Abstract
Staphylococcus aureus is a major human pathogen and a common cause of nosocomial infections. This facultative pathogen produces a large arsenal of virulence factors, including the haemolysins, which allow the bacterium to lyse erythrocytes and thereby release large amounts of the haem-containing haemoglobin. The released haem is thought to be the main iron source of this organism during the course of infection, and is considered to be crucial for bacterial proliferation in vivo. High concentrations of haem and its degradation products, on the other hand, are known to be toxic for S. aureus, making it essential for the pathogen to tightly control haem release from red blood cells. Here we show that S. aureus responds to haemin by downregulating the expression of haemolysins. Subinhibitory concentrations of haemin were found to significantly reduce transcription of the haemolysin genes hlb (encoding β-haemolysin) and hlgA (encoding the S-class component of γ-haemolysin), while hla (encoding α-haemolysin) and RNAIII (encoding δ-haemolysin) transcription did not appear to be affected. The presence of haemin also reduced the haemolytic potential of the supernatants of S. aureus LS1 cultures. Inactivation of the sae locus in LS1 abolished the haemin effect on the transcription of haemolysin genes, indicating that the two-component regulatory system is required for this regulatory effect. Iron limitation, on the other hand, was found to induce the expression of haemolysins, and this effect was again abolished in the sae mutant, indicating that S. aureus modulates its haemolysin production in response to iron and haem availability in an Sae-dependent manner.
Introduction
Iron is a key element for all living organisms due to its important role in enzyme processes and transport of electrons, mainly acting as a cofactor for other proteins or enzymes (Somerville & Proctor, 2009). In mammals, free iron is scarce, perhaps to prevent pathogenic growth and protect the organism itself from this reactive element in its ionic form, Fe2+ (Stojiljkovic & Perkins-Balding, 2002; Touati, 2000). Thus, 99.9 % of the iron in humans is located intracellularly (Reniere et al., 2007), and the largest amount (approx. 75 % of total iron) is complexed within the haem molecule inside erythrocytes (Stojiljkovic & Perkins-Balding, 2002).
Staphylococcus aureus is a major human pathogen and a frequent cause of life-threatening nosocomial and community-acquired infections (Diekema et al., 2001) such as sepsis, endocarditis, osteomyelitis and pneumonia (Lowy, 1998). An arsenal of virulence factors enables this bacterium to initiate colonization and to facilitate persistence within the host (Horsburgh et al., 2001). Among these factors, cytolysins, and specifically haemolysins, are secreted by S. aureus, leading to red blood cell lysis, release of iron complexes and a local increase in the available iron concentration of up to 100-fold (Rouault, 2004). The released haemoglobin is bound by the bacterium to special receptors, and the haem subunit is released and imported into the cytoplasm via two different systems called Isd (iron-regulated surface determinant system) and Hts (haem transport system) (Reniere et al., 2007).
The Isd system consists of 10 genes, expression of which is controlled by the iron-dependent ferric uptake regulator Fur (reviewed by Maresso & Schneewind, 2006). The isd locus encodes three cell wall-associated proteins (IsdA, B and C), one transpeptidase (SrtB), three membrane transport proteins (IsdD, E and F) and the haem oxygenase IsdG (Mazmanian et al., 2003), while IsdH (synonym HarA), another cell wall-anchored factor of this system, and IsdI, a second haem oxygenase, are located elsewhere on the chromosome (Dryla et al., 2003; Skaar et al., 2004). IsdB and IsdH bind haemoglobin on the cell surface (Dryla et al., 2003; Torres et al., 2006). The haem subunit is released from this complex via an as-yet-undefined mechanism, transported through the cell wall via IsdC, and passed through the cell membrane to the cytoplasm with IsdD, E and F and/or the Hts system (Mazmanian et al., 2003). The Hts system consists of a lipoprotein (HtsA) and two permeases (HtsB, HtsC) (Mason & Skaar, 2009), and also contributes to the uptake of iron–siderophore complexes (Beasley et al., 2009). Inside the cell, either IsdG or IsdI degrades the porphyrin ring and releases free iron, or the haem complex is bound to membrane-associated proteins to act as a cofactor in, e.g. electron transport (Thöny-Meyer, 1997).
However, as high intracellular concentrations of haem damage DNA and proteins due to mechanisms of an oxidative (Everse & Hsia, 1997) and/or non-oxygen-dependent nature (our own unpublished observations), S. aureus needs to tightly regulate the release and uptake of this iron-containing molecule. Consequently, S. aureus is equipped with a system, termed HrtAB (haem-regulated transport system) (Reniere et al., 2007), that is activated in the presence of increasing intracellular concentrations of haem. However, as in vitro studies have shown that increased concentrations of haemin (>10 µM) in the extracellular milieu suppress growth of this organism (Torres et al., 2007), it appears that the HrtAB export mechanism is not efficient enough to protect S. aureus fully from elevated concentrations of haem. This suggests that S. aureus would benefit from a regulatory mechanism to modulate haemolysin expression and reduce lysis of erythrocytes, and thus decrease the release of haemoglobin at the infection site(s).
Recently, we have shown that in S. aureus the iron status is sensed via Fur, and that Fur mediates the abundance of a large number of exoproteins secreted by the model strain Newman and a number of clinical laboratory S. aureus isolates. Moreover, it could be demonstrated that in Newman the haemolytic and cytotoxic activity is also affected by fur (Torres et al., 2010). Newman harbours a unique point mutation (Leu18→Pro18) in the sensor histidine kinase saeS that leads to a permanent activation of the sensor molecule of this two-component system (Geiger et al., 2008; Schäfer et al., 2009), yet this results in constitutive overexpression of class I sae target genes (coa, fnbA, eap, sib, efb, fib, sae) but not class II sae target genes, including the haemolysins hla and hlb (Mainiero et al., 2010). In addition to Fur, other regulators known to affect haemolysin expression and thus candidates for regulating red blood cell lysis are the Sae two-component regulatory system (a major regulator of haemolysin expression in S. aureus) (Giraudo et al., 1997; Goerke et al., 2005; Rogasch et al., 2006; Yamazaki et al., 2006), and the two-component system ArlRS, the SarA regulator, the agr system and the global stress component σB (reviewed by Bronner et al., 2004).
Thus, to characterize a putative regulatory network connecting haemin/iron availability with haemolysin expression/haemolytic function as well as the underlying regulators, a detailed analysis employing various media (iron-rich, iron-poor), media supplements (±haemin, iron sulphate or iron chelator) and S. aureus genetic backgrounds is needed. Using these tools, we show here that a functional S. aureus Sae system is a key switch regulating the production of haemolysins, suggesting that sae plays a major role in the protection of the microbial cell from exposure to cytotoxic concentrations of intracellular haem.
Methods
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37 °C and 150 r.p.m. in Luria–Bertani (LB) medium (Becton Dickinson) and RPMI 1640 (Invitrogen) supplemented with 1 % Casamino acids. Where required, media were supplemented with (per millilitre) 100 µg ampicillin, 7 µg chloramphenicol, 50 µg kanamycin or 10 µg tetracycline. The antibiotic resistance cassette-tagged hla, saePQRS and fur deletions of DU1090 (O’Reilly et al., 1986) and Newman 29 (Geiger et al., 2008) and MJH010 (Horsburgh et al., 2001) were transduced into S. aureus LS1 using phage 85. Haemin MICs were determined by broth microdilution as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2009).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype and phenotype* | Source or reference |
| E. coli strain | ||
| DH5α | F− ϕ80d/lacZΔM15 recA1 | Invitrogen |
| S. aureus strains | ||
| 8325-4 | NCTC8325 derivative lacking the prophages, rsbU | Novick (1967) |
| DU1090 | 8325-4 hla-erm1, pTS01, ErmR, TetR | O’Reilly et al. (1986) |
| COL | MRSA, highly meticillin-resistant clinical isolate, TetR | Dyke et al. (1966) |
| Cowan | ATCC12598, highly invasive clinical isolate | |
| LS1 | Murine arthritis isolate, saeSLeu allele, highly haemolytic on SBA | Bremell et al. (1992) |
| LS1 Δfur | LS1 Δfur, TetR, obtained by phage transduction from MJH010 | This study |
| LS1 Δhla | LS1 Δhla, ErmR, obtained by phage transduction from DU1090 | This study |
| LS1 Δsae | LS1 Δsae, KanR, obtained by phage transduction from New 29 | This study |
| MJH010 | 8325-4 Δfur, TetR | Horsburgh et al. (2001) |
| MSSA 1112 | Clinical isolate, bla+ | Entenza et al. (1997) |
| N315 | MRSA, clinical isolate | Kuroda et al. (2001) |
| Newman | ATCC25904, clinical isolate, ClfA/B and Eap hyperproducer, saeSPro allele | Duthie (1952) |
| New 29 | Newman ΔsaePQRS, KanR | Geiger et al. (2008) |
| New HG | Newman derivative with saeSLeu allele | Mainiero et al. (2010) |
| Plasmids | ||
| pSB2035 | E. coli–S. aureus shuttle plasmid, harbouring the cat gene conferring chloramphenicol resistance (CmR), and a gfp–lux dual reporter plasmid harbouring the agr P3 promoter | Qazi et al. (2001) |
| pSB2035/hlap | pSB2035 derivative, replacing the agr P3 promoter with an hla promoter fragment | This study |
| pSB2035/hlbp | pSB2035 derivative, replacing the agr P3 promoter with an hlb promoter fragment | This study |
| pSB2035/hlgAp | pSB2035 derivative, replacing the agr P3 promoter with an hlagA promoter fragment | This study |
CmR, chloramphenicol resistant; ErmR, erythromycin resistant; KanR, kanamycin resistant; TetR, tetracycline resistant. MRSA, meticillin-resistant Staphylococcus aureus.
