Abstract
Purpose
Cancer immunotherapy with adoptive transfer of tumor infiltrating lymphocytes (TILs) represents an effective treatment for patients with metastatic melanoma, with the objective regressions in up to 72% of patients in three clinical trials. However, the antigen targets recognized by these effective TILs remain largely unclear.
Experimental Design
Melanoma patients 2359 and 2591 both experienced durable complete regressions of metastases ongoing beyond five years following adoptive TIL transfer. Two conventional screening approaches were carried out to identify the antigens recognized by these clinically effective TILs. In addition, a novel approach was developed in this study to identify mutated T-cell antigens by screening a tandem minigene library, which comprised non-synonymous mutation sequences identified by whole-exome sequencing of autologous tumors.
Results
The autologous melanoma cDNA library screening led to the identification of previously undescribed non-mutated targets recognized by TIL 2359 or TIL 2591. On the other hand, the screening of tandem minigene libraries resulted in the identification of mutated kinesin family member 2C (KIF2C) antigen as a target of TIL 2359, and mutated DNA polymerase alpha subunit B (POLA2) antigen as a target of TIL 2591. Both KIF2C and POLA2 have been found to play important roles in cell proliferation.
Conclusions
These findings suggest that the minigene screening approach can facilitate the antigen repertoire analysis of tumor reactive T cells, and lead to the development of new adoptive cell therapies with purified T cells that recognize candidate mutated antigens derived from genes essential for the carcinogenesis.
Keywords: Melanoma/skin cancers, Phase I–III Trials, Clinical Immunology: Cellular immunotherapy, Tumor Immunobiology, Tumor antigens
Introduction
Adoptive cell therapy for patients with metastatic melanoma involves the infusion of autologous, in vitro expanded, tumor infiltrating lymphocytes (TILs) plus IL-2 into patients who have received a lymphodepleting preparative regimen (1, 2). In three sequential pilot clinical trials, adoptive TIL transfer mediated the objective regressions of metastatic melanoma in up to 72% of patients, including 36% with complete durable responses ongoing beyond five years (3). In spite of the strong anti-tumor activities of TILs ex vivo, not all of the patients receiving adoptive TIL transfer experienced durable regressions. One potential explanation for these observations is that many of the TILs used for treatment may predominantly recognize gene products that are dispensable for maintaining the tumorigenic phenotype and therefore subject to loss by down-regulation, mutation, or gene deletion. In contrast, TILs targeting antigens derived from indispensable genes may mediate long-term complete regressions more effectively (4–6). Many melanoma TILs have been discovered to recognize mutated gene products, consistent with the observation that tumor-reactive T cells can be isolated from melanoma. Nevertheless, most of the antigens recognized by adoptively transferred TILs that mediated long-term complete regressions remain elusive (7). To further test the hypothesis that mutated antigens frequently represent the targets of clinically effective TILs, we have begun to evaluate the use of whole-exome sequencing of tumor cell DNA to identify mutated antigens recognized by TILs.
In a recent murine tumor model system, mutated gene products were identified from a sarcoma cell line d42m1 by whole-exome sequencing, and were subjected to in silico analysis to identify candidate T cell epitopes that were predicted to bind to the MHC class I molecules H-2Db or H-2Kb with high affinity. Among these epitopes, R913L spectrin-β2 mutated epitope was predicted to be presented on tumor recognized by a T cell clone. Mutated spectrin-β2 was later confirmed to be served as a tumor-rejection antigen of d42m1 (8). In another study, candidate mutated epitopes identified by whole-exome sequencing of the B16F10 murine melanoma resulted in the identification of mutated peptides that could elicit protective and therapeutic T cell responses (9).
In our recent study, mutated genes from melanoma cell lines were identified by whole-exome sequencing, followed by in silico analysis to identify candidate peptides predicted to have high affinity to the autologous HLA molecules. Panels of synthetic peptides were then evaluated for their ability to stimulate autologous TILs. Using this approach, seven mutated epitopes were identified as the targets of three therapeutic TIL products (10). Two of the antigens that were identified using this approach, CSNK1A1 and PLEKHM2, were not identified in the initial cDNA library screening assay. However, this approach is limited by the accuracy of current HLA-binding prediction algorithms, which have not been thoroughly examined for infrequent HLA alleles. Furthermore, in some cases naturally processed epitopes may not correspond to those encoded in the genome due to the post-translational modifications (11, 12).
Previous studies have demonstrated that epitopes expressed from minigenes can be efficiently processed and presented by MHC class I molecules (13, 14). In addition, designs with tandem minigenes encoding multiple epitopes have been utilized for vaccine development (15). To overcome some of the limitations imposed by previous screening methods, tandem minigene constructs encoding mutated gene products identified by whole-exome sequencing were generated and transfected into target cell lines expressing each of the individual HLA class I gene products expressed by the autologous tumor. A single new mutated antigen target was identified from each of the two therapeutic TIL products by screening autologous tandem minigene libraries. These results indicate that this method may provide a valuable tool to identify potent antigens that may serve as the targets for future personalized therapies.
