Abstract
Oncogenic rearrangements of the TFE3 transcription factor gene are found in two distinct human cancers. These include ASPSCR1–TFE3 in all cases of alveolar soft part sarcoma (ASPS) and ASPSCR1–TFE3, PRCC-TFE3, SFPQ-TFE3 and others in a subset of paediatric and adult RCCs. Here we examined the functional properties of the ASPSCR1–TFE3 fusion oncoprotein, defined its target promoters on a genome-wide basis and performed a high-throughput RNA interference screen to identify which of its transcriptional targets contribute to cancer cell proliferation. We first confirmed that ASPSCR1–TFE3 has a predominantly nuclear localization and functions as a stronger transactivator than native TFE3. Genome-wide location analysis performed on the FU-UR-1 cell line, which expresses endogenous ASPSCR1–TFE3, identified 2193 genes bound by ASPSCR1–TFE3. Integration of these data with expression profiles of ASPS tumour samples and inducible cell lines expressing ASPSCR1–TFE3 defined a subset of 332 genes as putative up-regulated direct targets of ASPSCR1–TFE3, including MET (a previously known target gene) and 64 genes as down-regulated targets of ASPSCR1–TFE3. As validation of this approach to identify genuine ASPSCR1–TFE3 target genes, two up-regulated genes bound by ASPSCR1–TFE3, CYP17A1 and UPP1, were shown by multiple lines of evidence to be direct, endogenous targets of transactivation by ASPSCR1–TFE3. As the results indicated that ASPSCR1–TFE3 functions predominantly as a strong transcriptional activator, we hypothesized that a subset of its up-regulated direct targets mediate its oncogenic properties. We therefore chose 130 of these up-regulated direct target genes to study in high-throughput RNAi screens, using FU-UR-1 cells. In addition to MET, we provide evidence that 11 other ASPSCR1–TFE3 target genes contribute to the growth of ASPSCR1–TFE3-positive cells. Our data suggest new therapeutic possibilities for cancers driven by TFE3 fusions. More generally, this work establishes a combined integrated genomics/functional genomics strategy to dissect the biology of oncogenic, chimeric transcription factors.
Keywords: ASPSCR1, TFE3, CYP17A1, uridine phosphorylase, NAMPT, alveolar soft part sarcoma, renal carcinoma, chromosomal translocation
Introduction
The ASPSCR1–TFE3 chimeric transcription factor (TF), arising from a t(X;17)(p11.2;q25), was first identified in alveolar soft part sarcoma (ASPS) [1]. Subsequently, the same fusion was recognized in a subset of renal cell carcinomas (RCC) [2]. ASPS usually affects adolescents and young adults, with frequent metastases to the brain or lungs. Treatment options for this tumour are limited, given the disappointing experience with conventional chemotherapy and radiation therapy [3]. RCCs characterized by the ASPSCR1–TFE3 fusion are relatively over-represented in younger patients with RCC and they tend to present at more advanced stages [2,4]. Notably, the only generally available human cancer cell line endogenously expressing ASPSCR1–TFE3 is derived from such a kidney tumour (FU-UR-1) [5].
Transcription factor E3 (TFE3), along with TFEB, TFEC and MITF, forms the microphthalmia-TFE (MiT) subfamily of basic helix–loop–helix leucine zipper (bHLH-LZ) TFs [6,7] and binds the CANNTG motif recognized by all members of this group [8,9]. There are two forms of the ASPSCR1–TFE3 fusion, the type 2 variant including an additional TFE3 exon (see Supplementary material, Figure S1) [1]. Importantly, aside from ASPSCR1–TFE3 , TFE3 is also rearranged in several other oncogenic fusions in RCCs, including PRCC–TFE3 , SFPQ–TFE3 (a.k.a. PSF–TFE3 ), NONO–TFE3 and CLTC–TFE3 [10]. The involvement of TFE3 in five different gene fusions in RCCs (including ASPSCR1–TFE3 ) is consistent with a central role for TFE3-related transcriptional deregulation in these tumours. These fusions are all structurally similar, insofar as all contain the C-terminal portion of TFE3, including the TFE3 DNA-binding domain and nuclear localization signal. Native alveolar soft part sarcoma chromosome region candidate 1 (ASPSCR1, a.k.a. ASPL) is involved in intracellular regulation of the glucose transporter GLUT4, as established by studies of its mouse homologue, Aspscr1 [a.k.a. Tug (Tether containing a UBX domain for GLUT4)] [11–14].
We have reported on the central role of the MET receptor tyrosine kinase in TFE3 translocation tumours, both ASPS and RCC [15]. MET was found to be up-regulated in these tumours, due to direct transcriptional activation by TFE3 fusion oncoproteins, and this was associated with sensitivity to a MET kinase inhibitor [15]. This study supported the notion that candidate therapeutic targets may emerge from a more comprehensive understanding of the transcriptional target repertoire of these chimeric TFs. Here, we describe an integrative genomic analysis of expression profiles and genome-wide location analysis, followed by a functional genomics screen to characterize ASPSCR1–TFE3 target genes vital to its cellular growth effects.
Materials and methods
Cell lines
The following cell lines were used: 293 T; Cos-7; HeLa; MCF-7; and FU-UR-1 (gift of Dr M Ishiguro, Fukuoka University School of Medicine, Japan [5]).
Human promoter microarray analysis
DNA was hybridized for 40 h at 65 °C to the Agilent Human Promoter Array (Agilent Technologies). The probes represented sequences ranging from –5.5 to +2.5 kb within each promoter region and were spaced approximately every 195 bp. DNA labelling, array hybridization and scanning were performed at the Memorial Sloan-Kettering Cancer Center (MSKCC) Genomics Core Laboratory. Bound probes were identified using Tilemap. The ChIP-on-chip experiment was performed in triplicate to strengthen the validity of the results.
High-throughput RNAi
The MSKCC High-throughput Screening Core Facility obtained siRNAs specific for the selected genes from Ambion (Life Technologies, Grand Island, NY, USA). A minimum of three siRNAs/gene were used. FU-UR-1 cells were plated in 384-well plates at 1500 cells/well. Transfection of these cells with a single siRNA (100 nM)/well was performed using 0.5 μl HiPerFect (Qiagen) and incubation for 96 h. The experiment was performed in duplicate. A positive control (AllStars Cell death siRNA, Qiagen) and a negative control siRNA containing a GFP reporter were utilized to optimize the experiments. The effects of gene silencing were assessed by nuclear counts at 96 h following transfection, and by apoptosis at both 72 and 96 h. Hoechst stain was used to identify individual nuclei and NucView dye was added to capture caspase-3-dependent apoptosis. IN CELL Analyser (INCA 1000, GE Healthcare Biosciences) detected these markers and the data were analysed using IN CELL developer software. A t-like statistic (Ts) score was derived for each siRNA for both nuclear count and apoptosis screens; a Ts score >2 was considered significant for the first screen performed without doxorubicin and a Ts score of > 1.5 was considered significant for the screen performed with drug.