Construction of the reporter gene plasmids pSB2035/hlap, pSB2035/hlBp and pSB2035/hlgAp.
DNA fragments covering the hla, hlb and hlgA promoters were amplified by PCR using primer pairs hlap forward/reverse (for pSB2035/hlap), hlbp forward/reverse (for pSB2035/hlBp) and hlgAp forward/reverse (for pSB2035/hlgAp), respectively, and LS1 DNA as template (primer sequences are listed in Table S1 available with the online version of this paper). The resulting PCR products were EcoRI/SmaI-digested and cloned into pSB2035 (Qazi et al., 2001), thereby replacing the agr P3 promoter of pSB2035. Constructs harbouring the desired inserts were sequenced and used to electroporate RN4220, which served as donor for transducing the plasmids into LS1.
Quantitative real-time RT-PCR (qRT-PCR).
Bacteria were grown in LB for 6 h, and the cultures were split into equal parts and supplemented with different concentrations of haemin, 2,2′-bipyridine and FeSO4. The cultures were incubated for an additional 1 h at 37 °C and 150 r.p.m., and samples were removed at 15, 30 and 60 min after supplementation. RNA isolations and qRT-PCRs were carried out as described by Chatterjee et al. (2005). Levels of mRNA expression of the different genes were normalized against expression of the internal control gyrB. The amounts of different transcripts were expressed as the n-fold difference relative to the control gene (2−ΔCT, where ΔCT represents the difference in threshold cycle between the target and control genes).
Western blot analyses.
Bacteria were grown in LB supplemented as indicated, and samples were taken after 6 h of growth. To account for differences in growth, samples were diluted with fresh LB to a OD600 1, and equal volumes were centrifuged at 5000 g for 10 min at 4 °C. The supernatants were used for Hla determinations, and cell pellets were used to isolate Eap by the lithium chloride extraction method described by Sobke et al. (2006). Protein samples were separated by SDS-PAGE, electroblotted onto nitrocellulose membranes, and subjected to Western blot analyses with anti-Hla antibodies (Abcam) and anti-Eap antibodies (provided by J. Patti, Inhibitex), respectively.
Haemolytic activity assays.
Haemolytic activities for LS1 and its derivatives were assayed on LB agar plates supplemented with 5 % defibrinated sheep blood (SBA), human blood (HBA) and rabbit blood (RBA) after 24 h of growth at 37 °C followed by incubation at 4 °C for 24 h.
For determination of the haemolytic potential of S. aureus culture supernatants, overnight cultures were centrifuged at 5000 g for 10 min at 4 °C and the pellets resuspended in the same amount of fresh pre-heated medium. Cultures were then split into equal parts, and supplements added as indicated. Cultures were incubated for 2 h at 37 °C and 150 r.p.m., diluted with fresh LB to OD600 1, and centrifuged as described above. The supernatants were filter-sterilized and titrated for haemolytic activities as described by Nilsson et al. (1999). The highest dilution giving rise to visible lysis of erythrocytes was defined as the haemolytic titre.
Luciferase assay.
For assaying luciferase activities of S. aureus strains harbouring the pSB2035 derivatives, cells were grown in LB supplemented with chloramphenicol. After 6 h of growth, cultures were split into two, and one half was supplemented with haemin to a final concentration of 4 µM. Cultures were incubated for an additional 1 h. Then, 200 µl samples of the cell suspensions were removed at the time points indicated and transferred to wells of a 96-well clear-bottomed black plate (Greiner). The plate was placed in a Wallac Victor2 1420 Multilabel Counter (Perkin Elmer), and luminescence readings were taken for 5 s at 37 °C.
Statistical analyses.
The statistical significance of changes between supplemented and unsupplemented cultures, and wild-type and mutant strains was assessed with the Wilcoxon signed-rank test (for related samples), and the Mann–Whitney U test (for independent samples), respectively. P values <0.05 were considered significant.
Results and Discussion
Haemin affects growth of S. aureus in LB
Haemin is toxic for S. aureus if the concentration exceeds a certain value (Torres et al., 2006). To assess the cytotoxic impact of haemin on growth of the highly haemolytic S. aureus strain LS1 (Bremell et al., 1992), we determined its MIC and assayed growth in LB supplemented with different concentrations of haemin. Whereas LS1 was found to be relatively insensitive to haemin in the classical MIC assay (64 µM), its growth was clearly affected by much lower concentrations of this iron-containing molecule if LS1 was cultured as a batch culture in LB at 150 r.p.m. and 37 °C (Fig. 1a). Concentrations of haemin as low as 2 µM delayed the onset of growth of this isolate, and concentrations equal to or higher than 4 µM were sufficient to significantly delay the growth of LS1 under these conditions. Consequently, after 12 h of growth, the growth yields of the cultures supplemented with 4 and 8 µM haemin, respectively, were significantly reduced compared with the unsupplemented control (mean±sd OD600 values of 1.92±0.31 and 0.26±0.21 for the cultures challenged with 4 and 8 µM haemin, respectively, vs 3.56±0.43 for the untreated control; P<0.001 between treated and untreated samples). However, haemin concentrations up to 4 µM did not markedly affect the doubling times of the LS1 cultures during the exponential growth phase, which was significantly increased only in the LS1 culture challenged with 8 µM haemin (89.52±4.47 min for the haemin-supplemented culture vs 37.28±1.14 min for the control; P<0.01). Moreover, after 24 h of growth, all cultures showed comparable OD600 values, irrespective of the haemin addition, signalling that the haemin concentrations used here were not bacteriocidal for this strain (Table S2).
Fig. 1.
