Abstract
Anaplasma phagocytophilum is an obligately intracellular, tick-transmitted, bacterial pathogen of humans and other animals. In order to evade host immunity during the course of infection, A. phagocytophilum utilizes gene conversion to shuffle approximately 100 functional pseudogenes into a single expression cassette of the msp2(p44) gene, which encodes the major surface antigen, major surface protein 2 (Msp2). The role and extent of msp2(p44) recombination in a reservoir host for A. phagocytophilum have not been evaluated. In the current study, we explored patterns of recombination and expression site variability of the msp2(p44) gene in three chronically infected woodrats, a reservoir for the disease in the Western USA. All three woodrats developed persistent infection of at least 6 months duration; two of them maintained active infection for at least 8 months. In total, we detected the emergence of 60 unique msp2(p44) expression site variants with no common temporal patterns of expression site recombination among the three A. phagocytophilum populations. Both the strength of infection (i.e. pathogen load) and the genetic diversity of pseudogenes detected at the msp2(p44) expression site fluctuated periodically during the course of infection. An analysis of the genomic pseudogene exhaustion rate showed that the repertoire of pseudogenes available to the A. phagocytophilum population could in theory become depleted within a year. However, the apparent emergence of variant pseudogenes suggests that the pathogen could potentially evade host immunity indefinitely. Our findings suggest a tightly co-evolved relationship between A. phagocytophilum and woodrats in which the pathogen perpetually evades host immunity yet causes no detectable disease.
Introduction
The strategy of varying antigen presentation over time is found in diverse pathogens, including protozoa, viruses, helminths and bacteria (Barbour, 1987; Centurion-Lara et al., 2004; Diamond, 2003; Kline et al., 2003; Taylor & Rudenko, 2006; Zhang et al., 1997). In order to produce variant antigens, the pathogen typically can produce variant genes de novo (e.g. hypermutability due to faulty mismatch repair) or retains a library of DNA that can serve as a source for recombination into antigen expression sites. Because these mechanisms impose evolutionary cost, antigenic variation must also confer some selective advantage to the pathogen, and the most common explanation is that it allows the pathogen to evade host immunity. However, many pathogens persist during some part of their life history in a reservoir host that experiences little or no clinical disease. Such a pathogen–reservoir commensalism suggests some form of coevolved avirulence. Comparison of pathogenic interactions where one stage of the pathogen persists in a clinically affected, immunocompetent host while another persists in a reservoir would allow for better understanding of the antigenic variation strategy.
Anaplasma phagocytophilum is an intra-leukocytic rickettsial bacterium that infects a variety of animals, including humans, wildlife (such as rodents, carnivores and deer) and domestic animals (Bakken et al., 1996; Dumler et al., 2005; Foley et al., 2004; Nicholson et al., 2010). It is transmitted by the ticks Ixodes pacificus and Ixodes scapularis in North America, Ixodes ricinus in Europe and Ixodes persulcatus in Asia (Cao et al., 2003; Des Vignes & Fish, 1997; Macleod & Gordon, 1933; Ohashi et al., 2005; Richter et al., 1996). Infections with A. phagocytophilum are acute or persistent depending on host species. In humans, horses and some mouse models, acute infections self-limit concurrently with development of adaptive immunity (Dumler et al., 2005). Persistent infections may occur in sheep, some dogs, and numerous rodent species that serve as reservoir hosts, including dusky-footed woodrats (Neotoma fuscipes), western grey squirrels (Sciurus griseus) and redwood chipmunks (Tamias ochrogenys) in the western USA, white-footed mice (Peromyscus leucopus) in the eastern USA, and voles and wood mice (Myodes spp. and Apodemus spp.) in the Old World (Foley et al., 2002; Nicholson et al., 1999; Nieto & Foley, 2008, 2009; Scorpio et al., 2011; Stuen et al., 1998; Telford et al., 1996).
A. phagocytophilum, which resides in neutrophilic vacuoles, has several strategies for intracellular survival, including inhibition of phagosome–lysosome fusion to prevent cell-mediated destruction, and evasion of host adaptive immunity by serially presenting genetically variant antigens (Dumler et al., 2005). Gene conversion allows A. phagocytophilum to sequentially shuffle ~100 different antigen ‘functional pseudogenes’ into a hypervariable region of a single expression site of the msp2(p44) gene with conserved 5′ and 3′ ends (Barbet et al., 2006; Lin et al., 2006). The MSP2 protein is a surface antigen of A. phagocytophilum and is homologous with the MSP2 and MSP3 surface proteins of Anaplasma marginale, a related pathogen that infects only ungulates (Brayton et al., 2003; Dunning Hotopp et al., 2006).
