Abstract
Goat and sheep milk is consumed by human populations throughout the world; as a result, it has been proposed as an alternative, nutrient-rich milk to feed infants allergic to cow’s milk. Unfortunately, potentially harmful bacteria have not been thoroughly tested in goat or sheep milk. Listeria monocytogenes is a harmful bacterium that causes adverse health effects if ingested by humans. The purpose of this study was to estimate the prevalence and characterize the phenotype, genotype, virulence factors, biofilm formation, and antibiopotential of Listeria isolated from the milk of goat and sheep. Udder milk samples were collected from 107 goats and 102 sheep and screened for mastitis using the California mastitis test (CMT). Samples were then examined for the presence of pathogenic Listeria spp; if detected, the isolation of pathogenic Listeria (L. monocytogenes and Listeria ivanovii) was completed using isolation and identification techniques recommended by the International Organization for Standards (ISO 11290-1, 1996), in addition to serological, in vitro and in vivo pathogenicity tests. The isolates were subjected to PCR assay for virulence associated genes (hlyA, plcA, actA, and iap). Pathogenic Listeria spp. were isolated from 5.6% of goat and 3.9% sheep milk samples, with 33.3 and 25% of these selected samples respectively containing L. monocytogenes. The results of this study provide evidence of the low-likelihood of contamination leading to the presence of L. monocytogenes in raw goat and sheep milk; however, this study also confirmed a strong in vitro ability for biofilm formation and pathogenic capability of L. monocytogenes if discovered in the milk. L. monocytogenes may be present in goat and sheep milk and in order to reduce the exposure, hygienic milking conditions must be employed for the milk to be considered a safe alternative for human consumption.
Keywords: Listeria species, Goat, Ewe, Virulence genes, Biofilm formation, Antibiotic resistance
Introduction
For up to 6000 years, Egyptians have ranched goats and sheep and consumed their milk.1 The use of goat and sheep milk for medicinal purposes and household consumption is prominent throughout the world. Herd survivability, management ease, and affordability promote the ranching of goats primarily in low to middle-income countries.2 In locations with little availability to cattle or other alternate milk, populations with prominent milk allergies, or increased exposure to milk-borne pathogens such as Listeria, it is necessary to consider alternative milk. Goat and ewe milk has been proposed as an alternative, nutrient-rich milk to feed infants allergic to cow’s milk.3
Listeria species are ubiquitous; however, only two of 10 species, Listeria monocytogenes and Listeria ivanovii, are pathogenic in human populations.4 Listeria monocytogenes is a foodborne pathogen that may cause a severe, invasive illness with a corresponding mortality rate up to 30%;5 furthermore, exposure to L. monocytogenes is second only to salmonellosis among the most frequent causes of death due to a foodborne illness.6
Among animals, listeriosis may affect sheep and goats and the pathology may develop in different clinical forms and include mastitis.7 The excretion of Listeria in milk may persist throughout lactation and contribute to an increased risk of milk product contamination.6 Sheep milk has previously been described as a possible cause for listeriosis.8 Intermittent shedding of L. monocytogenes has been reported in goats and sheep without clinical signs of listeriosis.9,10 Therefore, the identification of infected animals was necessary due to the likely causal link to several outbreaks of listeriosis.8–12
Currently, studies confirming the virulence potential and polymorphism of L. monocytogenes strains prevalent in Egypt neither exist nor have been previously conducted. This study sought to compare milk related Listeria isolates of two domesticated animals, goats and sheep. Prevalence, biochemical and serological characterization, in vitro and in vivo pathogenicity, and antimicrobial resistance profiles were assessed. The plcB, actA, iap, and hlyA genes for L. monocytogenes were aptly chosen as target genes throughout this study.13
Materials and Methods
The study took place in private dairy farms surrounding Cairo, Egypt where 209 milk samples from the mammary glands of sheep (n = 102) and goats (n = 107) were collected. The animals had not been treated with an antibiotic for at least 30 days prior to collection. Milk samples were screened for mastitis using the California mastitis test (CMT) prior to the isolation of Listeria spp. Milk samples were taken under aseptic conditions for bacteriological examinations. A subsample of 15 ml of milk, taken from a composite milk sample, was collected in sterile universal bottles. The milk samples were quickly transported to the laboratory under chilled conditions and stored at 4°C till bacteriologically analyzed.
For the isolation and identification of Listeria species in the milk samples, the techniques recommended by the International Organization for Standards (ISO 11290-1, 1996) were implemented.
Isolation and identification of Listeria was performed using the double enrichment procedure, the first (Half Fraser) and the second (Fraser) enrichment broths. A loopful of growth from Fraser II was subcultured onto Palcam selective agar supplemented with SR 0150E (polymyxin B 5 mg, acriflavine HCl 2.5 mg) (Oxoid) and incubated micro-aerobically (5% O2, 10% CO2, and 85% N2). Colonies suspected to be Listeria were transferred onto tryptic soya yeast extract agar (TSYEA) (Difco, Bacton, PA, USA) and incubated at 30°C for 18–24 hours. Those putative Listeria colonies were tested for purity, in addition to morphological and biochemical characteristics (Table 1). Strains were also identified using the API® Listeria system (bioMe’rieux, Marcy l’Etoile, France) and the Oxoid Microbact™ Listeria 12L (MB1128A). Presumptive Listeria colonies were maintained at 4°C on Trypticase soy agar with 0.6% yeast extract slants, incubated at 37°C for 24 hours, and stored at 4°C for final serotyping and PCR confirmation.