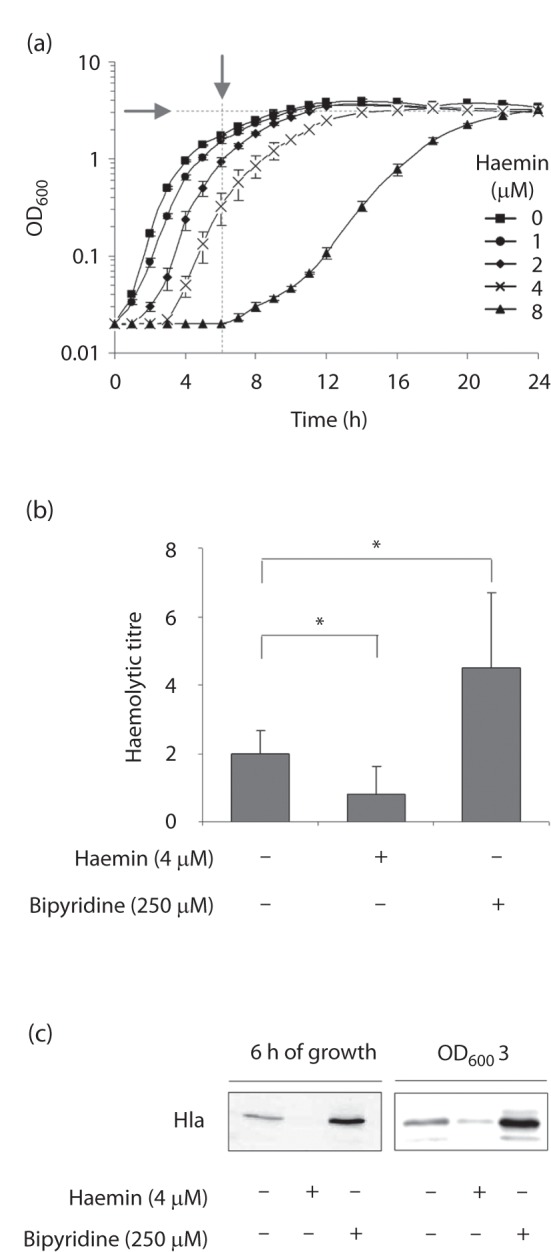
Impact of haemin on growth and haemolytic activity of S. aureus. (a) Growth characteristics of strain LS1 cultivated in LB supplemented with different concentrations of haemin. Data shown are the means±sd of four independent experiments done in duplicate. Arrows indicate the time points of sampling for the determination of Hla contents and haemolytic activities in supernatants. (b) Haemolytic activities of supernatants of LS1 cultures grown for 6 h in the absence and presence of haemin (4 µM) and 2,2′-bipyridine (250 µM). *P<0.05, for unsupplemented versus supplemented cultures (Wilcoxon signed-rank test). (c) Hla secretion in response to haemin (4 µM) and 2,2′-bipyridine (250 µM). Cultures were grown to the time points indicated, and filter-sterilized supernatants were subjected to Western blot analysis using anti-Hla antibodies. The results are representative of at least two independent experiments.
Haemin and 2,2′-bipyridine supplementation modulate α-haemolysin production and haemolytic activity of S. aureus LS1 in opposite directions
To study whether haemin might affect haemolysin production and its secretion in LS1, we tested the haemolytic activities and α-haemolysin (Hla) contents of supernatants of LS1 cells that were grown for 6 h either in the presence or in the absence of 4 µM haemin. To account for differences in growth yields, cultures were adjusted with fresh LB to OD600 1 prior to sampling the supernatants. The haemin-supplemented supernatants of LS1 cultures showed a significantly reduced capacity to lyse human erythrocytes, as compared with the control (Fig. 1b). Similarly, we detected a strong reduction in Hla in the haemin-supplemented culture supernatants after 6 h of growth (Fig. 1c). To rule out that growth phase-dependent differences between the haemin-supplemented culture and the untreated control (Fig. 1a) might account for this effect, we determined the Hla contents in the supernatants of early stationary phase cultures (OD600 3). We again found a decrease in Hla in the haemin-supplemented culture supernatants, although this was less pronounced than that observed after 6 h of growth (Fig. 1c). As we have recently observed that iron depletion increases the production of Hla and the haemolytic activity of S. aureus strain Newman (Torres et al., 2010), we also tested here whether and how the addition of the iron scavenger 2,2′-bipyridine (250 µM) affected the haemolytic activity and Hla expression of strain LS1. In line with our previous findings, we observed a significant increase in haemolytic activity in the supernatants of the 2,2′-bipyridine-challanged cultures (Fig. 1b), and detected elevated Hla signals in these culture supernatants (Fig. 1c). These data strongly suggest that subinhibitory amounts of haemin reduce the haemolytic activity and α-haemolysin production and/or secretion of S. aureus, while iron-limiting conditions trigger the opposite effects by upregulating Hla production/secretion and increasing the haemolytic activity.
Subinhibitory concentrations of haemin affect the transcription of haemolysins in S. aureus
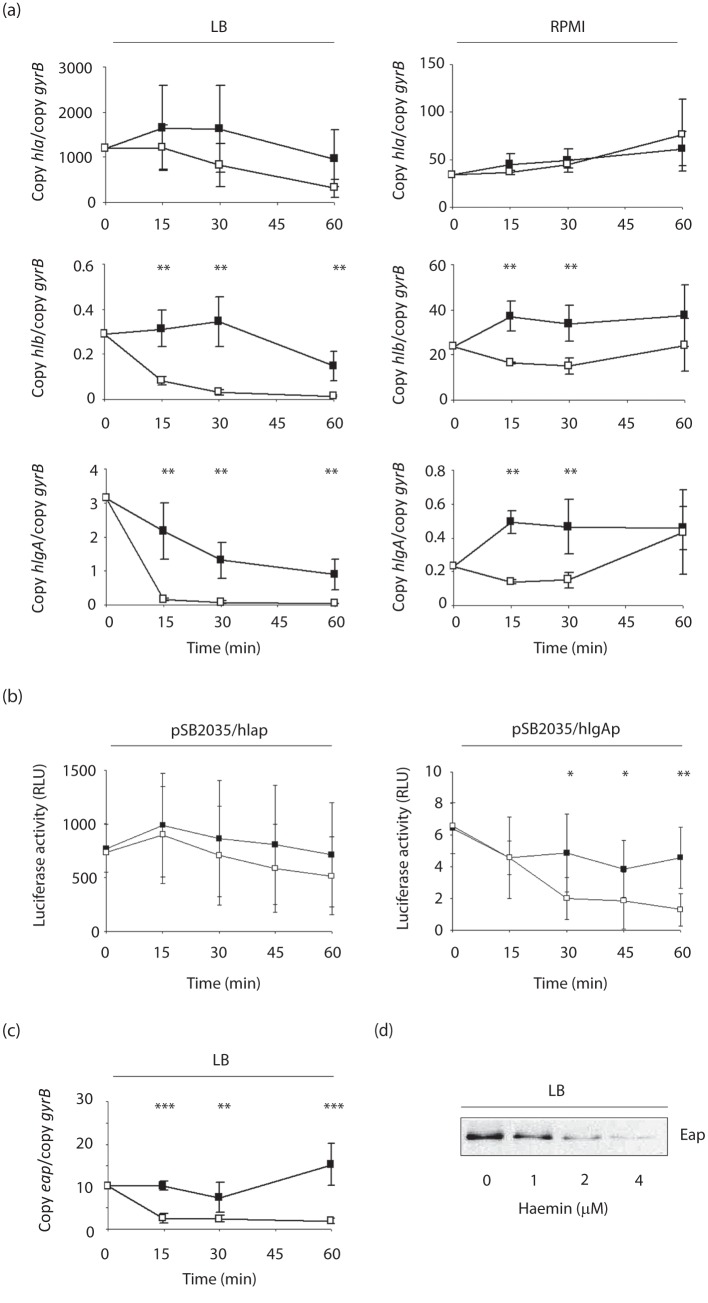
To determine how haemin affects the haemolysin production of S. aureus, we next analysed the transcription of four major haemolysins of this pathogen, hla (encoding α-haemolysin), hlb (encoding β-haemolysin), hlgA (encoding the S-class component of γ-haemolysin) and hld, which is encoded by RNAIII of the agr locus, in response to haemin. To eliminate potential growth phase-dependent differences in haemolysin transcription in this assay, LS1 cells were grown for 6 h in LB and split into two parts, and one part was supplemented with haemin (4 µM). Samples for qRT-PCR were taken immediately after addition of haemin (time point 0), and 15, 30 and 60 min later (Fig. 2a, c). Addition of subinhibitory concentrations of haemin resulted in significant reductions in transcription for hlb and hlgA as early as 15 min after the supplementation. A similar, but not significant, trend was observed for hla transcription. No such difference was observed for RNAIII transcription between the unsupplemented and the supplemented samples (data not shown).
Fig. 2.
