared the variants within each family. We observed that 25% of the variants on average (14% in Family I, 29% in Family II, 35% in Family III) were shared in multiple affected members in each family, whereas 75% on average (86% in Family I, 71% in Family II and 65% in Family III) were present only in single affected member in each family (Table 2). We then compared the shared variants between the three families, and found only 1 variant was shared between Family I and Family II, four variants were shared between Family I and Family III (Figure 2A). For the 689 variants identified in the probands, 82% were proband-specific, and only 18% were shared between probands at various frequencies (Figure 2B, Additional file 2: Table S2A, S2B). The results indicate that the majority of the novel, deleterious variants identified in the three families and probands are family-specific, i.e., present only in each family but not shared with other families.
Figure 2.
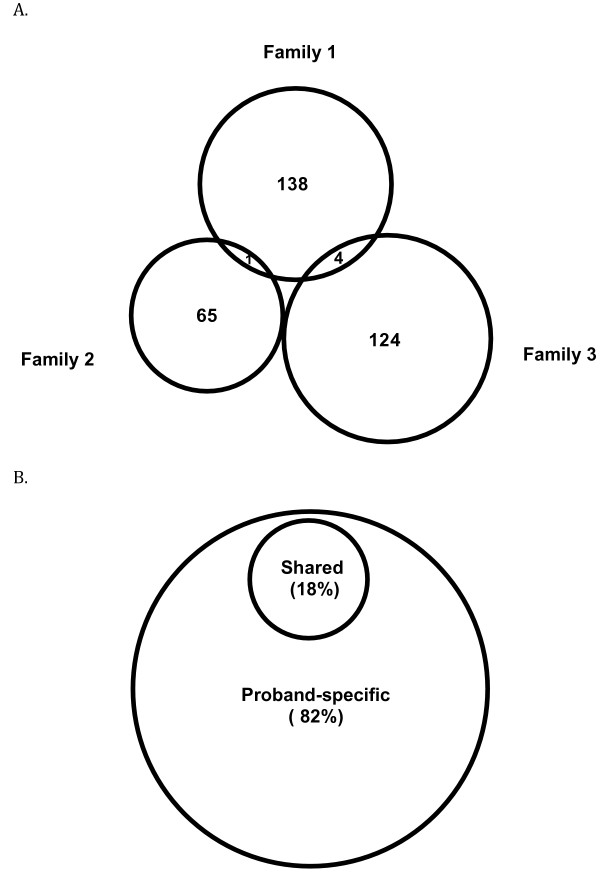
Comparison of the variants in BRCAx families and probands. A. Comparison in the three families. B. Comparison in the probands. The results show that the variants detected in the cancer-affected family members are highly family-specific. The higher rate (18%) of the shared variants in the probands are likely due to the remaining normal variants not filtered in the probands and the larger number of families represented by the probands than the three families.
Identification of putative genetic predispositions
We analyzed the shared mutations between the affected members of the same family, the functional class of the mutated genes, and existing evidence for their contribution to cancer. In doing so, we identified the variants as the putative predispositions in Family I, II, and III, and probands (Table 3, Additional file 1: Table S1A, S1B, S1C). For Family I, this was the PTEN-Induced Putative Kinase 1 (PINK1); for Family II, these were Lysine (K) Acetyltransferase 6B (KAT6B) and Neurogenic Locus Notch Homolog Protein 2 (NOTCH2); and for Family III, this was Phosphorylase Kinase Beta (PHKB).
Table 3.
