Abstract
Objective
We hypothesized that differential mRNA transcription between the sexes may be linked to the 9:1 female-to-male gender-related relative risk for the development of Sjögren's syndrome (SS), an autoimmune disease that leads to inflammation and dysfunction in the lachrymal and salivary glands.
Subjects and Methods
RNA from minor salivary glands was collected from nine healthy volunteers (four men and five women) and analyzed using the Agilent 4 × 44K human microarray platform. Differential expression was confirmed by qRT-PCR.
Results
Comparison of the transcriptome of minor salivary glands from normal male and female volunteers with that of salivary glands and secretory epithelia identified a number of gender, species, and tissue-specific gene expression patterns. These differences include, but are not limited to, a diverse set of genes involved in immune modulation, chemotactic control, inhibition of complement, metabolism, and neurogenesis.
Conclusion
Analysis of these changes provides insight into the protective and predisposing molecular factors that may be involved in the development of Sjögren's syndrome. Some of the gene changes observed in this study correlate with previously observed sexual dimorphisms in salivary gland function and also illustrate several new targets for further investigation.
Keywords: Genetics, microarray, salivary gland
Introduction
Sjögren's syndrome is a chronic autoimmune disease of unknown etiology. Although numerous organs can be involved, the focus of the disease is secretory epithelia such as the lacrimal and salivary glands, resulting in dry eyes and mouth. The estimated prevalence of SS is 0.5% or approximately 1.5 million Americans (Venables, 2004). It is strikingly more common in women than men, with a female-to-male ratio of about 9:1. This strong female predilection is very similar to some other autoimmune diseases, such as systemic lupus erythematosus and autoimmune thyroiditis. The reason for this discrepancy is not yet known, but it has been proposed that sex hormones or genes encoded on the sex chromosomes may predispose females to these diseases (Fish, 2008). Most of the studies looking at the mechanistic aspects of gender differences have focused on the effect of these factors on the immune system and inflammation. Little attention has been paid to gender-related differences in the target organs. We hypothesized that salivary glands from healthy females and males will have different gene expression profiles and that this sexual dimorphism may predispose females to chronic inflammation and / or exocrine dysfunction in response to hitherto unknown proinflammatory events.
Several studies have described sexual dimorphism in gene expression in the lung (Gruber et al, 2006), retina (Chowers et al, 2003), brain (Lu et al, 2004), and muscle (Roth et al, 2002) in humans as well as in the lacrimal and salivary glands in mice (Triester et al, 2005a,b; Richards et al, 2006). These changes in expression reflect a wide range of biological processes, molecular functions, and cellular components, including such activities as development, growth, transcription, metabolism, signal transduction, transport, receptor activity, and protein and nucleic acid binding.
Comparison of RNA isolated from different salivary glands in mice suggested that the nature of these sex-related variations in gene expression was unique to each tissue. For example, transforming growth factor β2 (TGFβ2) mRNA levels were increased in male submandibular and sublingual glands, but not in the parotid gland, whereas expression of interleukin 1 receptor type II was upregulated only in the male parotid gland (Triester et al, 2005a,b). Additionally, the gene for IL-3 was only upregulated in the female sublingual gland. These differences demonstrate not only the structural and functional differences among the major salivary glands but may also reflect different responsiveness to sex steroid hormones and other sex-related factors. Although 17β-estradiol and progesterone contribute little to the known sex-related differences in gene expression of the lacrimal gland (Suzuki et al, 2006), androgens are involved in more than 70% of these variations (Suzuki et al, 2006). This suggests that rather than females being more prone to developing autoimmune conditions, males may be more resistant.
A recent study showed significant differences in gene expression profiles between men and women in human parotid tissue obtained from the histologically normal part of benign parotid tumors (Srivastava et al, 2008). The same study also showed a marked difference in gene expressions between younger and older women. In this study, we have examined the transcriptome of minor salivary glands from normal male and female volunteers of similar ages to identify gender-related differences in gene expression profiles that may contribute to the higher risk of Sjögren's syndrome in women.
Materials and methods
Subjects
Human minor salivary glands were obtained from healthy volunteers enrolled in a study at the National Institutes of Health (NIH) Sjögren's syndrome clinic. The study was approved by the Institutional Review Board of the National Institute of Dental and Craniofacial Research at the NIH (NCT00001390) and is registered at clinicaltrials.gov. All subjects signed a written consent document prior to enrollment. The subjects presented with no signs of salivary dysfunction as measured by salivary flow. Minor salivary gland biopsies were negative for any histological abnormalities as scored by a clinically certified pathologist. Samples from nine subjects (four men and five women) were included in this study. (See Table 1 for ages of each).
Table 1. Patient age and gender.
| Patient number | Sex | Age |
|---|---|---|
| SG86 | Female | 53 |
| SG54 | Female | 49 |
| SG88 | Female | 34 |
| SG85 | Female | 42 |
| SG81 | Female | 54 |
| SG65 | Male | 49 |
| SG61 | Male | 62 |
| SG89 | Male | 44 |
| SG87 | Male | 53 |
RNA isolation and quality control
The work flow process is outlined in supplemental Figure 1. Total RNA from donor samples was isolated using the miRNeasy kit as per the manufacturer's standard protocols (Qiagen, Valencia, CA, USA). Following total RNA isolation, sample concentration was assayed using a Nanodrop-8000 (Thermo Scientific, Wilmington, DE, USA). RNA integrity was measured using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA), and samples containing low-quality RNA species were rejected from the study.
Microarray analysis
Total RNA samples (200 ng) were prepared and processed for microarray analysis using the Low RNA Input Linear Amplification Kit (Agilent) as per the manufacturer's standard protocol. Following labeled cRNA synthesis, samples were purified using an RNeasy kit (Qiagen) and quantified for dye integration and adequate amplification using a Nanodrop-8000 (Thermo Scientific). Following quantification, samples were hybridized overnight in a rotating hybridization oven and washed/scanned as per the standard Agilent Low RNA Input Linear Amplification Kit protocol.