Transcription of haemolysin genes and eap in LS1 in response to haemin. (a) Effect of haemin supplementation (4 µM) on the transcription of haemolysin genes in LS1 cells grown in LB and RPMI. Cells were grown for 6 h before haemin was added, and samples were taken at the time points indicated. Changes in transcription were assayed by qRT-PCR. Data shown are the means±sd of three independent experiments performed in duplicate. (b) Impact of haemin supplementation (4 µM) on luciferase activities of cultures of LS1 derivatives harbouring plasmids pSB2035/hlap and pSB2035/hlgAp. Cells were cultured in LB for 6 h before haemin (4 µM) was added. Luciferase activities were determined at the time points indicated. Data shown are the means±sd of seven independent experiments. (c) Effect of haemin supplementation (4 µM) on transcription of eap in LS1 cells grown in LB. *P<0.05, **P<0.01 for unsupplemented versus supplemented cultures (Wilcoxon signed-rank test). (d) Western blot analysis of Eap in cell-wall extracts of LS1 cells grown for 6 h in LB in the presence of different concentrations of haemin. The result is representative of at least two independent experiments.
The repressive effect of haemin on the transcription of haemolysins appears to be independent of growth media
To investigate the impact of the growth medium on the effect of haemin on haemolysin expression, we reproduced the experiment in a chemically defined medium, RPMI 1640 (hereafter RPMI), which contained smaller amounts of iron (approx. 0.5 µM) (Fenwick et al., 1996) than LB (approx. 20 µM) (Rayner & Sadler, 1990). In RPMI, strain LS1 produced nearly 60 times less hla transcripts than in LB after 6 h of growth, confirming recent findings that hla transcription is highly media-dependent (Ray et al., 2009). Transcription of hlb was also found to be affected by the growth medium, and was 120-fold increased in RPMI, while hlgA transcription varied only by a factor of 3 between LB and RPMI (Table 2). Addition of haemin to LS1 cells grown in RPMI for 6 h, however, again resulted in highly significant decreases in hlb and hlgA transcript levels when compared with the unsupplemented control (Fig. 2a). Transcription of hla, although strongly influenced by the growth medium in its transcription, was not affected by the addition of haemin.
Table 2. Effect of growth medium on transcription after 6 h of growth.
| Strain | Growth medium | Transcripts (copy/copy of gyrB)* | |||
| hla | hlb | hlgA | sae | ||
| LS1 | LB | 1631.9±964.1 | 0.3±0.1 | 1.3±0.5 | 696.8±371.4 |
| RPMI | 26.3±10.4 | 43.8±20.2 | 0.4±0.05 | 10.8±2.8 | |
| LS1 Δsae | LB | 0.003±0.0006 | 0.02±0.005 | 0.01±0.002 | − |
| RPMI | 0.07±0.03 | 0.06±0.02 | 0.01±0.004 | − | |
| LS1 Δfur | LB | 11.2±5.1 | 0.6±0.2 | 4.7±3.1 | 3.2±0.6 |
| RPMI | 115.63±99.75 | 66.3±61.9 | 0.2±0.1 | 4.8±0.9 | |
Transcript levels were determined by the 2–ΔCT method. Data are means±sd of at least three independent experiments performed in duplicate.
We confirmed the haemin effect on haemolysin transcription with promoter–reporter gene fusion experiments. We created a series of promoter–reporter gene fusion constructs based on agrP3-gfp-lux plasmid pSB2035 (Qazi et al., 2001), in which the agr P3 promoter was replaced by the hla, hlb and hlgA promoters. The plasmids were used to transform strain LS1, and the impact of haemin supplementation (4 µM) on the activities of the haemolysin promoters was determined as changes in luciferase activities. In line with our qRT-PCR data, we observed the strongest luciferase activity values with the LS1 derivative harbouring the hla promoter construct [773±224 relative light units (RLU) after 6 h of growth in LB]. Luciferase activity values with the LS1 derivative harbouring the hlgA promoter construct were more than 100-fold smaller than those found for the hla construct (6.4±1.6 RLU after 6 h of growth). However, we failed to detect luciferase activity with the LS1 derivative harbouring the hlb promoter construct, which was indistinguishable from the background luminescence of our test system. Addition of haemin to the 6 h LB cell cultures of LS1 derivatives harbouring the hla and hlgA promoter–reporter gene fusion constructs, resulted in a threefold reduction in luciferase activities for the strain harbouring the hlgA promoter construct, while luciferase activities were not affected by haemin in LS1 transformed with the hla promoter–reporter gene fusion construct (Fig. 2b). Although in line with our qRT-PCR data, the hla promoter–reporter gene fusion results are at first sight in conflict with our Western blot data demonstrating decreased Hla contents in haemin-supplemented culture supernatants (Fig. 1c). However, as Hla production and its stability are markedly affected by post-transcriptional and post-translational mechanisms (Morfeldt et al., 1995; Lindsay & Foster, 1999), we cannot exclude here that the observed decrease in Hla in the supernatants of haemolysin-challenged cultures might be due to haemin-induced alterations in hla translation or protease activities.
Iron sulphate affects haemolysin transcription in a media-dependent fashion
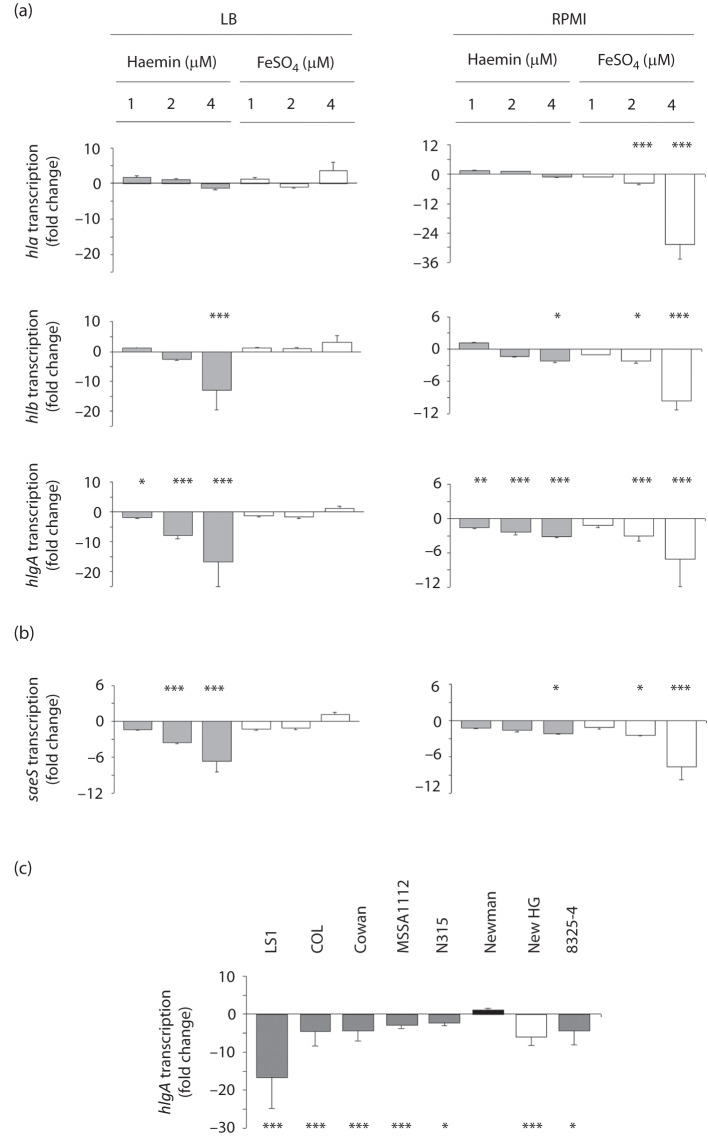
To test whether the observed effect on haemolysin transcription is haemin-dependent or could be induced by other iron sources as well, LS1 cells were grown for 6 h in either LB or RPMI. Subsequently, increasing amounts of haemin (1–4 µM) or FeSO4 (1–4 µM) were added to the cultures (Fig. 3). In the iron-rich medium LB, addition of FeSO4 did not significantly alter the transcription of hla, hlb or hlgA at any of the concentrations tested. Supplementation of LB with increasing amounts of haemin, on the other hand, decreased the transcription of hlb and hlgA. None of the conditions tested altered the transcription of hla significantly, although higher concentrations of FeSO4 seemed to foster transcription. In the iron-poor medium RPMI, the addition of either FeSO4 or haemin resulted in clear reductions in hlb and hlgA transcription (Fig. 3a), and FeSO4 seemed to repress the transcription of these two genes even more strongly than the equimolar amounts of haemin. However, as in LB, neither of the concentrations of haemin tested significantly altered the transcription of hla in RPMI. Interestingly, the haemin-induced repressive effect on the transcription of hlb and hlgA was still effective under growth conditions in which S. aureus suffered from iron-limitation, as addition of FeSO4 clearly increased the growth of the LS1 culture in RPMI (data not shown). This suggests that the repressive effect of haemin is independent of the iron content of the growth medium. The haemolysin gene transcription-regulating mechanisms that were effective under iron-limiting conditions, on the other hand, appeared to be insensitive to the iron source, as they were affected by both haemin and FeSO4 supplementation.