Putative predispositions in familial breast cancer families and probands
| |
Gene |
Description |
Position |
Nucleotide |
Amino acid |
Type |
PolyPhen2* |
Cancer-affected member |
Frequency |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Score prediction | ||||||||||||||||
| Family 1 |
|
|
|
|
|
|
|
|
1 |
2 |
3 |
4 |
6 |
7 |
8 |
|
| |
GPRIN1 |
G protein regulated inducer of neurite outgrowth 1 |
chr5:176026123 |
c.T713C |
p.L238S |
Exonic |
0.91 |
D |
- |
+ |
+ |
+ |
+ |
+ |
- |
5 |
| |
PINK1 |
PTEN induced putative kinase 1 |
chr1:20972051 |
c.960-2A > G |
|
Splicing |
NA |
NA |
- |
- |
+ |
+ |
- |
- |
- |
2 |
| |
POLK |
Polymerase (DNA directed) kappa |
chr5:74892737 |
c.A2219G |
p.H740R |
Exonic |
0.62 |
P |
- |
- |
- |
+ |
- |
- |
- |
1 |
| Family 2 |
|
|
|
|
|
|
|
|
1 |
2 |
3 |
4 |
5 |
6 |
|
|
| |
KAT6B |
K(lysine) acetyltransferase 6B |
chr10:76789128 |
c.G4546T |
p.D1516Y |
Exonic |
0.95 |
D |
- |
+ |
+ |
+ |
+ |
+ |
|
5 |
| |
KAT6B |
K(lysine) acetyltransferase 6B |
chr10:76789311 |
c.C4729T |
p.R1577C |
Exonic |
0.96 |
D |
- |
+ |
+ |
+ |
+ |
+ |
|
5 |
| |
NOTCH2 |
Notch 2 |
chr1:120459167 |
c.C6178T |
p.R2060C |
Exonic |
0.99 |
D |
- |
- |
+ |
- |
- |
+ |
|
2 |
| Family 3 |
|
|
|
|
|
|
|
|
1 |
2 |
4 |
5 |
6 |
|
|
|
| |
NANP |
N-acetylneuraminic acid phosphatase |
chr20:25596725 |
c.A583G |
p.I195V |
Exonic |
0.98 |
D |
+ |
- |
+ |
- |
- |
|
|
2 |
| |
PHKB |
phosphorylase kinase, beta |
chr16:47628126 |
c.1204 + 1G > T |
|
Splicing |
NA |
NA |
- |
+ |
- |
+ |
- |
|
|
2 |
| Proband |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
JAKMIP3 |
Janus kinase and microtubule interacting protein 3 |
chr10:133955524 |
c.G1574C |
p.G525A |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 2 |
POLQ |
Polymerase (DNA directed), theta |
chr3:121207798 |
c.A3980C |
p.Q1327P |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 3 |
DUX2 |
Double homeobox 2 |
chr10:135494906 |
|
|
Splicing |
NA |
NA |
|
|
|
|
|
|
|
|
| 4 |
UBE2L3 |
Ubiquitin-conjugating enzyme E2L 3 |
chr22:21975938 |
c.G349A |
p.E117K |
Exonic |
0.96 |
D |
. |
. |
|
|
|
|
|
|
| 5 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
chr9:110087260 |
c.C1028T |
p.P343L |
Exonic |
0.99 |
D |
. |
. |
|
|
|
|
|
|
| 7 |
GATA3 |
GATA binding protein 3 |
chr10:8100630 |
c.C604T |
p.R202C |
Exonic |
0.92 |
D |
|
|
|
|
|
|
|
|
| 8 |
KAT6B |
K(lysine) acetyltransferase 6B |
chr10:76744854 |
c.G2390A |
p.S797N |
Exonic |
0.98 |
D |
|
|
|
|
|
|
|
|
| 9 |
LIG1 |
Ligase I, DNA, ATP-dependent |
chr19:48637322 |
c.G1525A |
p.E509K |
Exonic |
0.95 |
D |
. |
. |
|
|
|
|
|
|
| 10 |
LIG4 |
Ligase IV, DNA, ATP-dependent |
chr13:108862463 |
c.G1154A |
p.R385K |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 14 |
NOTCH2 |
Notch 2 |
chr1:120529603 |
c.G854A |
p.R285H |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 15 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
chr9:133729493 |
c.G122A |
p.G41D |
Exonic |
0.92 |
D |
|
|
|
|
|
|
|
|
| 16 |
TNK2 |
Tyrosine kinase, non-receptor, 2 |
chr3:195596385 |
c.C1760T |
p.P587L |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 17 |
NFRKB |
Nuclear factor related to kappaB binding protein |
chr11:129755398 |
c.G611A |
p.R204H |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 18 |
NFKBIZ |
Nuclear factor of kappa light polypeptide gene enhancer |
chr3:101576029 |
|
|
Splicing |
NA |
NA |
|
|
|
|
|
|
|
|
| 19 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
chr16:18879624 |
c.C3083T |
p.T1028M |
Exonic |
0.99 |
D |
|
|
|
|
|
|
|
|
| 20 |
PRKCQ |
Protein kinase C, theta |
chr10:6528042 |
c.G855C |
p.Q285H |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 21 |
ADRA2A |
Adrenoceptor alpha 2A |
chr10:112838117 |
c.C363G |
p.C121W |
Exonic |
1.00 |
D |
|
|
|
|
|
|
|
|
| 22 | PPFIA4 | Protein tyrosine phosphatase, receptor type | chr1:203025582 | c.C668T | p.T223M | Exonic | 0.92 | D | ||||||||
D: Probably damaging (score: 0.909-1); P: Possibly damaging (score: 0.447 - 0.908).