Table 1. Cultural and biochemical differentiation between the Listeria species recovered from udder raw fresh whole milk of goat and sheep.
| Phenotypic characteristics | Listeria species | |||||
| Listeria monocytogenes; reference strain (ATCC 7494) | L. monocytogenes | L. seeligeri | L. innocua | L. welshimeri | L. grayi | |
| ALOA agar | Blue green with halo | Blue green with halo | Blue green without halo | Blue green without halo | Blue green without halo | Blue green without halo |
| Hemolysis | beta-hemolysis | beta-hemolysis | beta-hemolysis | Non-hemolytic | Non-hemolytic | Non-hemolytic |
| Motility | Motile at 22–25°C | Motile at 22–25°C | Motile at 37°C | Motile at 37°C | Motile at 37°C | Motile at 37°C |
| Nitrate reduction test | − | − | − | − | − | + |
| d-Xylose fermentation | − | − | + | − | + | − |
| l-Rhamnose fermentation | + | + | − | Variable | Variable | Variable |
| d-Mannitol | − | − | − | − | − | + |
| Alpha methyl d-mannoside | + | + | − | + | + | + |
| Genus Listeria distinguished on the basis of the 16S rRNA | + | + | + | + | + | + |
| Rapid detection of L. monocytogenes in milk using hlyA gene | + | + | + | − | − | − |
Serotyping was carried out on L. monocytogenes strains using commercial specific antisera (Behringwerke AG, Marburg, Germany) against the serovars 1and 4, following the manufacturer’s instructions. All purified isolates were tested by the standard disc diffusion method (CLSI, 2010) and were subjected to a susceptibility panel of 28 antibiotics (Oxoid) belonging to 11 drug classes (Table 6). Isolates were cultured in trypticase soy broth (TSB) supplemented with 0.6% yeast extract, and transferred to Mueller–Hinton agar (Oxoid). The plates were incubated at 37°C for 48 hours
Table 6. Result of antimicrobial susceptibility for Listeria species isolated from fresh udder milk.
| Antibiotics | Species of isolated Listeria | ||
| Listeria monocytogenes reference strain (ATCC7494) | L. monocytogenes | Other Listeria species | |
| Penicillins | |||
| Ampicillin (25 μg) | 1/1 | 3/3 | 1/7 |
| Penicillin G (10IU) | 1/1 | 3/3 | 3/7 |
| Amoxicillin/clavulanic acid (10 μg) | 1/1 | 3/3 | 1/7 |
| Cloxacillin (5 μg) | 0/1 | 0/3 | 0/7 |
| Oxacillin (1 μg) | 0/1 | 0/3 | 0/7 |
| Amoxicillin (25 μg) | 1/1 | 3/3 | 1/7 |
| Fluoroquinolones | |||
| Ofloxacin (10 μg) | 1/1 | 3/3 | 7/7 |
| Enrofloxacin (10 μg) | 1/1 | 3/3 | 7/7 |
| Ciprofloxacin (5 μg) | 1/1 | 3/3 | 1/7 |
| Flumequine (30 μg) | 0/1 | 0/3 | 0/7 |
| Pefloxacin (30 μg) | 0/1 | 0/3 | 0/ |
| Aminoglycosides | |||
| Amikacin (30 μg) | 1/1 | 3/3 | 7/7 |
| Gentamicin (10 μg) | 1/1 | 3/3 | 7/7 |
| Kanamycin (30 μg) | 1/1 | 3/3 | 7/7 |
| Neomycin (10 μg) | 1/1 | 3/3 | 7/7 |
| Streptomycin (10 μg) | 1/1 | 3/3 | 1/7 |
| Phenicol | |||
| Chloramphenicol (30 μg) | 1/1 | 2/3 | 7/7 |
| Tetracycline | |||
| Tetracycline (30 μg) | 1/1 | 2/3 | 7/7 |
| Sulfonamide | |||
| Trimethoprim-sulfamethoxazole 1∶19 (25 μg) | 1/1 | 2/3 | 1/7 |
| Cephalosporins | |||
| Cefotaxime (30 μg) | 0/1 | 0/3 | 0/7 |
| Cephalothin (30 μg) | 0/1 | 0/3 | 0/7 |
| Lincosamides | |||
| Lincomycin (2 μg) | 0/1 | 0/3 | 0/7 |
| Clindamycin (2 μg) | 0/1 | 0/3 | 0/7 |
| Polypeptides | |||
| Bacitracin (10 units) | 0/1 | 0/3 | 0/7 |
| Glycopeptide | |||
| Vancomycin (30 μg) | 1/1 | 3/3 | 7/7 |
| Macrolide | |||
| Erythromycin (15 μg) | 1/1 | 3/3 | 7/7 |
| Spiramycin (100 μg) | 1/1 | 3/3 | 7/7 |
| Rifamycin | |||
| Rifampicin (5 μg) | 1/1 | 3/3 | 2/7 |
All the biochemically characterized Listeria isolates were assayed for phosphatidylinositol-specific phospholipase C (PI-PLC) activity,14 and DL-alanine-beta-naphthylamide HCl (DLABN) and d-alanine-p-nitroanilide (DAPN) tests further differentiated between L. monocytogenes and other species of Listeria Clark and McLauchlin (1997).15 The classical tests for Listeria pathogenicity implemented were the Anton conjunctivitis test (rabbits), inoculation of mice, inoculation of embryonated eggs, and ingestion assay on Vero cells.16,17 A qualitative assessment of the biofilm formation assay was developed according to Giacaman et al. (2010) through the Christensen's tube method and by the tissue culture plate assay, as described by Borucki et al. (2003).18,19
The PCR was used for the detection of the genus Listeria. Freshly grown typical Listeria-like colonies (black colonies) from the surfaces of Palcam plates were boiled in 400 μl of 1× Tris-EDTA buffer (pH 8.0) for 10 minutes and centrifuged at 16 873 g for 10 minutes to remove denatured proteins and bacterial membranes. Listeria was distinguished on the basis of the 16S rRNA that can be revealed by PCR (U1forward primer 5-CAG-CMG-CCG-CGG-TAA-TWC-3, where M denotes A or C and W denotes A or T and LI1 reverse primer 5-CTC-CAT-AAA-GGT-GAC-CCT-3) to target a 938-bp 16S rRNA sequence in members of the genus Listeria.20,21 Amplification was performed in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, CT, USA). The samples were subjected to an initial denaturation step of 94°C for 4 minutes, followed by 25 amplification cycles of 1 minute at 94°C (denaturation), 1 minute at 60°C (primer annealing), and 1 minute at 72°C (primer extension) followed by a final extension step of 72°C for 5 minutes. PCR reaction products were separated on 1.5% agarose gels, stained with ethidium bromide and visualized. An L. monocytogenes strain (ATCC 7494) and an E. coli strain (ATCC 25922) were included as positive and negative controls respectively.