Microarray data analysis
Microarray datasets were imported into the Genespring GX 9 (Agilent) software suite for analysis. Following 75th percentile shift normalization, low intensity probes subject to high degrees of noise were removed by filtering the lowest 20% of each sample. Sexually dimorphic genes were then identified by grouping the datasets based upon gender and filtering for genes at least twofold differentially transcribed between the sexes. Differential gene expression was tested by Student's t-statistic. Statistical significance of differential expression was assessed by calculating two-tailed P-values from the t-distribution (degrees of freedom = number of samples − 1). Only genes that passed the above criteria with a P-value < 0.05 were included in the high-confidence cutoff list and used for real-time validation and comparison to lung tissue. To minimize the false discovery rate and relax parametric assumptions about the distribution of gene expression, exact P-values were calculated for each gene using permutations of genders. The result of this second analysis was termed the moderate confidence list and used for gene ontology and Ingenuity pathway analysis.
Real-time PCR
To validate microarray results, a subset of genes identified as sexually dimorphic by microarray analysis were analyzed for gender-related differential expression by quantitative real-time PCR (qRT-PCR). Total RNA for qRT-PCR analysis was isolated from male (n = 5) and female (n = 5) healthy volunteers meeting the same criteria as required for inclusion in the microarray study. One microgram of the isolated total RNA was then reverse transcribed into cDNA for qRT-PCR analysis using the Superscript III Reverse Transcriptase system (Invitrogen, Carlsbad, CA, USA) according to manufacturer's standard protocols. Quantification of genes indicated to be differentially transcribed was performed using validated Taqman probes and the Taqman Gene Expression Master Mix (Applied Biosystems, Carlsbad, CA, USA).
Ingenuity pathway analysis
The list of differentially expressed genes was entered into Ingenuity pathway analysis (IPA) and filtered for curated interactions with estrogen receptors (ER; both alpha-and beta-subunits) using the Grow function of Pathway Assist. The pathway was expanded (using the same Grow function) to show relationships to the androgen receptor (AR), as well as interactions (using Path Explorer) between ER or AR and the molecule TGFβ2. Interactions reported in the literature as well as protein–protein interactions were reported in the final pathway map. The differential expression (relative to females) of genes associated with the analysis was color-coded to show higher expression in red and lower expression in green.
Gender-related organ-specific changes
Differentially identified genes from the conservative cutoff were then compared with changes in other tissue in two ways. The first approach involved comparing the differentially expressed salivary gland genes to a list of differentially transcribed genes identified from the Gruber et al (2006) dataset of sexually dimorphic genes in the human lung (GEO GDS1673). All significant genes from the lung dataset were at least twofold differentially regulated between the sexes and had P-values < 0.05.
The second approach used the IPA organ system filter to compare the differentially expressed salivary gland genes to genes associated with other secretory tissue, such as pancreas and lung.
Results
To analyze sexually dimorphic transcription that may be associated with the increased risk of Sjögren's syndrome in women, we analyzed nine human minor salivary glands via Agilent 4 × 44K microarrays, quantitative reverse transcriptase PCR, and computational methods (Figure S1).
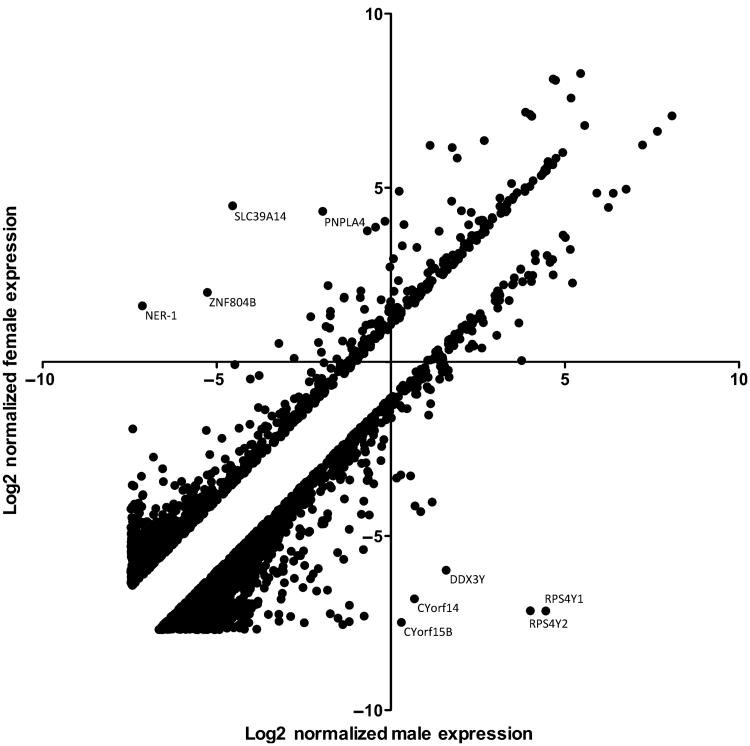
Genes that were differentially expressed between genders were separated based on statistical significance (P < 0.05, following 75th percentile shift normalization). The resultant gene signature efficiently clustered into two groups, separating males from females using unsupervised clustering techniques (Figure 1). From the 33717 probes detected, 360 genes were differentially transcribed and met the moderate cutoff criteria (Table S1). Consistent with earlier reports (Srivastava et al, 2008), the majority of changes associated with gender-specific differential transcription are minor in nature, with 68% of the 360 statistically significant changes being less than twofold between the sexes. A few examples of highly differentially expressed genes are indicated by arrows in Figure 2. Interestingly, only 13 genes are differentially expressed at greater than tenfold levels between the sexes, and all but one (XIST) of these 13 genes are upregulated in males over females (Table 2). The expression intensity of the mRNA transcripts detected as sexually dimorphic varied widely, with some transcripts detected at high intensity levels (e.g. RPS4Y1: mean expression level of 1.53-fold below GAPDH in males and mean expression of 5284-fold below GAPDH in females) and at lower levels (e.g. COL10A1: mean 92.1-fold expression below GAPDH in males and mean expression 209-fold below GAPDH in females).
