Abstract
The Par polarity proteins play key roles in asymmetric division of Drosophila stem cells; however, whether the same mechanisms control stem cells in mammals is controversial. Although necessary for mammary gland morphogenesis, Par3 is not essential for mammary stem cell function. We discovered that, instead, a previously uncharacterized protein, Par3-Like (Par3L), is vital for mammary gland stem cell maintenance. Par3L function has been mysterious because unlike Par3 it does not interact with atypical protein kinase C or the Par6 polarity protein. We found that Par3L is expressed by multipotent stem cells in the terminal end buds of murine mammary glands. Ablation of Par3L resulted in rapid and profound stem cell loss. Unexpectedly, Par3L, but not Par3, binds to the tumor suppressor protein LKB1 and inhibits its kinase activity. This interaction is key for the function of Par3L in mammary stem cell maintenance. Our data reveal insights into a linkage between cell polarity proteins and stem cell survival, and uncover the first known biological function for Par3L.
The Par3 protein sits at the apex of a signaling network that controls apical/basal polarity and spindle orientation not only in differentiated epithelial cells but also in the Drosophila neuroblast, intestinal, and sensory organ precursor stem cells [1,2]. Loss of spatially organized Par signaling through atypical protein kinase C (aPKC) in these stem cells disrupts asymmetric cell division and perturbs self-renewal and the differentiation of daughter cells. Although the Par proteins are conserved throughout the Metazoa it remains unclear whether their functions have been retained in mammalian stem cells. For example, although the Par3 pathway does appear to play a role in the asymmetric divisions of radial glial progenitors [3], it is not essential for mammary gland regeneration from stem cells [4], and aPKC is completely dispensable for primitive and adult hematopoietic stem cell polarization, activity, and blood formation [5].
While in lower animals the Par proteins and aPKC are each encoded only by single genes, diversification has occurred in the vertebrate lineage, and the various isoforms can have distinct biological functions. For instance, isoforms of Par6 are not interchangeable in zebrafish [6]. In 2002 we cloned a gene (pard3b) closely related to the Par3 gene (pard3) and that expresses a protein we named Par3-like (Par3L) [7]. The function of Par3L has been enigmatic because, unlike Par3, it does not bind to either Par6 or aPKC, but exogenous Par3L can colocalize with these proteins and with Par3 at epithelial tight junctions [7,8]. Nothing has been known about any biological roles for Par3L. It has not been shown to participate in cell polarization or in stem cell functions. Here we identify Par3L as an essential factor for mouse mammary stem cell maintenance, and show that it acts by suppressing the kinase activity of the Par4/LKB1 polarity protein.
RESULTS
Par3L is expressed in cap cells and is required for mammary ductal outgrowth
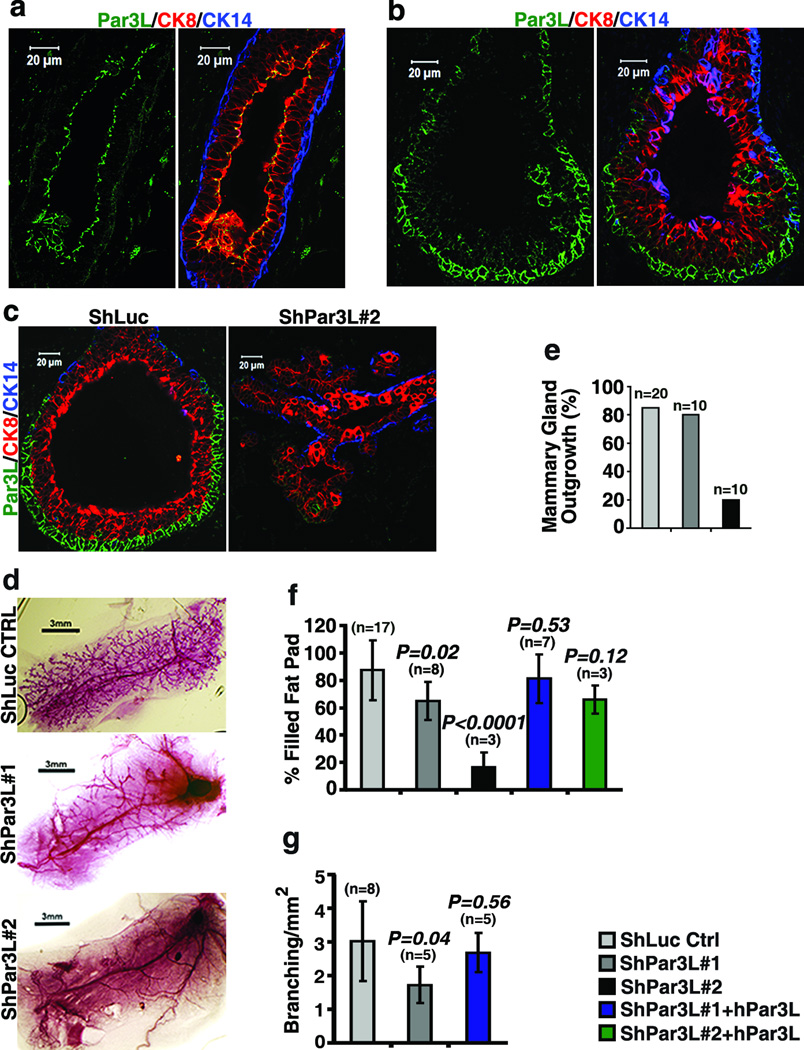
To test for a function of Par3L in mammary gland development we first developed Par3L-specific antibodies (Supplementary Figure 1a, b) and stained sections from mature and developing murine mammary glands. In mature glands Par3L staining was restricted to the tight junctions of luminal epithelial cells (Figure 1a), as predicted from previous studies [8,9]. Strikingly, however, in the developing glands Par3L was detected at the cell cortex in the cap cells of terminal end buds (Figure 1b). Cap cells express s-SHIP and, using a transgenic mouse model (Tg11.5kb-GFP) that expresses GFP from the s-SHP promoter [9], they have been shown to be active, multipotent stem cells that can regenerate a functional mammary gland upon transplantation into cleared fat pads of recipient mice [10]. We refer to these cells as mammary repopulating units (MRUs). They are negative for cytokeratin 14 (CK14) and cytokeratin 8 (CK8), but are CD49f and CD24 positive [10]. Notably, cap cells that express GFP from the s-SHIP promoter were also strongly positive for Par3L, while the majority of the terminal end bud body cells, which lack GFP expression, were negative for Par3L (Supplementary Figure 1c).
Figure 1.