PINK1 is a mitochondrial serine/threonine-protein kinase. Mutation in PINK1 causes autosomal recessive Parkinson’s disease [22]. KAT6B is a histone acetyl transferase involved in DNA replication, gene expression and regulation, and epigenetic modification of chromosomal structure [23]. Mutations in KAT6B cause multiple neurological diseases [24]. NOTCH2 is a member of the Notch family involved in controlling cell fate decision. Low Notch activity leads to hyperproliferative activity in breast cancer [25] and mutation in NOTCH2 causes Hajdu-Cheney syndrome [26]. PHKB regulates the function of phosphorylase kinase [27]. Mutation in PHKB causes glycogen storage disease type 9B [28]. Interestingly, a variant in Polymerase (DNA-Directed) Kappa (POLK) was present in Family I member #4. POLK is a member of Y family DNA polymerases, and functions by repairing the replication fork passing through DNA lesions [29]. Although we are not able to validate it due to the lack of DNA from the subject’s parents, it raises a possibility that this variant could be a de novo mutation in this individual. Multiple transcriptional factors were also affected by the mutations in each family. For example, the following transcriptional factors were mutated in Family I: ZNF335, LRRC66, ZNF417, ZNF587, GTF2I, ZFAND4, EIF4G2, GZF1, CCDC86, ZSCAN18, ZNF546, TAF1L, and LRIG3 (Additional file 1: Table S1A).
The variant data from probands show similar patterns as those of the three families (Table 3). In the 22 probands, four carried variants affecting the genes involved in DNA replication and damaging repair. Those include Polymerase (DNA-directed) Theta (POLQ) in Proband #2, RAD23 Homolog B (S. cerevisiae) (RAD23B) in Proband #3, Ligase I DNA, ATP-dependent (LIG1) in Proband #9, and Ligase IV DNA, ATP-dependent (LIG4) in Proband #10. POLQ repairs the apurinic sites [30]. RAD23B plays a role in nucleotide excision repair [31]. LIG1 ligates nascent DNA of the lagging strand, and a mutation in LIG1 causes replication errors, genome instability, and cancer [32]. LIG4 catalyzes double-strand break repair by joining non-homologous ends, and mutation in LIG4 causes LIG4 syndrome [33]. Several variants are found in well-known oncogenes and tumor suppressor genes, such as GATA Binding Protein 3 (GATA3) in Proband #7 and Abelson Murine Leukemia Viral Oncogene Homolog 1 (ABL1) in Proband #18. GATA3 regulates luminal epithelial cell differentiation in the mammary gland [34,35]. The abnormal expression of GATA3 causes luminal A-type breast cancer [36-38]. ABL1 is a tyrosine kinase that controls cell differentiation and division. It is involved in (9, 22) translocation, forming BCR-ABL fusion gene in chronic myelogenous leukemia (CML) [39]. Several individual variants in different cases affect the same genes but at different positions. For example, in Proband #8, a variant in KAT6B (c.G1841A/p.S614N) affects the HAT domain at the N terminal, whereas two variants in KAT6B in Family II (c.G3997T/p.D1333Y and c.C4180T/p.R1394C) affect the Met-rich domain at the C-terminal. In Proband #14 and Family II, two different NOTCH2 variants (c.G854A/p.R285H, c.C6178T/p.R2060C) were present. Multiple variants affect the genes involved in phosphorylation. These include Tyrosine Kinase Non-Receptor 2 (TNK2) in proband #16, Phosphatidylinositol 3 Kinase-Related Kinase (SMG1) in Proband #19, Protein Kinase C Theta (PRKCQ) in Proband #20, and Protein Tyrosine Phosphatase, Receptor Type F (PPFIZ4) in Proband #22.