Figure 1.
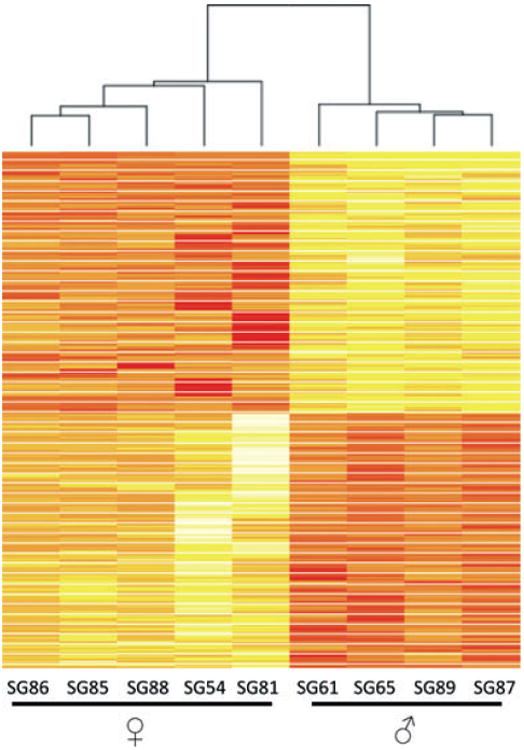
Heat map of differentially expressed genes meeting the moderate statistical threshold. Color intensity correlates with the degree of gene expression. Rows are normalized to set the mean at zero, with the standard deviation set at ±1. Rows colored in red are repressed while yellow represents upregulated transcripts
Figure 2.
Scatter plot analysis of 2637 genes that are at least twofold differentially expressed between male and female normal volunteers. Expression values were normalized to the 75th percentile of expression and transformed into log2space for visualization prior to scatter plot analysis
Table 2.
Genes meeting the conservative statistical threshold are primarily male biased. Genes with large fold changes are primarily male biased and present on the Y chromosome. Female-biased genes are present on a number of chromosomes and are maximally upregulated at lower levels than those detected in males
| Fold change | GeneName | Description | Chromosome |
|---|---|---|---|
| Up-regulated in Males | |||
| 3604.8 | RPS4Y1 | Ribosomal protein S4, Y-linked 1 | Y |
| 2543.6 | RPS4Y2 | Ribosomal protein S4, Y-linked 2 | Y |
| 109.5 | CYorfl4 | Chromosome Y open reading frame 14 | Y |
| 78.6 | CYorfl5A | Chromosome Y open reading frame 15A | Y |
| 66.0 | AL713714 | mRNA; cDNA DKFZp667C0715 (from clone DKFZp667C0715) | X |
| 44.6 | DDX3Y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | Y |
| 37.0 | JARID1D | Jumonji, AT rich interactive domain ID (JARID1D) | Y |
| 32.7 | TTTY15 | Testis-specific transcript, Y-linked 15 | Y |
| 23.1 | CYorfl5B | Lipopolysaccaride-Specific Response 5-Like Protein | Y |
| 20.9 | TTTY2 | Testis-specific transcript, Y-linked 2 | Y |
| 16.0 | USP9Y | Ubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) | Y |
| 13.9 | UTY | Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked, transcript variant 3 | Y |
| 9.4 | TTTY14 | Testis-specific transcript, Y-linked 14 on chromosome Y | Y |
| 9.2 | NLGN4Y | Neuroligin 4, Y-linked | Y |
| 8.1 | EIF1AY | Eukaryotic translation initiation factor 1A, Y-linked | Y |
| 7.7 | ZFY | Zinc finger protein, Y-linked | Y |
| 6.3 | UTY | Ubiquitously transcribed tetratricopeptide repeat gene, Y-linked, transcript variant 1 | Y |
| 5.2 | FLJ43390 | cDNA FU39142 fis, clone OCBBF1000049. | Y |
| 3.2 | AW976081 | EST3SS190 MAGE resequences, MAGN Homo sapiens cDNA | 7 |
| 3.2 | ZSCAN4 | Zinc finger and SCAN domain containing 4 | 19 |
| 3.0 | PRKY | Protein kinase, Y-linked | Y |
| 2.9 | ALOX12P2 | Arachidonate 12-lipoxygenase pseudogene 2 | 17 |
| 2.8 | BX094246 | Soares testis NHTcDNA clone | 4 |
| 2.5 | SV2B | Synaptic vesicle glycoprotein 2B | 15 |
| 2.3 | THC2674900 | Unknown | 6 |
| 2.3 | PTPRG | Protein tyrosine phosphatase, receptor type, G | 3 |
| 2.3 | CYP1A2 | Cytochrome P450, family 1, subfamily A, polypeptide 2 | 15 |
| 2.2 | THC2621536 | Unknown | 1 |
| 2.1 | KLHL34 | Kelch-like 34 (Drosophila) | X |
| 2.0 | COL10A1 | Collagen, type X, alpha KSchmid metaphyseal chondrodysplasia) | 6 |
| Up-regulated in Females | |||
| 17.7 | XIST | X (inactive)-specific transcript (XIST) | X |
| 3.2 | ARSD | Arylsulfatase D, transcript variant 2 | X |
| 2.7 | TSPAN13 | Tetraspanin 13 | 7 |
| 2.6 | CHST2 | Carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 | 3 |
| 2.1 | PEX7 | Peroxisomal biogenesis factor 7 | 6 |
| 2.1 | TXNDC12 | Thioredoxin domain containing 12 | 1 |
To determine potentially relevant biological pathways linked to possible disease predisposition or physiological differences, we performed gene ontology analysis (GO analysis) (Table S2) and IPA on the differentially transcribed genes (Table 3). IPA indicates that sexually dimorphic gene transcription is associated with a diverse set of cellular processes. Genes upregulated in female salivary glands were primarily associated with metabolic pathways but also contained transcripts whose roles are attributed to chromatin modification and electron transport functions. In contrast to the female glands, a diverse set of transcripts were upregulated in male salivary glands. These upregulated genes are involved in immune regulation, chemotaxis, neuronal development and metabolism. Consistent with previous reports in mice, TGFβ2 levels are 1.9-fold upregulated in male minor salivary glands (Triester et al, 2005a,b). TGFβ2 is expressed at a mean expression level 9.98-fold lower than GAPDH in males and mean 21.29-fold lower expression than GAPDH in females. Additionally, complement factor I (CFI), an inhibitor of complement, is upregulated roughly 2.5-fold in male minor salivary glands. Mean CFI expressioninmalesis12.02-fold lower than GAPDH and 33.84-fold lower than GAPDH in females.