The kinetics and mechanisms of msp2(p44) pseudogene recombination in A. phagocytophilum have been investigated in mice and horses, which experience acute, self-limiting infection (Lin & Rikihisa, 2005; Rejmanek et al., 2012b; Scorpio et al., 2008; Wang et al., 2004). Based on the detection of expression site variants, even during short-lived infections there is a high rate of expressed pseudogene turnover with no clear patterns of expression shared between individuals, although some pseudogenes are expressed more frequently than others. In contrast, in chronically infected sheep, infections are detectable in cyclical waves, 2 or 3 weeks apart, suggesting that msp2(p44) recombination may allow the pathogen to temporarily escape cellular and humoral immune responses by antigenic variation (Granquist et al., 2008, 2010). In one particular study, pseudogenes were detected in different sheep at similar times post-infection, indicating a loosely programmed order of expression of certain pseudogenes (Granquist et al., 2010). Although sheep are a good model for studying persistent A. phagocytophilum infections, they are not natural reservoirs for this bacterium.
Dusky-footed woodrats (hereafter referred to as woodrats) are a common reservoir host for A. phagocytophilum throughout the Coast Range and parts of the Sierra Nevada of California. Chronic infection of at least 8–14 months was documented in field studies (Castro et al., 2001; Nicholson, 1998), and in one laboratory study, naturally infected woodrats remained A. phagocytophilum PCR-positive for up to 59 days, while woodrats experimentally infected with an equine-origin A. phagocytophilum strain remained positive for up to 90 days (Foley et al., 2002). In a related study, woodrats infected with one of three different strains (woodrat, equine or canine origin) remained PCR-positive from 35 to 62 days (Nieto et al., 2010). In all reports of anaplasmosis in woodrats, no evidence of clinical disease has been observed.
Little is known about how infection kinetics and patterns of msp2(p44) recombination differ among acutely infected, chronically infected but sick, and chronically infected natural reservoir hosts for A. phagocytophilum. In the current study we sought to describe the infection kinetics of A. phagocytophilum and, in particular, the patterns of pseudogene recombination in a reservoir host by infecting wild-caught woodrats with a woodrat-origin strain of A. phagocytophilum and monitoring the course of infection for up to 8 months.
Methods
Woodrat inoculation and sampling.
Three dusky-footed adult woodrats [one male (W9), and two females (W12 and W16)] were live-trapped in a mixed conifer/oak woodland near Bonny Doon, CA (N 37° 2′ 29.76′′, W 122° 9′ 1.95′′) and transported to the University of California, Davis. The woodrats were maintained in a barrier facility and received rodent chow and water ad libitum as well as occasional apples, peanuts and fresh oak leaves. All animal experiments were conducted with approval of the Institutional Animal Care and Use Committee at UC Davis, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. Prior to inoculation, the woodrats were held for at least 3 weeks and tested weekly for A. phagocytophilum infection by real-time PCR and once by serology using an indirect immunofluorescence antibody test (IFAT) following established protocols (Nicholson et al., 1999). When negative infection status was established the woodrats were inoculated intraperitoneally (i.p.) with 0.5 ml each of freshly collected woodrat blood from a naturally infected woodrat (DU-1), with a real-time PCR CT value of 28.2, trapped at Hendy Woods State Park in Northern California. One week post-inoculation (p.i.), woodrats were anaesthetized with ketamine (20 mg kg−1) and xylazine (4 mg kg−1), and 50 µl of blood was obtained via the retro-orbital sinus and placed in EDTA tubes (BD). Woodrats were then similarly bled once a week for the first 6 weeks and then every other week for the duration of the study. At the end of the experiment woodrats were euthanized by ketamine/xylazine overdose followed by cervical dislocation.
DNA extraction and PCR.
DNA from all blood samples including the DU-1 inoculum was extracted using a Qiagen Blood & Tissue kit (Qiagen) per manufacturer’s instructions with the following modifications. For each extraction, 50 µl blood was used as a template and the DNA was eluted in 50 µl water. Extracted DNA samples were screened for the presence of A. phagocytophilum DNA using a real-time PCR assay targeting the msp2(p44) gene following a previously established protocol (Drazenovich et al., 2006), using Maxima qPCR Master Mix (Fermentas). A subset of real-time PCR-positive DNA samples corresponding to a range of different time points throughout the course of infection for each of the three woodrats were chosen for amplification of the entire msp2(p44) expression site by conventional PCR methods. The msp2(p44) expression site was amplified from DNA extracts using a nested PCR assay. In the first round of PCR, primers AB 1058 (5′-GAACCATCCCCTTAAAATACTTTC-3′) and AB 1207 (5′-GGGAGTGCTCTGGTTAGATTTAGG-3′), which generate a fragment of approximately 3 kb containing P44Sup1/omp-1n, msp2(p44), and truncated recA, were used (Barbet et al., 2006). In the second round of PCR, primer MSP2iF (5′-GCTGAAGTGAGGAGACGAAG-3′), which anneals in the 5′ region flanking the msp2(p44) gene, and MSP2iR (5′-AATGGTAGCAGAACCAGAAG-3′), which anneals just 3′ to the truncated recA gene, were used to generate a fragment of approximately 1.5 kb (Rejmanek et al., 2012b). The PCR conditions were: an initial denaturation cycle of 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 1 min at 55 °C and 4 min at 72 °C, and a final extension of 10 min at 72 °C. Products were prepared for cloning using the Qiagen Gel Extraction kit.