For PCR detection of virulence genes among Listeria species, the Listeria pellet collected from the surfaces of Palcam plates was dispersed in 400 μl 10 mM Tris-1 mM EDTA buffer containing 0.1% Tween 80 and 150 μl (2 mg/ml) lysozyme and incubated for 15 minutes on ice. The pellet was lysed by redissolving in TE buffer containing 40 μl (100 μg of Proteinase K/ml) Proteinase K and 60 μl (10% (w/v)) sodium dodecyl sulfate and incubated for 30 minutes at 37°C. DNA was extracted by adding an equal volume of TE saturated phenol: chloroform: iso-amyl alcohol (25∶24∶1 v/v/v) and precipitated by the addition of 20 μg of acrylamide/ml, 0.05 volumes of 3M sodium acetate and 2.5 volumes of ethanol.
The 10 goat and ewe Listeria isolates were screened by PCR for the presence of the virulence genes using the primers: hlyA (for.) 5′-GCA GTT GCA AGC GCT TGG AGT GAA-3′, (rev.) 5′- GCA ACG TAT CCT CCA GAG TGA TCG-3′ to amplify the 456-bp fragment;22 plcB (for.) 5′-CTG CTT GAG CGT TCA TGTCTC ATC CCC C-3′, (rev.) 5′-ATG GGT TTC ACT CTCCTT CTA C-3′ to amplify the 1484-bp fragment;23 actA (for.) 5′-CGC CGC GGA AATTAA AAA AAG A-3′, (rev.) 5′- ACG AAG GAA CCG GGCTGC TAG - 3′ to amplify the 839-bp fragment; and Iap (for.) 5′-ACA AGC TGC ACC TGT TGC AG-3′, (rev.) 5′-TGACAG CGT GTG TAG TAG CA-3′ to amplify the 131-bp fragment.24 Amplification was performed in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, CT, USA). The samples were subjected to an initial denaturation step of 95°C for 2 minutes, followed by 35 amplification cycles of 15 seconds at 95°C (denaturation), 30 seconds at 60°C (annealing), and 90 seconds at 72°C (primer extension), followed by a final extension step of 72°C for 10 minutes. Visualization of the PCR products was carried out as previously indicated (Fig. 1).
Figure 1.
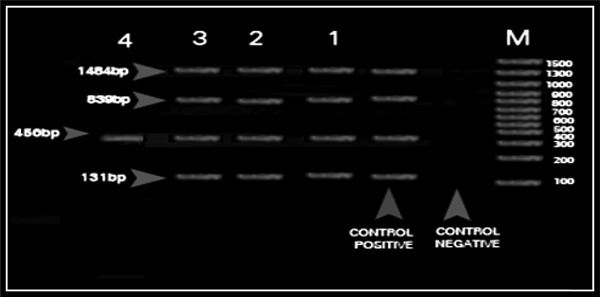
Multiplex PCR of four virulence associated genes for standard Listeria monocytogenes (ATCC 7494) and isolated Listeria species. M: DNA marker (100–1500 bp); control negative: E. coli strain ATCC 25922; control positive: amplified products of four virulence associated genes of standard L. monocytogenes (ATCC 7494) – plcB gene (1484 bp); actA gene (839 bp); hlyA gene (456 bp); iap gene (131 bp); Lane 1: amplified products of four virulence associated genes of the isolated L. monocytogenes; Lane 2: amplified products of one virulence associated gene hlyA gene (456 bp) of the isolated Listeria seeligeri.
The study was performed at the Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, Egypt in accordance with the recommendations in the updated Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NRC, 2010).
Results
A total of 209 milk samples collected from goats (107) and sheep (102) were screened for Listeria species (Table 4). The microbiological analysis revealed an overall prevalence of 10 (goat, 6/107; sheep, 4/102) putative Listeria spp isolates (10/209; 4.8%). Out of these, three isolates (goats, 2/6; sheep 1/4) were identified as L. monocytogenes (3/10; 30.0%). The remaining seven were considered Listeria species. In the 107 goat udder milk samples, two were positive for L. monocytogenes (1.9%); while in the 102 ewe udder milk samples, only one sample was positive for L. monocytogenes (1.0%). The overall prevalence of Listeria species were found to be: Listeria innocua (2/209; 1.0%), L. welshimeri (1/209; 0.5%), L. seeligari (2/209; 1,0%), and Listeria grayi (2/209; 1.0%). The three L. monocytogenes isolated from 209 milk samples (3/209; 1.4%) were serologically typed as either identified L. monocytogenes serotype Type 1 (1/1) isolated from the goat sample or Type 4 isolated from samples obtained from both goat (1/2) and sheep (1/1) milk samples (Table 5). The L. monocytogenes isolate from all three samples (goat, 2/6; sheep, 1/4) possessed all the four virulence associated genes while L. seeligeri carried the hlyA gene only.