Additional details are provided in Supplementary methods (see Supplementary material), including cell lines, plasmids and antibodies and more standard experimental methods, such as RNA extraction and cDNA microarray analysis, immunoblotting, transactivation assays, nuclear extract preparation, electrophoretic mobility shift assays, quantitative real-time RT–PCR assays, chromatin immunoprecipitation (ChIP) assays, RNAi target validation studies and cell growth assays.
Results
ASPSCR1–TFE3 shows nuclear localization
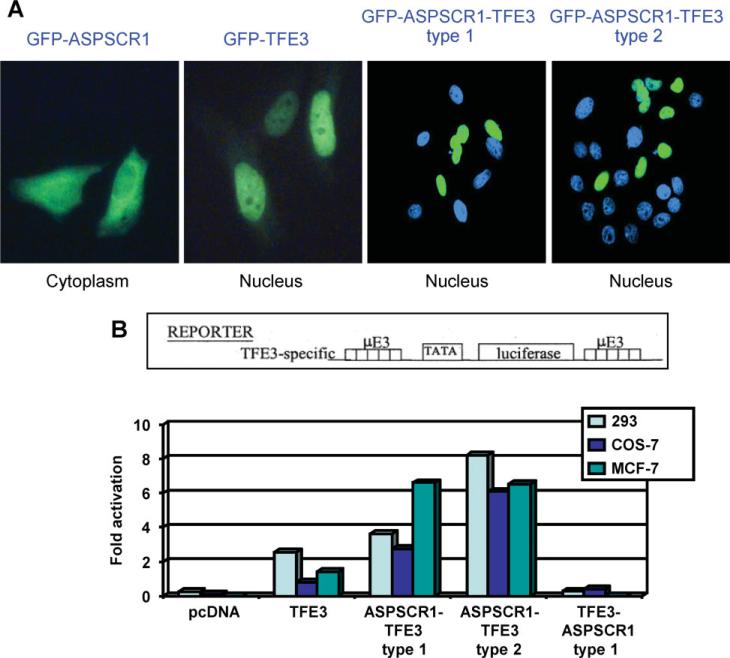
TFE3 contains a nuclear localization signal [16,17] in a region retained in both types of the ASPSCR1–TFE3 fusion protein [1] (see Supplementary material, Figure S1). To study subcellular localization, we cloned ASPSCR1, ASPSCR1–TFE3 type1, ASPSCR1–TFE3 type2 and TFE3 cDNAs into GFP expression vector pEGFR-C1 (BD Biosciences Clontech). ASPSCR1–TFE3 fusion proteins showed diffuse nuclear localization with nucleolar exclusion, indistinguishable from that of native TFE3. In contrast, native ASPSCR1 had a primarily cytoplasmic localization (Figure 1A), consistent with published data for Tug, the murine counterpart of ASPSCR1 [11–13].
Figure 1.
(A) ASPSCR1, TFE3, ASPSCR1–TFE3 type1 and ASPSCR1–TFE3 type2 were cloned into a GFP vector and over-expressed in HeLa cells. Both types of ASPSCR1–TFE3 and native TFE3 showed predominantly nuclear localization, whereas native ASPSCR1 had a primarily cytoplasmic distribution. (B) To assess the activity of ASPSCR1–TFE3 as a TF, transactivation assays were performed using the µE3-luciferase reporter known to be bound by native TFE3. Both types of ASPSCR1–TFE3 were stronger activators of the μE3 reporter relative to TFE3. ASPSCR1–TFE3 type 2 was a stronger activator than ASPSCR1–TFE3 type 1 in 293 and Cos7 cells, but not in MCF-7 cells
ASPSCR1–TFE3 is a stronger activator than native TFE3
To examine transactivation by ASPSCR1–TFE3, we used the uE3 reporter known to be bound by TFE3 [9] driven by five copies of the prototypical TFE3 binding sequence, CATGTG. ASPSCR1–TFE3 was a stronger activator of the uE3 reporter compared to TFE3 in the 293 T, COS7 and MCF7 cell lines (Figure 1B). In 293 T and COS7 cell lines, the ASPSCR1–TFE3 type 2 fusion protein appeared more active than ASPSCR1–TFE3 type 1. The difference between type 1 and type 2 ASPSCR1–TFE3 fusions is the presence in the latter of an additional exon encoding the TFE3 activation domain (see Supplementary material, Figure S1), which may contribute to stronger activation by ASPSCR1–TFE3 type 2.
Expression profiling of the ASPSCR1–TFE3-associated transcriptome
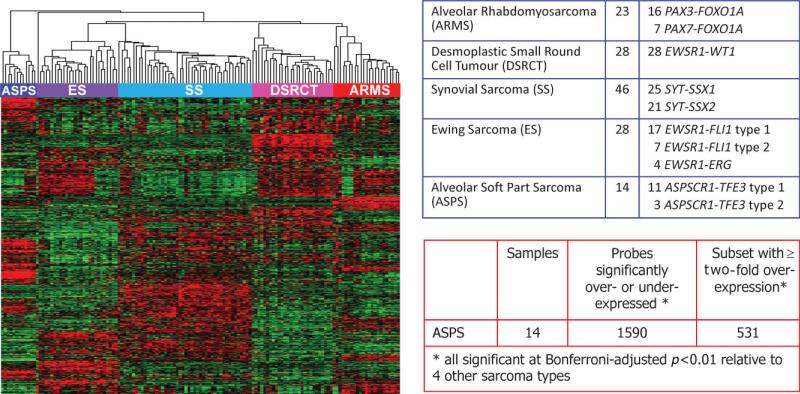
To better understand the significance of ASPSCR1–TFE3 in vivo and to define its global transcriptomic effects, we used two complementary approaches to identify genes whose expression is altered in the presence of this chimeric TF. The first approach is based on the fact that ASPS is one of a group of primitive sarcomas defined by unique chimeric TFs formed through type-specific chromosomal translocations [18]. Thus, the distinctive expression profile of these sarcomas is thought to reflect in part the direct and indirect effects of the respective chimeric TFs [18]. We therefore derived an ASPS expression profile from a large microarray analysis of five major translocation sarcomas based on the Affymetrix HG-U133A GeneChip, previously described in the context of other studies [19,20]. Tumour samples were procured under clinical protocols approved by the Institutional Review Board of Memorial Sloan-Kettering Cancer Center. Specifically, we compared the expression profiles of 14 cases of ASPS (11 type 1 and three type 2 ASPSCR1–TFE3 variants) against 125 tumour samples from four other translocation sarcomas: 28 Ewing sarcoma, 23 alveolar rhabdomyosarcoma, 46 synovial sarcoma and 28 desmoplastic small round cell tumour. Based on a stringent Bonferroni-corrected p < 0.01 cut-off, we identified 1531 array probes for genes significantly altered in expression in ASPS compared to the other four sarcoma types. Of these 1531 significantly dysregulated probes, 531 demonstrated > two-fold over-expression relative to the other four sarcoma types. These 531 probes corresponded to 415 unique genes that were highly and significantly up-regulated in ASPS compared to the other four sarcomas (Figure 2).
Figure 2.