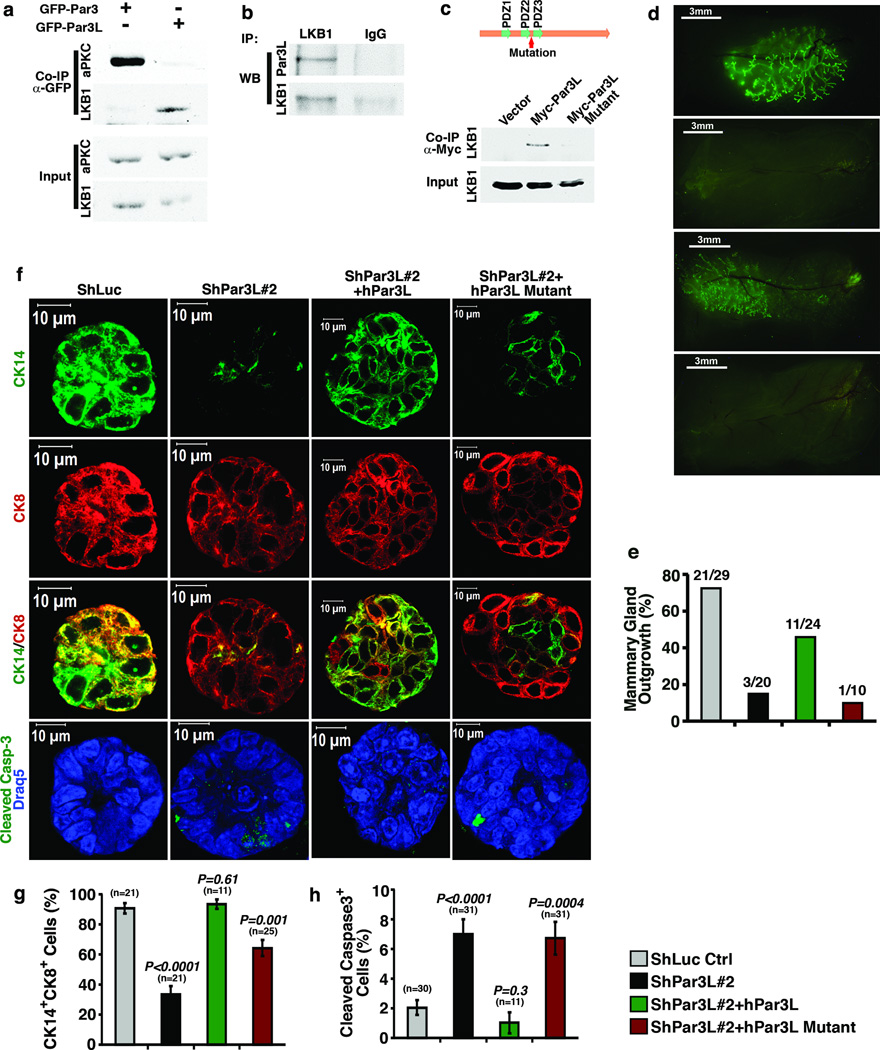
Par3L is required for mammary gland regeneration. (a, b) immuno-staining shows that Par3L is expressed specifically at the apical junctions of luminal epithelial cells (marked with CK8, red) in mature mammary ducts and in cap cells of the terminal end buds of developing mammary glands. (c) Par3L depletion results in abnormal terminal end bud morphology. (d) Carmine alum staining of whole mammary glands shows that Par3L depletion impairs mammary gland regeneration (e) quantification of regeneration, as % of fat pads showing any ductal outgrowth. (f) depletion of Par3L significantly reduces the extent of outgrowth into the fat pads. This phenotype could be reversed by over-expression of human Par3L, which is resistant to the Par3L shRNAs. (g) partial depletion of Par3L by shPar3L#1 significantly reduces mammary ductal tree side branching. This phenotype could be rescued by over-expression of human Par3L. Graph shows mean ± SD. n represents the number of mammary glands examined in each experiment. Statistical significance was calculated for each experimental group compared to the shLuc control group; p values were calculated by the Student t test.
To test the function of Par3L in these mammary gland cells two lentiviral shRNAs were developed to target the murine Par3L, with ~70% knock down by shPar3L#1 and 99% knock down by shPar3L#2 compared to the luciferase shRNA (shLuc) control (Supplementary Figure 1d, e). Par3L was silenced by these two shRNAs in mammary gland stem/progenitor cells, which were subsequently transplanted into clear fat pads for mammary gland regeneration. Whole mount staining of these fat pads showed that shPar3L#2 drastically reduced mammary gland take rate as compared to the controls (Figure 1d, e). Both shRNAs targeting Par3L significantly reduced fat pad filling for those glands that did regenerate (Figure 1d, f). Ductal branching was measured in the fat pads transplanted by mammary cells with shPar3L#1, which was permissive for mammary gland outgrowth. The number of ductal braching points per unit area was significantly reduced (Figure 1d, g). Importantly, these phenotypes were reversed by the expression of human Par3L, which is resistant to shRNA-mediated silencing, demonstrating that the defect is specific to Par3L knockdown rather than to off-target effects of the shRNAs. Since Par3L is expressed at either the cortex of cap cells in developing mammary glands or the tight junction of luminal epithelial cells in mature mammary glands, the phenotype caused by Par3L depletion could be due to defects in either type of cells. To further address this issue we stained mammary gland sections to examine the structure of mammary glands. Silencing of Par3L substantially reduced anti-Par3L antibody staining of mammary glands (Figure 1c). Moreover, the normal structure of terminal end buds was severely disrupted in glands that grew out from mammary stem cells transduced with shPar3L#2. All the terminal end bud cells were CK8 positive and there was no distinct cap cell layer (Figure 1c). This experiment demonstrates that Par3L silencing causes defects in ductal outgrowth and terminal end bud structures, most likely through effects on the cap cell MRUs.
Par3L is essential for mammary stem cell maintenance
To further investigate whether Par3L affects MRUs, we quantified the in vivo regeneration capacity of isolated mammary cells, after implantation into the cleared fat pads of syngeneic recipient mice, using limiting dilution assays [11,12]. Based on positive outgrowths scored for each dilution, the MRU frequency was 1/2,100 for wild type cells, in line with previous results [11,12], but only 1/215,000 for cells expressing shPar3L#2 (Table 1). These data indicate that silencing of Par3L causes a highly significant ~100-fold decrease in functional MRUs. By contrast, no significant decrease in MRUs was observed upon depletion of the closely related Par3 gene [4].
Table 1.