We also performed an analysis at the pathway level by annotating the mutation-affected genes in the three families using KEGG database (http://www.genome.jp/kegg/pathway.html). Certain mutations were identified to affect several functional pathways. For example, the genes mutated in Family I (ACADVL, AHCY, ALDOA, SGPL1, MAT1A, GALNT8, GGT1) are involved in metabolic pathways. The genes mutated in Family 2 (NOTCH2, DUSP16) are involved in Notch signaling pathway and MAPK signaling pathway; genes mutated in Family III (SLC9A1, ITGAX, ITGAD) are involved in regulation of actin cytoskeleton.
Discussion
The majority of families with familial breast cancer lack evidence for their genetic predispositions. Efforts in past decade have made slow progress in determining the unknown genetic predispositions. Currently, population-based approach is adapted as the major promising tool to reach the goal [40]. One weakness of this approach is that it can “dilute out the effects of a very strong association in a small subset of the study population” [41]. It requires a large-size disease population of over tens of thousands but the predispositions identified will likely remain very rare in the disease population. Due to the extreme rarity, such genetic predispositions are often difficult to confirm in different disease populations and to distinguish from normal polymorphisms [5,10]. Our study observed the presence of family-specific, novel, deleterious variants, and putative predispositions in the families and probands analyzed. The information implies that, in addition to the population-based approach, a family-based approach provides another option to determine the genetic predisposition.
Based on the higher frequencies of well-known predispositions identified by traditional approaches, the rarity of the predispositions recently identified by population-based approach, and the presence of family-specific, novel, deleterious variants in disease families revealed in our study, we propose a model to explain the genetic predispositions in familial breast cancer (Figure 3). In this model, the predisposition in BRCA1 has the highest frequency in the familial breast cancer population, other known predispositions gradually decrease their frequencies to insignificant levels, and the predispositions for many BRCAx familial breast cancers are family-specific. The model explains the difficulty in using traditional and population-based approaches to determine the unknown predispositions, and highlights that applying family-focused approach will be able to determine the genetic predispositions for many BRCAx disease families. This model can be further tested in larger number of BRCAx familial breast cancer families.
Figure 3.
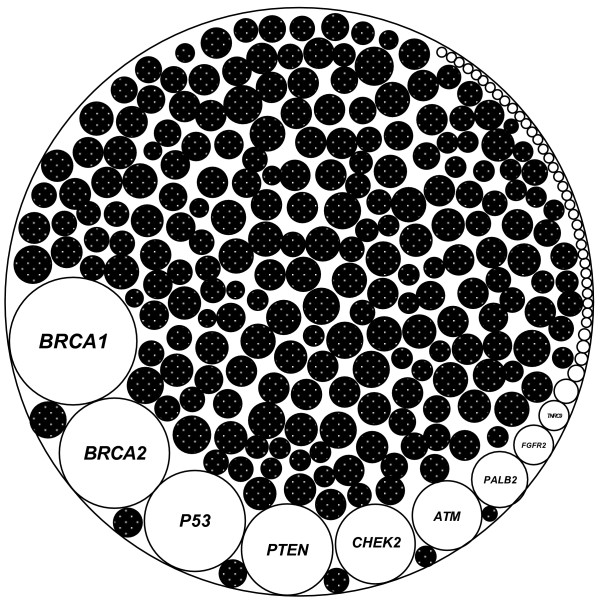
A model for the genetic predispositions in familial breast cancer. The known predisposition in BRCA1 has the highest sharing frequency in the disease population, other known predispositions decrease their frequencies towards extreme rarity in the disease populations, and the family-specific predispositions are enriched in many disease families without known predispositions. The biggest circle represents the entire genetic predispositions in familial breast cancer. The open circles represent the shared, known predispositions, and the black circles represent the family-specific predispositions.