Table 4. Prevalence of Listeria species recovered from udder raw fresh whole milk of goat and sheep.
| Animal species | Health condition of the animals | Number of examined samples | Isolated Listeria species | |||||||||||
| L. monocytogenes | L. innocua | L. welshimeri | L. seeligeri | L. grayi | Total | |||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | |||
| Goat | Apparent Healthy animal | 33 | 0 | 0 | 0 | 0 | 1 | 3.0 | 0 | 0 | 0 | 0 | 1 | 3.0 |
| Subclinical Mastitic animal | 35 | 1 | 2.9 | 1 | 2.9 | 0 | 0 | 1 | 2.9 | 0 | 0 | 3 | 8.6 | |
| Clinical Mastitic animal | 39 | 1 | 2.6 | 0 | 0 | 0 | 0 | 1 | 2.6 | 0 | 0 | 2 | 5.1 | |
| Total | 107 | 2 | 1.9 | 1 | 0.9 | 1 | 0.9 | 2 | 1.9 | 0 | 0 | 6 | 5.6 | |
| Sheep | Apparent Healthy animal | 38 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2.6 | 1 | 2.6 |
| Subclinical Mastitic animal | 41 | 1 | 2.4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2.4 | 2 | 4.9 | |
| Clinical Mastitic animal | 23 | 0 | 0 | 1 | 4.4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4.4 | |
| Total | 102 | 1 | 1.0 | 1 | 1.0 | 0 | 0 | 0 | 0 | 2 | 2.0 | 4 | 3.9 | |
| Total | 209 | 3 | 1.4 | 2 | 1.0 | 1 | 0.5 | 2 | 1.0 | 2 | 1.0 | 10 | 4.8 | |
Table 5. Serotyping of Listeria monocytogenes recovered from udder raw fresh whole milk of goat and sheep.
| Animal species | Number of L. monocytogenes isolates | L. monocytogenes serotypes | |||||
| Type 1 | Type 4 | Untyped | |||||
| n | % | n | % | n | % | ||
| Goat milk | 2 | 1 | 50 | 1 | 50 | 0 | 0 |
| Sheep milk | 1 | 0 | 0 | 1 | 100 | 0 | 0 |
| Total | 3 | 1 | 33.3 | 2 | 66.7 | 0 | 0 |
The milk samples did not appear visibly abnormal throughout the course of the study. In total, 107 goat milk samples were collected; 33 were normal, 35 were subclinical mastitic, and 33 were clinically mastitic. The 102 milk samples collected from sheep were comprised of 23 clinically mastitic, 41 subclinically mastitic, and 38 normal milk samples.
All tested isolates were small, Gram-positive rods, negative for indole, oxidase, urease, and H2S production from organic sulfur compounds and test-positive for methyl red and Voges–Proskauer, had the ability to grow at 35°C, were catalase-positive and motile in wet mounts indicative for all Listeria species. They utilized dextrose, esculin, maltose, and some species utilized rhamnose and xylose with production of acid. The only isolate that utilized mannitol with acid production was L. grayi, while L. monocytogenes and L. seeligeri produced hemolysis in sheep blood stabs and consequently were CAMP test-positive (Table 1). Of the two, only L. monocytogenes failed to utilize xylose and was positive for rhamnose utilization. The researchers observed that the non-hemolytic species L. innocua and L. grayi provided the same xylose reactions as L. monocytogenes but were negative in the CAMP test. Listeria innocua gave a variable result for the utilization of rhamnose, an observation that was also recorded in the present investigation with L. welshimeri and L. grayi (Table 1).
The pathogenicity testing of the 10 Listeria isolates via the PI-PLC, DLABN, and DAPN assays, as well as in vivo tests, namely chick embryo, mice inoculation tests, Anton’s eye test, and Vero ingestion assay, indicated that three hemolytic L. monocytogenes isolates were determined to be pathogenic (Table 2). Splenic samples from deceased mice of the L. monocytogenes challenge contained viable L. monocytogenes that were recovered on BHI agar and confirmed by PCR; conversely, splenic samples from surviving mice had no viable L. monocytogenes detectable on BHI agar. The three hemolytic isolates of L. monocytogenes showed the characteristic enhancement of hemolytic zone with Staphylococcus aureus (Tables 1 and 2). All other Listeria spp. isolates were non-pathogenic (Table 3).