Limiting dilution mammary regeneration assay
| Experiment | Cells/Injection into fat pad |
Outgrowths (number/total) |
Stem cell frequency (Upper and lower limit) |
|---|---|---|---|
| shLuc control | 1,000 | 7/16 | |
| 2,000 | 4/6 | ||
| 4,000 | 6/6 | 1/2,062 | |
| 8,000 | 5/6 | (1/1,205 – 1/3,400) | |
| 16,000 | 6/6 | ||
| shPar3L#2 | 16,000 | 1/6 | |
| 32,000 | 2/6 | ||
| 64,000 | 1/6 | 1/214,960 | |
| 128,000 | 2/6 | (1/112,340 – 1/411,330) | |
| 256,000 | 4/6 | ||
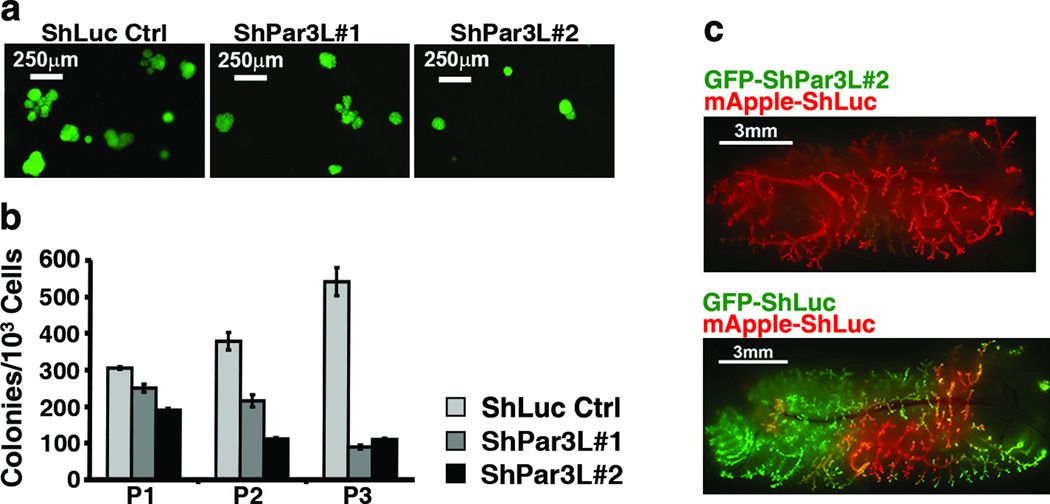
As an in vitro assay for stem cell survival we used a serial colony-forming assay, a schematic for which is shown in Supplementary Figure 2a. Single mammary cell suspensions were prepared from mammary cells transduced with lentivector shRNAs against Par3L or shLuc control, and seeded into Matrigel. Colonies formed (Figure 2a) after one week and were scored, then trypsinized and replated as single cells into fresh Matrigel. The number of colony forming cells (CFCs) increases in the second and third passages of the wild type control (Figure 2b). Importantly, this serial passage procedure strongly enriches for MRUs, as shown by the very high efficiency with which the control cells from tertiary colonies could repopulate fat pads in vivo (Supplementary Figure 2b). When Par3L was depleted, however, the number of CFCs was significantly lower, even in the primary culture, and dropped substantially in subsequent passages, for two independent shRNAs (Figure 2a,b). These results are consistent with a rapid depletion of functional stem cells in the absence of Par3L expression.
Figure 2.
Par3L is essential for mammary gland stem cell maintenance. (a) Lentivirus-transduced (GFP+) colonies grown from single cells in Matrigel, first passage. (b) serial colony-forming assay shows that Par3L depletion decreases the number of colony forming cells. Graph shows mean ± SD for 3 experimental replicates (independent transductions). (c) competition assay demonstrates that Par3L depletion impairs MRU maintenance in a cell autonomous manner. Equal numbers of red (control) and green (shPar3, top panel, or control, bottom panel) were mixed, injected into cleared fat pads and allowed to regenerate for 6 weeks.
We reasoned that Par3L might be required cell-autonomously for MRU survival, or might alternatively support the stem cell micro-environment in a cell non-autonomous fashion. To distinguish these two possibilities, we used a competition assay to compare the efficiencies of ductal outgrowth by shLuc control MRUs versus Par3L-depleted cells. Cells expressing the shPar3L#2 were marked green with GFP, and the shLuc were marked red with mApple. Equal numbers of cells were then mixed and implanted into cleared fat pads, as diagrammed in Supplementary Figure 2b. If Par3L were necessary for niche maintenance, or for secretion of factors that support stem cells, then one would predict that green, Par3L-depleted cells would be unable to form a niche and negatively impact the outgrowth of neighboring control red cells. Conversely, the wild type cells should provide a functional niche for green Par3L-depleted stem cells, enabling their outgrowth. Reproducibly, however, only the red control cells populated the entire fat pad, and the green Par3L-depleted MRUs contributed negligibly to ductal outgrowth (Figure 2c). When, instead, an equal mix of red and green control cells was implanted into the fat pads, both populations were able to contribute efficiently to the regenerated ductal trees. We conclude from these data that Par3L is essential for murine mammary gland stem cell function, and that it functions cell autonomously.
Depletion of Par3 promotes apoptosis and differentiation into CK8+ cells
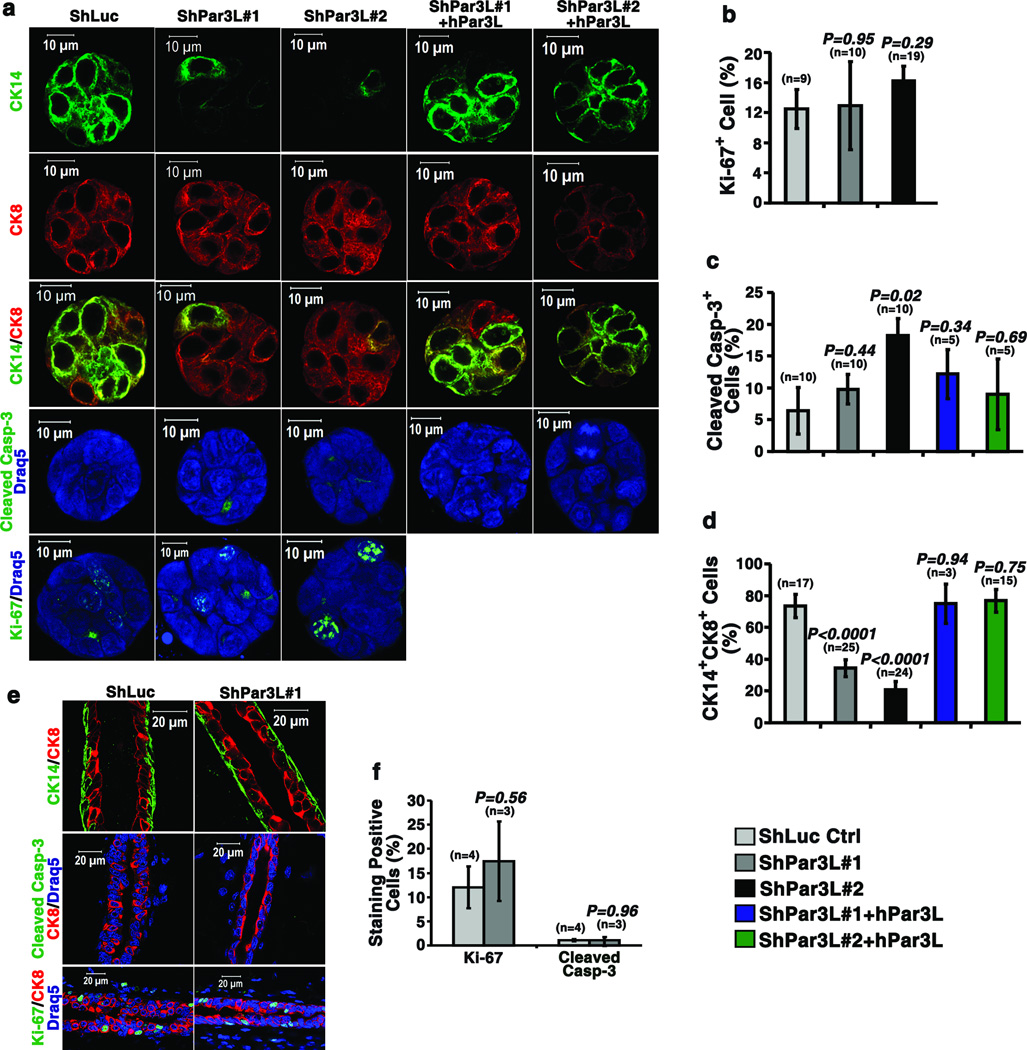
Some MRUs in young mice are actively cycling [9]. Therefore, a decrease in relative MRU number could arise from either reduced proliferation or increased cell death. To distinguish these possibilities we stained the colonies grown in Matrigel for Ki67 or cleaved Caspase-3, and for CK8 as a luminal epithelial marker and CK14 as a myoepithelial marker. Proliferation, as marked by Ki67+ cells, was not affected by expression of Par3L shRNAs (Figure 3a, b), but cleaved Caspase-3 staining was significantly increased with shPar3L#2, and this was reduced to control levels by hPar3L expression (Figure 3a, c). Moreover, the number of basal-like (CK14+) and dual-positive (CK8+/CK14+) cells, which are possible progenitors, was substantially reduced by both Par3L shRNAs, an effect that was efficiently reversed by hPar3L expression (Figure 3a, d). In contrast, we could detect no difference in either Ki67 or Caspase-3 staining for mature ducts arising from cells expressing shPar3L#1 as compared to shLuc controls, and there were no detectable CK8+/CK14+ cells (Figure 3e, f). These data suggest that loss of Par3L specifically depletes MRUs by promoting apoptosis and/or preventing their self-renewal, but has no effect on the survival of differentiated mammary cells.