Expression profiling data set on 139 sarcomas with chimeric TFs using Affymetrix U133A microarrays. Left panel shows unsupervised clustering of the expression data, demonstrating complete separation of the five sarcoma types in the dataset. The five sarcoma types and the corresponding gene fusions documented in all the cases are listed in the top right table. The bottom left table shows the analysis for differentially expressed genes in ASPS samples, all of which contained the ASPCR1–TFE3 fusion
In a second approach to the ASPSCR1–TFE3-associated transcriptome, we profiled gene expression in 293 T cells transfected with tetracycline-inducible plasmids containing either ASPSCR1–TFE3 type 1 or type 2 constructs. RNA from time points 0, 12, 24, 36 and 48 h were analysed on Affymetrix U133 Plus 2.0 arrays. Moderated t-statistics were used to test whether genes were differentially expressed between the time points of interest. We identified the subset of genes from the 36 and 48 h time points that were significantly dysregulated compared to time 0 (see Supplementary material, Table S1) and further cross-referenced them with our initial lists of differentially expressed genes in ASPS from the tumour profiling data described above. There were 103 genes that were up-regulated in both sets of profiles (Table 1). By combining genes significantly altered in one or both of these two expression profiles, we generated a list of 1116 genes that are up-regulated and 951 genes that are down-regulated in the presence of ASPSCR1–TFE3.
Table 1.
Genes (n = 103) that were both differentially over-expressed in ASPSCR1 -TFE3-positive ASPS relative to other sarcomas and up-regulated upon expression of ASPSCR1 -TFE3 in inducible 293 cells
| ABCB9 | CTTN | IFI30 | RHBDF1 |
| ABHD2 | CUGBP2 | IGF2R | RRAGD |
| ANGPTL2 | CYP17A1 | ITGB1BP3 | SCARB1 |
| APOE | DEXI | KCNJ4 | SCPEP1 |
| APOL2 | DST | KLHL21 | SFXN3 |
| ASAH1 | EPAS1 | LAMP1 | SLC19A2 |
| ATP6V0C | EPOR | MAFF | SLC9A1 |
| ATP6V0E1 | EPS15L1 | MET | SNCB |
| ATP6V1B2 | EZR/VIL2 | MFSD1 | SOD2 |
| ATP6V1D | FAS | MFSD5 | STOM |
| ATP6V1E1 | FER1L3 | MREG | STX3 |
| ATP6V1H | FLJ10815 | NAPA | SULT1C2 |
| AVPI1 | G0S2 | NPC2 | SUV39H1 |
| BHLHB2 | GABARAPL1 | OLFML2A | SV2B |
| BHLHB3 | GDF15 | OSTM1 | TACC1 |
| BIN1 | GLB1 | P4HA2 | TACC2 |
| C12orf49 | GM2A | PACSIN2 | TCEB1 |
| C1orf38 | GNS | PEPD | TCN2 |
| C2orf18 | GPNMB | PMP22 | TMEM70 |
| C8orf55 | GPR56 | POPDC2 | TMEM8 |
| CAST | GPRC5B | PPARGC1A | UAP1L1 |
| CD59 | GRN | PRODH | UPP1 |
| CIB1 | HLA-E | PXN | VAT1 |
| CRYAB | HMOX1 | RAB32 | WBP2 |
| CTSA/PPGB | HSD17B14 | RAB7A | WIPI1 |
| CTSD | HTATIP2 | RALGDS |
Genome-wide location analysis of ASPSCR1–TFE3
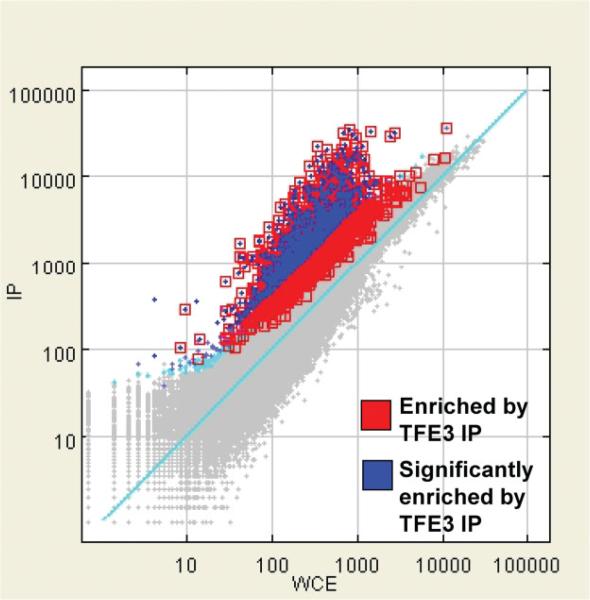
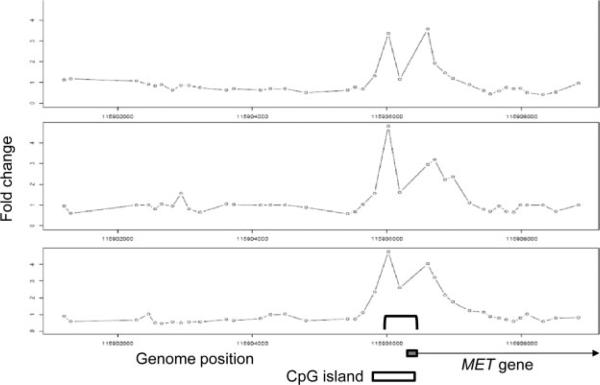
Having defined the repertoire of genes whose expression is altered in the presence of ASPSCR1–TFE3, we set out to delineate the subset of those genes that are its direct transcriptional targets. We performed genome-wide ChIP assays (ChIP-on-chip) on the FU-UR-1 cell line to identify the direct targets of ASPSCR1–TFE3. The absence of expression of native TFE3 in this cell line, confirmed using both RT–PCR and western blots (results not shown), allowed the use a TFE3 antibody for the selective immunoprecipitation of ASPSCR1–TFE3. We identified 2193 genes bound by ASPSCR1–TFE3, representing approximately 11.5% of the genes queried by the Agilent promoter microarray (Figure 3). An initial validation of the results was provided by the identification of a previously described direct target of ASPSCR1–TFE3, the MET gene [15] (Figure 4).
Figure 3.
ChIP-on-chip results for ASPSCR1–TFE3-bound regions in the FU-UR-1 cell line. There were 2193 genes (11.5%) showing significant enrichment following IP for ASPSCR1–TFE3 using a TFE3 antibody in this cell line lacking native TFE3 expression
Figure 4.