Materials and Methods
Patient materials and cell lines
All patient materials were obtained in the course of a National Cancer Institute Institutional Review Board approved clinical trial. Patient 2359 and Patient 2591 were enrolled on clinical trials (Trial registration ID: NCT00096382 and ID: NCT00335127, respectively) that have been described in detail previously (16). The patients underwent resections from which both a TIL line and a tumor cell line were established. TILs used for this study were generated by methods described previously (17). Briefly, tumor fragments were excised and cultured in media containing IL-2. TIL cultures that expanded were screened for recognition of autologous or HLA-matched tumor, and reactive TILs were expanded using a rapid expansion protocol (REP) with IL-2, anti-CD3 antibody and irradiated feeder cells to large numbers for patient infusion (18). A small portion of TILs underwent a second REP for the experiments shown in this report. For co-culture assays, T cells and tumor cells were cultured at 1:1 ratio in a 96-well plate with 200 μL medium (AIM-V medium supplemented with 5% human serum) for 16 hr. In antibody blocking experiments, melanoma cells were pre-incubated with HLA-A*02 (SB-02), HLA-B,C (B1.23.2) or HLA-A,B,C (W6/32) blocking antibodies (40 μg/mL) for 3 hr, followed by co-cultured with T cells. The concentration of IFN-γ in the supernatant was determined by ELISA (Thermo Scientific).
Whole-exome sequencing
The method has been described previously (10). Genomic DNA purification, library construction, exome capture of approximately 20,000 coding genes and next-generation sequencing of tumor and normal samples were performed at Personal Genome Diagnostics (Baltimore, MD). In brief, genomic DNA from tumor and normal samples was fragmented and used for Illumina TruSeq library construction (Illumina). Exonic regions were captured in solution using the Agilent SureSelect 50 Mb kit (version 3) according to the manufacturer’s instructions (Agilent). Paired-end sequencing, resulting in 100 bases from each end of each fragment, was performed using a HiSeq 2000 Genome Analyzer (Illumina). Sequence data were mapped to the reference human genome sequence, and sequence alterations were determined by comparison of over 50 million bases of tumor and normal DNA. Over 8 billion bases of sequence data were obtained for each sample, and a high fraction of the bases were from the captured coding regions. Over 43 million bases of target DNA were analyzed in the tumor and normal samples, and an average of 42–51 reads were obtained at each base in the normal and tumor DNA samples.
Bioinformatic analyses were carried out by Personal Genome Diagnostics and the Genome Technology Access Center, Genomics and Pathology Services of the Washington University School of Medicine. The tags were aligned to the human genome reference sequence (hg18) using the Eland algorithm of the CASAVA 1.6 software (Illumina). The chastity filter of the BaseCall software of Illumina was used to select sequence reads for subsequent analyses. The ELANDv2 algorithm of the CASAVA 1.6 software was then applied to identify point mutations and small insertions and deletions. Known polymorphisms recorded in dbSNP were removed from the analysis. Potential somatic mutations were filtered and visually inspected as described previously (19).
The construction of tandem minigene library and cDNA library
Non-synonymous mutations from melanoma samples were identified from whole-exome sequencing data. Tandem minigene constructs that encode polypeptides containing 6 identified mutated amino acid residues flanked on their N- and C- termini, 12 amino acids on both sides, were synthesized (Integrated DNA Technologies, Coralville, Iowa), and then cloned into pcDNA3.1 expression vector using the In-Fusion Advantage PCR Cloning Kit (Clontech), according to the manufacture’s instruction.
The cDNA libraries were generated from Mel 2359 and Mel 2591 mRNA using the SMARTer RACE cDNA Amplification Kit (Clontech), and the cDNA libraries were cloned into pCMV6 expression vector using the In-Fusion Advantage PCR Cloning Kit (Clontech), as described previously (6).
PCR amplification and quantitative PCR
The following primer sets were used for PCR amplification: 5′-GCG GCG TTA AGA CTT CGT AGG GT-3′ and 5′-GCT CCT CAC CAT TAC TGC GTT GG-3′ for the amplification of Exon 1 in KIF2C ORF from genomic DNA and KIF2C cDNA. 5′-TGT CCA GAT GGA TGG GTG TGA AG-3′ and 5′-TCA AGG AGT GGG TGC TGC TAC CT-3′ for the amplification of Exon 14 in POLA2 ORF from genomic DNA. 5′-CAG TGG ATT TAT CTG AGC TTA AGG-3′ and 5′-TTA AGG GCC CAC ACA GCA GAG AA-3′ for the amplification of POLA2 cDNA. SLC24A5 WT TaqMan primer/probe set (Hs01385410_m1) was obtained from Applied Biosystems.
IFN-γ ELISPOT assay
The responses directed against tumor cell lines and peptide-pulsed target cells were quantified in an IFN-γ ELISPOT assay using 96-well PVDF-membrane filter plates (EMD Millipore, Billerica, MA) coated with 15 μg/ml of the monoclonal anti–IFN-γ antibody 1D1K (Mabtech, Inc., Cincinnati, OH). Bound cytokine was detected using 1 μg/ml of the biotinylated anti–IFN-γ antibody 7-B6-1 (Mabtech). HEK293 cells expressing HLA-A*0201, HLA-A*0205 or HLA-C*0701 were pulsed with peptides for 2 h at 37 °C. The following peptides were used: MART-1: AAGIGILTV, DUSP12: VSCAGQMLEV, SERPINE2: AVYFKGLWKS, SLC24A5: RRDGGIIIYF, mutated KIF2C: RLFPGLTIKI, mutated POLA2: TRSSGSHFVF. T cells were co-cultured overnight with target cells or medium containing 50 ng/ml PMA plus 1 μM ionomycin (PMA/I). The numbers of spots per 105 T cells were calculated.