Figure 3.
Loss of Par3L causes mammary stem cell loss through apoptosis and/or failure of self-renewal. (a) representative images of the staining of tertiary colonies. Par3L depletion decreases the percentage of CK14+CK8+ double positive cells, increased cleave Caspase-3+ cells, but does not change the Ki67+ cell ratio. (b to d) Quantification of Ki67+ cells,(b), cleaved Caspase-3+ cells (c), and CK14+CK8+ cells, (d). n represents the number of colonies scored. Original source data for (c) is provided in Supplementary Table 1. (e) Staining of mature mammary gland ducts regenerated from mammary cells expressing shPar3L#1 or shLuc control. The ducts have normal myoepithelial and luminal epithelial organization. Partial depletion of Par3L has no effects on cleaved Caspase-3 or Ki67 staining in the ducts. (f) Quantification of Ki67 and cleaved Caspase-3 staining in the mammary gland ducts. Graph shows mean ± SEM. n represents the number of mammary glands that were scored. Statistical significance was calculated for each experimental group compared to the shLuc control group. p values were calculated by Student t test.
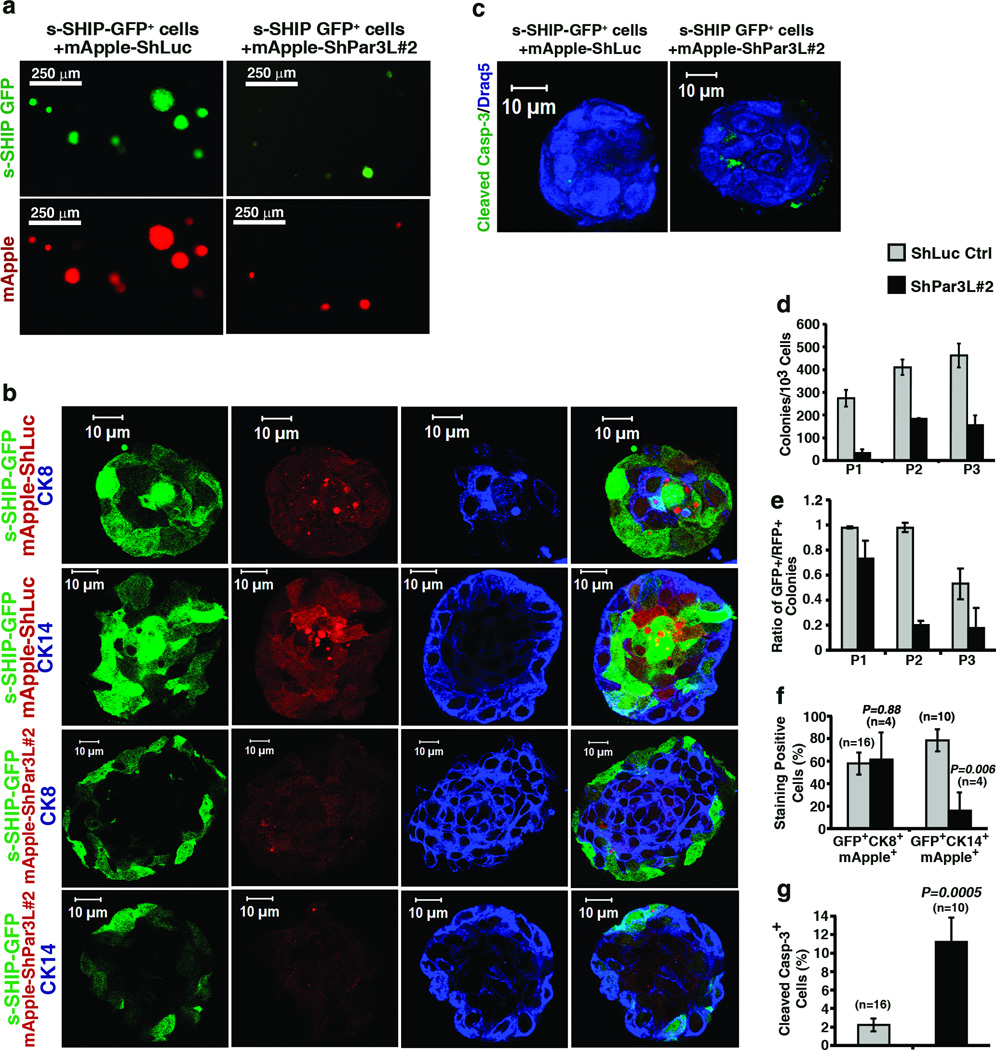
Par3L expression is not entirely specific to cap cells, since it was also detectable at the tight junctions of luminal cells in mature ducts (Figure 1a). Therefore, to test more definitively for a stem cell-specific function of Par3L, we used FACS to purify GFP+ MRUs to homogeneity from the Tg11.5kb-GFP mice (Supplementary Figure 4 a,b). The GFP+ cells were transduced with lentivectors expressing mApple plus shLuc (control) or shPar3L#2 hairpin RNAs. The cells were then grown into colonies in Matrigel, and passaged as described above (Supplementary Figure 2). Most of the control colonies continued to express GFP, and were positive for the lentiviral marker (Figure 4 a,d). However, very few cells transduced with shPar3L#2 formed colonies (Figure 4 a,d). Additionally, those colonies that grew out were generally smaller, and less positive for mApple (Figure 4a, b), suggesting that there was selection against cells containing multiple copies of the shRNA/mApple vector. The effect of Par3L depletion is even more striking when GFP+ colonies are scored. After the third passage >50% of the colonies in the shLuc control group were GFP+, while only 17% of surviving colonies in the shPar3L#2 group were GFP+ (Figure 4e). This result demonstrates that Par3L depletion causes rapid loss of GFP+ mammary stem cells. We next focused on the GFP+mApple+ colonies to understand what effects Par3L depletion might have on these GFP+ MRUs. In surviving colonies from cells transduced with shPar3L#2, over 60% cells were CK8+, which is not different than the shLuc control. However, only 16% cells were CK14+ in the shPar3L#2 group, a significant decrease compared to 79% CK14+ cells for the shLuc control (Figure 4b, f). This result is consistent with our earlier observation that Par3L depletion causes loss of basal CK14+ or CK8+CK14+ dual-positive cells (Figure 3a, d). Moreover, the surviving Par3L-depleted colonies had a substantially higher apoptotic index (Figure 4c, f). We conclude that Par3L possesses a MRU-specific function that is essential for stem cell maintenance.
Figure 4.