Cloning and sequencing of expression site variants.
PCR-amplified fragments were cloned into the pGEM-T Easy vector (Promega) followed by transformation into Escherichia coli DH5α cells, and plated onto LB agar containing 100 µg ampicillin ml−1. Individual colonies were grown overnight in LB broth containing 100 µg ampicillin ml−1 and plasmids were purified using a Quantum Prep Plasmid Miniprep kit (Bio-Rad). Plasmids were assessed for appropriate insert size following EcoRI digestion. In order to evaluate the diversity of expressed msp2(p44) pseudogenes, 10 clones from each time point were randomly chosen for sequencing. In addition, 40 msp2(p44) expression site clones from the initial DU-1 inoculum were also sequenced. Sequencing was performed using an ABI 3730 sequencer (Davis Sequencing). Expression site sequences were manually trimmed to the nucleotides encoding the LAKT amino acid residues present on both sides of the hypervariable region. Nucleotide and amino acid sequences were aligned using the clustal_x sequence alignment program (Larkin et al., 2007). All unique msp2(p44) pseudogene sequences were compared with known A. phagocytophilum HZ-strain pseudogenes by searching the A. phagocytophilum HZ genome using the Comprehensive Microbial Resource (CMR) website (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi). A comparison of the entire complement of expression site msp2/p44 pseudogenes with each other was conducted by sequence alignment using mafft (version 6.0).
Data analysis.
Data were maintained in Excel (Microsoft) and analysed with the statistical package ‘R’ (R-Development Core Team, http://www.r-project.org). For all tests, a value of P≤0.05 was considered evidence of statistical significance. For each woodrat at each time, we calculated the gene diversity H (often called ‘expected heterozygosity’) using the maximum-likelihood estimator H = 1−Σp2 (Nei & Kumar, 2000; Weir, 1990), where p is the frequency of the allele in the population. The number of alleles, A, is affected by the sample size more than the effective number of alleles, Ae = 1/Σp2, or than H (Crow & Kimura, 1970). A more exhaustive justification of these methods is described in our previous paper (Rejmanek et al., 2012b).
The msp2 pseudogene library includes approximately 100 functional pseudogenes (Foley et al., 2009). This library was assumed to be exhausted as a Poisson process with an exponential decay. Thus we log-transformed the number of unused pseudogenes, psleft, in order to linearize its relationship with time. In R we used the function call:
to run an analysis of covariance to determine whether different woodrat lines lost their library at different rates.
Results
Duration and kinetics of infection
All three woodrats developed real-time PCR-detectable infections by 1 week post-inoculation (p.i.). Although their CT values fluctuated over time, the woodrats continued to have PCR-detectable infections at all time points (except W12 week 5 and W16 week 20) for the duration of the study (Fig. 1). After several weeks of high CT values, woodrat W9 was euthanized at 26 weeks p.i. Woodrats W12 and W16 were held until 34 weeks p.i., at which point they both still had moderately strong A. phagocytophilum infections (CT values of 30.0 and 32.6, respectively). Throughout the sampling period, mean CT values for W9 (29.2±4.0), W12 (29.9±3.5) and W16 (31.3±2.9) were in close agreement with each other. However, as can be observed in Fig. 1, CT values for W9 cycled twice at approximately 2 week intervals and then trended upwards over time, while CT values for W12 and W16 fluctuated in 4–6 week intervals throughout the study period.
Fig. 1.
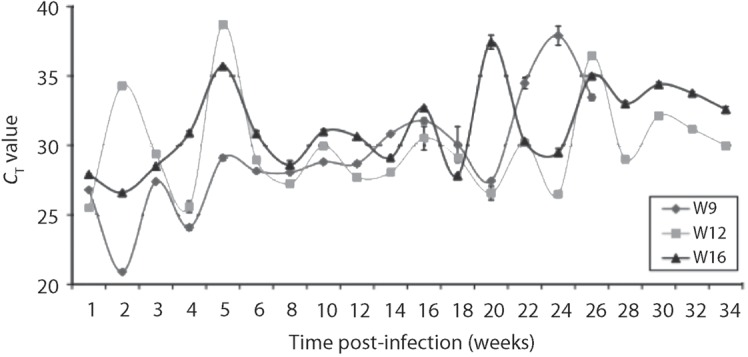
Real-time TaqMan PCR results charting the course of A. phagocytophilum DU-1 strain infections in three woodrats (W9, W12 and W16) over time. Each data point represents the mean CT value of three replicate PCRs. Error bars, sem.