Results
To evaluate the antigen reactivity of TIL with clinical activity, we focused on two metastatic melanoma patients who experienced durable complete responses to adoptive TIL therapy. Patient 2359 had a primary cutaneous melanoma at the right knee that metastasized to the thigh, iliac and inguinal lymph nodes. This individual experienced a complete regression of all metastatic lesions in response to autologous TIL transfer that is ongoing for over eight years following treatment. Patient 2591 had a primary back melanoma that metastasized to the abdominal wall, mesenteric lymph nodes, right colon, and supraclavicular lymph nodes. This individual experienced a complete regression of all metastatic lesions in response to autologous TIL transfer and remains disease free nine years after treatment.
TIL 2591 recognized previously described non-mutated self-antigens
Experiments were first carried out to identify the predominant HLA-restriction elements for the recognition of autologous melanoma by TIL 2359 and TIL 2591. HLA genotyping of PBMC indicated that patient 2359 expressed A*0205, A*1101, B*4403, B*4901, C*0701, and C*1601, while patient 2591 expressed A*0101, A*0201, B*0801, B*3501, C*0401, C*0701. TIL 2359 T cells induced the release of IFN-γ following co-culture with autologous tumor cells, and was completely blocked by the HLA-A*02 blocking antibody SB-02 (Supplementary Fig. S1A). The results indicated that HLA-A*0205 represented the predominant restriction element mediating tumor recognition of TIL 2359. The release of IFN-γ from a co-culture of TIL 2591 with autologous tumor cells that were pre-incubated with the HLA-A*02 blocking antibody SB-02 was inhibited by approximately 50%, while incubation with the pan-HLA-B/C antibody B1.23.2 also partially inhibited the release of IFN-γ in response to autologous tumor cells (Supplementary Fig. S1B). These results suggested that TIL 2591 recognized autologous melanoma through multiple HLA-restriction elements.
The ability of TIL 2359 and 2591 to recognize shared antigens was initially evaluated by co-transfecting COS-7 cells with each of the autologous HLA class I alleles expressed by these patients with a panel of 30 non-mutated melanoma antigens that included members of the melanocyte differentiation family of genes, cancer germ-line genes, as well as a set of additional genes previously identified as targets of melanoma-reactive T cells. As shown in Supplementary Fig. S2A, TIL 2359 failed to recognize HLA-A*0205-transfected target cells that were co-transfected with any of the candidate antigens that were tested. In contrast, TIL 2591 recognized HLA-A*0101+ target cells that were transfected with tyrosinase (Supplementary Fig. S2B), as well as HLA-A*0201+ target cells transfected with MART-1, NA-17A and tyrosinase (Supplementary Fig. S2C).
TIL 2359 and TIL 2591 recognized new non-mutated self-antigens
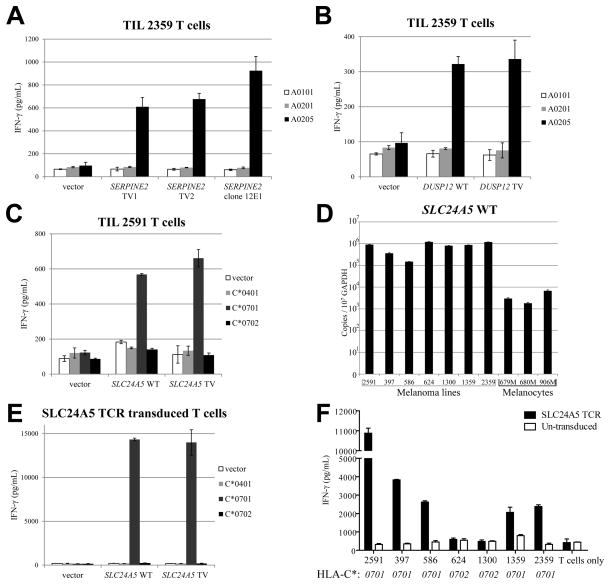
In an attempt to identify the antigen or antigens recognized by TIL 2359, HEK293 cells that stably expressed the dominant restriction element HLA-A*0205 were transiently transfected individually with approximately 2,000 pools consisting of 50 cDNA clones generated from the autologous Mel 2359 cell line. These transfectants were screened for their ability to stimulate autologous TIL, as previously described (6). Three positive pools were identified and confirmed, and all the positive clones isolated from the 1st pool corresponded to the truncated transcript of SERPINE2 [serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2]. The sequence of the SERPINE2 transcript (clone 12E1) appeared to represent a partial cDNA transcript that lacked the first 434 b.p. of SERPINE2 transcript variant 2 (TV2) (Supplementary Fig. S3A). TIL 2359 demonstrated similar recognition of COS-7 cells that were transiently transfected with either the truncated SERPINE2 transcript (clone 12E1) or full-length SERPINE2 TV1 or TV2 cDNA, together with HLA-A*0205 (Fig. 1A). Therefore, TIL 2359 T cells recognized COS-7 cells transfected with each of the SERPINE2 transcripts in an HLA-A*0205-restricted manner. The positive clones isolated from the 2nd and 3rd pool corresponded to a transcript variant of DUSP12 (dual specificity phosphatase 12), which is identical to the sequence CR603104.1 in GenBank database (Supplementary Fig. S3B). Similarly, COS-7 cells were transfected with either WT or TV of DUSP12 cDNA, together with HLA-A*0205 cDNA. TIL 2359 T cells recognized both DUSP12 WT and DUSP12 TV gene product, in an HLA-A*0205-restricted manner (Fig. 1B).
Figure 1.