Table 3.
Sexually dimorphic expression is detected within a diverse set of genes. Differentially transcribed genes between the genders that met the moderate statistical cutoff for significance were analyzed using Ingenuity pathway analysis bioinformatics database for shared ontological terminology. Sexually dimorphic gene expression was detected in a wide variety of molecular function, cellular location, and biological processes. The top five pathways are listed
| Genes in network | Function |
|---|---|
| ↑ACACA, AKR1B10, AKT1, ↑ALDH9Al, ARL4D, ↑C20ORF30, CASP3, ↓CFI, COL4A1, ↓COL7A1, ↓ COX4Il,FNl, ↑IFT20, INSIG1, LPIN1, NCSTN, ONECUT1, PPARG, PSENEN, PTPN12,↑RAPIA. RAPGEF5, ↓RBM5, RGS14, ↑RHEB, ↑RRAGD, SCD, ↓TGFB2, ↑TMBIM6, ↑TMED2, ↑TMEDIO, TNF, ↑TNFRSF19, TRAF6, UCP1 | Connective Tissue Development and Function, Organismal Functions, Lipid Metabolism |
| ABCB7, ↑ALG2, APP, ↑CDK2AP1, CHRM4, COX5B, ↓EFHC2, ↑ERP29, EXOC8, ↑FECH, ↑GNB1,HGS, HTT, IL6, ↑LAMP2, MME.MYC, ↑MYOSB, MY05C, ↑NDUFA9, NDUFS3, NDUFV2, RTN2, ↑RTN4, RTN4R, SEC61A1, ↓SLIT2, ↓SV2B, TPD52, ↑TPD52Ll, UQCRC1, UQCRC2. VAMP4, VTI1B, XBP1 | Neurological Disease, Organismal Injury and Abnormalities, Cellular Assembly and Organization |
| ABCF3, ACINI, ATF4, CDK5RAP3, CEBPA, CHMP1B, DISCI, ↓ERMP1, ↑FOXA1, G6PC, HNF4A. KLF5, MID1IP1, ↑MOCS2, ↑MRPS23, ↓NEK3, NR0B2, ONCUT1, PCK1, ↑PPC6C, PRLR, ↑PYCRl. RORA, RORC, ↓SFRS8,↑SNX3, SPP1, STAM, ↑STAMBP, TAGLN, TAT, ↑TMEM59, ↑TRMT12, UFC1, TTR | Carbohydrate Metabolism, Gene Expression, Amino Acid Metabolism |
| ↑BTG1, ↑C220RF28, COL4A1, ↑COMMDIO, DNM1L, DTX3, DTX3L, ↑FUCA1, ↓GLS, GSK3B, HEX A, ↑HEXB, ↑HMBS, ↑PARK7, ↑PRUNE, RELA, RNF10, ↑RNF14, RNF114, RNF128, RNF166, RNF185, SUMOl, SYT1, ↓TLK2, TNF, TP53, TPD52, TRIM2, TRIM32, UBA2, ↑UBE2E1, ↑UBE2H. UBOX5, YWHAZ | Decreased Levels of Albumin, Antimicrobial Response, Lipid Metabolism |
| ANXA2, ATPA1A, CTNNB1, ↑CTNNBIPl, ↑DNAJA2, E2F2, E2F5, EDN1, ↑EFS, EGFR, EZH2. FAS, ↑HADH, KIT, KLF5, NCOR1, NDFIP1, NDFIP2, ↑PCBP2, ↑PDCD2, PHB2, ↑PHB, PIK3R1. PSEN1, ↓PTPRG, RBL2, ↓SEMA5A, ↑SETD8, ↑SLCHA2, ↑SLC5A1, SMARCA2, SPP1, TCF7L2, VDR, XPOl | Cell Cycle, Gene Expression, Cellular Growth and Proliferation |
Genes in bold are those differentially expressed in the pathway, direction of arrow indicates up or down regulation.
To validate the microarray results, we performed quantitative reverse transcriptase PCR (qRT-PCR; Figure 3). Genes chosen for qRT-PCR analysis were TSPAN13, COL10, LSG1, TGFβ2, CFI, and CSIG. To assess the capacity of the microarray to correctly detect differential transcription in closely expressed genes, targets with small fold changes between the sexes were selected (COL10A1, LSG1, TSPAN13). CFI and TGFB2 expression levels were also confirmed by qRT-PCR owing to the potential clinical relevance of these molecules. All of the genes assayed using qRT-PCR presented with similar fold changes to those detected by microarray.
Figure 3.