Fig. 3.
Effect of haemin and FeSO4 supplementation on transcription of haemolysin genes and of saeS in S. aureus. Cells were grown for 6 h in LB and RPMI, before the haemin or FeSO4 concentrations indicated were added, and samples were taken 30 min after addition of the substances. (a) Effects on haemolysin gene transcription in LS1. (b) Effects on saeS transcription in LS1. (c) Effect of haemin supplementation (4 µM) on hlgA transcription in different S. aureus lineages. Changes in transcription were assayed by qRT-PCR. Data shown are the means±sd of fold changes between supplemented and unsupplemented controls of three independent experiments done in duplicate. *P<0.05, **P<0.01, ***P<0.001 for unsupplemented versus supplemented cultures (Wilcoxon signed-rank test).
Effect of haemin supplementation on the activity of selected global regulators
To identify regulatory molecules that might exert the haemin effect on haemolysin transcription, we tested the impact of haemin supplementation on the transcription of selected regulators known to affect haemolysin expression, such as ArlRS (Liang et al., 2005), SaePQRS (Giraudo et al., 1997; Goerke et al., 2001, 2005; Li & Cheung, 2008; Xiong et al., 2006), SarA (Goerke et al., 2001; Xiong et al., 2006) and σB (Bischoff et al., 2004; Cheung et al., 1999; Horsburgh et al., 2002; Goerke et al., 2005). The addition of haemin to LS1 cells grown in LB did not affect the transcription of arlRS, sarA or sigB (data not shown). However, in LB a strong repression in sae transcription was observed after the addition of haemin (Fig. 3b), indicating that this iron-containing porphyrin molecule might affect the activity of the Sae two-component regulatory system. Moreover, similarly to hla and hlb, sae expression was found to be greatly affected by the medium: when LS1 cells were grown in RPMI, sae transcription was found to be ~60-fold decreased when compared with LB (Table 2). Supplementing RPMI with haemin resulted in a slight (and significant only at 4 µM) reduction in sae transcription, whereas FeSO4 had a stronger effect (Fig. 3b). However, the expression patterns of sae upon haemin and FeSO4 supplementation in both media were similar to those found for hlb and hlgA (Fig. 3a).
The sae locus is required for the effect of haemin on the transcription of haemolysins
We and others have reported recently that the ferric uptake regulator Fur modulates haemolysin production and Sae activity in S. aureus strain Newman under iron-limiting conditions (Johnson et al., 2011; Torres et al., 2010). To test whether Sae and/or Fur are required for mediating the haemin effect on haemolysin transcription, we inactivated the sae and fur loci in LS1. We first determined the impact of the mutations on haemolytic activities after growth on SBA, RBA and HBA plates (Fig. 4a). While the wild-type showed clear haemolytic zones surrounding the colonies on all plate types tested, this was not the case with the mutants. Inactivation of the sae locus in LS1 resulted in a marked decrease of the haemolytic areas surrounding the mutant colonies, and this phenotype was observed on all blood agar plate types. The LS1 Δfur mutant, on the other hand, produced haemolytic zones on SBA and RBA plates that were comparable with those found with the wild-type. On agar plates supplemented with human blood, however, LS1 Δfur produced almost no haemolytic areas, and this phenotype was associated with a clearly decreased colony size of the mutant. Interestingly, an LS1 Δhla mutant, incapable of producing α-haemolysin, formed smaller haemolytic areas on RBA but not on SBA or HBA plates, indicating that Hla, although the predominantly expressed haemolysin of this strain, was not essential for the haemolytic activity of LS1 on human red blood cells.
Fig. 4.
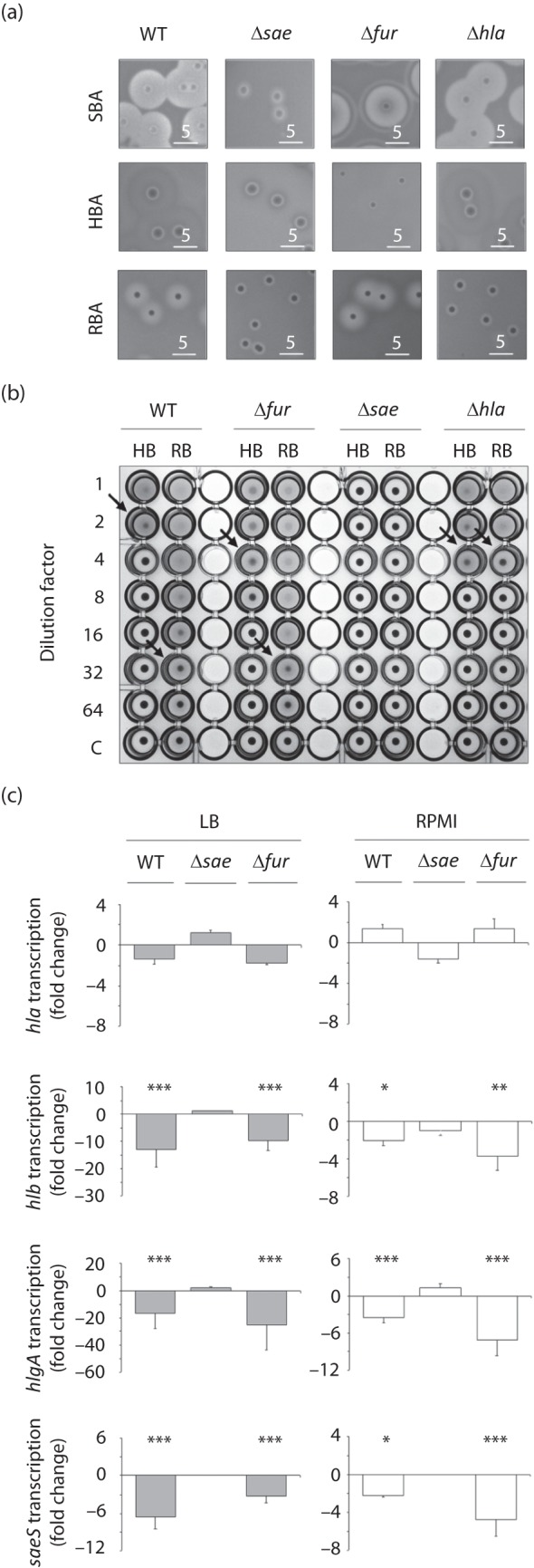
Effects of fur, hla and sae deletions on the haemolytic capacity of LS1. (a) Cells were grown for 24 h at 37 °C on agar plates supplemented with sheep blood (SBA), human blood (HBA) and rabbit blood (RBA), followed by 24 h incubation at 4 °C. (b) Haemolytic activities of supernatants of LS1 and its derivatives on human (HB) and rabbit erythrocytes (RB). (c) Effect of haemin supplementation (4 µM) on transcription of hla, hlb, hlgA and saeS of strain LS1 and its derivatives grown in LB and RPMI. Cells were grown for 6 h before haemin was added, and samples were taken 30 min after addition of the substances. Changes in transcription were assayed by qRT-PCR. Data shown are the means±sd of fold changes between supplemented and unsupplemented controls of three independent experiments performed in duplicate. *P<0.05, **P<0.01, ***P<0.001 for unsupplemented versus supplemented cultures (Wilcoxon signed-rank test). WT, wild-type.
Second, we assayed the haemolytic potentials of supernatants of OD600-adjusted overnight cultures of LS1 and its derivatives on human and rabbit erythrocytes (Fig. 4b). These analyses essentially confirmed our findings described above: whereas supernatants of LS1 Δsae produced no visible haemolytic activities in these assays, LS1 Δfur supernatants showed haemolytic activities that were comparable with those obtained with the wild-type supernatants. Interestingly, supernatants of LS1 Δhla showed haemolytic activities on human erythrocytes that were in the range of those found with the wild-type and LS1 Δfur supernatants, while haemolysis of rabbit erythrocytes was clearly decreased with the LS1 Δhla supernatants when compared with the wild-type and LS1 Δfur.