Expression of msp2(p44) pseudogenes
The msp2(p44) expression site from individual woodrats was successfully amplified from blood samples taken at time points corresponding to low CT values (i.e. <30). In total, five time points (weeks 1, 4, 6, 10 and 14) from woodrat W9, nine time points (weeks 1, 3, 8, 12, 16, 20, 24, 28 and 32) from woodrat W12, and seven time points (weeks 1, 3, 8, 14, 18, 24 and 32) from woodrat W16 were analysed. For each time point, 10 randomly chosen clones were sequenced. In addition, 40 clones from the original DU-1 inoculum were also sequenced.
Sixty unique pseudogenes were detected among the three woodrats and from the inoculum. As it is not possible to conclusively identify all truly unique pseudogenes because the DU-1 genome has not been sequenced, small base-pair differences amounting to less than 3 % of the hypervariable region (~12 bp) between any two different pseudogene clones were not analysed as unique genes. The 60 pseudogenes could be classified as having either low (63–77 %, n = 21), medium (85–90 %, n = 11) or high (91–100 %, n = 28) nucleotide identity to known pseudogenes present in the A. phagocytophilum HZ strain genome (Table 1); those with >90 % homology with an HZ pseudogene are named according to their position in the HZ genome and denoted in this paper with an asterisk.
Table 1. Description of expressed msp2(p44) pseudogenes detected in all three woodrats and the DU-1 inoculum.
The similarity of each pseudogene is compared with known pseudogenes within the A. phagocytophilum HZ strain. Similarity type (low, medium or high) is based on percentage identity to the HZ strain.
| Woodrat pseudogene | Most similar HZ pseudogene | Percentage identity | Similarity type |
| D-6 | P44-27 | 63 | Low |
| D-4 | P44-43 | 67 | |
| D-7 | P44-54 | 69 | |
| D-2 | P44-43 | 70 | |
| D-13 | P44-27 | 70 | |
| D-15 | P44-40 | 70 | |
| D-19 | P44-52 | 70 | |
| D-22 | P44-51 | 70 | |
| D-12 | P44-56 | 71 | |
| D-14 | P44-15b | 71 | |
| D-16 | P44-56 | 71 | |
| D-18 | P44-52 | 71 | |
| D-21 | P44-32 | 71 | |
| D-3 | P44-44 | 72 | |
| D-17 | P44-37 | 72 | |
| D-20 | P44-44 | 72 | |
| D-9 | P44-48 | 73 | |
| D-10 | P44-29 | 74 | |
| D-11 | P44-27 | 74 | |
| D-5 | P44-73 | 76 | |
| D-1 | P44-44 | 77 | |
| D-25 | P44-8 | 85 | Medium |
| D-28 | P44-16b | 87 | |
| D-8 | P44-55 | 88 | |
| D-32 | P44-64 | 88 | |
| D-23 | P44-44 | 89 | |
| D-24 | P44-14 | 89 | |
| D-27 | P44-16 | 89 | |
| D-29 | P44-16 | 90 | |
| D-30 | P44-16 | 90 | |
| D-31 | P44-64 | 90 | |
| D-33 | P44-11 | 90 | |
| P44-16* | P44-16 | 91 | High |
| P44-49* | P44-49 | 91 | |
| P44-50* | P44-50 | 91 | |
| P44-53* | P44-53 | 91 | |
| P44-7* | P44-7 | 92 | |
| P44-39* | P44-39 | 92 | |
| P44-18* | P44-18 | 93 | |
| P44-23* | P44-23 | 93 | |
| P44-43* | P44-43 | 93 | |
| P44-6a* | P44-6 | 93 | |
| P44-2b* | P44-2b | 94 | |
| P44-30a* | P44-30 | 94 | |
| P44-34* | P44-34 | 94 | |
| P44-52* | P44-52 | 94 | |
| P44-6* | P44-6 | 95 | |
| P44-24* | P44-24 | 95 | |
| P44-37* | P44-37 | 95 | |
| P44-44* | P44-44 | 95 | |
| P44-30* | P44-30 | 96 | |
| P44-10* | P44-10 | 97 | |
| P44-57* | P44-57 | 97 | High |
| P44-65* | P44-65 | 97 | |
| P44-69* | P44-69 | 98 | |
| P44-75* | P44-75 | 98 | |
| P44-5* | P44-5 | 99 | |
| P44-55* | P44-55 | 99 | |
| P44-68* | P44-68 | 99 | |
| P44-11* | P44-11 | 100 |
Among the 40 inoculum clones analysed, 15 unique pseudogenes were detected. However, a single pseudogene (P44-6*) accounted for nearly 40 % of the expression site pseudogene population. For all time points analysed, the mean number of different pseudogenes (out of 10 randomly sampled clones) ranged from one to seven (mean = 4.4±2.4) for W9, one to five (mean = 3.0±1.8) for W12, and one to four (mean = 2.5±1.2) for W16. There were no apparent patterns in the order or variety of pseudogenes detected at the expression site among the three woodrats. In fact, excluding the inoculum pseudogenes, only five pseudogenes (D-15, D-29, P44-23*, P44-44* and P44-52*) were detected in more than one woodrat. Even among those five pseudogenes there was not a common temporal pattern detected at the expression site. For example, P44-23* was detected at the expression site on week 1 in W9 and on week 12 in W12. Similarly, P44-44* was expressed on week 6 in W9 and on week 32 in W12.