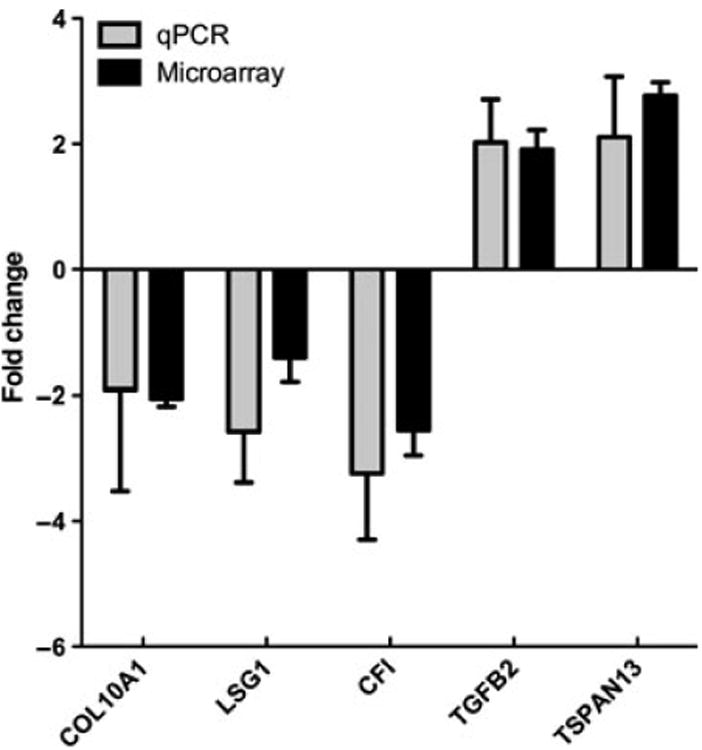
Microarray data closely correlated with quantitative reverse transcription PCR. A subset of genes flagged as differentially transcribed by the microarray analysis were selected for confirmation via qRT-PCR. Microarray fold change values correspond well with data obtained using the Taqman qPCR system. Positive fold change indicates female-biased expression. Standard error of the mean (s.e.m.) is plotted for each
To find differentially expressed genes that may be unique to the salivary glands, we compared our results to microarray datasets taken from male and female, non-smoking, human lungs (Table 4). As expected, genes associated with dosage compensation such as XIST are shared between the two datasets. However, 29 of the 34 genes meeting the conservative statistical cutoff are differentially transcribed between the two genders in the salivary glands but not in lung tissue. Of the 29 genes uniquely transcribed within the salivary gland but not the lung, 24 are upregulated in males over females. The five genes upregulated in women are done so at a low level, ranging from two- to threefold over male levels. Functional classification of the 24 genes upregulated in males indicates that the transcripts are present in five clusters associated with nucleic acid binding, ion binding, metabolic processes, gene expression, and membrane association (Table S3). Given the dominant involvement of the lachrymal and salivary glands in the majority of Sjögren's patients, we compared the differentially expressed genes to gene expression profiles of other secretory organs, such as lung and pancreas. A total of eight genes from the list of gender differentially expressed genes were unique to the salivary gland and not expressed in pancreas tissue based on the IPA database (Table 5). Most were related to metabolic processes and transport with two of these genes reported to be involved in calcium signaling (RRAGD, TPD52L1), a critical part of salivary gland secretory activity. TNFRSF19, a molecule involved in caspase-independent apoptosis, was not expressed in the pancreas but was present in the lung and upregulated in the minor salivary glands of females. TNFRSF19 is expressed at a mean expression level 762-fold below GADPH in males and mean expression 405-fold below GAPDH in females.
Table 4.
Comparison of sexually dimorphic gene expression in human minor salivary glands against genes differentially transcribed in human lung. Sexually dimorphic genes meeting the conservative cutoff were compared against dimorphic genes detected in human lung. Although a core subset of genes are shared between the two tissue types, many differentially transcribed genes are uniquely dimorphic within the minor salivary gland
| Gene | Name | Ratio | GO biologic process |
|---|---|---|---|
| XIST | Inactive X Specific Transcripts | Female:17 | Dosage compensation, regulation of gene expres sion, epigenetic |
| CYorfl5A | Chromosome Y Open Reading Frame 15a | Female:78.6 | G-protein mediated signaling, intracellular protein traffic, MHCI-mediated immunity, cell proliferation and differentiation, general vesicle transport |
| PTPRG | Protein Tyrosine Phosphatase, Receptor Type G | Female:2.4 | Phosphate metabolic process, cell surface receptor linked signal transduction, biopolymer modification, cellular protein metabolic process |
| UTY | Ubiquitously Transcribed Tetratricopeptide Re peat Gene, Y-linked | Male:13.9 | Transport, establishment of localization |
| EIF1AY | Eukaryotic Translation Initiation Factor la, Y-linked | Male:8.2 | Translation, protein-RNA complex assembly, cellular protein metabolic process, macromolec ular complex assembly |
| ZFY | Zinc finger protein, Y-linked | Male:7.7 | Regulation of transcription |
| JARID1D | SMCY Homolog, Y-linked (Mouse) | Male:37 | DNA metabolic process, male gamete generation, chromosome organization and biogenesis |
Table 5.