Third, we assayed the impact of the mutations on the transcription of the haemolysin genes. Inactivation of both sae and fur clearly affected the transcription of haemolysin genes, although this effect was highly media-dependent (Table 2). However, addition of subinhibitory concentrations of haemin to LS1 Δfur cells grown in LB and RPMI yielded similar reductions in hlb, hlgA and sae transcript levels as in LS1 wild-type cells, while inactivation of sae in LS1 abolished the haemin-induced reduction of haemolysin transcription (Fig. 4c). Transcription of hla was apparently not affected by the addition of haemin to either of the mutant cultures.
These findings suggest that Hla is not a major player in the haemolysis phenotype observed with LS1, and indicate that Hla is not essential to the effect of haemin on haemolysis of human erythrocytes. Interestingly, although hla transcription was severely downregulated in saeRS deletion strains (Giraudo et al., 1997; Goerke et al., 2001, 2005; Xiong et al., 2006), and direct binding of the response regulator SaeR to the hla promoter has been recently demonstrated (Nygaard et al., 2010; Sun et al., 2010), hla transcription was not significantly affected by haemin-mediated sae repression in LS1, while hlb, hlgA and saeS responded to this stimulus in an Sae-dependent manner. It has been shown that sae target genes differ significantly regarding SaeRS dosage and SaeS-sensing mechanisms. For instance, hla expression is not affected by mutations in SaeS as present in strain Newman and is rather insensitive towards SaeRS dosage (Mainiero et al., 2010). The data presented here suggest that the Sae-dependent modulation of haemolysin genes in S. aureus might be even more complicated.
S. aureus strain Newman differs in its haemin effect on hlgA transcription
To test whether the repressive effect of haemin on haemolysin transcription is restricted to S. aureus strain LS1, we analysed the impact of haemin supplementation on the transcription of hlgA in a number of laboratory strains (Fig. 3c). With the exception of wild-type strain Newman (harbouring a Pro18 mutation in saeS), all other isolates tested, including the Newman derivative New HG, which contains a repaired saeS gene (saeSLeu18) (Mainiero et al., 2010), showed the same effect as LS1, although the repression intensities differed from strain to strain.
In the context of our previous findings (Torres et al., 2010), our new observations underline the importance of strain background and media when assessing the consequences of haemin, iron exposure or iron restriction on regulation and target gene expression in S. aureus. In our previous study, a fur mutant of strain Newman displayed not only elevated total exoprotein production but also increased haemolysis and hla expression upon cultivation in an iron-rich medium. In the fur mutant of strain LS1, we also observed increased hla transcription, but only in the low iron-containing medium RPMI. Cultivation of LS1 Δfur cells in the iron-rich medium LB, on the other hand, resulted in a strong decrease in hla transcription, contrary to our previous findings and those of Johnson et al. (2011) with strain Newman. An apparent explanation for this discrepancy might be the leucine to proline mutation at position 18 in SaeS of strain Newman, which leads to a constitutive activation of the Sae system in this genetic lineage (Adhikari & Novick, 2008; Mainiero et al., 2010). We and others have shown that the Newman sae system responds differently to environmental stresses (Geiger et al., 2008; Schäfer et al., 2009) from a Newman derivative harbouring a leucine at position 18 of SaeS (SaeSLeu), and that a Newman SaeSPro derivative expresses exoproteins differently from Newman wild-type (Adhikari & Novick, 2008; Luong et al., 2011; Mainiero et al., 2010). In line with this, we found that a Newman derivative harbouring an saeSLeu allele, New HG, responded to haemin exposure similarly to strain LS1 and a number of other S. aureus genetic backgrounds, while strain Newman did not (Fig. 3c), strongly suggesting that the response of the Sae system to environmental signals is severely altered by the saeS mutation of strain Newman.
Subinhibitory concentrations of haemin and 2,2′-bipyridine affect Sae activity
Although Sae is known to affect its own expression (Adhikari & Novick, 2008), differences observed in sae transcription do not necessarily reflect Sae activity, as other regulators have been reported to modulate the transcription of this regulatory locus too (Giraudo et al., 2003; Li & Cheung, 2008; Novick & Jiang, 2003). To test whether haemin and 2,2′-bipyridine affect not only sae transcription but also Sae activity, we analysed the impact of haemin and 2,2′-bipyridine supplementation, on the expression of the extracellular adherence protein (Eap), which was previously shown to be dependent on Sae activity in its expression (Harraghy et al., 2005). Similar to our observations made for hlb and hlgA, addition of 4 µM haemin to an LS1 culture grown for 6 h in LB resulted in an immediate reduction in eap transcription, suggesting that Sae activity is altered by the presence of subinhibitory concentrations of this molecule (Fig. 2c). This hypothesis is further strengthened by findings obtained from Western blot analyses performed with cell-wall extracts of LS1 cells that were cultured in the absence and presence of increasing concentrations of haemin. Haemin concentrations as low as 1 µM were sufficient to reduce the amount of this cell wall-associated adhesin, while 4 µM haemin nearly abolished the production of Eap (Fig. 2d). Addition of the iron scavenger 2,2′-bipyridine (250 µM) to a 6 h LB culture of LS1, on the other hand, yielded an immediate and significant increase in sae and hlgA transcription (Fig. 5a). Similarly, growth of LS1 in LB supplemented with 2,2′-bipyridine increased Eap quantity on the bacterial cell surface markedly (Fig. 5b), in line with recent observations showing that low iron conditions affect Sae activity in a Fur-dependent manner (Johnson et al., 2011).
Fig. 5.
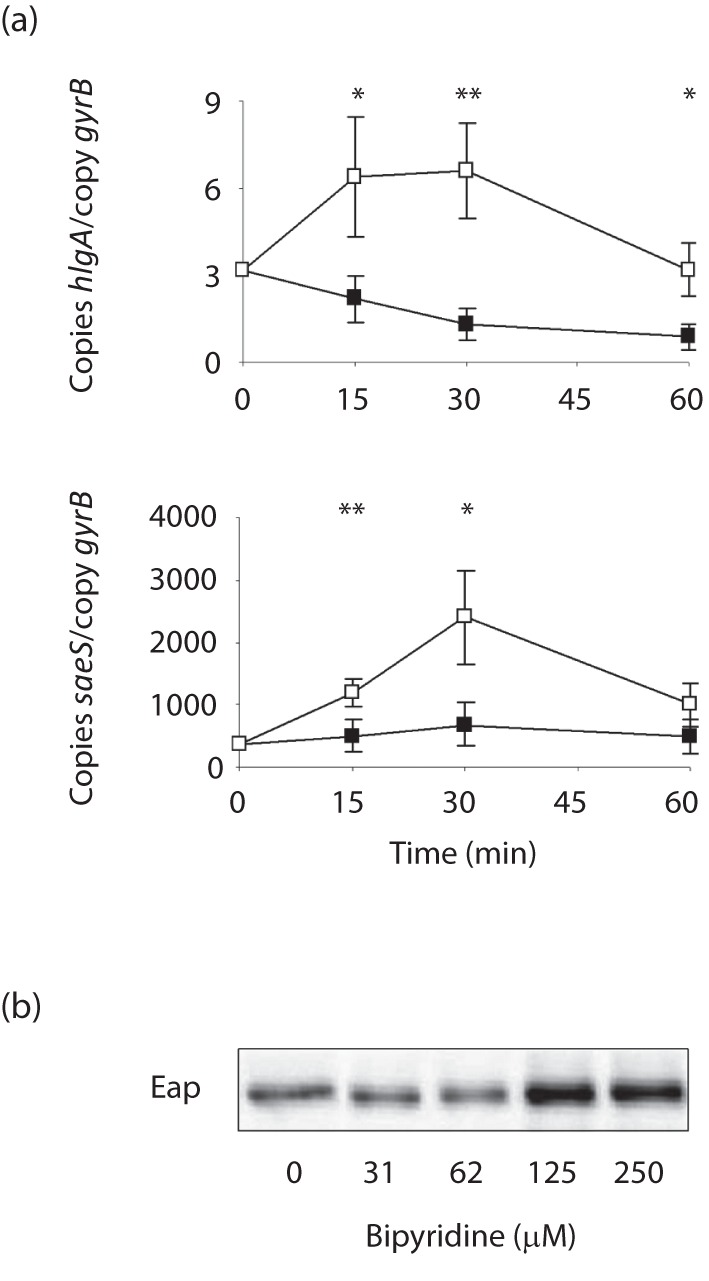
Effect of 2,2′-bipyridine supplementation on the transcription of saeS and hlgA, and on Eap production in LS1. (a) LS1 cells were grown for 6 h in LB before 2,2′-bipyridine (250 µM) was added, and samples were taken at the time points indicated. Changes in transcription were assayed by qRT-PCR. Data shown are the means±sd of three independent experiments performed in duplicate. *P<0.05, **P<0.01 for unsupplemented versus supplemented cultures (Wilcoxon signed-rank test). (b) Western blot analysis of Eap in cell-wall extracts of LS1 cells grown for 6 h in LB in the presence of the 2,2′-bipyridine concentrations indicated. The result is representative of at least two independent experiments.