Table 2. Pathogenicity profiles of Listeria monocytogenes recovered from udder raw fresh whole milk of goat and sheep.
| Source of milk samples | Serotype | Pathogenicity profile | PCR profile of virulence associated genes | ||||||||||||
| CAMP (±) | PI-PLC | DLABN | DAPN | Anton's eye test | Mice lethality | Chick-embryo lethality | Vero cell ingestion assay | Biofilm | hlyA | plcB | actA | iap | |||
| Christensen's tube | Microtiter plate assay | + | + | + | + | ||||||||||
| Reference strain (ATCC 7494) | 4 | ± | + | − | − | + | + | + | ++ | Strong | Strong (OD = 0.158) | + | + | + | + |
| Goat | 1 | +/+ | + | − | − | + | + | + | ++ | Moderate | Very strong (OD = 0.201) | + | + | + | + |
| 4 | ± | + | − | − | + | + | + | ++ | Strong | Strong (OD = 0.104) | + | + | + | + | |
| Ewe | 4 | ± | + | − | − | + | + | + | ++ | Strong | Strong (OD = 0.117) | + | + | + | + |
CAMP: Christie, Atkins, Munch-Petersen test; S/R: Staphylococcus aureus/Rhodococcus equi; PI-PLC: phosphatidylinositol-specific phospholipase C; DLABN: DL-alanine- beta-naphthylamide HCl; DAPN: d-alanine-p-nitroanilide; OD: optical density; OD595 < 0.1 = weak; OD595 ≤ 0.1 = strong; OD595 > 1 = very strong.
Table 3. Pathogenicity profiles of Listeria species recovered from udder raw fresh whole milk of goat and sheep.
| Source of milk samples | Listeria species | Pathogenicity profile | PCR profile of virulence associated genes | ||||||||||||
| CAMP (±) with S/R | PI-PLC | DLABN | DAPN | Anton's eye test | Mice lethality | Chick-embryo lethality | Vero cell ingestion assay | Biofilm | |||||||
| Christensen's tube | Microtiter plate assay | hlyA | plc</emph>B | actA | iap | ||||||||||
| L. monocytogenes (ATCC 7494) | ± | + | − | − | + | + | + | + | Strong | Strong (OD = 0.158) | + | + | + | + | |
| Goat | L. innocua | − | − | + | + | − | − | − | − | Weak | Strong (OD = 0.159) | _ | _ | _ | _ |
| L. seeligeri | − | − | + | + | − | − | − | − | Moderate | Strong (OD = 0.221) | + | _ | _ | _ | |
| L. welshimeri | − | − | + | + | − | − | − | − | Moderate | Very strong OD = 0.112) | _ | _ | _ | _ | |
| Sheep | L. innocua | − | − | + | + | − | − | − | − | Moderate | Strong (OD = 0.107) | _ | _ | _ | _ |
| L. grayi | − | − | + | + | − | − | − | − | Weak | Strong (OD = 0.127) | _ | _ | _ | _ | |
| L. grayi | − | − | + | + | − | − | − | − | Moderate | Strong (OD = 0.126) | _ | _ | _ | _ | |
CAMP: Christie, Atkins, Munch-Petersen test; S/R: Staphylococcus aureus/Rhodococcus equi; PI-PLC: phosphatidylinositol-specific phospholipase C; DLABN: DL-alanine-beta-naphthylamide HCl; DAPN: d-alanine-p-nitroanilide; OD: optical density; OD < 0.1 = weak; OD 0.1 = strong; OD > 0.1 = very strong.
The results of adherence assay to test glass tube assessed by 0.1% Crystal Violet stain showed that the three (1/2 goat and 1/1 sheep) L. monocytogenes isolates were able to strongly form a biofilm on a glass surface, while one of the two isolates from the goat was moderately adherent (Table 2). L. innocua, L. welshimeri, L. seeligari, and L. grayi varied in their adherence and biofilm formation (Table 3). The three L. monocytogenes strains were screened for their adherence to polystyrene 96-well microtiter plates at different degrees; the results conclude L. monocytogenes was able to form a strong biofilm on polystyrene (OD570 > 1) (Table 2). The same strong affinity was also observed with the seven (goats, 4/6; sheep, 3/4) Listeria species, L. innocua (goat 1/6; sheep, 1/4), L. welshimeri (goat, 1/6), L. seeligari (goat, 2/6), and L. grayi (sheep, 2/4) (Table 3).
Discussion
From the results, Listeria was distinguished from Brochothrix, which is of concern as a food spoilage organism, by its inability to grow at 35°C and its lack of motility.17 From the five species isolated, the only isolate that utilized mannitol with acid production was L. grayi.17 It was also confirmed that L. monocytogenes and L. seeligeri produced hemolysis in sheep blood stabs, as also determined by Hitchins and Jinneman (2013).17 Prior research has determined that L. innocua may produce negative results for the utilization of rhamnose;17 this observation was also recorded in the present investigation with L. welshimeri and L. grayi (Table 1). After confirming negative results for the utilization of rhamnose, serotyping of Listeria isolates was differentiated. Listeriolysin O (LLO) is the main virulence factor and exists in all of the pathogenic L. monocytogenes coded by hlyA gene; according to a study by Choi and Hong,51 the hlyA gene of L. monocytogenes would be able to determine two cells in milk through a PCR assay (Tables 1–3). Although L. ivanovii is predominantly an animal pathogen affecting ruminants and sheep,20,25 it was not isolated in this study.