Par3L is necessary for colony forming activity by s-SHIP GFP+ cells. GFP+ cells from 6 week-old s-SHIP GFP transgenic mice were FACS sorted to homogeneity and infected by mApple-shLuc control or mApple-shPar3L#2 virus. (a, d) serial colony-forming assay shows that Par3L depletion decreases the number of colony forming cells. All mApple-positive colonies were counted and presented in the bar graph. Graph represents mean ± SD for 3 technical replicates (independent transductions of GFP+ cells from a single mouse). Original source data provided in Supplementary Table 1. (b) representative images of the staining of tertiary colonies for CK8, CK14. (c) staining for cleaved Caspase-3. (e) the ratio of GFP+ colonies over mApple positive colonies indicates shPar3L#2 over-expression causes GFP+ stem cell loss. Graph represents mean ± SD for 3 technical replicates (independent transductions of GFP+ cells from a single mouse). (f) Quantification of GFP, mApple, and CK8 triple-positive or GFP, mApple, and CK14 triple-positive cells. Par3L depletion significantly decreases the number of CK14 positive cells but not CK8 positive cells. Error bars represent mean ± SEM (n shown on figure). Statistical significance was calculated for each experimental group compared to the shLuc control group. p values were calculated by Student t test. (g) Quantification of cleaved Caspase-3+ cells. Par3L depletion significantly increased cleaved Caspase-3+ cells. Graph shows mean ± SEM. n represents the number of colonies that were scored. Statistical significance was calculated for ShPar3L#2 compared to the shLuc control group. p values were calculated by Student t test.
Par3L, but not Par3, associates with LKB1
How might Par3L act to maintain MRU survival? The Par3L protein does not bind to aPKC or Par6, so we tested other polarity proteins, and identified an unanticipated interaction with LKB1, the mammalian homologue of the C. elegans Par4 polarity protein (Figure 5a). The endogenous association of the two proteins was confirmed using lysates from mouse kidneys, which express Par3L at relatively high levels (Figure 5b). This interaction is specific, because LKB1 does not bind the Par3 protein (which was able to bind aPKC, Figure 5a). We next identified the region of Par3L required for LKB1 binding. As shown in Supplementary Figure 5 and Figure 5c, the interaction involves the PDZ2-PDZ3 region of Par3L, and specifically requires a small region lying between the 2 PDZ domains, of no known function, and which shows no sequence similarity to the same region in Par3 (or to any other sequence in the Genbank database). Replacement of the Par3L sequence in this region with that from Par3 abolished LKB1 binding by full-length hPar3L (Figure 5c). Therefore, this short Par3L sequence is required for LKB1 binding. To determine if the interaction has biological relevance we asked if the mutant could rescue mammary outgrowth in the fat pad repopulation assay. Importantly, although the wild type hPar3L efficiently rescued ductal outgrowth, the LKB1 binding-defective mutant did not (Figure 5d, e). Moreover, only the wild type hPar3L rescued colony formation in vitro by Par3L-depleted mammary cells, reducing cleaved Caspase-3 staining and increasing to control levels the number of CK14+/CK8+ dual-positive cells present in each colony. The mutant Par3L was unable to suppress apoptosis and only partially reversed the effects on CK14+/CK8+ abundance (Figure 5 f – h). Together, these data strongly suggest that LKB1 binding to Par3L is crucial for the maintenance of murine mammary stem cells.
Figure 5.
Par3L maintains mammary stem cells through interaction with LKB1. (a) Par3L interacts with LKB1 but not aPKC; while Par3 interacts with aPKC, but not LKB1. About 2% of LKB1 was captured by Par3L while only ~0.02% LKB1 precipitated with Par3. (b) The endogenous interaction between Par3L and LKB1 was confirmed by co-immunoprecipitation using mouse kidney lysates, which express relatively high levels of Par3L. (c) The region on Par3L required for LKB1 binding was mapped between PDZ2 and PDZ3 domains. Replacement of this region of Par3L with the corresponding region of Par3 disrupts the interaction between LKB1 and Par3L. All full scan images are presented in supplementary figure 6. (d, e) Mammary gland regeneration assay. Wild type human Par3L can rescue mammary gland regeneration from Par3L-depleted mammary cells, while the LKB1 binding-deficient Par3L mutant is unable to do so. (f) representative images of the staining of tertiary colonies for CK8, CK14, and cleaved Caspase-3. (g, h) quantification of CK14+CK8+ dual-positive (g) and cleaved Caspase-3+ (h) cells in in vitro serial colony forming assay. Wild type human Par3L, but not the mutant Par3L, was able to reverse the Par3L depletion effects. Graph shows mean ± SEM. n represents the number of colonies that were scored. Statistical significance was calculated for each experimental group compared to the shLuc control group. p values were calculated by Student t test.