ChIP-on-chip results for MET, demonstrating the reproducibility of the results in the triplicate assays and providing basic validation of the overall experiment as the location of the four significant probes in ChIP-on-chip data coincides with the location of ASPSCR1–TFE3 binding (bracket in bottom panel), previously shown by simple ChIP analyses of the MET promoter region [15]
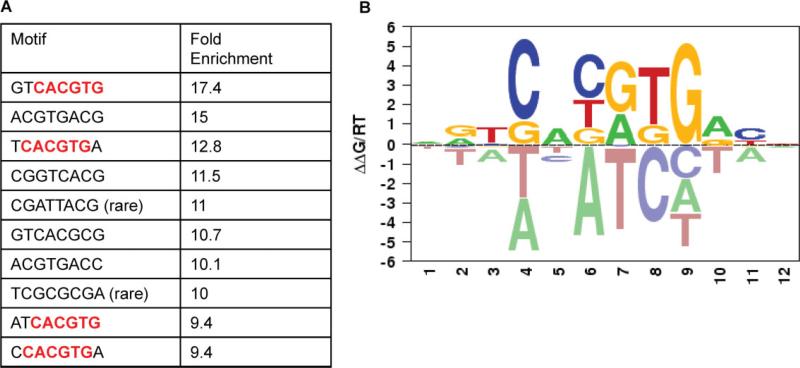
Next, we analysed the regions of the promoters that were bound by ASPSCR1–TFE3 to confirm enrichment for the previously described motif bound by native TFE3 (CANNTG) and possibly refine the consensus binding sequence. We defined regions of bound promoters that were within a 700 bp region surrounding a significant probe (−350 bp to 350 bp). An unbiased search of all possible 4–8 bp sequences was performed in these regions and compared to 10 000 unbound regions. The ASPSCR1–TFE3-bound promoters were significantly enriched for CACGTG. When the motif was analysed in the context of 8 bp sequences, the most common patterns appeared to favour a 5 T and a 3 A (Figure 5A). Separately, we also utilized the MatrixRE-DUCE algorithm to define the preferred binding motifs for ASPSCR1–TFE3. The regions investigated were 50 bp upstream and 250 bp downstream from the significantly bound probes and again we identified CACGTG as a top motif, with a preference for a 5'T and a 3'A [21] (Figure 5B).
Figure 5.
A. An analysis of over-represented 8 bp sequences in promoter regions bound by ASPSCR1–TFE3 rediscovered the CACGTG binding sequence which was previously described for TFE3, present here in four of the 10 top-scoring 8 bp motifs (boldface, red). In addition, it appeared to favour a 5'T and a 3'A. (B) Using MatrixREDUCE analysis, again the CACGTG consensus binding sequence was identified and favoured 5'T and 3'A. In this data visualization, the size and vertical order of the bases above the horizontal axis indicates the consensus and its strength at that position (see text for details)
Integrated analysis of expression profiles and genome-wide location analysis
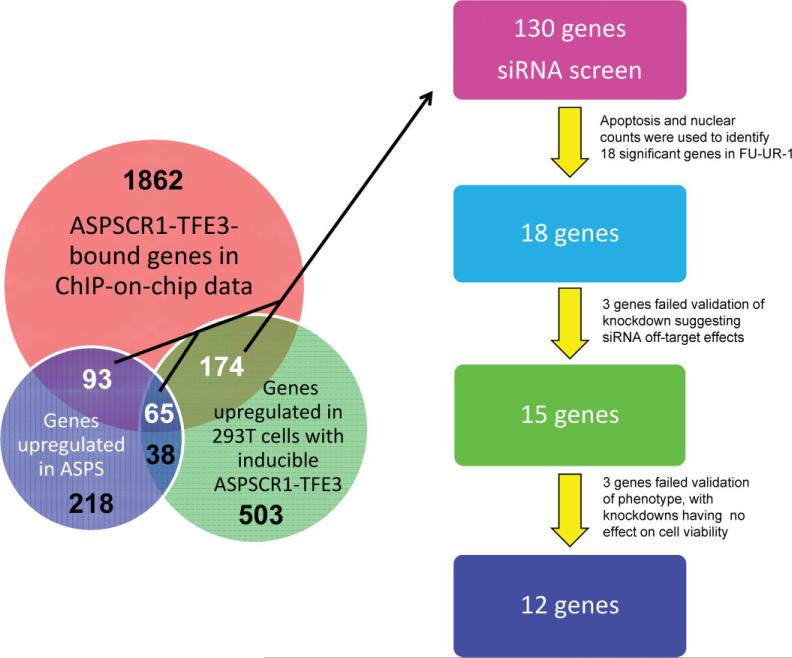
The bound promoter regions identified in the ASPSCR1–TFE3 ChIP-on-chip experiments were cross-referenced with the genes identified through the expression profiling studies described above as being dysregulated in the presence of ASPSCR1–TFE3. This analysis showed a highly significant enrichment for genes up-regulated in the presence of ASPSCR1–TFE3 among the genes bound by it (332 genes; Fisher exact test, p < 10–3). In contrast, there was a substantially less significant enrichment for genes down-regulated in the presence of ASPSCR1–TFE3 among genes bound by it in the ChIP-on-chip experiments (64 genes; Fisher exact test, p 0.002). This integrated analysis suggests that ASPSCR1–TFE3 functions primarily as a transcriptional activator.
Validation of novel direct transcriptional targets of ASPSCR1–TFE3
To further validate the above integrated genomics strategy and to highlight the significance of up-regulated genes that are under direct transcriptional control of ASPSCR1–TFE3, we present here a more detailed analysis of two highly up-regulated genes, CYP17A1 and UPP1 , that were also significantly bound by ASPSCR1–TFE3 in the ChIP-on-chip data. The CYP17A1 gene, encoding cytochrome P450 sub-family XVII (steroid 17α-hydroxylase) had the highest average fold change (121-fold over-expression; Student's t-test, p = 10–11) in ASPS relative to the four other sarcomas in the tumour expression profiling dataset. UPP1 , encoding uridine phosphorylase, showed a 47-fold over-expression in ASPS (Student's t-test, p = 10–10) relative to the four other sarcomas.
As presented in detail in the Validation data (see Supplementary material), co-transfection assays showed that a reporter plasmid driven by the CYP17A1 promoter was strongly transactivated by ASPSCR1–TFE3 (see Supplementary material, Figure S2). Quantitative real-time RT–PCR for CYP17A1 in 293 T cells expressing ASPSCR1–TFE3 confirmed transcriptional up-regulation (see Supplementary material, Figure S3). Mutagenesis studies defined the TFE3 binding-site required for transcriptional activation by ASPSCR1–TFE3 (see Supplementary material, Figure S2). Electrophoretic mobility shift assays and conventional ChIP analysis confirmed our ChIP-on-chip data showing that ASPSCR1–TFE3 binds the CYP17A1 promoter (see Supplementary material, Figure S3) and suggested that the type 2 form of the fusion may show stronger DNA binding (see Supplementary material, Figure S4). For UPP1 , multiple lines of evidence likewise supported its direct transcriptional control by ASPSCR1–TFE3 (see Supplementary material, Validation data and Figures S5 and S6). Thus, these independent single gene data for CYP17A1 and UPP1 , along with our previous data on MET, confirm that genes identified by our integrated genomics strategy described above are indeed likely to be directly trans-activated by the ASPSCR1–TFE3 fusion oncoprotein.