One common trend among all three woodrats was that few pseudogenes were detected at the expression site during subsequent time points. Overall, 13 pseudogenes were detected during multiple time points of infection. However, seven of those pseudogenes, detected on weeks 1 or 3 p.i., were identical to pseudogenes expressed in the inoculum. Additionally, two similar pseudogenes detected later on during the course of infection varied from the originally detected pseudogenes by 3–10 nt translating to 2–4 amino acid differences across the hypervariable region. This left four pseudogenes that were essentially identical (i.e. 1 or 2 nt differences with no amino acid changes) to those found at the expression site earlier in the course of infection. In contrast, 51 pseudogenes were detected at only a single time point.
A comparison of the entire complement of amino acid sequences from expression site msp2 pseudogenes with each other and with previously expressed woodrat pseudogenes revealed four unique groups of two or three sequences each containing shared central hypervariable regions but diverse N and/or C ends (Fig. 2).
Fig. 2.

Amino acid alignments of the msp2(p44) hypervariable region among expression site variants containing identical or nearly identical central portions with variable N and C ends. The start and end of each hypervariable region are denoted by the amino acids LAKT. Grey shading indicates areas of amino acid substitutions, insertions and/or deletions, and a white background denotes areas of amino acid identity. These pseudogenes represent possible recombination breakpoint variants.
Genetic diversity and pseudogene exhaustion
Table 2 gives three measures of the msp2 genetic diversity for each woodrat line over time. Also shown is the number of remaining pseudogenes (of approximately 100) left in each A. phagocytophilum line. Although still infected, as determined by real-time PCR, we were unable to amplify and sequence expressed pseudogenes in the W9 line beyond 14 weeks, at which point about one-third of the available pseudogenes had been expressed. The other two lines persisted past 30 weeks with a slower rate of pseudogene exhaustion. Analysis of covariance showed that the three lines differed significantly in pseudogene library exhaustion rate (P = 1.3×10−7). The W9 pseudogene library was exhausted at a rate of approximately 2 % per week, while the W12 and W16 libraries were exhausted at rates of 1 and 0.4 %, respectively. The mean pseudogene exhaustion rate among the three woodrat A. phagocytophilum lines was 1.1 % per week. Under these conditions it would take over 2 years of continual infection to use up the entire pseudogene repertoire. Also presented in Table 2 is the gene diversity, H, of expression site msp2/p44 pseudogenes in each of the three A. phagocytophilum populations over the course of 34 weeks. Expressed gene diversity in each population fluctuated widely across successive time points, and no discernible pattern between individual populations was observed.
Table 2. Expressed msp2 allelic diversity over time (in weeks) in three woodrat chronic infections of A. phagocytophilum.
Gene diversity, H, fluctuates over time, as does the observed number of alleles, A, and the effective number of alleles, Ae. The size of the pseudogene library remaining, psleft, reflects the starting state of about 100 pseudogenes and the number of distinct pseudogenes already expressed.
| Woodrat | Time | H | A | Ae | psleft |
| W9 | 0 | 0.81 | 15 | 5.27 | 85 |
| 1 | 0.58 | 3 | 2.38 | 84 | |
| 4 | 0.84 | 7 | 6.25 | 77 | |
| 6 | 0.82 | 7 | 5.56 | 70 | |
| 10 | 0.48 | 2 | 1.92 | 68 | |
| 14 | 0.56 | 3 | 2.27 | 65 | |
| W12 | 0 | 0.81 | 15 | 5.27 | 85 |
| 1 | 0 | 1 | 1 | 84 | |
| 3 | 0.18 | 2 | 1.22 | 84 | |
| 8 | 0.66 | 4 | 2.94 | 81 | |
| 12 | 0.76 | 5 | 4.17 | 77 | |
| 16 | 0 | 1 | 1 | 76 | |
| 20 | 0.74 | 5 | 3.85 | 71 | |
| 24 | 0.74 | 5 | 3.85 | 68 | |
| 28 | 0.66 | 3 | 2.94 | 65 | |
| 32 | 0 | 1 | 1 | 64 | |
| W16 | 0 | 0.81 | 15 | 5.27 | 85 |
| 1 | 0.62 | 4 | 2.61 | 84 | |
| 3 | 0.62 | 3 | 2.63 | 83 | |
| 8 | 0.66 | 4 | 2.94 | 79 | |
| 14 | 0.18 | 2 | 1.22 | 78 | |
| 18 | 0.56 | 3 | 2.27 | 77 | |
| 24 | 0 | 1 | 1 | 76 | |
| 34 | 0 | 1 | 1 | 75 |
Discussion
Few good model systems exist for studying the mechanisms and kinetics of chronic A. phagocytophilum infection. Although horse and mouse models for granulocytic anaplasmosis have been employed (Scorpio et al., 2008; Zhi et al., 1999), infections in these hosts are generally short-lived. And while sheep tend to develop persistent infections (Granquist et al., 2010), they develop clinical disease and are not true reservoir hosts for A. phagocytophilum. In the current study we infected natural reservoir dusky-footed woodrats with a woodrat-origin strain of A. phagocytophilum in order to describe the infection kinetics and patterns of msp2(p44) pseudogene recombination during a persistent infection.