Par3L suppresses LKB1 kinase activity
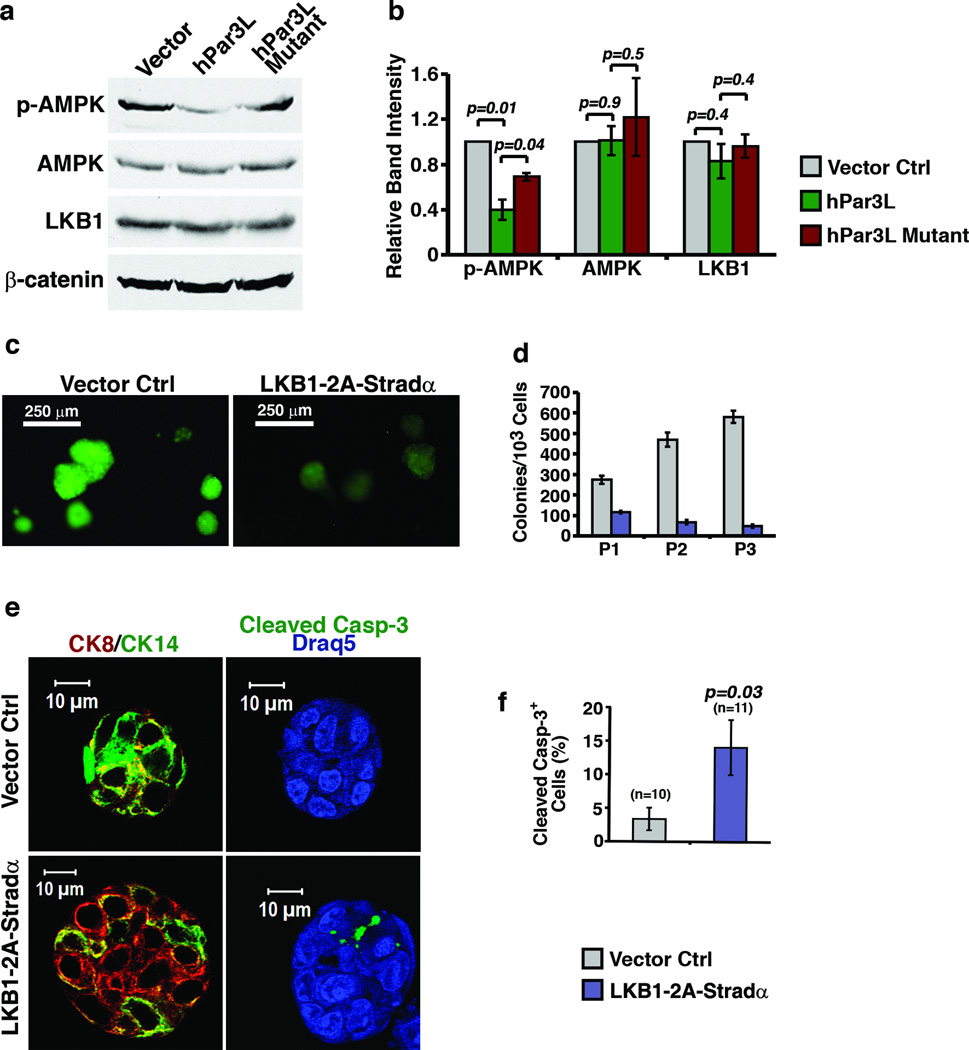
We next tested whether Par3L modulates LKB1 activity. To do so we created stably transduced HEK293T cells expressing either wild type or the mutant Par3L, and probed lysates for phosphorylated AMPK, a downstream target of LKB1. Notably, the expression of wild type Par3L in HEK293T cells inhibited AMPK phosphorylation, with no change in expression levels of either LKB1 or AMPK, while the mutant had significantly less inhibitory effect on phospho-AMPK (Figure 6a, b). We were unable to test this effect in mammary stem cells because we could not purify them in sufficient numbers for the biochemical experiments. Instead, we performed a serial colony-forming assay to determine if an increase in LKB1 activity in the stem cells would alter their behavior. Strikingly, co-expression of LKB1 and its activator, Stradα, strongly suppressed colony outgrowth in the serial colony-forming assay. The few colonies that grew in the presence of LKB1+Stradα had a significantly higher apoptotic index, as measured by staining for cleaved Caspase-3 (Figure 6e, f). We conclude that Par3L can inhibit LKB1, and that uncontrolled LKB1 activity severely compromises mammary stem cell survival.
Figure 6.
Par3L maintains mammary stem cells by suppressing LKB1 activity. (a) Par3L inhibits LKB1 activity in HEK293T cells. Wild type or mutant Par3L was over-expressed in HEK293T cells and phospho-AMPK was blotted as an indicator for LKB1 activity. The full scan images are presented in supplementary figure 6. (b) Wild type Par3L reduced the phospho-AMPK level to about 40% of the control level, while mutant Par3L reduced phospho-AMPK level to about 70% of the control level. Graph shows means and ± SEM for 3 independent experiments.. p values were calculated by paired Student t test. (c) mammary cells from female C3H mice were infected by vector control or LKB1-2A-Stradα polycistronic expression virus and subjected to the serial colony-forming assay. (d) LKB1 over-expression decreases the number of colony forming cells. Graph shows mean ± SD for 3 experimental replicates (independent transductions). Original source data provided in Supplementary Table 1. (e) representative images of the staining of tertiary colonies for CK8, CK14, and cleaved Caspase-3. (f) Quantification of cleaved Caspase-3+ cells in the in vitro serial colony-forming assay. LKB1 over-expression significantly increases cleaved Caspase-3+ cells comparing to the vector control group. Graphs show mean ± SEM. n represents the number of colonies that were scored. Statistical significance was calculated for each experimental group compared to the vector control group. p values were calculated by unpaired Student t test.
DISCUSSION
Taken together, these data reveal the first defined biological function for Par3L, a previously uncharacterized protein that is closely related to the Par3 polarity protein [7,8]. Both proteins possess 3 PDZ domains and a conserved N-terminus that in Par3 homodimerizes. Par3 also associates with other components of the polarity machinery, including Par6 and aPKC, and functions in many cellular contexts to create spatial asymmetries [2]. Pertinently, the Par polarity machinery is required for asymmetric cell divisions of many stem cells, including the C. elegans zygote, Drosophila neuroblasts, and mammalian radial glial progenitors [1–3]. However, it is dispensable in other situations, including hematopoietic stem cell and mammary stem cell maintenance [4, 5]. Whether the Par3L isoform, which does not bind to Par6 or aPKC, plays any role either in cell polarity or in stem cell function has not been previously addressed. We now show that Par3L is expressed in one important subset of mammary stem cells, the cap cells of the terminal end buds [9], and is essential for their maintenance. Moreover, Par3L but not Par3 binds to a distinct polarity protein called Par4 or LKB1.