Functional genomics screen of direct targets of ASPSCR1–TFE3
The bioinformatic rediscovery of the consensus TFE3 binding sequence and single gene validation analyses described above supported the robustness of the overall approach to identify direct targets of transactivation by ASPSCR1–TFE3. We hypothesized that a subset of these ASPSCR1–TFE3 target genes mediate its oncogenic effects, as we had previously shown individually for MET [15]. Based on biological plausibility, we chose 130 of the 332 up-regulated direct target genes of ASPSCR1–TFE3 to study in a functional genomics screen. In addition, we included siRNAs to the portion of TFE3 included in the ASPSCR1–TFE3 fusion transcript as a tumour-specific positive control. We performed arrayed RNAi in 384-well plates in the FUUR-1 cell line. A total of 404 siRNAs were used to analyse the cellular effects of knockdown of these 130 target genes, with a minimum of three siRNAs/gene (used separately). The cellular phenotypes that were scored were apoptosis using NucView, a marker of caspase 3-dependent apoptosis, and cell proliferation based on nuclear counts. We performed the screen in duplicate and each siRNA was given a t-like statistic (Ts) score for both the NucView and the cell proliferation assay; genes were considered significant if two siRNAs attained a Ts score of >2. The initial screen identified a small number of genes whose knockdown resulted in increased apoptosis and/or decreased proliferation. We performed an additional screen in the presence of doxorubicin to potentiate the cellular effects of the siRNAs. In total, we chose to further investigate 18 of the genes from the 130 genes analysed in the high-throughput RNAi screen that had significant Ts scores (Figure 6). As a positive control, TFE3 itself was a strong hit based on decreased proliferation in the RNAi screen and the specificity of the knockdown was confirmed by RT–PCR (results not shown).
Figure 6.
There were a total of 332 genes (numbers in white) that were bound targets of ASPSCR1–TFE3 in the ChIP-on-chip analysis and up-regulated in either the ASPS sarcoma profiling signature (n = 93), the inducible 293 T cell signature (n = 174) or both (n = 65). We chose 130 of these 332 up-regulated ASPSCR1–TFE3 target genes to study in a high-throughput RNAi screen, based on biological plausibility and therapeutic potential. The final 12 genes emerging from this functional genomics screen are listed in Table 2. In addition, the 103 genes (65 + 38) that are in common between the ASPS sarcoma profiling signature and the inducible 293 T cell signature are listed in Table 1
Validation of hits from the functional genomics screen
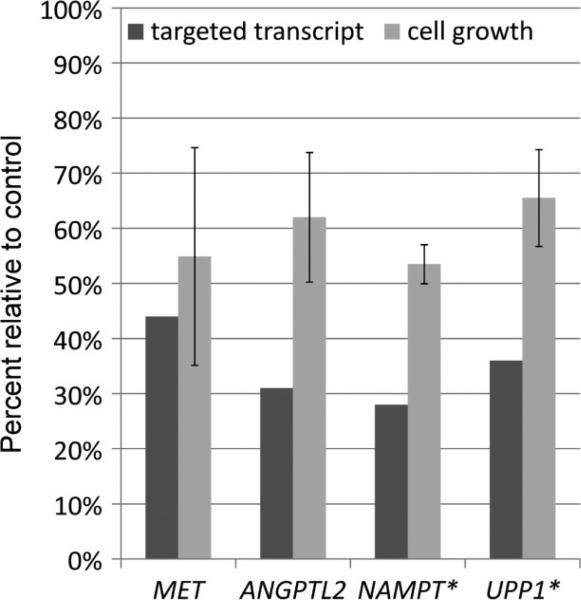
We first sought to confirm that the hits in the RNAi screen were due to on-target effects of the specific siRNAs. Indeed, siRNAs targeting three genes (GLB1 , NR1D1 , SLC38A7 ) could not be validated as on-target at the RNA or protein level and were assumed to have scored positive due to off-target effects (Figure 6). For the remaining 15 siRNAs whose knockdown of the expected target genes was validated at the RNA or protein level, cell proliferation assays were used to independently confirm, in six-well plates, their cellular effects in the same FU-UR-1 cell line used in the original high-throughput screen in 384-well plates (as the FU-UR-1 cell line is the only readily available cell line with endogenous ASPSCR1–TFE3 ). For 12 genes, we confirmed the phenotypic changes caused by their knockdown; three genes showed no observable changes in this secondary screen, BACH1 , SHB and SS18L1 . We confirmed a decrease in cell proliferation when the following genes were knocked down: ANGPTL2 , LGALS3BP, NAMPT, PCGF1 , PRICKLE3 , PTPRF, SLC29A1 , SOCS3 , SV2B, TYRO3 and UPP1 (Table 2, Figure 7). MET, whose inhibition has previously been reported to decrease cell proliferation [15], was rediscovered in this screen, serving as a quality control (Figure 7).
Table 2.
Up-regulated direct targets of ASPSCR1 –TFE3 confirmed to contribute to proliferation and survival of ASPSCR1 –TFE3-positive cells (see text for details)
| Expression profiles |
Primary screen* without Doxo |
Primary screen with Doxo |
Secondary screen | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | ChIP-on-CHIP | ASPS | Inducible 293 | Stockwin [28] | Apoptosis | Decrease in cell number | Apoptosis | Decrease in cell number | Decrease in cell number | |
| 1 | ANGPTL2 | + | + | + | + | + | - | + | - | + |
| 2 | LGALS3BP | + | + | - | + | + | - | + | + | + |
| 3 | MET | + | + | + | + | + | + | + | + | + |
| 4 | NAMPT | + | - | + | + | + | - | + | + | + |
| 5 | PCGF1 | + | - | + | - | - | - | - | + | + |
| 6 | PRICKLE3 | + | -** | + | - | + | - | + | + | + |
| 7 | PTPRF | + | - | + | - | - | + | + | + | + |
| 8 | SLC29A1 | + | + | - | + | + | - | + | + | + |
| 9 | SOCS3 | + | - | + | - | + | - | - | + | + |
| 10 | SV2B | + | + | + | + | - | - | + | + | + |
| 11 | 7YRO3 | + | - | + | - | - | - | + | + | + |
| 12 | UPP1 | + | + | + | + | - | - | - | + | + |
Numbers refer to quantity of siRNAs/gene that resulted in a changed phenotype.
This gene was not on the Affymetrix U133A array.
Figure 7.
Individual validation of RNAi-mediated gene silencing of selected genes identified in the high-throughput primary screens. For each RNAi target, the percentage of transcript detected following knockdown is shown by the dark bar and the percentage of growth of FU-UR-1 cells following knockdown compared to mock-transfected control is shown by the light bar. *Cell viability assays following knockdowns of NAMPT and UPP1 were performed in the presence of 10 nM doxorubicin
To validate the RNAi data for NAMPT pharmacologically and to explore it as a potential therapeutic target, we studied the effects of a NAMPT inhibitor, FK866 [22], on FU-UR-1 cells. FK866 decreased viability in FU-UR-1 in a dose-dependent manner, and its IC50 in this cell line was 20 nM at 96 h (results not shown), which is within the range of FK866 concentrations (3–50 nM) that have demonstrated activity in cell lines considered sensitive to NAMPT inhibition [23–26].
Discussion
Tumours expressing ASPSCR1–TFE3 are resistant to conventional therapies [3]. Although targeting the unique chromosomal aberration in these tumours would be a logical approach, pharmacological inhibition of TFs (other than nuclear hormone receptors) remains quite challenging [18]. In order to discover novel therapeutic approaches to these tumours, we sought to enhance the understanding of this chimeric TF and the downstream pathways under its direct transcriptional control. Specifically, we performed an integrated analysis of expression profiles and genome-wide TF binding followed by a functional genomics screen to characterize ASPSCR1–TFE3 target genes that contribute to neoplastic cell proliferation.