New non-mutated T-cell antigens are identified by cDNA library screening. (A) COS-7 cells were transfected with HLA cDNA constructs, together with full-length SERPINE2 transcription variant 1 (TV1), variant 2 (TV2) or clone 12E1 isolated from Mel 2359 cDNA library. These transfected cells were co-cultured with TIL 2359 T cells overnight. (B) COS-7 cells were transfected with HLA cDNA constructs, together with full-length WT DUSP12 or its transcription variant (DUSP12 TV). These transfected cells were co-cultured overnight with TIL 2359 T cells, and the release of IFN-γ was determined by ELISA. (C) COS-7 cells were transfected with HLA cDNA constructs, together with full-length WT SLC24A5 or its transcription variant (SLC24A5 TV). These transfected cells were co-cultured with TIL 2591 T cells overnight. (D) The copy numbers of SLC24A5 WT or SLC24A5 TV in normal melanocyte samples or melanoma lines were determined by quantitative PCR. (E) Healthy donor T cells were transduced with SLC24A5 TCR isolated from TIL 2591. These T cells were co-cultured with COS-7 cells transfected with HLA cDNA constructs, together with SLC24A5 WT or SLC24A5 TV. (F) SLC24A5 TCR transduced T cells were co-cultured overnight with various melanoma cells lines. The secretion of IFN-γ was determined by ELISA. The types of HLA-C are indicated below.
Using a similar approach, we attempted to determine whether or not TIL 2591 recognized any antigens in addition to MART-1, NA-17A and tyrosinase (Supplementary Fig. S2). Pools of 50 cDNA clones generated from Mel 2591 were then transiently transfected into HEK293 cell lines that were stably transfected with either HLA-A*0101, A*0201, B*0801, B*3501, C*0401, C*0701. Following transient transfection of each of the six HLA class I-transfected cell lines with approximately 2000 cDNA library pools, a single pool was identified that stimulated the release of significant levels of IFN-γ from TIL 2591 when transfected into HEK293 cells that expressed HLA-C*0701. Positive clones isolated from this pool corresponded to a transcript of the SLC24A5 [solute carrier family 24 (sodium/potassium/calcium exchanger), member 5] containing intronic sequences that were spliced from the majority of SLC24A5 transcripts (Supplementary Fig. S3C). TIL 2591 cells recognized COS-7 cells that were co-transfected with HLA-C*0701 and either the full length SLC24A5 transcript or the partial cDNA isolated by expression cloning, but failed to recognize cells that were co-transfected with HLA-C*0702 or C*0401 (Fig. 1C).
SLC24A5 contains the predicted sodium-calcium exchanger activity, and SLC24A5 knockdown disrupts the melanin pigment production in normal human melanocytes (20). Additional experiments were carried out to further characterize this antigen. As shown in Fig. 1D, similar levels of expression of SLC24A5 were found in all melanoma lines that were tested, while melanocytes expressed SLC24A5 at approximately 1% of the levels observed in melanoma lines. While the majority of normal tissues expressed low levels of SLC24A5 expression, retina, skin and testis expressed levels of SLC24A5 that were similar to those observed in melanocytes (Supplementary Fig. S4). A T cell receptor was then isolated from an SLC24A5-reactive T cell clone derived from TIL 2591, and introduced into a recombinant retroviral construct. Activated T cells obtained from the peripheral blood of healthy donors transduced with SLC24A5 TCR recognized SLC24A5 association in a C*0701-restricted manner (Fig. 1E). SLC24A5 TCR transduced T cells recognized all melanoma lines expressing HLA-C*0701, but not Mel 624 and 1300, which expressed HLA-C*0702 (Fig. 1F).
TIL 2359 recognized a mutated antigen as assessed by minigene library screening
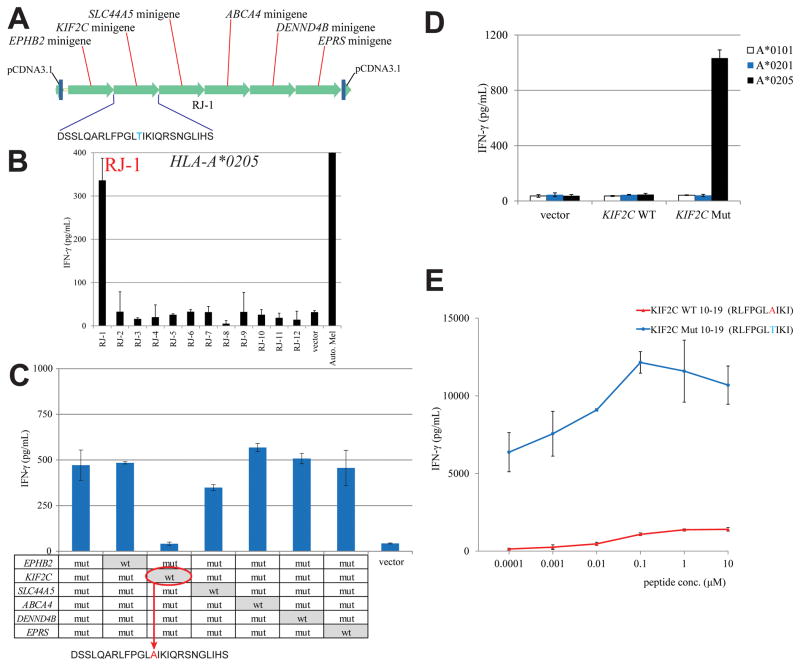
The reactivity of TIL 2359 was then further evaluated using a novel approach. In this approach, tandem minigene constructs were generated based on the non-synonymous mutations identified by exomic analysis of tumor and normal DNA. Each tandem minigene constructs encoded up to six individual minigene fragments that corresponded to the mutated codon flanked on either side by the 12 additional codons present in the normal gene product. One example is illustrated in Fig. 2A.