Our study aimed to determine if there are germline mutations present, rather than reach for comprehensive coverage of germline mutations in each family. We achieved this by eliminating all variants matched in population-derived variant databases (i.e., dbSNP137, ESP6500, 1000 genomes) to maximally avoid the variants representing normal polymorphism. Inclusion of such variants as the predisposition candidates, even with the use of certain cut-off such as minor allele frequency (MAF) <0.01, can increase the sensitivity but decrease the specificity of the variants referred to as putative predispositions.
Assignment of a specific mutation as a true predisposition to a disease family requires solid phenotypic evidence from in vitro analysis, cell line tests, search of the literature, bioinformatics data analysis, and animal models. This is best evidenced by determining the BRCA1 germline mutations as genetic predispositions in breast cancer, in which the definitive conclusion for its contribution to breast cancer is based on the mouse models showing development of breast cancer with the germline mutated BRCA1[42]. Our current study aims to provide evidence that the BRCAx disease families are enriched with germline damaging mutations, such that focusing on each disease family will be required to determine the genetic predisposition in each family. Indeed, even under strict mapping conditions, large numbers of mutations have been detected in each disease family and probands. While the data provide rich resources to identify the true predisposition for the disease family, the data cannot be considered as true predisposition without further phenotypic and functional evidences.
Conclusions
Our study shows that genetic predispositions in many BRCAx familial breast cancer families can be family-specific.
Abbreviations
BRCAx: Familial breast cancer without known mutations in BRCA1 and BRCA2; Proband: the first affected family member seeking medical attention; Exome sequencing: Sequencing the entire coding region in a genome using the next generation DNA sequencing technology; SAM: Sequence Alignment/Map format used for storing sequence data in a series of tab delimited ASCII columns; BAM: A binary format for storing sequence data in a compressed, indexed, binary form; GATK: Genome Analysis Toolkit. It is a software package to analyse next-generation resequencing data; VarScan 2: a software package to detect variants in next-generation resequencing data; PolyPhen-2: a software to predict possible impact of an amino acid substitution on the structure and function of a protein; Primer3: a software for designing PCR primers; NCBI: The National Center for Biotechnology Information; dbSNP: Single Nucleotide Polymorphism Database; ESP: Exome Sequencing Project; MAF: Minor Allele Frequency.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FX, HW, BD performed experiments. YK performed bioinformatics data analysis. CS, DB performed pedigree analysis, identified the study subjects, and prepared DNA samples. JL performed statistical analysis. EAF, SS, KC developed the UNMC Breast Cancer Collaborative Register used in the study [43]. HL and SMW conceived the study. SMW designed the experiment and wrote the paper. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Variants detected in breast cancer-affected members in three BRCAx familial breast cancer families. Table S1A. Family 1; Table S1B. Family 2; Table S1C. Family 3; Table S1D. Variants shared among the three families; Table S1E. Variants validated by Sanger sequencing.
Variants identified in 22 probands. Table S2A. Variants only in single proband; Table S2B. Variants shared among probands.
Contributor Information
Hongxiu Wen, Email: wenhongxiu@gmail.com.
Yeong C Kim, Email: yeong.kim@unmc.edu.
Carrie Snyder, Email: csnyder@creighton.edu.
Fengxia Xiao, Email: fengxia.xiao@unmc.edu.
Elizabeth A Fleissner, Email: eafleissner@unmc.edu.
Dina Becirovic, Email: DinaBecirovic@creighton.edu.
Jiangtao Luo, Email: jiangtao.luo@unmc.edu.
Bradley Downs, Email: bradley.downs@unmc.edu.
Simon Sherman, Email: ssherm@unmc.edu.
Kenneth H Cowan, Email: kcowan@unmc.edu.
Henry T Lynch, Email: htlynch@creighton.edu.
San Ming Wang, Email: sanming.wang@unmc.edu.