To our knowledge, the meta-analysis of expression profiles reported here represents the largest description of genes dysregulated in the presence of ASPSCR1–TFE3, including both tumour samples and in vitro data. We compared 14 tumour samples of ASPS against 125 samples from four other sarcoma types. In addition, using inducible 293 T cells, we identified genes significantly dysregulated upon induction of ASPSCR1–TFE3 expression. Other groups have recently analysed the expression profiles of ASPS to identify possible therapeutic approaches. Lazar et al. reported the up-regulation of 18 angiogenesis-associated genes in three ASPS tumours [27]; of these 18 genes, four (MDK , EPSAS1 , HIF1A and TIMP2 ) were identified as up-regulated in our expression profiling data and our ChIP-on-chip analysis found three of the 18 genes to be bound by ASPSCR1–TFE3 (MDK , JAG1 and LAMA5 ). Stockwin et al. [28] identified differentially expressed genes in seven ASPS tumours, relative to a universal reference RNA; comparing our up-regulated genes with those described in that study, we found a 22% (359/1634) overlap. Additionally, 79 genes were up-regulated in all three expression profiles (see Supplementary material, Table S2); no genes were found to be down-regulated in all three profiles. Ultimately, we identified 51 genes that were up-regulated in our analysis (sarcoma profiling and 293 T-inducible cell line) and the Stockwin et al. study that were also targets of ASPSCR1–TFE3 based on our ChIP-on-chip data (see Supplementary material, Table S2).
The effective integration of expression profiles with genome-wide TF-binding site data depended on the quality of our ChIP-on-chip data for ASPSCR1–TFE3. The ChIP-on-chip data quality was validated in two ways. First, we were able to bioinformatically redis-cover the known CACGTG recognition sequence for the TFE3 DNA-binding domain [29]. Additionally, this approach supported TCACGTGA as the preferred sequence context for the CACGTG binding motif for ASPSCR1–TFE3, based on two different analyses of the ChIP-on-chip data. Secondly, independent experimental data validating selected genes as direct targets of ASPSCR1–TFE3 further confirmed the quality of the ChIP-on-chip data. As mentioned above, we had previously identified MET as a direct transcriptional target of ASPSCR1–TFE3 [15] and, indeed, scored as significantly bound by ASPSCR1–TFE3 in our ChIP-on-chip data. Previously known or proposed transcriptional targets of native TFE3 in other cell types were also validated, but less consistently. These included genes encoding plasminogen activator inhibitor-1 (PAI-1 ), SMAD7 [30,31], cathepsin K [32,33], NPT2 [29], tyrosine hydroxylase [34], tyrosinase and TYRPI [35]. Of these genes, three (PAI-1 , cathepsin K and NPT2) were found to be significantly bound by ASPSCR1–TFE3 in our ChIP-on-chip data. To supplement this, we therefore provide independent experimental data validating two genes in the ASPSCR1–TFE3 ChIP-on-chip data as novel direct targets of ASPSCR1–TFE3, CYP17A1 and UPP1 (see Supplementary material, Validation data).
CYP17A1 encodes a single cytochrome P450 17-hydroxylase with both 17α-hydroxylase and C17,20-lyase activities [36,37]. In addition to the gonadal and adrenal tissues, both major sites for CYP17A1 expression [38,39], CYP17A1 is also expressed in a variety of other non-steroidal tissues, suggesting broader steroidogenic activities. Levels of CYP17A1 in patients with prostate cancer were correlated with clinical progression and metastasis [40]. Although it is unclear that CYP17A1 is contributing to tumourigenesis in ASPS, and indeed its RNAi-mediated silencing did not cause apoptosis or a notable decrease in proliferation in our high-throughput screen, it may nonetheless prove to be a useful marker for ASPS, as it has been repeatedly discovered to be highly up-regulated in ASPS expression profiles [28].
UPP1 encodes uridine phosphorylase, an enzyme of pharmacological interest because it can convert the pyrimidine analogue, 5'-deoxy-5'-fluorouridine, to 5-fluoruracil (5-FU), allowing administration of the former as a prodrug, the latter having low toxicity for non-neoplastic cells expressing only basal levels of the enzyme [41]. Our findings therefore suggest 5' -deoxy-5' -fluorouridine as a novel therapeutic approach in ASPS. Cells with higher expression of UPP1 may also display enhanced sensitivity to 5-FU itself [42], providing a rationale to investigate 5-FU activity in ASPS patients. UPP1 has been shown to be a target gene of EWS-FLI1 in Ewing sarcoma cells [41], but we observed substantially higher expression of UPP1 in ASPS compared to Ewing sarcoma (analysis not shown).
As these data confirmed the quality of the ChIP-on-chip analyses, we cross-referenced these genome-wide TF binding site data with the ASPSCR1–TFE3-associated expression profiles, in order to define a list of presumed or confirmed direct transcriptional targets of ASPSCR1–TFE3 that we wished to query functionally as potential therapeutic targets using a high-throughput RNAi screen. Our previous identification of MET as both an up-regulated target of ASPSCR1–TFE3 and a potential therapeutic target [15] provided the rationale for this high-throughput RNAi screen and for a recent clinical trial of ARQ197, a selective MET inhibitor, which reported disease stabilization in 15/17 patients with ASPS [43]. Furthermore, rediscovering MET as a hit in our high-throughput RNAi screen served to validate our overall strategy.
Aside from MET and UPP1 , we found that 10 other ASPSCR1–TFE3 target genes, ANGPTL2 , LGALS3BP; NAMPT, PCGF1 , PRICKLE3 , PTPRF, SLC29A1 , SOCS3 , SV2B and TYRO3 , appear to have a significant role in the survival of ASPSCR1–TFE3-driven cells, based on phenotype changes in the high-throughput RNAi screens (Table 2). Two genes with potential clinical implications, ANGPLT2 and NAMPT, are discussed in more detail below. The remaining eight genes are described in detail in the Gene descriptions (see Supplementary material).
ANGPTL2 encodes Angiopoietin-like 2 protein, a member of the vascular endothelial growth factor family involved in the formation of blood vessels [44]. ANGPLT2 has been reported, in the context of a mouse knockout, to contribute to metastasis of squamous cell carcinoma [45]. Over-expression of ANGPLT2 was previously reported in ASPS [28]. We found ANGPLT2 to be dramatically up-regulated in both of our ASPSCR1–TFE3-associated expression profiles (ASPS tumours, 293 T cells expressing inducible APSL–TFE3) and also directly bound by ASPSCR1–TFE3 in our ChIP-on-chip data. Furthermore, we provide the first evidence that silencing of ANGPTL2 by RNAi results in an apoptotic response and a decrease in cell proliferation. Thus, ANGPTL2 blockade may have therapeutic potential in tumours expressing ASPSCR1–TFE3.