Differentially expressed salivary gland genes unique to salivary glands compared with lung and pancreas. Ingenuity pathway analysis organ system filter was used to identify unique genes in the salivary gland that were not expressed in pancreas. Gene summary as reported by Genecards (http://www.genecards.org)
|
Fold change F vs M |
Gene | SG | Lung | Proposed function |
|---|---|---|---|---|
| 1.86 | RRAGD | X | RRAGD is a monomeric guanine nucleotide-binding protein, or G protein. By binding GTP or GDP, small G proteins act as molecular switches in numerous cell processes and signaling pathways and may form homo- or hetero-dimer with TPD52 family members. The protein is reported to be involved in cell proliferation and calcium signaling. It also interacts with the mitogen-activated protein kinase 5 | |
| 2.27 | SLC5A1 | X | This gene encodes a member of the sodium-dependent glucose transporter (SGLT) family. The encoded integral membrane protein is the primary mediator of dietary glucose and galactose uptake from the intestinal lumen. Mutations in this gene have been associated with glucose-galactose malabsorption | |
| 1.67 | TPD52L1 | x | This gene encodes a member of the tumor protein D52 (TPD52) family. The encoded protein contains a coiled-coil domain and may form homo- or hetero-dimer with TPD52 family members. The protein is reported to be involved in cell proliferation and calcium signaling | |
| −2.21 | CYP4F12 | X | Catalyzes leukotriene B4 omega hydroxylation and arachidonic acid omega-hydroxylation but with an activity much lower than that of CYP4F2. Catalyzes the hydroxylation of the antihistamine ebastine | |
| −2.39 | PTPRG | X | X | The protein encoded by this gene is a member of the protein tyrosine phosphatase (PTP) family. PTPs are known to be signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. This PTP possesses an extracellular region, a single transmembrane region, and two tandem intracytoplasmic catalytic domains, and thus represents a receptor-type PTP. The extracellular region of this PTP contains a carbonic anhydrase-like (CAH) domain, which is also found in the extracellular region of PTPRBETA/ ZETA. This gene is located in a chromosomal region that is frequently deleted in renal cell carcinoma and lung carcinoma, thus is thought to be a candidate tumor suppressor gene |
| 2.02 | TNFRSF19 | X | X | The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptor is highly expressed during embryonic development. It has been shown to interact with TRAF family members, and to activate JNK signaling pathway when overexpressed in cells. This receptor is capable of inducing apoptosis by a caspase-independent mechanism, and it is thought to play an essential role in embryonic development |
| −1.43 | GLS | X | X | Sahai (1983, PubMed 6825316) demonstrated phosphate-activated glutaminase (EC 3.5.1.2) in human platelets. It is the major enzyme yielding glutamate from glutamine. Significance of the enzyme derives from its possible implication in behavior disturbances in which glutamate acts as a neurotransmitter (Prusiner, 1981). High heritability of platelet glutaminase was indicated by studies of Sahai and Vogel (1983, PubMed 6682827) who found an intraclass correlation coefficient of 0.96 for monozygotic twins and 0.53 for dizygotic twins |
| 1.38 | HMBS | X | X | This gene encodes a member of the hydroxymethylbilane synthase superfamily. The encoded protein is the third enzyme of the heme biosynthetic pathway and catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane. Mutations in this gene are associated with the autosomal dominant disease acute intermittent porphyria. Alternatively spliced transcript variants encoding different isoforms have been described |
Further analysis suggested that multiple sexually dimorphic genes were found to interact with the androgen and estrogen receptors (Figure 4). Four genes upregulated in males, CYP1A2, NRIP2, OXTR, and TGFβ2, were involved in androgen or estrogen receptor signaling. (Mean expression levels: CYP1A2: 3141-fold below GAPDH in males, 3553-fold below GAPDH in females. NRIP2: 324-fold below GAPDH in males, 1050-fold below GAPDH in females. OXTR: 433-fold below GAPDH in males, 1361-fold below GAPDH in females. TGFβ2: 9.9-fold below GAPDH in males, 21-fold below GAPDH in females.) The six genes that were detected as upregulated in females, PARK7, FOXA1, PCBP2, GNB1, CTNNBIP1, and RNF17, all interact with the androgen receptor in a stimulatory fashion. (Mean expression levels: PARK7: 1.4-fold above GAPDH in males, 1.9-fold above in females. FOXA1: 6.4-fold below GAPDH in males, 4.5-fold below GAPDH in females. PCBP2: 49-fold below GAPDH in males, 37-fold below GAPDH in females. GNB1: 13-fold below GAPDH in males, 9.5-fold below GAPDH in females. CTNNBIP1: 1.28-fold below GAPDH in males, 1.1-fold above GAPDH in females. RNF17: 4847-fold below GAPDH in males, 4814-fold below GAPDH in females.)
Figure 4.
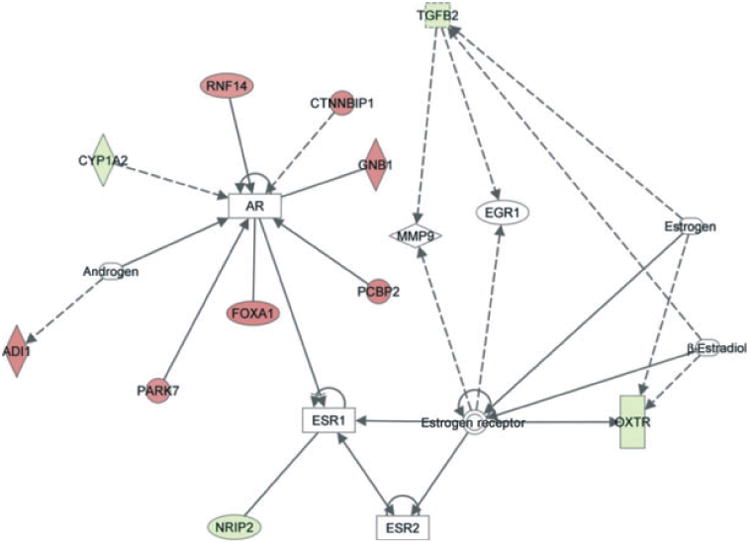
Androgen as well as estrogen receptors have known interactions with several differentially expressed genes. Ingenuity pathway analysis was used to explore interactions between ER and AR and statistically significant, differentially expressed genes. Genes colored in red have higher expression in women with respect to men, whereas genes colored in green show lower expression. Colorless molecules were not found in the gene expression data
Discussion
Our data indicate that despite gross morphological similarity, large differences exist within the transcrip-tome of male and female minor salivary glands. Although minor salivary glands contribute only a small proportion to total saliva production, the degree of inflammation is similar to that seen in major glands; therefore, it represents a good model to assess the potential impact of gender-related differences on the risk for Sjögren's syndrome. The low-level expression of potentially anti-inflammatory molecules, such as TGFβ2 and CFI, and the concomitant upregulation of TNFRSF19 may enhance apoptosis and directly contribute to the increased vulnerability of female salivary glands. Interestingly, TGFB2 has been demonstrated to be involved in both salivary development and the regulation of important immune processes, such as the differentiation of FoxP3+ T-regulatory cells (Fu et al, 2004; Lourenco et al, 2008; Hall et al, 2010). This finding provides support for the hypothesis that many differentially expressed molecular processes may contribute to salivary gland dysfunction and the progression of chronic inflammation and autoimmunity. The observation that complement factor I (CFI) was 2.5-fold upregulated in male mSGs is worth noting in light of reports highlighting the potential importance of the complement system in SS progression (Nguyen et al, 2007). Upregulation of CFI expression, a serine proteinase that regulates the formation of the C3 / C5 convertase enzymes through cleavage of C4B /C3, may play a role in the prevention of SS within males via inhibition of complement-mediated inflammation.