Listeria monocytogenes isolated from sheep milk in animals suffering from subclinical mastitis was characterized by a persistent shedding of Listeria and an apparently normal milk consistency and appearance.26–29 Listeria spp. was never found in sheep subclinical mastitis without obvious inflammation. L. seeligeri was isolated from 1.67% of healthy-appearing goat fecal samples.29–31
Animals may become infected through the ingestion of Listeria-contaminated food, poor-quality silage, pathogen inhalation, direct contact to the source, drinking water, feed components, soil in which fodder plants were grown, silage feeding, sawdust bedding, and farm yard manure;32–38,39 therefore, distributing untreated manure for agricultural purposes is regarded as a primary risk factor for the transmission of Listeria, thereby contributing to the spread of foodborne disease.37
Literature confirming the virulence of associated Listeria genes isolated from sheep and goats currently does not exist. With the exception of L. seeligeri being hemolytic, though non-pathogenic, the only pathogenic strain of L. monocytogenes is hemolytic. Variations in the degree of hemolysis among the bacteria have also been reported by Rawool et al. (2007).12 It has also been demonstrated that the L. monocytogenes phospholipases are essential determinants of pathogenicity.38,39 These studies provided evidence for the virulence factor activity of PI-PLC, expressed only by pathogenic spp. of Listeria i.e., L. monocytogenes.40,41 This expression remains a reliable marker for the discrimination between pathogenic and non-pathogenic Listeria species.40,41 The positivity of the L. monocytogenes in PI-PLC assay may be explained by the common regulation of the hlyA, actA, and plcB genes.42
Genetic tests that have been used for the detection and/or identification of Listeria species have exploited the molecular differences of the 16S and 23S rRNA genes.43 Polymerase chain reaction assays have also been developed to target specific virulence genes such as the hlyA gene (part of LIPI-1), which are present in specific Listeria species alongside the virulence gene, iap, that is common to all Listeria species.44,45 Listeria monocytogenes is pathogenic at the species level, but various strains display varied virulence and pathogenic potential. Some strains may be virulent and cause disease while other strains may remain non-pathogenic and produce no apparent malaise.44,46,47,48 The difference between these virulent and non-virulent strains is likely minimal, as they possess the same virulence gene cluster LIPI-1 (prfA, hlyA, plcA, mpI, actA, and plcB) and major virulence proteins involved in L. monocytogenes pathogenesis, but may differ in the expression of unique genes.44 LIPI-1 includes the genes prfA (encodes positive regulatory factor A), plcA (PI-PLC), hlyA (hemolysin LLO), mpl (zinc metalloprotease), actA (surface actin polymerization protein), plcB (phosphatidylcholine phospholipase C (PC-PLC)), and the iap (invasion associated protein) encodes the protein p60. The latter gene can be used to distinguish between the different Listeria species as it is only present in L. ivanovii, L. seeligeri, and L. monocytogenes.49 The gene plcA, which encodes PI-PLC, however, is only present in L. ivanovii and L. monocytogenes. Phosphatidylinositol-specific phospholipase C and LLO acts in synergy to destroy the phagosome in which the bacterium is trapped during infection. This is one of four major steps involved in the cellular mechanism of Listeria pathogenesis. The absence of the plcA gene in L. seeligeri is, therefore, a contributing factor to its non-pathogenic status.49 The hlyA gene, is responsible for the hemolysis of blood cells48 and is only found in virulent strains of L. monocytogenes. The PCR employed in the present study was specific for individual detection of the four virulence associated genes plcB, actA, hlyA, and iap found in L. monocytogenes while L. seeligeri had an intact hlyA gene only. These findings commensurate with the former scientific research for the detection of hlyA gene,22 plcB gene,14 iap gene, and actA gene.24 Although Gouin et al. (1994) and Bhunia (2008) assumed that the plcB could be used to distinguish between the different Listeria species in L. ivanovii, L. seeligeri, and L. monocytogenes, it was not detected in the two isolated strains of L. seeligeri in the present investigation.50 As LLO is a secreted protein, the detection of it in a food sample can also be an indicator of the presence of L. monocytogenes cells.20
Conclusion
This study concludes a low, though viable, prevalence of L. monocytogenes in both sheep and goat milk. Human exposure can occur through the consumption of raw, unpasteurized milk. Pregnant women should undergo health and safety precautions in milking goats and sheep, especially those animals that have recently given birth; pregnant women should also avoid coming into contact with the afterbirth or with newborn lambs. Immunosuppressed individuals are advised to undertake similar precautions. It is important for human populations to maintain strict hygiene and practice pasteurization of raw milk in order to minimize the risks of human infection with L. monocytogenes.
Conflict of Interest
The authors declare no personal or financial conflicts of interests in the development, writing, and submission of this study.
Funding
This research was funded and supported by Cairo University and the researchers.