NAMPT encodes nicotinamide phosphoribosyltransferase, the enzyme involved in the synthesis of nicotinamide mononucleotide from nicotinamide [46,47]. NAMPT was up-regulated in 293 T cells upon induction of ASPSCR1–TFE3 in the present study and has been reported to be over-expressed in ASPS cells [28]. The NAMPT promoter region was also bound by ASPSCR1–TFE3 in our ChIP-on-chip experiments. Two separate siRNAs for NAMPT caused apoptosis in FU-UR-1 cells and blocking NAMPT, either by RNAi or by chemical inhibition (using FK866), reduced cell viability. NAMPT has been implicated in tumourigenesis, poor response to therapy and higher stage in a variety of tumours [48–50]. NAMPT inhibitors, including FK866 (APO866) and CHS828, are in clinical trials. Although single-agent trials of NAMPT inhibitors have not been successful, combining these inhibitors with conventional chemotherapy or radiation therapy may enhance responses [24–26,47,51]. Interestingly, the inhibition of NAMPT in gastric cancer cells may lead to increased sensitivity to fluorouracil [24]. Given the significance of UPP1 in ASPSCR1–TFE3-expressing cells, the therapeutic potential of combining a NAMPT inhibitor with 5-fluorouracil (or other conventional chemotherapy agents) in these patients may warrant further investigation.
In conclusion, our results support a gain-of-function role for ASPSCR1–TFE3. We provide comprehensive data on the repertoire of direct transcriptional targets of ASPSCR1–TFE3 and used a high-throughput functional genomics strategy to define a subset of these deregulated transcriptional target genes that contribute to proliferation and survival of these cancer cells. Several of these genes open new therapeutic opportunities for patients with ASPSCR1–TFE3 -positve tumours. More generally, this study demonstrates a robust approach to identifying biologically significant, up-regulated direct targets of chimeric TFs, using high-throughput tailored siRNA screens, that should be readily applicable to other translocation-associated cancers driven by specific chimeric TFs.
Supplementary Material
Acknowledgements
This study was supported in part by NIH/NCI Grant R01 CA95785 (to M Ladanyi) and by the Alliance Against Alveolar Soft Part Sarcoma. We thank Zhen Lu for technical guidance with ChIP-on-chip assays. We thank Agnes Viale and the staff of the MSKCC Genomics Core laboratory for all microarray data generation. We also thank Yufang Shao, Zhengdeng Lei, Paul A Calder, Bhavneet Bhinder and Constantin Radu for all their help during the course of the siRNA screening project in the MSKCC HTS Core Facility, which is partially supported by Mr William H Goodwin and Mrs Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, the William Randolph Hearst Fund in Experimental Therapeutics, the Lillian S Wells Foundation, and by a NIH/NCI Cancer Center Support Grant (No. 5 P30 CA008748-44).
Footnotes
Microarray data are available at: http://cbio.mskcc.org/public/sarcoma_array_data/
No conflicts of interest were declared.
Author contributions
M Ladanyi conceived and supervised the analyses and experiments; RK, MN, MT, MYM, TS and M Laé performed the experiments; QM, AO, SL and CL performed the bioinformatic analyses; CA, IO and HD performed the high-throughput RNAi experiments and analysed the RNAi screening data; and M Ladanyi and RK assembled and finalized the manuscript.
References
- 1.Ladanyi M, Lui MY, Antonescu CR, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57. doi: 10.1038/sj.onc.1204074. [DOI] [PubMed] [Google Scholar]
- 2.Argani P, Antonescu CR, Illei PB, et al. Primary renal neoplasms with the ASPL–TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kayton ML, Meyers P, Wexler LH, et al. Clinical presentation, treatment, and outcome of alveolar soft part sarcoma in children, adolescents, and young adults. J Pediatr Surg. 2006;41:187–193. doi: 10.1016/j.jpedsurg.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Argani P, Olgac S, Tickoo SK, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–1160. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
- 5.Ishiguro M, Iwasaki H, Ohjimi Y, et al. Establishment and characterization of a renal cell carcinoma cell line (FU-UR-1) with the reciprocal ASPL–TFE3 fusion transcript. Oncol Rep. 2004;11:1169–1175. [PubMed] [Google Scholar]
- 6.Hemesath TJ, Steingrimsson E, McGill G, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 7.Rehli M, Den Elzen N, Cassady AI, et al. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- 8.Zhao GQ, Zhao Q, Zhou X, et al. TFEC, a basic helix–loop–helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol Cell Biol. 1993;13:4505–4512. doi: 10.1128/mcb.13.8.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckmann H, Su LK, Kadesch T. TFE3: a helix–loop–helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 10.Haq R, Fisher DE. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J Clin Oncol. 2011;29:3474–3482. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- 11.Yu C, Cresswell J, Loffler MG, et al. The glucose transporter 4-regulating protein TUG is essential for highly insulin-responsive glucose uptake in 3 T3-L1 adipocytes. J Biol Chem. 2007;282:7710–7722. doi: 10.1074/jbc.M610824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogan JS, Hendon N, McKee AE, et al. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature. 2003;425:727–733. doi: 10.1038/nature01989. [DOI] [PubMed] [Google Scholar]
- 13.Rubin BR, Bogan JS. Intracellular retention and insulin-stimulated mobilization of GLUT4 glucose transporters. Vitam Horm. 2009;80:155–192. doi: 10.1016/S0083-6729(08)00607-9. [DOI] [PubMed] [Google Scholar]
- 14.Bogan JS, Kandror KV. Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr Opin Cell Biol. 2010;22:506–512. doi: 10.1016/j.ceb.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuda M, Davis IJ, Argani P, et al. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 16.Weterman MJ, van Groningen JJ, Jansen A, et al. Nuclear localization and transactivating capacities of the papillary renal cell carcinoma-associated TFE3 and PRCC (fusion) proteins. Oncogene. 2000;19:69–74. doi: 10.1038/sj.onc.1203255. [DOI] [PubMed] [Google Scholar]
- 17.Takebayashi K, Chida K, Tsukamoto I, et al. The recessive phenotype displayed by a dominant negative microphthalmia-associated transcription factor mutant is a result of impaired nucleation potential. Mol Cell Biol. 1996;16:1203–1211. doi: 10.1128/mcb.16.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BS, Barretina J, Maki RG, et al. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11:541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filion C, Motoi T, Olshen AB, et al. The EWSR1/NR4A3 fusion protein of extraskeletal myxoid chondrosarcoma activates the PPARG nuclear receptor gene. J Pathol. 2009;217:83–93. doi: 10.1002/path.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho AL, Deraje Vasudeva S, Lae M, et al. PDGF receptor-α is an alternative mediator of rapamycin-induced Akt activation: implications for combination targeted therapy of synovial sarcoma. Cancer Res. 2012;72:4515–4525. doi: 10.1158/0008-5472.CAN-12-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foat BC, Morozov AV, Bussemaker HJ. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics. 2006;22:e141–149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- 22.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 23.Thakur BK, Dittrich T, Chandra P, et al. Inhibition of NAMPT pathway by FK866 activates the function of p53 in HEK293T cells. Biochem Biophys Res Commun. 2012;424:371–377. doi: 10.1016/j.bbrc.2012.06.075. [DOI] [PubMed] [Google Scholar]