Conclusions
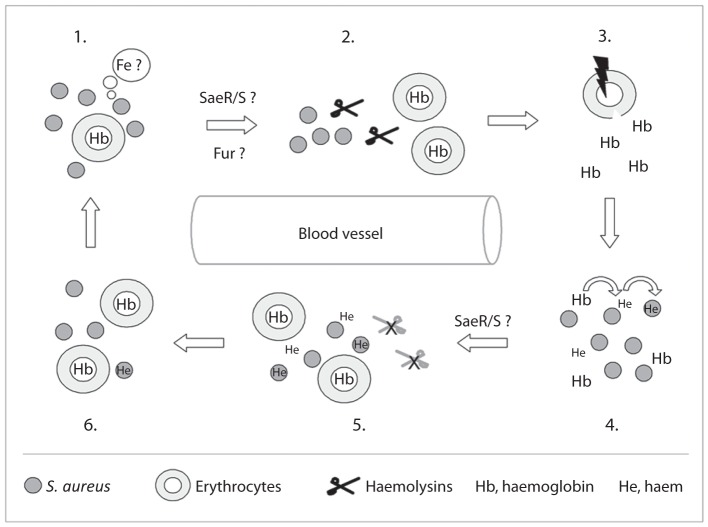
Haemoglobin is considered to be the major iron source for S. aureus during infection (Lee et al., 2008). Accordingly, the pathogen is equipped with a number of factors, allowing it (i) to release haemoglobin from red blood cells, (ii) to bind haemoglobin and to import the iron-containing porphyrin (haem), (iii) to degrade the imported haem and to release the iron, and (iv) to protect itself from the cytotoxic effects that are associated with this molecule. Here, we show that the sae system is likely to contribute to this complex regulatory circuit by modulating the haemolysin production of S. aureus in response to iron limitation and haem availability. Based on our transcriptional data, we propose that S. aureus regulates its haemolytic potential in an Sae- and Fur-dependent manner (Fig. 6). Under iron-limiting conditions, S. aureus activates the production and secretion of haemolysins via the Sae and Fur regulatory systems, while higher (subinhibitory) concentrations of haemin in the extracellular environment of S. aureus – probably signalling to the bacterium that iron is available in sufficient amounts – lead to a downregulation of haemolysin production that is due to a repression of Sae activity. However, the molecular mechanism(s) by which haemin can interact with the Sae regulatory system to modulate its activity still need to be identified.
Fig. 6.
Proposed model for the regulation of S. aureus haemolytic activity in response to iron availability and haem. Iron limitation (1) leads in S. aureus to an increased production and secretion of haemolysins via Fur and SaeR/S (2), which disintegrate erythrocytes to release haemoglobin (3). The released iron-storage protein is bound by the bacterium to particular receptors, and the haem subunit is released and imported into the cytoplasm (4). The increased intracellular/extracellular haem concentration is sensed by the SaeR/S system to decrease SaeS activity, which leads to a downregulation of haemolysin production (5) to prevent intoxication of the bacterium by this molecule. The ongoing uptake/consumption of extracellular haemoglobin/haem by S. aureus (6) depletes the iron in the extracellular milieu, which restarts the cycle (1).
Acknowledgements
This study was supported by the Deutsche Forschungsgemeinschaft (DFG; grant He 1850/7-3), the German Federal Ministry for Education and Science (BMBF) (grant no. 01 KI 07103 Skinstaph to M. H.) and through the HOMFOR programme of the University of Saarland Hospitals. E. P. S. was supported by the National Institutes of Health, USA (AI069233 and AI073843). We thank Karin Hilgert and Torsten Hartmann for technical assistance, and Dr Joe Patti for providing the anti-Eap antibody.
Abbreviations:
- qRT-PCR
quantitative real-time RT-PCR
- RLU
relative light units
Footnotes
Two supplementary tables are available with the online version of this paper.
References
- Adhikari R. P., Novick R. P. (2008). Regulatory organization of the staphylococcal sae locus. Microbiology 154, 949–959. 10.1099/mic.0.2007/012245-0 [DOI] [PubMed] [Google Scholar]
- Beasley F. C., Vinés E. D., Grigg J. C., Zheng Q., Liu S., Lajoie G. A., Murphy M. E., Heinrichs D. E. (2009). Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol 72, 947–963. 10.1111/j.1365-2958.2009.06698.x [DOI] [PubMed] [Google Scholar]
- Bischoff M., Dunman P., Kormanec J., Macapagal D., Murphy E., Mounts W., Berger-Bächi B., Projan S. (2004). Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186, 4085–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremell T., Abdelnour A., Tarkowski A. (1992). Histopathological and serological progression of experimental Staphylococcus aureus arthritis. Infect Immun 60, 2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner S., Monteil H., Prévost G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28, 183–200. [DOI] [PubMed] [Google Scholar]
- Chatterjee I., Becker P., Grundmeier M., Bischoff M., Somerville G. A., Peters G., Sinha B., Harraghy N., Proctor R. A., Herrmann M. (2005). Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol 187, 4488–4496. 10.1128/JB.187.13.4488-4496.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. L., Chien Y. T., Bayer A. S. (1999). Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun 67, 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2009). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved standard, 8th edn, CLSI document M07–A8. Wayne, PA: Clinical and Laboratory Standards Institute.