Acknowledgments
The study was supported by a pilot grant from Fred & Pamela Buffett Cancer Center, University of Nebraska Medical Center (SMW), and a NIH grant 1R21CA180008 (SMW). The funding bodies play no roles in design, collection, analysis, and interpretation of data. We also wish to thank for Melody A. Montgomery at the UNMC Research Editorial Office for her professional assistance in editing this manuscript.
References
- American Cancer Society. Cancer Facts & Figures – 2013. 2013.
- Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, Yates LR, Papaemmanuil E, Beare D, Butler A, Cheverton A, Gamble J, Hinton J, Jia M, Jayakumar A, Jones D, Latimer C, Lau KW, McLaren S, McBride DJ, Menzies A, Mudie L, Raine K, Rad R, Chapman MS, Teague J. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, Neuhausen SL, John EM, Andrulis IL, Terry MB, Daly M, Buys S, Le Calvez-Kelm F, Lonie A, Pope BJ, Tsimiklis H, Voegele C, Hilbers FM, Hoogerbrugge N, Barroso A, Osorio A, Giles GG, Devilee P, Benitez J, Hopper JL, Tavtigian SV, Goldgar DE, Southey MC. Breast Cancer Family Registry; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, Thorne H, Barnes DR, Li J, Ellul J, Philip GK, Antill YC, James PA, Trainer AH, Mitchell G, Campbell IG. kConFab. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape K, Ruark E, Tarpey P, Renwick A, Turnbull C, Seal S, Murray A, Hanks S, Douglas J, Stratton MR, Rahman N. Predisposition gene identification in common cancers by exome sequencing: insights from familial breast cancer. Breast Cancer Res Treat. 2012;134:429–433. doi: 10.1007/s10549-012-2057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia-Aznarez FJ, Fernandez V, Pita G, Peterlongo P, Dominguez O, de la Hoya M, Duran M, Osorio A, Moreno L, Gonzalez-Neira A, Rosa-Rosa JM, Sinilnikova O, Mazoyer S, Hopper J, Lazaro C, Southey M, Odefrey F, Manoukian S, Catucci I, Caldes T, Lynch HT, Hilbers FS, van Asperen CJ, Vasen HF, Goldgar D, Radice P, Devilee P, Benitez J. Whole exome sequencing suggests much of non-BRCA1/BRCA2 familial breast cancer is due to moderate and low penetrance susceptibility alleles. PLoS One. 2013;8:e55681. doi: 10.1371/journal.pone.0055681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers FS, Meijers CM, Laros JF, van Galen M, Hoogerbrugge N, Vasen HF, Nederlof PM, Wijnen JT, van Asperen CJ, Devilee P. Exome sequencing of germline DNA from non-BRCA1/2 familial breast cancer cases selected on the basis of aCGH tumor profiling. PLoS One. 2013;8:e55734. doi: 10.1371/journal.pone.0055734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers FS, Meijers CM, Laros JF, van Galen M, Hoogerbrugge N, Vasen HF, Nederlof PM, Wijnen JT, van Asperen CJ, Devilee P. Rare variants in XRCC2 as breast cancer susceptibility alleles. J Med Genet. 2012;49:618–620. doi: 10.1136/jmedgenet-2012-101191. [DOI] [PubMed] [Google Scholar]
- Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, Duarte Sdel V, Rivas MA, Warren-Perry M, Zachariou A, Campion-Flora A, Hanks S, Murray A, Ansari Pour N, Douglas J, Gregory L, Rimmer A, Walker NM, Yang TP, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C. et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Krush AJ, Lemon HM, Kaplan AR, Condit PT, Bottomley RH. Tumor variation in families with breast cancer. JAMA. 1972;222:1631–1635. [PubMed] [Google Scholar]
- Lynch H, Wen H, Kim YC, Snyder C, Kinarsky Y, Chen PX, Xiao F, Goldgar D, Cowan KH, Wang SM. Can unknown predisposition in familial breast cancer be family-specific? Breast J. 2013;19:520–528. doi: 10.1111/tbj.12145. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup, 1000 Genome Project Data. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonita-Laza I, Ottman R. Study designs for identification of rare disease variants in complex diseases: the utility of family-based designs. Genetics. 2011;189(3):1061–1068. doi: 10.1534/genetics.111.131813. PMID: 21840850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin D, Campbell M, Fayers P, Pinol A. Sample Size Tables for Clinical Studies. 