All three woodrats developed persistent infection of 6 months duration (at least 8 months for two of the woodrats) with strongly positive test results for those two even on the last time point of the experiment. This finding corroborates field data demonstrating long-term infections in woodrats (Castro et al., 2001; Nicholson et al., 1999); given that the average life span of dusky-footed woodrats is only 1 to 2 years, a single exposure could result in life-long infection (Linsdale & Tevis, 1951). During this chronic infection, our data hint at infection cycles of strong followed by weak infection with approximately 4 to 6 week periods. While these observations were based on a limited set of expression site clones, we did not detect similar expression site variants during corresponding peaks of infection between any of the woodrats. In addition, the observed pseudogenes did not correspond to pseudogenes expressed in other animal studies at similar times of infection (Granquist et al., 2008; Wang et al., 2004). Two to three week cycles have been observed in sheep that are chronically infected with A. phagocytophilum and cattle infected with A. marginale, associated with sequential expression of antigenically variant MSP2 proteins encoded by msp2 pseudogenes (French et al., 1999; Granquist et al., 2008). In the current study, unique msp2(p44) pseudogenes were also observed at the expression site during each rickettsial cycle. In future studies, it would be interesting to look more closely at possible associations between specific expression site variants and corresponding serological responses over time. This could be accomplished by using synthetic peptides corresponding to the hypervariable regions of specific DU-1 pseudogenes. Such a technique was successfully used to track levels of serological responses to specific msp2/p44 pseudogenes over time in a sheep model of A. phagocytophilum infection (Granquist et al., 2008).
During the course of infection of the three woodrats evaluated in the present study, 60 unique pseudogenes were detected at the expression site. Although a genome sequence for a woodrat strain of A. phagocytophilum is not yet available, analysis of genome sequences from seven strains of Anaplasma (two of A. marginale, one of A. marginale subsp. centrale and four of A. phagocytophilum) shows that the overall number and synteny of msp2 paralogues within a species is maintained. In A. marginale and A. marginale subsp. centrale there are either six or seven msp2 paralogues. In four strains of A. phagocytophilum (HZ, two strains isolated from infected dogs, and one strain from a rodent reservoir; B. Al-Khedery and A. Barbet, unpublished data) there are 83–85 msp2/p44 pseudogenes in each strain, conservatively defined as containing both the 5′ and 3′ conserved elements of msp2/p44 on the same strand and separated by <500 bp. Allowing for an additional 15 or so msp2/p44 pseudogenes with either 5′ or 3′ conserved elements brings the total number of potentially functional pseudogenes in each of the different A. phagocytophilum strains to approximately 100. Therefore, we consider it unlikely that a woodrat strain would have a significantly greater number of msp2/p44 pseudogenes than other strains of A. phagocytophilum. Not surprisingly, nucleotide identity between the hypervariable regions of expression site variants detected in the current study and those in the HZ strain varied considerably, from 63 to 100 %. Similar between-strain variation across the msp2(p44) hypervariable region has been reported among other A. phagocytophilum strains (Barbet et al., 2006). Interestingly, three of the DU-1 strain pseudogenes were identical or nearly identical (1 nt difference) to those in the HZ strain even though these two strains have been shown to be dissimilar across multiple genes, consistently falling into separate phylogenetic clades (Rejmanek et al., 2012a). It may be that these particular pseudogenes play an especially important role in cellular adhesion or host immune evasion and are thus less likely to acquire mutations.
In the present study, pseudogenes detected at the expression site showed no apparent temporal patterns among individual animals. That is, the same pseudogenes were never detected at the same or even similar time points between any of the woodrats. This is similar to what has been reported in other studies in acute hosts, including mice and horses (Rejmanek et al., 2012b; Scorpio et al., 2008; Wang et al., 2004). In contrast, infection in chronically infected sheep has shown that certain pseudogenes are detected at the expression site at similar time points, suggesting at least a loose order of expression in this host (Granquist et al., 2008). Why chronically infected sheep and woodrats differ with respect to pseudogene expression is not clear, although it may be due to differential immune responses elicited by these two hosts towards the pathogen. However, our findings regarding temporal pseudogene diversity must be tempered, as our analysis was restricted to 10 expression site clones per time point, allowing potential low-frequency variants to go unnoticed.