We propose that Par3L maintains mammary stem cells through suppression of LKB1 kinase activity, and that loss of this restraint results in a failure of self-renewal and increased apoptosis. This model is in accord with known functions of LKB1, which is capable of driving p53-dependent apoptosis when over-expressed [13,14], and which has been recently identified as an essential factor in a Caspase-independent cell death and extrusion pathway in C. elegan [15]. LKB1 is also required for the maintenance of hematopoietic stem cell dormancy [16–18]. Consistent with our model, the conditional deletion of LKB1 in mammary glands promotes ductal hyperbranching, which is the opposite of the phenotype caused by loss of Par3L, and promotes tumorigenesis [19]. It will be of interest to determine if Par3L functions to restrain LKB1 activity in other mammalian stem cell populations and perhaps acts as a tumor promoter through suppression of LKB1 activity. LKB1 is a master kinase that regulates 14 downstream protein kinases, and an important question for the future concerns which of these kinases mediates the effects of LKB1 on mammary stem cell maintenance. We note that SIK1 couples LKB1 to p53-dependent apoptosis triggered by cell detachment [13]. Finally, we speculate that in Drosophila - which does not contain a Par3L gene – the negative regulation of LKB1 is occupied by Bazooka, the fly homologue of Par3.
Supplementary Material
(a, b) Specificity of the Par3L antibody. Par3L antibody recognizes over-expressed human YFP-tagged Par3L in HEK293T cells and endogenous murine Par3L in mouse kidney lysates. (c) Par3L staining co-localizes with GFP staining in the mammary glands of 6-week old s-Ship-GFP transgenic mice. (d, e) Immunoblot to test the efficiency of shPar3L hairpin RNAs. HEK293T cells over-expressing mouse Par3L gene were infected by Par3L hairpin virus. shPar3L#1 reduced Par3L expression by 70%, and shPar3L#2 completely depleted Par3L protein. Graph represents mean ± SEM for n= 3 independent experiments. All full scan images of western blots can be found in supplementary figure 6.
(a) Scheme of serial colony-forming assay. Mammary gland cells were isolated from adult female C3H or s-SHIP-GFP transgenic mice. Cells were cultured for two days in suspension after virus infection to allow recovery. Before Matrigel culture cells were dissociated in fresh 0.05% trypsin for 45 min at 37°C to obtained single cells. Three hundred to 1,000 cells in 5µl of medium were mixed with 45µl growth factor reduced phenol-red free Matrigel. The mixture were laid on the bottom of ultra-low attachment 24-well cluster (Corning) as a drop and solidified at 37C for 30 min. The solidified Matrigel drops were covered by 500µl of mammosphere medium. The cultures were fed every other day. Colonies were scored and imaged using an Olympus SZX16 microscope. For passage, colonies were recovered in 0.5 ml Cell Recovery Solution (BD) at 4°C for 30 min. Single cells were obtained by trypsinization and plated in Matrigel culture as described previously. After the third passage, colonies were analyzed by immuno-staining in vitro or by transplantation. (b) Summary of limiting dilution assay using control tertiary colonies and a representative image of a GFP+ mammary gland regenerated in the limiting dilution assay from these control cells. Mammary gland cells expressing shLuc were used for the serial colony-forming assay. After the third passage, colonies of the indicated numbers were injected into cleared fat pad for the limiting dilution assay. Mammary gland regeneration was tested in 8 cleared mammary fat pads. (c) Scheme of competition assay. Mammary gland cells were isolated from adult female C3H mice. The cells were infected with mApple-ShLuc virus, EGFP-ShLuc virus or EGFP-ShPar3L#2 virus. After 2 days recovery in suspension culture, 50,000 mApple-ShLuc virus infected cells were combined with an equal number of EGFP-ShLuc virus infected cells or EGFP-ShPar3L#2 virus infected cells and injected into cleared fat pads of 3 week old C3H mice. Mammary glands were scored for outgrowth after 6 weeks.
Representative images of mature mammary ducts stained for CK8, CK14, or cleaved Caspase-3. Those mammary ducts that grew out from cells depleted of Par3L show a normal morphology with a single layer of luminal epithelial cells surrounded by a single layer of myoepithelial cells. The number of cleaved Caspase-3+ cells is very low and not significantly different from the ShLuc control.
(a) Scheme of mammary stem cell colony-forming assay. Mammary gland cells were isolated from 6-week old female s-SHIP-GFP transgenic mice. GFP+ mammary gland stem cells were purified by FACS sorting as described in METHODS. The sorted cells were then infected with mApple-ShLuc virus or mApple-ShPar3L#2 virus. After recovery for 2 days in suspension culture the cells were plated in Matrigel. (b) Scheme of FACS for the GFP+ mammary gland stem cells. Mammary gland cells from s-SHIP-GFP transgenic mice were stained by the following lineage antibodies: PE-CD31 (MEC13.3, BD), PE-CD45 (30-F11, BD), and PE-Ter119 (TER-119, BD). 7AAD (BD) was added before sorting. GFP+PE-7AAD- single cells were collected for the experiments.
The region between PDZ2 and PDZ3 on Par3L protein is necessary for the interaction between Par3L and LKB1. (a) schematic view of the Par3L truncation mutants used for co-immunoprecipitation with LKB1. (b) co-precipitation of LKB1 and Par3L truncation mutants group #1. LKB1 binding to Par3L protein required the region from PDZ2 to PDZ3 domain. (c) co-precipitation of LKB1 and Par3L truncation mutants group #2. LKB1 was unable to bind to any one of the 3 isolated PDZ domains. (d) co-precipitation of LKB1 and Par3L truncation mutants group #3. LKB1 binding to Par3L protein requires PDZ domains 2 and 3 plus the intervening region. All experiments were conducted 3× independently. All full scan images of western blots can be found in supplementary figure 6.
Original data for the immunoblots presented in the other figures.
Acknowledgements
This study was supported by NIH grant GM070902 (I.G.M.) and DoD postdoctoral fellowship W81XWH-11-1-0083 (Y.H.)
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. Y.H. performed the experiments; Y.H. and I.G.M. analyzed the data and prepared the manuscript.
The authors declare no competing financial interests.