2. Malden, MA: Blackwell Science; 1997. [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y, Heng HH, Yang XJ. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- Kraft M, Cirstea IC, Voss AK, Thomas T, Goehring I, Sheikh BN, Gordon L, Scott H, Smyth GK, Ahmadian MR, Trautmann U, Zenker M, Tartaglia M, Ekici A, Reis A, Dörr HG, Rauch A, Thiel CT. Disruption of the histone acetyltransferase MYST4 leads to a Noonan syndrome-like phenotype and hyperactivated MAPK signaling in humans and mice. J Clin Invest. 2011;121:3479–3491. doi: 10.1172/JCI43428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, Artavanis-Tsakonas S, Brugge JS. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci U S A. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet. 2011;43:303–305. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Mol Cell Biol. 2010;30:1368–1381. doi: 10.1128/MCB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg IE, van Beurden EA, de Klerk JB, van Diggelen OP, Malingré HE, Boer MM, Berger R. Autosomal recessive phosphorylase kinase deficiency in liver, caused by mutations in the gene encoding the beta subunit (PHKB) Am J Hum Genet. 1997;61:539–546. doi: 10.1086/515502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2005;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spek PJ, Smit EM, Beverloo HB, Sugasawa K, Masutani C, Hanaoka F, Hoeijmakers JH, Hagemeijer A. Chromosomal localization of three repair genes: the xeroderma pigmentosum group C gene and two human homologs of yeast RAD23. Genomics. 1995;23:651–658. doi: 10.1006/geno.1994.1554. [DOI] [PubMed] [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, Gatti RA, Varon R, Oettinger MA, Neitzel H, Jeggo PA, Concannon P. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Kim JW, Bechis SK, Werb Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol. 2008;20:164–170. doi: 10.1016/j.ceb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BJ, Giguere V. Meta-analysis of human cancer microarrays reveals that GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;7:49. doi: 10.1186/1476-4598-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dydensborg AB, Rose AA, Wilson BJ, Grote D, Paquet M, Giguère V, Siegel PM, Bouchard M. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene. 2009;28:2634–2642. doi: 10.1038/onc.2009.126. [DOI] [PubMed] [Google Scholar]
- Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Southey MC, Park DJ, Nguyen-Dumont T, Campbell I, Thompson E, Trainer AH, Chenevix-Trench G, Simard J, Dumont M, Soucy P, Thomassen M, Jønson L, Pedersen IS, Hansen TV, Nevanlinna H, Khan S, Sinilnikova O, Mazoyer S, Lesueur F, Damiola F, Schmutzler R, Meindl A, Hahnen E, Dufault MR, Chris Chan T, Kwong A, Barkardóttir R, Radice P, Peterlongo P. COMPLEXO et al. COMPLEXO: identifying the missing heritability of breast cancer via next generation collaboration. Breast Cancer Res. 2013;15:402. doi: 10.1186/bcr3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SM, Klotz DM, Cohn CS. In: Hormone and Cancer. Vedeckis, Wayne V, editor. Boston: Birkhauser; 1996. Genetics of Breast cancer; p. 199. [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Sherman S, Shats O, Fleissner E, Bascom G, Yiee K, Copur M, Crow K, Rooney J, Mateen Z, Ketcham MA, Feng J, Sherman A, Gleason M, Kinarsky L, Silva-Lopez E, Edney J, Reed E, Berger A, Cowan K. Multicenter breast cancer collaborative registry. Cancer Inform. 2011;10:217–226. doi: 10.4137/CIN.S7845. PMID: 21918596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variants detected in breast cancer-affected members in three BRCAx familial breast cancer families. Table S1A. Family 1; Table S1B. Family 2; Table S1C. Family 3; Table S1D. Variants shared among the three families; Table S1E. Variants validated by Sanger sequencing.
Variants identified in 22 probands. Table S2A. Variants only in single proband; Table S2B. Variants shared among probands.