The present study showed the diversity, H, of pseudogenes at the expression site fluctuating without any obvious approach to stationarity. This differs from the self-limiting infection of laboratory mice, in which expression site pseudogene diversity increases over two or three passes until reaching stationarity (Rejmanek et al., 2012b). In that study, DNA was harvested during peak rickettsemia, i.e. before the development of a strong adaptive immunity response, differing from the methods used in the present study to evaluate chronic infection. Thus, selection on the expression site msp2/p44 alleles took a different form in the woodrats. Each sample was a snapshot of a dynamic micro-coevolutionary race between the adaptive woodrat immune system and the A. phagocytophilum msp2/p44 library deployment system. Fluctuations in the number of expression site pseudogenes, or even periodic cycles, are to be expected in such an evolutionary race.
The rate of A. phagocytophilum pseudogene turnover (i.e. expression of previously unused pseudogenes) in woodrat infection, which was approximately 1 % per week, was lower than the 2 % per week reported earlier in serially passed infection in laboratory mice (Rejmanek et al., 2012b), although estimates in both studies may have been underestimates of the true exhaustion rate. In particular, the highest apparent exhaustion rate was detected in W9, which was strongly PCR-positive (and thus allowed for msp2 sequencing) approximately every 3 weeks, while W16, with the slowest rate of pseudogene exhaustion, could be sampled only every 5 weeks, and W12 with an intermediate rate of exhaustion was sampled every 3.5 weeks. Because it takes approximately 10–14 days to mount a considerable IgG antibody response to a novel antigen, the expression of new pseudogenes would be expected to follow a similar time frame. If this is the case, the exhaustion rate of approximately 2 % observed in W9 might represent the most realistic exhaustion rate among the woodrat lines. However, we must be cautious in making any broad generalizations about the duration of infections in woodrats given our low sample size. In addition, it is important to keep in mind the natural heterogeneity inherent in wild populations, which could result in a wide range of exhaustion rates among individuals.
A 2 % exhaustion rate would result in the depletion of the entire pseudogene repertoire available in the genome in approximately 1 year. Because woodrats commonly live longer than 1 year and their infections can readily last at least 34 weeks, a scenario in which all genome-encoded pseudogenes are exhausted is certainly plausible. At this point, once all available pseudogenes have been expressed and resulted in the development of corresponding host adaptive immunity, the woodrat could potentially clear the infection. Another possibility is that host antibodies targeting pseudogene antigens expressed early during the course of infection could wane to the point where re-expression of those pseudogenes allows the bacterium to continue evading the host immune response. Such a diminishing antibody response to particular MSP2(P44) antigens has been demonstrated in chronically infected sheep (Granquist et al., 2010). In the present study, at least four pseudogenes were subsequently detected at the expression site at later time points during the course of infection. Another possibility is that mutated pseudogene variants or mosaics could be expressed, which would allow the bacterial population to perpetuate even longer. In the current study, we observed pseudogene variants with shared central hypervariable regions but diverse 5′ and/or 3′ ends. Sometimes, the different ends extended quite far into the hypervariable region, similar to three variants of the HZ strain that we observed previously (Rejmanek et al., 2012b). These pseudogenes may represent recombination breakpoint variants similar to those observed in another study (Lin & Rikihisa, 2005). If these 5′ and 3′ changes produce new antigenic specificities, the antigenic repertoire would be larger than the genomic pseudogene repertoire. As presently understood, these variant expressed pseudogenes differ from the more extensive mosaic sequences that are commonly observed during persistent A. marginale infections (Brayton et al., 2003). It is possible that they represent an early stage in diversification that could give rise later in infection to the more extensive mosaic sequence blocks observed previously in natural infections of woodrats (Barbet et al., 2006). To define this will require genome sequencing of a woodrat strain and comparison with the expressed variants in long-term infections.
In conclusion, we have demonstrated that woodrats infected with a woodrat-origin A. phagocytophilum strain can maintain detectable infection for at least 8 months, confirming their role as competent reservoir hosts for this zoonotic pathogen. Both the strength of infection (i.e. pathogen load) and the genetic diversity of msp2(p44) pseudogenes detected at the expression site were shown to fluctuate periodically over time, and while the repertoire of pseuodgenes available to the infective A. phagocytophilum population could in theory become depleted within a year, the emergence of variant pseudogenes suggests that the pathogen likely continues evading host immunity indefinitely. Maintenance of such chronic infections without clinical disease indicates a stable co-evolutionary relationship between woodrats and their associated A. phagocytophilum strains.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases Evolution of Infectious Disease program (RO1 GM081714). We thank Nate Nieto for contributions to the study design, members of the Foley laboratory for helpful discussions, and Joy Worth, Susan Wang, Julia Harper, Katie Azervand and Jennifer Truong for laboratory assistance.