Figure 2.
TIL 2359 T cells recognize mutated KIF2C gene product. (A) An example of tandem minigene construct, which encoded polypeptides containing 6 identified mutated amino acid residues flanked on their N- and C- termini, 12 amino acids on both sides. These tandem minigenes were synthesized and cloned into pcDNA3.1 vector. (B) A screening assay was carried out to co-transfect HLA-A*0205 with individual tandem minigene construct into COS-7 cells. TIL 2359 T cells were co-cultured with these transfected COS-7 cells overnight, and the release of IFN-γ from TIL 2359 was evaluated by ELISA. The structure of the tandem minigene construct RJ-1 has been shown in (A). (C) Each point mutation site at the minigenes of RJ-1 construct was converted back to WT sequence individually, indicated on the table. TIL 2359 were co-cultured with COS-7 cells transfected with individual RJ-1 variant and HLA-A*0205. The secretion of IFN-γ by TIL 2359 was determined by ELISA. (D) COS-7 cells were transfected with WT or mutated KIF2C cDNA construct, together with HLA cDNA constructs. These transfected cells were co-cultured with TIL 2359 T cells overnight. (E) HEK293 cells stably expressed HLA-A*0205 were pulsed with WT or mutated KIF2C peptides, followed by co-cultured with TIL 2359 T cells overnight. The release of IFN-γ was determined by ELISA.
COS-7 cells were transiently transfected individually with twelve tandem minigenes encoding the 71 minigenes based on exomic DNA sequences containing non-synonymous point mutations identified from Mel 2359 (Supplementary Table S1). These COS-7 cells were also co-transfected with HLA-A*0205, the dominant HLA restriction element used for autologous tumor cell recognition by this TIL. Co-culture of these transfectants with TIL 2359 resulted in the recognition of one of the 12 tandem minigene constructs, RJ-1 (Fig. 2B). RJ-1 encoded mutated fragments of the EPHB2, KIF2C, SLC44A5, ABCA4, DENND4B, and EPRS genes, as shown in Fig. 2A. Subsequently, six RJ-1 variant constructs were generated, each of which encoded the WT rather than the mutated residue present in one of the six minigenes (Fig. 2C). TIL 2359 recognized COS-7 cells co-transfected with HLA-A*0205 plus five of the six individually transfected RJ-1 variants, but failed to recognize the variant encoding the WT KIF2C sequence, indicating that this minigene encoded a mutated epitope recognized by TIL 2359 (Fig. 2C). To further test this observation, COS-7 cells were co-transfected with either WT or mutated full-length KIF2C cDNA transcripts that were amplified from Mel 2359, together with either HLA-A*0101, HLA-A*0201 or HLA-A*0205 cDNA. The co-culture experiment indicated that TIL 2359 T cells recognized COS-7 cells co-transfected with the mutated but not WT KIF2C gene product, in a HLA-A*0205-restricted manner (Fig. 2D).
The mutated KIF2C coding region contained a single C to A transversion at nucleotide 46 that resulted in a substitution of threonine for alanine at position 16 in the native KIF2C protein (Supplementary Fig. S5A). Exomic sequencing results indicated that DNA from Mel 2359 exclusively corresponded to the mutated but not the normal residue at position 46, results confirmed by direct Sanger sequencing of Mel 2359 DNA, indicating the loss of heterozygosity at this locus (Supplementary Fig. S5B). In an attempt to identify the mutated KIF2C epitope recognized by TIL 2359, peptides encompassing the KIF2C mutation that were predicted to bind with high affinity to HLA-A*0205 were synthesized (21), and pulsed on HEK293 cells that stably expressed HLA-A*0205 (Supplementary Table S2). HEK293-A*0205 cells pulsed with a decamer corresponding to residues 10–19 stimulated the release of high levels of IFN-γ from TIL 2359 T cells, and the peptide was recognized at a minimum concentration of 0.1 nM. In contrast, the corresponding WT peptide did not induce significant IFN-γ release at a concentration as high as 10 μM (Fig. 2E).