- Diekema D. J., Pfaller M. A., Schmitz F. J., Smayevsky J., Bell J., Jones R. N., Beach M., SENTRY Partcipants Group (2001). Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis 32 (Suppl 2), S114–S132. 10.1086/320184 [DOI] [PubMed] [Google Scholar]
- Dryla A., Gelbmann D., von Gabain A., Nagy E. (2003). Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol 49, 37–53. 10.1046/j.1365-2958.2003.03542.x [DOI] [PubMed] [Google Scholar]
- Duthie E. S. (1952). Variation in the antigenic composition of staphylococcal coagulase. J Gen Microbiol 7, 320–326. 10.1099/00221287-7-3-4-320 [DOI] [PubMed] [Google Scholar]
- Dyke K. G., Jevons M. P., Parker M. T. (1966). Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aureus. Lancet 287, 835–838. 10.1016/S0140-6736(66)90182-6 [DOI] [PubMed] [Google Scholar]
- Entenza J. M., Vouillamoz J., Glauser M. P., Moreillon P. (1997). Levofloxacin versus ciprofloxacin, flucloxacillin, or vancomycin for treatment of experimental endocarditis due to methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob Agents Chemother 41, 1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everse J., Hsia N. (1997). The toxicities of native and modified hemoglobins. Free Radic Biol Med 22, 1075–1099. 10.1016/S0891-5849(96)00499-6 [DOI] [PubMed] [Google Scholar]
- Fenwick B., Rider M., Liang J., Brightman A. (1996). Iron repressible outer membrane proteins of Moraxella bovis and demonstration of siderophore-like activity. Vet Microbiol 48, 315–324. 10.1016/0378-1135(95)00155-7 [DOI] [PubMed] [Google Scholar]
- Geiger T., Goerke C., Mainiero M., Kraus D., Wolz C. (2008). The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol 190, 3419–3428. 10.1128/JB.01927-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo A. T., Cheung A. L., Nagel R. (1997). The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch Microbiol 168, 53–58. 10.1007/s002030050469 [DOI] [PubMed] [Google Scholar]
- Giraudo A. T., Mansilla C., Chan A., Raspanti C., Nagel R. (2003). Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr Microbiol 46, 246–250. [DOI] [PubMed] [Google Scholar]
- Goerke C., Fluckiger U., Steinhuber A., Zimmerli W., Wolz C. (2001). Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol 40, 1439–1447. 10.1046/j.1365-2958.2001.02494.x [DOI] [PubMed] [Google Scholar]
- Goerke C., Fluckiger U., Steinhuber A., Bisanzio V., Ulrich M., Bischoff M., Patti J. M., Wolz C. (2005). Role of Staphylococcus aureus global regulators sae and sigmaB in virulence gene expression during device-related infection. Infect Immun 73, 3415–3421. 10.1128/IAI.73.6.3415-3421.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraghy N., Kormanec J., Wolz C., Homerova D., Goerke C., Ohlsen K., Qazi S., Hill P., Herrmann M. (2005). sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 151, 1789–1800. 10.1099/mic.0.27902-0 [DOI] [PubMed] [Google Scholar]
- Horsburgh M. J., Ingham E., Foster S. J. (2001). In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183, 468–475. 10.1128/JB.183.2.468-475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. (2002). sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184, 5457–5467. 10.1128/JB.184.19.5457-5467.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Sengupta M., Purves J., Tarrant E., Williams P. H., Cockayne A., Muthaiyan A., Stephenson R., Ledala N. & other authors (2011). Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int J Med Microbiol 301, 44–52. 10.1016/j.ijmm.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Ohta T., Uchiyama I., Baba T., Yuzawa H., Kobayashi I., Cui L., Oguchi A., Aoki K. & other authors (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357, 1225–1240. 10.1016/S0140-6736(00)04403-2 [DOI] [PubMed] [Google Scholar]
- Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. P. (2008). Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem 283, 30957–30963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Cheung A. (2008). Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun 76, 1068–1075. 10.1128/IAI.01069-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Zheng L., Landwehr C., Lunsford D., Holmes D., Ji Y. (2005). Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol 187, 5486–5492. 10.1128/JB.187.15.5486-5492.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay J. A., Foster S. J. (1999). Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol Gen Genet 262, 323–331. 10.1007/s004380051090 [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N Engl J Med 339, 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Luong T. T., Sau K., Roux C., Sau S., Dunman P. M., Lee C. Y. (2011). Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol 193, 686–694. 10.1128/JB.00987-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero M., Goerke C., Geiger T., Gonser C., Herbert S., Wolz C. (2010). Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol 192, 613–623. 10.1128/JB.01242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso A. W., Schneewind O. (2006). Iron acquisition and transport in Staphylococcus aureus. Biometals 19, 193–203. [DOI] [PubMed] [Google Scholar]
- Mason W. J., Skaar E. P. (2009). Assessing the contribution of heme-iron acquisition to Staphylococcus aureus pneumonia using computed tomography. PLoS ONE 4, e6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., Schneewind O. (2003). Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909. 10.1126/science.1081147 [DOI] [PubMed] [Google Scholar]
- Morfeldt E., Taylor D., von Gabain A., Arvidson S. (1995). Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14, 4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. M., Hartford O., Foster T., Tarkowski A. (1999). Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect Immun 67, 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. (1967). Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33, 155–166. 10.1016/0042-6822(67)90105-5 [DOI] [PubMed] [Google Scholar]
- Novick R. P., Jiang D. (2003). The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149, 2709–2717. [DOI] [PubMed] [Google Scholar]
- Nygaard T. K., Pallister K. B., Ruzevich P., Griffith S., Vuong C., Voyich J. M. (2010). SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis 201, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly M., de Azavedo J. C. S., Kennedy S., Foster T. J. (1986). Inactivation of the alpha-haemolysin gene of Staphylococcus aureus 8325-4 by site-directed mutagenesis and studies on the expression of its haemolysins. Microb Pathog 1, 125–138. 10.1016/0882-4010(86)90015-X [DOI] [PubMed] [Google Scholar]
- Qazi S. N., Counil E., Morrissey J., Rees C. E., Cockayne A., Winzer K., Chan W. C., Williams P., Hill P. J. (2001). agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect Immun 69, 7074–7082. 10.1128/IAI.69.11.7074-7082.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B., Ballal A., Manna A. C. (2009). Transcriptional variation of regulatory and virulence genes due to different media in Staphylococcus aureus. Microb Pathog 47, 94–100. 10.1016/j.micpath.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Rayner M. H., Sadler P. J. (1990). Precipitation of cadmium in a bacterial culture medium: Luria-Bertani broth. FEMS Microbiol Lett 71, 253–257. 10.1111/j.1574-6968.1990.tb03832.x [DOI] [PubMed] [Google Scholar]
- Reniere M. L., Torres V. J., Skaar E. P. (2007). Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20, 333–345. 10.1007/s10534-006-9032-0 [DOI] [PubMed] [Google Scholar]
- Rogasch K., Rühmling V., Pané-Farré J., Höper D., Weinberg C., Fuchs S., Schmudde M., Bröker B. M., Wolz C. & other authors (2006). Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol 188, 7742–7758. 10.1128/JB.00555-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A. (2004). Microbiology. Pathogenic bacteria prefer heme. Science 305, 1577–1578. 10.1126/science.1102975 [DOI] [PubMed] [Google Scholar]
- Schäfer D., Lâm T. T., Geiger T., Mainiero M., Engelmann S., Hussain M., Bosserhoff A., Frosch M., Bischoff M. & other authors (2009). A point mutation in the sensor histidine kinase SaeS of Staphylococcus aureus strain Newman alters the response to biocide exposure. J Bacteriol 191, 7306–7314. 10.1128/JB.00630-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar E. P., Gaspar A. H., Schneewind O. (2004). IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem 279, 436–443. 10.1074/jbc.M307952200 [DOI] [PubMed] [Google Scholar]
- Sobke A. C., Selimovic D., Orlova V., Hassan M., Chavakis T., Athanasopoulos A. N., Schubert U., Hussain M., Thiel G. & other authors (2006). The extracellular adherence protein from Staphylococcus aureus abrogates angiogenic responses of endothelial cells by blocking Ras activation. FASEB J 20, 2621–2623. 10.1096/fj.06-5764fje [DOI] [PubMed] [Google Scholar]
- Somerville G. A., Proctor R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev 73, 233–248. 10.1128/MMBR.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I., Perkins-Balding D. (2002). Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol 21, 281–295. 10.1089/104454902753759708 [DOI] [PubMed] [Google Scholar]
- Sun F., Li C., Jeong D., Sohn C., He C., Bae T. (2010). In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol 192, 2111–2127. 10.1128/JB.01524-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny-Meyer L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev 61, 337–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. J., Pishchany G., Humayun M., Schneewind O., Skaar E. P. (2006). Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol 188, 8421–8429. 10.1128/JB.01335-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. J., Stauff D. L., Pishchany G., Bezbradica J. S., Gordy L. E., Iturregui J., Anderson K. L., Dunman P. M., Joyce S., Skaar E. P. (2007). A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1, 109–119. 10.1016/j.chom.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. J., Attia A. S., Mason W. J., Hood M. I., Corbin B. D., Beasley F. C., Anderson K. L., Stauff D. L., McDonald W. H. & other authors (2010). Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect Immun 78, 1618–1628. 10.1128/IAI.01423-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. (2000). Iron and oxidative stress in bacteria. Arch Biochem Biophys 373, 1–6. 10.1006/abbi.1999.1518 [DOI] [PubMed] [Google Scholar]
- Xiong Y. Q., Willard J., Yeaman M. R., Cheung A. L., Bayer A. S. (2006). Regulation of Staphylococcus aureus α-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis 194, 1267–1275. 10.1086/508210 [DOI] [PubMed] [Google Scholar]
- Yamazaki K., Kato F., Kamio Y., Kaneko J. (2006). Expression of γ-hemolysin regulated by sae in Staphylococcus aureus strain Smith 5R. FEMS Microbiol Lett 259, 174–180. 10.1111/j.1574-6968.2006.00236.x [DOI] [PubMed] [Google Scholar]