- 24.Bi TQ, Che XM, Liao XH, et al. Overexpression of Nampt in gastric cancer and chemopotentiating effects of the Nampt inhibitor FK866 in combination with fluorouracil. Oncol Rep. 2011;26:1251–1257. doi: 10.3892/or.2011.1378. [DOI] [PubMed] [Google Scholar]
- 25.Travelli C, Drago V, Maldi E, et al. Reciprocal potentiation of the antitumoral activities of FK866, an inhibitor of nicotinamide phosphoribosyltransferase, and etoposide or cisplatin in neuroblastoma cells. J Pharmacol Exp Ther. 2011;338:829–840. doi: 10.1124/jpet.111.184630. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Hasan MK, Alvarado E, et al. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–921. doi: 10.1038/onc.2010.468. [DOI] [PubMed] [Google Scholar]
- 27.Lazar AJ, Das P, Tuvin D, et al. Angiogenesis-promoting gene patterns in alveolar soft part sarcoma. Clin Cancer Res. 2007;13:7314–7321. doi: 10.1158/1078-0432.CCR-07-0174. [DOI] [PubMed] [Google Scholar]
- 28.Stockwin LH, Vistica DT, Kenney S, et al. Gene expression profiling of alveolar soft-part sarcoma (ASPS). BMC Cancer. 2009;9:22. doi: 10.1186/1471-2407-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kido S, Miyamoto K, Mizobuchi H, et al. Identification of regulatory sequences and binding proteins in the type II sodium/phosphate cotransporter NPT2 gene responsive to dietary phosphate. J Biol Chem. 1999;274:28256–28263. doi: 10.1074/jbc.274.40.28256. [DOI] [PubMed] [Google Scholar]
- 30.Hua X, Miller ZA, Wu G, et al. Specificity in transforming growth factor-β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor muE3, and Smad proteins. Proc Natl Acad Sci USA. 1999;96:13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua X, Miller ZA, Benchabane H, et al. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor-β-induced transcription of the Smad7 gene. J Biol Chem. 2000;275:33205–33208. doi: 10.1074/jbc.C000568200. [DOI] [PubMed] [Google Scholar]
- 32.Motyckova G, Weilbaecher KN, Horstmann M, et al. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci USA. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martignoni G, Gobbo S, Camparo P, et al. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod Pathol. 2011;24:1313–1319. doi: 10.1038/modpathol.2011.93. [DOI] [PubMed] [Google Scholar]
- 34.Wong SC, Moffat MA, Coker GT, et al. The 3′ flanking region of the human tyrosine hydroxylase gene directs reporter gene expression in peripheral neuroendocrine tissues. J Neurochem. 1995;65:23–31. doi: 10.1046/j.1471-4159.1995.65010023.x. [DOI] [PubMed] [Google Scholar]
- 35.Verastegui C, Bertolotto C, Bille K, et al. TFE3, a transcription factor homologous to microphthalmia, is a potential transcriptional activator of tyrosinase and TyrpI genes. Mol Endocrinol. 2000;14:449–456. doi: 10.1210/mend.14.3.0428. [DOI] [PubMed] [Google Scholar]
- 36.Conley AJ, Bird IM. The role of cytochrome P450 17 α-hydroxylase and 3β-hydroxysteroid dehydrogenase in the integration of gonadal and adrenal steroidogenesis via the δ5 and δ4 pathways of steroidogenesis in mammals. Biol Reprod. 1997;56:789–799. doi: 10.1095/biolreprod56.4.789. [DOI] [PubMed] [Google Scholar]
- 37.Payne AH. Hormonal regulation of cytochrome P450 enzymes, cholesterol side-chain cleavage and 17α-hydroxylase/C17–20 lyase in Leydig cells. Biol Reprod. 1990;42:399–404. doi: 10.1095/biolreprod42.3.399. [DOI] [PubMed] [Google Scholar]
- 38.Voutilainen R, Tapanainen J, Chung BC, et al. Hormonal regulation of P450scc (20,22-desmolase) and P450c17 (17α-hydroxylase/17,20-lyase) in cultured human granulosa cells. J Clin Endocrinol Metab. 1986;63:202–207. doi: 10.1210/jcem-63-1-202. [DOI] [PubMed] [Google Scholar]
- 39.Chung BC, Picado-Leonard J, Haniu M, et al. Cytochrome P450c17 (steroid 17α-hydroxylase/17,20 lyase): cloning of human adrenal and testis cDNAs indicates the same gene is expressed in both tissues. Proc Natl Acad Sci USA. 1987;84:407–411. doi: 10.1073/pnas.84.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locke JA, Fazli L, Adomat H, et al. A novel communication role for CYP17A1 in the progression of castration-resistant prostate cancer. Prostate. 2009;69:928–937. doi: 10.1002/pros.20940. [DOI] [PubMed] [Google Scholar]
- 41.Deneen B, Hamidi H, Denny CT. Functional analysis of the EWS/ETS target gene uridine phosphorylase. Cancer Res. 2003;63:4268–4274. [PubMed] [Google Scholar]
- 42.Im YS, Shin HK, Kim HR, et al. Enhanced cytotoxicity of 5-FU by bFGF through up-regulation of uridine phosphorylase 1. Mol Cell. 2009;28:119–124. doi: 10.1007/s10059-009-0116-x. [DOI] [PubMed] [Google Scholar]
- 43.Adjei AA, Schwartz B, Garmey E. Early clinical development of ARQ 197, a selective, non-ATP-competitive inhibitor targeting MET tyrosine kinase for the treatment of advanced cancers. Oncologist. 2011;16:788–799. doi: 10.1634/theoncologist.2010-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oike Y, Yasunaga K, Suda T. Angiopoietin-related/angiopoietin-like proteins regulate angiogenesis. Int J Hematol. 2004;80:21–28. doi: 10.1532/ijh97.04034. [DOI] [PubMed] [Google Scholar]
- 45.Aoi J, Endo M, Kadomatsu T, et al. Angiopoietin-like protein 2 is an important facilitator of inflammatory carcinogenesis and metastasis. Cancer Res. 2011;71:7502–7512. doi: 10.1158/0008-5472.CAN-11-1758. [DOI] [PubMed] [Google Scholar]
- 46.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 47.Bi TQ, Che XM. Nampt/PBEF/visfatin and cancer. Cancer Biol Ther. 2010;10:119–125. doi: 10.4161/cbt.10.2.12581. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima TE, Yamada Y, Hamano T, et al. Adipocytokines as new promising markers of colorectal tumors: adiponectin for colorectal adenoma, and resistin and visfatin for colorectal cancer. Cancer Sci. 2010;101:1286–1291. doi: 10.1111/j.1349-7006.2010.01518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Folgueira MA, Carraro DM, Brentani H, et al. Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin Cancer Res. 2005;11:7434–7443. doi: 10.1158/1078-0432.CCR-04-0548. [DOI] [PubMed] [Google Scholar]
- 50.Shackelford RE, Bui MM, Coppola D, et al. Over-expression of nicotinamide phosphoribosyltransferase in ovarian cancers. Int J Clin Exp Pathol. 2010;3:522–527. [PMC free article] [PubMed] [Google Scholar]
- 51.von Heideman A, Berglund A, Larsson R, et al. Safety and efficacy of NAD depleting cancer drugs: results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother Pharmacol. 2010;65:1165–1172. doi: 10.1007/s00280-009-1125-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.