TIL 2591 recognized a mutated antigen identified by minigene library screening
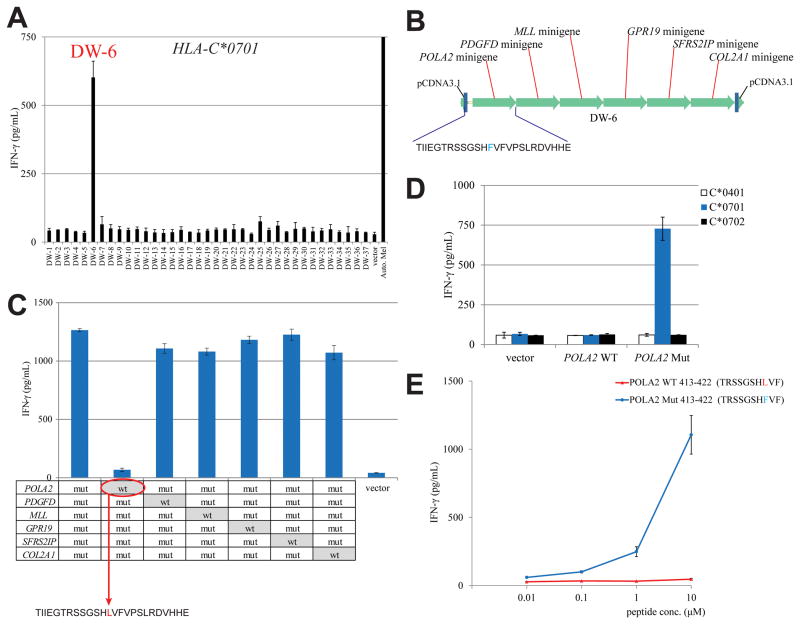
We then attempted to identify mutated T-cell antigen recognized by TIL 2591 by synthesizing 37 tandem minigene constructs encoding the 217 minigenes based on exomic DNA sequences containing non-synonymous point mutations identified from Mel 2591 (Supplementary Table S3). As noted above, TIL 2591 appeared to recognize autologous tumor cells in the context of multiple HLA restriction elements (Supplementary Fig. S1). Therefore, HEK293 cell lines stably expressing each of the six MHC class I HLA molecules isolated from Mel 2591 were transiently transfected individually with the 37 tandem minigene constructs, followed by an overnight co-culture with TIL 2591. Initial results indicated that TIL 2591 recognized HLA-C*0701+ HEK293 cells (HEK293-C*0701) cells that were transiently transfected with minigene DW-6, but failed to respond significantly to the other minigene constructs (Fig. 3A). Each of the six individual mutated minigenes in the DW-6 tandem construct (Fig. 3B) were then individually reverted to the WT sequence (Fig. 3C). Evaluation of responses to the WT variants indicated that TIL 2591 recognized COS-7 cells transfected with each of the DW-6 variants, with the exception of a construct encoding the WT POLA2 fragment (Fig. 3C). To test these findings, COS-7 cells were transfected with either a WT or mutated full-length POLA2 cDNA construct, together with HLA-C*0401, HLA-C*0701 or HLA-C*0702 cDNA. TIL 2591 T cells only recognized target cells transfected with HLA-C*0701 plus the mutated POLA2 construct, but not the corresponding WT transcript (Fig. 3D). The single C to T transition at nucleotide 1258 of the POLA2 coding region resulted in a substitution of leucine for phenylalanine at position 420 of the WT POLA2 protein (Supplementary Fig. S5C). Sanger sequencing indicated that both genomic DNA and cDNA derived from Mel 2591 RNA contained both the WT and mutated nucleotide at position 1258, whereas genomic DNA isolated from PBMC of patient 2591 corresponded to the WT sequence, indicating that this represented a heterozygous somatic mutation in Mel 2591 cells (Supplementary Fig. S5D).
Figure 3.
Mutated POLA2 gene product is recognized by TIL 2591 T cells. (A) Individual tandem minigene construct was transfected in HEK293 cells stably expressed HLA-C*0701. TIL 2591 T cells were co-cultured with these transfected cells overnight, and the release of IFN-γ was determined by ELISA. (B) The structure of tandem minigene construct DW-6. (C) Each point mutation site at the minigenes of DW-6 construct was converted back to WT sequence individually, indicated on the table. TIL 2591 T cells were co-cultured with COS-7 cells transfected with individual DW-6 variant and HLA-C*0701. The secretion of IFN-γ by TIL 2591 was determined by ELISA. (D) COS-7 cells were transfected with WT or mutated POLA2 cDNA construct, together with HLA cDNA constructs. These transfected cells were co-cultured with TIL 2359 T cells overnight. (E) HEK293 cells stably expressed HLA-C*0701 were pulsed with WT or mutated POLA2 peptides, followed by co-cultured with TIL 2591 T cells overnight. The release of IFN-γ was determined by ELISA.
An HLA-C*0701 binding algorithm was then used to identify candidate POLA2 peptides overlapping with the mutated leucine residue at position 420 (Supplementary Table S4). Co-culture results indicated that HLA-C*0701+ HEK293 cells pulsed with a decamer corresponding to residues 413–422 of mutated POLA2 stimulated the release of IFN-γ from TIL 2591 T cells at a minimum concentration of 10 nM. In contrast, the corresponding WT peptide did not induce significant IFN-γ release at a concentration as high as 10 μM (Fig. 3E).
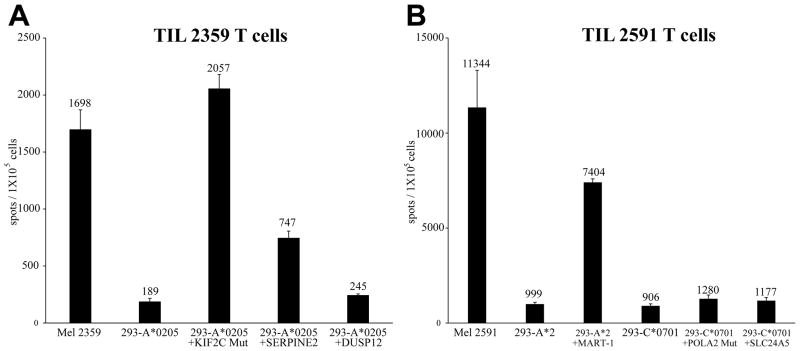
We then estimated the proportion of T cells in TIL 2359 and 2591 recognizing the mutated KIF2C and POLA2, respectively, as well as a panel of commonly recognized non-mutated melanoma T cell epitopes using IFN-γ enzyme-linked immunosorbent spot (ELISPOT) assays. TIL 2359 generated approximately 2,000 spots per 100,000 T cells in response to HLA-A*0205+ cells pulsed with the mutated KIF2C epitope, similar to that observed in response to the autologous melanoma (Fig. 4A). TIL 2591 generated greater than 7,000 spots in response to the HLA-A2 restricted MART-1 epitope, while only small fractions of T cells reacted against the HLA-C*0701-restricted mutated POLA2 epitope and non-mutated SLC24A5 (Fig. 4B). The results are consistent with the TIL 2591 co-culture experiments, which showed a high level of IFN-γ secretion following stimulation by MART-1 and low levels of IFN-γ secretion following stimulation by SLC24A5 and mutated POLA2 (Fig. 1, 3 and Supplementary Fig. S2C). We did not consistently detect tyrosinase or NA-17A reactive T cells using ELISPOT assays (data not shown).