References
- 1.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCaffrey LM, Macara IG. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. doi: 10.1101/gad.1795909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sengupta A, Duran A, Ishikawa E, Florian MC, Dunn SK, et al. Atypical protein kinase C (aPKCzeta and aPKClambda) is dispensable for mammalian hematopoietic stem cell activity and blood formation. Proc Natl Acad Sci U S A. 2011;108:9957–9962. doi: 10.1073/pnas.1103132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munson C, Huisken J, Bit-Avragim N, Kuo T, Dong PD, et al. Regulation of neurocoel morphogenesis by Pard6 gamma b. Dev Biol. 2008;324:41–54. doi: 10.1016/j.ydbio.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao L, Macara IG, Joberty G. Multiple splice variants of Par3 and of a novel related gene, Par3L, produce proteins with different binding properties. Gene. 2002;294:99–107. doi: 10.1016/s0378-1119(02)00681-9. [DOI] [PubMed] [Google Scholar]
- 8.Kohjima M, Noda Y, Takeya R, Saito N, Takeuchi K, Sumimoto H. PAR3beta, a novel homologue of the cell polarity protein PAR3, localizes to tight junctions. Biochem Biophys Res Commun. 2002;299:641–646. doi: 10.1016/s0006-291x(02)02698-0. [DOI] [PubMed] [Google Scholar]
- 9.Rohrschneider LR, Custodio JM, Anderson TA, Miller CP, Gu H. The intron 5/6 promoter region of the ship1 gene regulates expression in stem/progenitor cells of the mouse embryo. Dev Biol. 2005;283:503–521. doi: 10.1016/j.ydbio.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 10.Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev. 2010;24:1882–1892. doi: 10.1101/gad.1932810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 12.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H, Liu P, Wang ZC, Zou L, Santiago S, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2:ra35. doi: 10.1126/scisignal.2000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karuman P, Gozani O, Odze RD, Zhou XC, Zhu H, et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001;7:1307–1319. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 15.Denning DP, Hatch V, Horvitz HR. Programmed elimination of cells by caspase-independent cell extrusion in C. elegans. Nature. 2012;488:226–230. doi: 10.1038/nature11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gan B, Hu J, Jiang S, Liu Y, Sahin E, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partanen JI, Tervonen TA, Myllynen M, Lind E, Imai M, et al. Tumor suppressor function of Liver kinase B1 (Lkb1) is linked to regulation of epithelial integrity. Proc Natl Acad Sci U S A. 2012;109:E388–E397. doi: 10.1073/pnas.1120421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a, b) Specificity of the Par3L antibody. Par3L antibody recognizes over-expressed human YFP-tagged Par3L in HEK293T cells and endogenous murine Par3L in mouse kidney lysates. (c) Par3L staining co-localizes with GFP staining in the mammary glands of 6-week old s-Ship-GFP transgenic mice. (d, e) Immunoblot to test the efficiency of shPar3L hairpin RNAs. HEK293T cells over-expressing mouse Par3L gene were infected by Par3L hairpin virus. shPar3L#1 reduced Par3L expression by 70%, and shPar3L#2 completely depleted Par3L protein. Graph represents mean ± SEM for n= 3 independent experiments. All full scan images of western blots can be found in supplementary figure 6.
(a) Scheme of serial colony-forming assay. Mammary gland cells were isolated from adult female C3H or s-SHIP-GFP transgenic mice. Cells were cultured for two days in suspension after virus infection to allow recovery. Before Matrigel culture cells were dissociated in fresh 0.05% trypsin for 45 min at 37°C to obtained single cells. Three hundred to 1,000 cells in 5µl of medium were mixed with 45µl growth factor reduced phenol-red free Matrigel. The mixture were laid on the bottom of ultra-low attachment 24-well cluster (Corning) as a drop and solidified at 37C for 30 min. The solidified Matrigel drops were covered by 500µl of mammosphere medium. The cultures were fed every other day. Colonies were scored and imaged using an Olympus SZX16 microscope. For passage, colonies were recovered in 0.5 ml Cell Recovery Solution (BD) at 4°C for 30 min. Single cells were obtained by trypsinization and plated in Matrigel culture as described previously. After the third passage, colonies were analyzed by immuno-staining in vitro or by transplantation. (b) Summary of limiting dilution assay using control tertiary colonies and a representative image of a GFP+ mammary gland regenerated in the limiting dilution assay from these control cells. Mammary gland cells expressing shLuc were used for the serial colony-forming assay. After the third passage, colonies of the indicated numbers were injected into cleared fat pad for the limiting dilution assay. Mammary gland regeneration was tested in 8 cleared mammary fat pads. (c) Scheme of competition assay. Mammary gland cells were isolated from adult female C3H mice. The cells were infected with mApple-ShLuc virus, EGFP-ShLuc virus or EGFP-ShPar3L#2 virus. After 2 days recovery in suspension culture, 50,000 mApple-ShLuc virus infected cells were combined with an equal number of EGFP-ShLuc virus infected cells or EGFP-ShPar3L#2 virus infected cells and injected into cleared fat pads of 3 week old C3H mice. Mammary glands were scored for outgrowth after 6 weeks.
Representative images of mature mammary ducts stained for CK8, CK14, or cleaved Caspase-3. Those mammary ducts that grew out from cells depleted of Par3L show a normal morphology with a single layer of luminal epithelial cells surrounded by a single layer of myoepithelial cells. The number of cleaved Caspase-3+ cells is very low and not significantly different from the ShLuc control.
(a) Scheme of mammary stem cell colony-forming assay. Mammary gland cells were isolated from 6-week old female s-SHIP-GFP transgenic mice. GFP+ mammary gland stem cells were purified by FACS sorting as described in METHODS. The sorted cells were then infected with mApple-ShLuc virus or mApple-ShPar3L#2 virus. After recovery for 2 days in suspension culture the cells were plated in Matrigel. (b) Scheme of FACS for the GFP+ mammary gland stem cells. Mammary gland cells from s-SHIP-GFP transgenic mice were stained by the following lineage antibodies: PE-CD31 (MEC13.3, BD), PE-CD45 (30-F11, BD), and PE-Ter119 (TER-119, BD). 7AAD (BD) was added before sorting. GFP+PE-7AAD- single cells were collected for the experiments.
The region between PDZ2 and PDZ3 on Par3L protein is necessary for the interaction between Par3L and LKB1. (a) schematic view of the Par3L truncation mutants used for co-immunoprecipitation with LKB1. (b) co-precipitation of LKB1 and Par3L truncation mutants group #1. LKB1 binding to Par3L protein required the region from PDZ2 to PDZ3 domain. (c) co-precipitation of LKB1 and Par3L truncation mutants group #2. LKB1 was unable to bind to any one of the 3 isolated PDZ domains. (d) co-precipitation of LKB1 and Par3L truncation mutants group #3. LKB1 binding to Par3L protein requires PDZ domains 2 and 3 plus the intervening region. All experiments were conducted 3× independently. All full scan images of western blots can be found in supplementary figure 6.
Original data for the immunoblots presented in the other figures.