References
- 1. Herodotus 440 B.C. The History of Herodotus By Herodotus Written 440 B.C.E Translated by George Rawlinson Book II Provided by The Internet Classics Archive. Available http://classics.mit.edu//Herodotus/history.html. [Google Scholar]
- 2.Haenlein GFW. Goat milk in human nutrition. Small Ruminant Res. 2004;51:155–63. [Google Scholar]
- 3.Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–94. doi: 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- 4.Rebagliati V, Philippi R, Rossi M, Troncoso A. Prevention of foodborne listeriosis. Indian J Pathol Microbiol. 2009;52:145–9. doi: 10.4103/0377-4929.48903. [DOI] [PubMed] [Google Scholar]
- 5.Kasalica A, Vuković V, Vranješ A, Memiši N. Listeria monocytogenes in milk and dairy products. Biotechnol Anim Husb. 2011;27:1067–82. [Google Scholar]
- 6.Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fthenakis GC, Saratsis Ph, Tzora A, Linde K. Naturally occurring subclinical ovine mastitis associated with Listeria monocytogenes. Small Ruminant Res. 1998;31:23–7. [Google Scholar]
- 8.Gronstol H. Listeriosis in sheep. Listeria monocytogenes excretion and immunological state in healthy sheep. Acta Vet Scand. 1979;20:168–79. doi: 10.1186/BF03546609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp MW. Bovine mastitis and Listeria monocytogenes. Vet Rec. 1989;125:512–3. doi: 10.1136/vr.125.20.512-b. [DOI] [PubMed] [Google Scholar]
- 10.Conly JM, Johnston BL. Listeria: a persistent food-borne pathogen. Can J Infect Dis Med Microbiol. 2008;9:327–8. doi: 10.1155/2008/702565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MMWR. Outbreak of Listeria monocytogenes infections associated with pasteurized milk from a local dairy – Massachusetts, 2007. MMWR Morb Mortal Wkly Rep. 2008;57:1097–100. [PubMed] [Google Scholar]
- 12.Rawool DB, Malik SVS, Shakuntala I, Sahare AM, Barbuddhe SB. Detection of multiple virulence-associated genes in Listeria monocytogenes isolated from bovine mastitis cases. Int J Food Microbiol. 2007;113:201–7. doi: 10.1016/j.ijfoodmicro.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 13.Notermans SHW, Dufrenne J, Leimeister-Wachter M, Domann E, Chakraborty T. Phosphatidylinositol-specific phospholipase C activity as a marker to distinguish between pathogenic and non-pathogenic Listeria species. Appl. Environ. Microbiol. 1991;57:2666–70. doi: 10.1128/aem.57.9.2666-2670.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark AG, McLauchlin J. Simple color tests based on an alanyl peptidase reaction which differentiate Listeria monocytogenes from other Listeria species. J Clin Microbiol. 1997;35:2155–6. doi: 10.1128/jcm.35.8.2155-2156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raja RK, Ramesh N, Maripandi A. Invasion and interaction studies of Salmonella Typhimurium sub sp enteritis in vero and MDCK cell lines. Adv Biol Res. 2010;4:86–91. [Google Scholar]
- 16.Hitchins AS, Jinneman K. 2013. Bacteriological analytical manual, chapter 10 detection and enumeration of Listeria monocytogenes in foods. Silver Spring, MD, USA: U.S. Food and Drug Administration, Page Last Updated; 2013 February 14. [Google Scholar]
- 17.Giacaman RA, Araneda E, Padilla C. Association between biofilm forming isolates of mutans Streptococci and caries experience in adults. Arch Oral Biol. 2010;55:550–4. doi: 10.1016/j.archoralbio.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Borucki MK, Peppin JD, White D, Loge F, Call DR. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol. 2003;69:7336–42. doi: 10.1128/AEM.69.12.7336-7342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, et al. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wesley IV, Harmon KM, Dickson JS, Schwartz AR. Application of a multiplex polymerase chain reaction assay for the simultaneous confirmation of Listeria monocytogenes and other Listeria species in turkey sample surveillance. J Food Prot. 2002;65:780–5. doi: 10.4315/0362-028x-65.5.780. [DOI] [PubMed] [Google Scholar]
- 21.Paziak-Domanska B, Bogulawska E, Wiekowska-Szakiel M, Kotlowski R, Rozalska B, Chmiela M, et al. Evaluation of the API test, phosphatidylinositol-specific phospholipase C activity and PCR method in identification of Listeria monocytogenes in meat foods. FEMS Microbiol Lett. 1999;171:209–14. doi: 10.1111/j.1574-6968.1999.tb13434.x. [DOI] [PubMed] [Google Scholar]
- 22.Ramaswamy V, Cresence VM, Rejitha JS, Lekshmi MU, Dharsana KS, Prasad SP, et al. Listeria: review of epidemiology and pathogenesis. J Microbiol Immunol Infect. 2007;40:4–13. [PubMed] [Google Scholar]
- 23.Barza M. Listeriosis and milk. N Engl J Med. 1985;312:438–40. doi: 10.1056/NEJM198502143120710. [DOI] [PubMed] [Google Scholar]
- 24.Schoder D, Winter P, Kareem A, Baumgartner W, Wagner M. A case of sporadic ovine mastitis caused by Listeria monocytogenes and its effect on contamination of raw milk and raw-milk cheeses produced in the on-farm dairy. J Dairy Res. 2003;70:395–401. doi: 10.1017/s0022029903006277. [DOI] [PubMed] [Google Scholar]
- 25.Winter P, Schilcher F, Bagò Z, Schoder D, Egerbacher M, Baumgartner W, et al. Clinical and histopathological aspects of naturally occurring mastitis caused by Listeria monocytogenes in cattle and ewes. J Vet Med B Infect Dis Vet Public Health. 2004;51:176–9. doi: 10.1111/j.1439-0450.2004.00751.x. [DOI] [PubMed] [Google Scholar]
- 26.Pintado CM, Grant KA, Halford-Maw R, Hampton MD, Ferreira MA, McLauchlin J. Association between a case study of asymptomatic ovine listerial mastitis and the contamination of soft cheese and cheese processing environment with Listeria monocytogenes in Portugal. Foodborne Pathog Dis. 2009;6:569–75. doi: 10.1089/fpd.2008.0246. [DOI] [PubMed] [Google Scholar]