Figure 4.
IFN-γ ELISPOT responses of TILs. (A) IFN-γ spots per 105 cells of TIL 2359. TIL2359 T cells were co-cultured with autologous Mel 2359 or HLA-A*0205-positive HEK293 cells pulsed with peptides. Additionally, TIL 2591 T cells were cultured with autologous Mel 2591 in the ELISPOT assay. TIL 2359 T cells stimulated with the polyclonal activator PMA plus ionomycin generated 59,385 spots per 105 T cells in this assay. (B) IFN-γ spots per 105 cells of TIL 2591 T cells. These T cells were co-cultured HLA-A*02-positive or HLA-C*07-positive HEK293 cells pulsed with peptides. In addition, TIL 2591 T cells were cultured with autologous Mel 2591 in the ELISPOT assay. TIL 2591 T cells stimulated with the polyclonal activator PMA plus ionomycin generated 60,162 spots per 105 T cells in this assay.
In summary, our results provide evidence for the utility of multiple screening approaches for evaluating the antigen reactivity of polyclonal T cell populations. These methods include conventional cDNA library screening approaches as well as a novel method involving the transfection of minigene libraries encoding mutated candidate antigens generated based on tumor whole-exome sequencing data. This minigene approach provides an important tool that should facilitate the identification of T-cell antigens.
Discussion
The identification of novel antigens presented by tumors and recognized by T cells has been a bottleneck in the development of effective cancer immunotherapies. It has primarily been carried out using approaches that are based upon the transfection of COS-7 or HEK293 cells with cDNA library pools. In this study, a conventional cDNA library screening approach was used to identify the antigens recognized by therapeutic TIL 2359 and 2591. Screening of an autologous cDNA library prepared from Mel 2359 led to the identification of two non-mutated antigens, SERPINE2 and DUSP12, as HLA-A*0205-restricted targets of TIL 2359. On the other hand, TIL 2591 recognized several previously described epitopes, including HLA-A*0201-restricted epitopes of MART-1, tyrosinase and NA17-A, as well as an HLA-A*01 restricted epitope of tyrosinase. Similarly, screening of an autologous cDNA library prepared from Mel 2591 led to the identification of a previously undescribed antigen, SLC24A5, that was recognized in the context of HLA-C*0701 by TIL 2591.
Further attempts were then made to characterize the response of these TILs to mutated cancer antigens through the use of whole-exome sequencing. A new approach was developed based upon the transfection of HLA-matched targets with mutated minigene constructs. Using this approach, we identified two mutated T-cell antigens, KIF2C and POLA2, as targets of TIL 2359 and 2591, respectively. KIF2C, together with KIF2A and KIF2B, comprise the kinesin-13 family of microtubule motor proteins, which are defined by the localization of the conserved kinesin motor domain in the middle of the polypeptide (22). Kinesin-13 proteins are non-motile and induce microtubule de-polymerization by disassembling tubulin subunits from the polymer end (23). Each member of kinesin-13 family has distinct roles during mitosis (24). KIF2C, also known as mitotic centromere-associated kinesin (MCAK), uses microtubule depolymerizing activity to correct improper microtubule attachments at kinetochores (25). KIF2C promotes rapid restructuring of the microtubule cytoskeleton by making catastrophe, an event which microtubule plus ends abruptly switch from growth to de-polymerization, to become a random single-step process (26, 27). POLA2, also known as p70 α2 subunit, is one of the 4 subunits of the highly conserved DNA polymerase α (28). This polymerase is unique among DNA polymerase family member, because of its primase activity to initiate de novo DNA synthesis. The primase synthesizes short RNA-DNA primers, ~40 nucleotides in length, for the leading strand synthesis or for the Okazaki fragment on the lagging strand (29). The POLA2 subunit interacts directly with the p180 catalytic subunit, but does not possess any enzymatic activity. However, it does play an essential role at the early stage of chromosomal DNA replication. Studies in S. cerevisiae demonstrate that the POLA2 subunit is required for in vivo DNA synthesis and correct progression through S phase (30). Taken together, both KIF2C and POLA2 play important roles during the cell proliferation. They are likely essential genes for carcinogenesis, and may explain the ability of these TILs that recognize the mutated epitopes to mediate the complete durable regressions of cancer in these patients.