Our data showing that expression levels of hundreds of genes are sexually dimorphic within the human minor salivary gland are consistent with the report by Srivastava et al (2008), who detected 787 genes that were at least 1.5-fold differentially expressed between the sexes in parotid glands. Despite some similarities, there are some important differences between the studies. First, in our study, minor salivary glands showed a 4.5:1 ratio of male-biased differential transcription, which closely resembled the difference seen in mouse submandibular glands (Triester et al, 2005a,b). In contrast, microarray analysis performed by Srivastava et al (2008) on human parotid gland tissue indicated that 89 genes were differentially transcribed by at least twofold, with 40 genes upregulated in men and 49 genes upregulated in women. Comparison of expression levels for individual transcripts between the two studies indicates that a core subgroup of genes such as XIST, RPS4Y1, DDX3Y, and other genes located primarily on the sex chromosomes are consistently differentially transcribed between the sexes. Outside of this core group, many genes were sexually dimorphic in one but not the other study. This could be attributed to specific expression of selected genes in various salivary glands, differences in the matching of males and females for age between the two studies or the microarray platforms as previously reported by Suzuki et al (2009). The Srivastava et al (2008) study on parotid gene expression clearly demonstrated that age has a significant effect on gene expression. Comparing a group of young and old women revealed differential expression of 228 genes. As in the previous study of parotid gene expression males and females were not matched for age, it is possible that potential gender-related differences in the transcriptome were masked by changes related to age. To avoid this possibility, we included subjects with similar ages in our study.
The significant degree of sexually dimorphic gene expression observed in our study suggests differential activity of sex steroid hormone receptors present within the glands. Comparison of the sexually dimorphic genes detected within this report and genes demonstrated to be upregulated upon the administration of testosterone to orchiectomized male BALB / c mice indicates that androgens are likely to play an important role in human salivary dimorphic gene expression (Triester et al, 2005a,b). An alternative area not explored in this manuscript is differential transcription related to hypothalamic-produced neuropeptides such as gonadotropin-releasing hormone that can act on pituitary gland activity related to X-inactivation, a significant aspect of sexually dimorphic gene expression (Suzuki et al, 2009).
In addition to the direct effect of sex hormones on their receptors, other differentially expressed genes may also modify both estrogen and androgen receptor signaling. IPA indicates that both TGFβ2 and OXTR are estrogen responsive genes and both are downregulated in females compared with males (Takahashi et al, 1994; Chang et al, 1999; Bale et al, 2001, Larcher et al, 1995, Triester et al, 2005a,b). In contrast, many genes such as RNF14 and PARK7 are upregulated in females compared with males and have a stimulatory effect on the AR (Yang et al, 2007; Takahashi et al, 2001). The pathway analysis further shows an inhibitory relationship between AR and the estrogen receptors. Together, this data support a difference in signaling in male and female salivary glands with ER signaling likely decreased (hence the decrease in TGFB2 and OXTR) in the salivary glands of women compared with men. This is particularly interesting because most women in this study were in the perimenopausal years, which is the most likely period to develop Sjögren's syndrome. One can hypothesize that a loss of this delicate balance can lead to an increased risk of Sjögren's syndrome. This theory is consistent with observations that the intracrine conversion of dehydroepeandrosterone to androgen, which is the main source of androgens in the salivary glands in postmenopausal women, is impaired in women with Sjögren's compared to age-matched control women (Laine et al, 1995; Porola et al, 2008).
Microarray technology can provide a wealth of information and help formulate new concepts related to molecular mechanisms underlying human disease. In this study, we have made use of the highly parallelized nature of microarray technology to characterize the sexually dimorphic nature of gene expression in healthy male and female minor salivary glands at the RNA level. Further studies confirming these observations of differential expression between men and women at the protein level in healthy humans and patients with Sjögren's syndrome are necessary to better understand the clinical significance of these findings. In parallel, the hypothesis that the biological processes defined by these differentially expressed genes contribute to the risk of Sjögren's syndrome could also be tested in animal models, leading to a better understanding of the physiologic changes associated with the disease.
Supplementary Material
Supporting Information
Additional Supporting information may be found in the online version of this article:
Figure S1 Process workflow. RNA isolated from human minor salivary glands using the Qiagen miRNeasy kit. Samples that met quality control metrics for purity, concentration and integrity were accepted for study and analyzed via two statistical thresholds to maximize the amount of data available for analysis.
Table S1 List of 361 genes that are differentially expressed between male and female salivary glands.
Table S2 GO Moderate Cutoff Data.
Table S3 Functional Clusters present in genes uniquely differently expressed in mSG.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Acknowledgments
The research in this manuscript was supported by an NIH NIDCR intramural research grant to JAC.
Footnotes
Author contributions: D.G.M., G.G.I. and J.A.C. designed the study. S.A, I.A. and G.G.I. managed the clinical aspects of sample acquisition and assessment. D.G.M. and M.W. performed the experiments. D.G.M., S.S., J.C. and J.A.C. analyzed the data. D.G.M., J.A.C. and G.G.I. wrote the paper.