- 27.Elezebeth G, Malik SVS, Chaudhari SP, Barbuddhe SB. The occurrence of Listeria species and antibodies against listeriolysin-O in naturally infected goats. Small Ruminant Res. 2007;67:173–8. [Google Scholar]
- 28.Fotou K, Tzora A, Voidarou Ch, Alexopoulos A, Plessas S, Avgeris I, et al. Isolation of microbial pathogens of subclinical mastitis from raw sheep's milk of Epirus (Greece) and their role in its hygiene. Anaerobe. 2011;17:315–9. doi: 10.1016/j.anaerobe.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 29.De Luca G, Zanetti F, Fateh-Moghadm P, Stampi S. Occurrence of Listeria monocytogenes in sewage sludge. Zentralbl Hyg Umweltmed. 1998;201:269–77. [PubMed] [Google Scholar]
- 30.Yoshida T, Kato Y, Sato M, Hirai K. Sources and routes of contamination of raw milk with Listeria monocytogenes and its control. J Vet Med Sci. 1998;60:1165–8. doi: 10.1292/jvms.60.1165. [DOI] [PubMed] [Google Scholar]
- 31.Ryser ET, Marth EH.(editors)Listeria, listeriosis, and food safety. CRC Press2007 [Google Scholar]
- 32.Zundel E, Bernard S. Listeria monocytogenes translocates throughout the digestive tract in asymptomatic sheep. J Med Microbiol. 2006;55:1717–23. doi: 10.1099/jmm.0.46709-0. [DOI] [PubMed] [Google Scholar]
- 33.Brugère-Picoux J. Ovine listeriosis. Small Ruminant Res. 2008;76:12–20. [Google Scholar]
- 34.Hellstrom S. Contamination routes and control of Listeria monocytogenes in food production. Finland: Department of Food Hygiene and Environmental Health, Faculty of Veterinary Medicine, University of Helsinki; 2011. [Helsinki University Print] [Google Scholar]
- 35.Latorre AA, Van Kessel JAS, Karns JS, Zurakowski MJ, Pradhan AK, Boor KJ, et al. Increased In vitro adherence and on-farm persistence of predominant and persistent Listeria monocytogenes strains in the milking system. Appl Environ Microbiol. 2011;77:3676–84. doi: 10.1128/AEM.02441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behravesh CB, Williams IT, Tauxe RV. Emerging foodborne pathogens and problems: expanding prevention efforts before slaughter or harvest. In: Institute of Medicine (US). Improving food safety through a one health approach: workshop summary. Washington, DC: National Academies Press; 2012. p. A14. Available from: http://www.ncbi.nlm.nih.gov/books/NBK114501/ [PubMed] [Google Scholar]
- 37.Oliver SP, Jayarao BM, Almeida RA. 2005. Foodborne pathogens, mastitis, milk quality, and dairy food safety. NMC Annual Meeting Proceedings, Orlando, Florida, USA. [DOI] [PubMed] [Google Scholar]
- 38.Marquis H, Doshi V, Portnoy DA. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect Immun. 1995;63:4531–4. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith GA, Marquis H, Jones S, Johnston NC, Portnoy DA, Goldfine H. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect Immun. 1995;63:4231–7. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu D. Listeria monocytogenes: comparative interpretation of mouse virulence assay. FEMS Microbiol Lett. 2004;233:159–64. doi: 10.1016/j.femsle.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Graham T, Golsteyn-Thomas EJ, Gannon VP, Thomas JE. Genus- and species-specific detection of Listeria monocytogenes using polymerase chain reaction assays targeting the 16S/23S intergenic spacer region of the rRNA operon. Can J Microbiol. 1996;42:1155–62. doi: 10.1139/m96-147. [DOI] [PubMed] [Google Scholar]
- 42.Graham TA, Golsteyn-Thomas EJ, Thomas JE, Gannon VP. Inter- and intraspecies comparison of the 16S–23S rRNA operon intergenic spacer regions of six Listeria spp. Int J Syst Bacteriol. 1997;47:863–9. doi: 10.1099/00207713-47-3-863. [DOI] [PubMed] [Google Scholar]
- 43.Sallen BA, Rajoharison S, Desverenne S, Quinn F, Mabilat C. Comparative analysis of 16S and 23S rRNA sequences of Listeria species. Int J Syst Bacteriol. 1996;46:669–74. doi: 10.1099/00207713-46-3-669. [DOI] [PubMed] [Google Scholar]
- 44.Blais BW, Turner G, Sooknanan R, Malek LT. A nucleic acid sequence based amplification system for detection of Listeria monocytogenes hlyA sequences. Appl Environ Microbiol. 1997;63:310–3. doi: 10.1128/aem.63.1.310-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bubert A, Hein I, Raunch M, Lehner A, Yoon B, Goebel W, et al. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl Environ Microbiol. 1999;65:4688–92. doi: 10.1128/aem.65.10.4688-4692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erdenlig S, Ainsworth AJ, Austin FW. Pathogenicity and production of virulence factors by Listeria monocytogenes isolates from channel catfish. J Food Prot. 2000;63:613–9. doi: 10.4315/0362-028x-63.5.613. [DOI] [PubMed] [Google Scholar]
- 47.Gracieux P, Roche SM, Pardon P, Velge P. Hypovirulent Listeria monocytogenes strains are less frequently recovered than virulent strains on PALCAM and Rapid' L. mono media. Int J Food Microbiol. 2003;83:133–45. doi: 10.1016/s0168-1605(02)00321-5. [DOI] [PubMed] [Google Scholar]
- 48.Doumith M, Cazalet C, Simoes N, Frangeul L, Jacquet C, Kunst F, et al. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect Immun. 2004;72:1072–83. doi: 10.1128/IAI.72.2.1072-1083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kingdon GC, Sword P. Effect of Listeria monocytogenes hemolysin on phagocytic cells and lysosomes. Infect Immun. 1970;1:356–62. doi: 10.1128/iai.1.4.356-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhunia AK. Listeria monocytogenes. In: Foodborne microbial pathogens. New York: Springer Science+Business Media; 2008. p. 165–82. [Google Scholar]
- 51.Choi WS, Hong C-H. Rapid enumeration of Listeria monocytogenes in milk using competitive PCR. Int J Food Microbiol. 2003;84:79–85. doi: 10.1016/s0168-1605(02)00401-4. [DOI] [PubMed] [Google Scholar]