Our previous efforts have been focused on identifying widely shared target antigens. However, targeting antigens that are expressed in normal tissues can induce severe autoimmune toxicity because of the gene expression in normal tissues (31–33), which could be avoided by targeting patient-specific, tumor-specific mutated antigens. The minigene approach demonstrated in this study can facilitate the identification of mutated antigens, and allow more effective cancer immunotherapy by developing methods to specifically target the mutated antigens. Currently, TILs are grown from individual tumor fragments to allow the selection of cultures with anti-tumor reactivity for further expansion and subsequent administration to patients. Some TIL populations, such as the two TILs evaluated in this report, recognize mutated as well as non-mutated antigens, whereas additional TILs appear to predominantly or exclusively recognize mutated antigens (10). Use of the minigene screening approach will facilitate the identification of individual mutation-reactive T cell cultures. This approach may be further enhanced by the development of techniques such as sorting with specific MHC tetramers to increase the frequency of mutation-reactive T cells from bulk TILs that may contain significant numbers of non-tumor reactive cells (34). Alternatively, TCRs isolated from cells reactive with mutated epitopes can be isolated and introduced into autologous peripheral blood T cells for the subsequent adoptive T-cell transfer (35, 36).
In conclusion, this study suggests that the identification of antigens using an approach based upon the screening of minigenes that encode mutated candidate antigens complements the use of conventional cDNA library screening methods to evaluate the antigen repertoire of tumor reactive T cells. Targeting mutated antigens, in particular those derived from gene products that are involved with carcinogenesis, can potentially enhance the effectiveness of cancer immunotherapy using adoptive transfer of TILs.
Supplementary Material
Translational Relevance.
To develop next-generation adoptive cell therapies using purified TILs with defined specificity, the identification of T-cell antigens recognized by TILs is one of the major bottlenecks. Previous studies for antigen identification have primarily involved screening cDNA libraries, which is time-consuming and low-throughput. The minigene approach demonstrated in this study can significantly facilitate the identification of T-cell antigens, and it will pave the way for new adoptive cell therapies targeting tumor-specific, mutated antigens.
Acknowledgments
Grand Support: This research is supported by the NCI Director’s Innovation Award (Y.L.), by the Intramural Research Program of NCI, by Adelson Medical Research Foundation (AMRF), by Milstein Family Foundation, and by European Research Council (StG-335377) (Y.S.).
We are grateful to John R. Wunderlich, Kenichi Hanada, Qiong J. Wang, and Shigui Zhu for suggestions and technical support.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: Y. Lu, P. F. Robbins
Development of methodology: Y. Lu, P. F. Robbins
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Y. Lu, X. Yao, J. S. Crystal, Y. F. Li, M. El-Gamil, L. Davis
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Lu, X. Yao, J. S. Crystal, C. Gross,
Writing, review, and/or revision of the manuscript: Y. Lu, S. A. Rosenberg, P. F. Robbins
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. E. Dudley, J. C. Yang, Y. Samuels
Study supervision: S. A. Rosenberg, P. F. Robbins
References
- 1.Rosenberg SA. Raising the bar: the curative potential of human cancer immunotherapy. Sci Transl Med. 2012;4:127–8. doi: 10.1126/scitranslmed.3003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–4. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res. 2011;17:4550–7. doi: 10.1158/1078-0432.CCR-11-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 5.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 6.Lu YC, Yao X, Li YF, El-Gamil M, Dudley ME, Yang JC, et al. Mutated PPP1R3B Is Recognized by T Cells Used To Treat a Melanoma Patient Who Experienced a Durable Complete Tumor Regression. J Immunol. 2013;190:6034–42. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–81. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 10.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004;427:252–6. doi: 10.1038/nature02240. [DOI] [PubMed] [Google Scholar]
- 12.Dalet A, Robbins PF, Stroobant V, Vigneron N, Li YF, El-Gamil M, et al. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc Natl Acad Sci U S A. 2011;108:E323–31. doi: 10.1073/pnas.1101892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–26. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 14.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–54. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 15.Martin P, Simon B, Lone YC, Chatel L, Barry R, Inchauspe G, et al. A vector-based minigene vaccine approach results in strong induction of T-cell responses specific of hepatitis C virus. Vaccine. 2008;26:2471–81. doi: 10.1016/j.vaccine.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Dudley ME, Yang JC, Sherry R, Hughes MS, Royal R, Kammula U, et al. Adoptive cell therapy for patients with metastatic melanoma: evaluation of intensive myeloablative chemoradiation preparative regimens. J Clin Oncol. 2008;26:5233–9. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26:332–42. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–41. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 19.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 21.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 24.Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–9. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–68. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 26.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 2011;147:1092–103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Akhmanova A, Dogterom M. Kinesins lead aging microtubules to catastrophe. Cell. 2011;147:966–8. doi: 10.1016/j.cell.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Wang TS. Eukaryotic DNA polymerases. Annual review of biochemistry. 1991;60:513–52. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 29.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annual review of biochemistry. 1998;67:721–51. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 30.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Molecular and cellular biology. 1994;14:923–33. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–46. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhurst MR, Yang JC, Langan RC, Dudley ME, Nathan DA, Feldman SA, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19:620–6. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36:133–51. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, et al. Design and use of conditional MHC class I ligands. Nature medicine. 2006;12:246–51. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 35.Linnemann C, Heemskerk B, Kvistborg P, Kluin RJ, Bolotin DA, Chen X, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nature medicine. 2013;19:1534–41. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi E, Mizukoshi E, Kishi H, Ozawa T, Hamana H, Nagai T, et al. A new cloning and expression system yields and validates TCRs from blood lymphocytes of patients with cancer within 10 days. Nature medicine. 2013;19:1542–6. doi: 10.1038/nm.3358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.