References
- Bale TL, Davis AM, Auger AP, Dorsa DM, McCarthy MM. CNS region-specific oxytocin receptor expression: importance in regulation of anxiety and sex behavior. J Neurosci. 2001;21:2546–2552. doi: 10.1523/JNEUROSCI.21-07-02546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WY, Birch L, Woodham C, Gold LI, Prins GS. Neonatal estrogen exposure alters the transforming growth factor-beta signaling system in the developing rat prostate and blocks the transient p21(cip1 / waf1) expression associated with epithelial differentiation. Endocrinology. 1999;140:2801–2813. doi: 10.1210/endo.140.6.6833. [DOI] [PubMed] [Google Scholar]
- Chowers I, Liu D, Farkas RH, et al. Gene expression variation in the adult human retina. Hum Mol Genet. 2003;12:2881–2893. doi: 10.1093/hmg/ddg326. [DOI] [PubMed] [Google Scholar]
- Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;9:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Zhang N, Yopp AC, et al. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 - precursors. Am J Transplant. 2004;4:1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- Gruber MP, Coldren CD, Woolum MD, et al. Human lung project: evaluating variance of gene expression in the human lung. Am J Respir Cell Mol Biol. 2006;35:65–71. doi: 10.1165/rcmb.2004-0261OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BE, Zheng C, Swaim WD, et al. Conditional overexpression of TGFB1 disrupts mouse salivary gland development and function. Lab Invest. 2010;90:543–555. doi: 10.1038/labinvest.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher A, Neculcea J, Breton C, et al. Oxytocin receptor gene expression in the rat uterus during pregnancy and the estrous cycle and in response to gonadal steroid treatment. Endocrinology. 1995;136:5350–5356. doi: 10.1210/endo.136.12.7588281. [DOI] [PubMed] [Google Scholar]
- Lourenco SV, Uyekita SH, Lima DMC, Soares FA. Developing human minor salivary glands: morphological parallel relation between the expression of TGF-beta isoforms and cytoskeletal markers of glandular maturation. Virchows Arch. 2008;452:427–434. doi: 10.1007/s00428-007-0552-y. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SH, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Nguyen CQ, Kim H, Cornelius JG, Peck AB. Development of Sjogren's syndrome in nonobese diabetic-derived autoimmune-prone C57BL /6.NOD-Aec1Aec2 mice is dependent on complement component 3. J Immunol. 2007;179:2318–2329. doi: 10.4049/jimmunol.179.4.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porola P, Virkki L, Przybyla BD, et al. Androgen deficiency and defective intracrine processing of dehydroepiandrosterone in salivary glands in Sjögren's syndrome. J Rheumatol. 2008;35:2229–2235. doi: 10.3899/jrheum.080220. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Disorders of glutamate metabolism and neurological dysfunction. Annu Rev Med. 1981;32:521–542. doi: 10.1146/annurev.me.32.020181.002513. Review. [DOI] [PubMed] [Google Scholar]
- Richards SM, Jensen RV, Liu M, et al. Influence of sex on gene expression in the mouse lacrimal gland. Exp Eye Res. 2006;82:13–23. doi: 10.1016/j.exer.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Roth SM, Ferrel RE, Peters DG, Metter EJ, Hurley BF, Rogers MA. Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genomics. 2002;10:191–197. doi: 10.1152/physiolgenomics.00028.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai S. Glutaminase in human platelets. Clin Chim Acta. 1983;127:197–203. doi: 10.1016/s0009-8981(83)80004-7. [DOI] [PubMed] [Google Scholar]
- Sahai S, Vogel F. Genetic control of platelet glutaminase: a twin study. Hum Genet. 1983;63:292–293. doi: 10.1007/BF00284668. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Wang J, Zhou H, Melvin JE, Wong DT. Age and gender related differences in human parotid gland gene expression. Arch Oral Biol. 2008;53:1058–1070. doi: 10.1016/j.archoralbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Schirra F, Richards SM, et al. Estrogen's and progesterone's impact on gene expression in the mouse lacrimal gland. Invest Ophthalmol Vis Sci. 2006;47:158–168. doi: 10.1167/iovs.05-1003. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Richards SM, Liu S, Jensen R, Sullivan DA. Influence of sex on gene expression in human corneal epithelial cells. Mol Vis. 2009;15:2554–2569. [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Eitzman B, Bossert NL, et al. Transforming growth factors beta 1, beta 2, and beta 3 messenger RNA and protein expression in mouse uterus and vagina during estrogen-induced growth: a comparison to other estrogen-regulated genes. Cell Growth Differ. 1994;5:919–935. [PubMed] [Google Scholar]
- Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J Biol Chem. 2001;276:37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- Triester NS, Richards SM, Lombardi MJ, Rowley P, Jensen RV, Sullivan DA. Sex-related differences in gene expression in salivary glands of BALB /c mice. J Dent Res. 2005a;84:160–165. doi: 10.1177/154405910508400210. [DOI] [PubMed] [Google Scholar]
- Triester NS, Richards SM, Suzuki T, Jensen RV, Sullivan DA. Influence of androgens on gene expression in the BALB/ c mouse submandibular gland. J Dent Res. 2005b;84:1187–1192. doi: 10.1177/154405910508401218. [DOI] [PubMed] [Google Scholar]
- Venables PJ. Sjogrens syndrome. Best Pract Res Clin Rheumatol. 2004;18:313–329. doi: 10.1016/j.berh.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Yang Z, Chang YJ, Miyamoto H, et al. Transgelin functions as a suppressor via inhibition of ARA54-enhanced androgen receptor transactivation and prostate cancer cell growth. Mol Endocrinol. 2007;21:343–358. doi: 10.1210/me.2006-0104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Additional Supporting information may be found in the online version of this article:
Figure S1 Process workflow. RNA isolated from human minor salivary glands using the Qiagen miRNeasy kit. Samples that met quality control metrics for purity, concentration and integrity were accepted for study and analyzed via two statistical thresholds to maximize the amount of data available for analysis.
Table S1 List of 361 genes that are differentially expressed between male and female salivary glands.
Table S2 GO Moderate Cutoff Data.
Table S3 Functional Clusters present in genes uniquely differently expressed in mSG.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.