Abbreviations:
- p.i.
post-inoculation
References
- Bakken J. S., Krueth J., Tilden R. L., Dumler J. S., Kristiansen B. E. (1996). Serological evidence of human granulocytic ehrlichiosis in Norway. Eur J Clin Microbiol Infect Dis 15, 829–832. 10.1007/BF01701530 [DOI] [PubMed] [Google Scholar]
- Barbet A. F., Lundgren A. M., Alleman A. R., Stuen S., Bjöersdorff A., Brown R. N., Drazenovich N. L., Foley J. E. (2006). Structure of the expression site reveals global diversity in MSP2 (P44) variants in Anaplasma phagocytophilum. Infect Immun 74, 6429–6437. 10.1128/IAI.00809-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A. G. (1987). Immunobiology of relapsing fever. Contrib Microbiol Immunol 8, 125–137. [PubMed] [Google Scholar]
- Brayton K. A., Meeus P. F., Barbet A. F., Palmer G. H. (2003). Simultaneous variation of the immunodominant outer membrane proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect Immun 71, 6627–6632. 10.1128/IAI.71.11.6627-6632.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. C., Zhao Q. M., Zhang P. H., Yang H., Wu X. M., Wen B. H., Zhang X. T., Habbema J. D. (2003). Prevalence of Anaplasma phagocytophila and Borrelia burgdorferi in Ixodes persulcatus ticks from northeastern China. Am J Trop Med Hyg 68, 547–550. [DOI] [PubMed] [Google Scholar]
- Castro M. B., Nicholson W. L., Kramer V. L., Childs J. E. (2001). Persistent infection in Neotoma fuscipes (Muridae: Sigmodontinae) with Ehrlichia phagocytophila sensu lato. Am J Trop Med Hyg 65, 261–267. [DOI] [PubMed] [Google Scholar]
- Centurion-Lara A., LaFond R. E., Hevner K., Godornes C., Molini B. J., Van Voorhis W. C., Lukehart S. A. (2004). Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol Microbiol 52, 1579–1596. 10.1111/j.1365-2958.2004.04086.x [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura M. (1970). An Introduction to Population Genetics Theory. New York: Burgess Publishing Company. [Google Scholar]
- Des Vignes F., Fish D. (1997). Transmission of the agent of human granulocytic ehrlichiosis by host-seeking Ixodus scapularis (Acari:Ixodidae) in southern New York state. J Med Entomol 34, 379–382. [DOI] [PubMed] [Google Scholar]
- Diamond M. S. (2003). Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol 81, 196–206. 10.1046/j.1440-1711.2003.01157.x [DOI] [PubMed] [Google Scholar]
- Drazenovich N., Foley J., Brown R. N. (2006). Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic Anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis 6, 83–90. 10.1089/vbz.2006.6.83 [DOI] [PubMed] [Google Scholar]
- Dumler J. S., Choi K. S., Garcia-Garcia J. C., Barat N. S., Scorpio D. G., Garyu J. W., Grab D. J., Bakken J. S. (2005). Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis 11, 1828–1834. 10.3201/eid1112.050898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp J. C., Lin M., Madupu R., Crabtree J., Angiuoli S. V., Eisen J. A., Seshadri R., Ren Q., Wu M. & other authors (2006). Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2, e21. 10.1371/journal.pgen.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. E., Kramer V., Weber D. (2002). Experimental infection of dusky-footed wood rats (Neotoma fuscipes) with Ehrlichia phagocytophila sensu lato. J Wildl Dis 38, 194–198. [DOI] [PubMed] [Google Scholar]
- Foley J. E., Foley P., Brown R. N., Lane R. S., Dumlers J. S., Madigan J. E. (2004). Ecology of Anaplasma phagocytophilum and Borrelia burgdorferi in the western United States. J Vector Ecol 29, 41–50. [PubMed] [Google Scholar]
- Foley J. E., Nieto N. C., Barbet A., Foley P. (2009). Antigen diversity in the parasitic bacterium Anaplasma phagocytophilum arises from selectively-represented, spatially clustered functional pseudogenes. PLoS ONE 4, e8265. 10.1371/journal.pone.0008265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French D. M., Brown W. C., Palmer G. H. (1999). Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect Immun 67, 5834–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granquist E. G., Stuen S., Lundgren A. M., Bråten M., Barbet A. F. (2008). Outer membrane protein sequence variation in lambs experimentally infected with Anaplasma phagocytophilum. Infect Immun 76, 120–126. 10.1128/IAI.01206-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granquist E. G., Stuen S., Crosby L., Lundgren A. M., Alleman A. R., Barbet A. F. (2010). Variant-specific and diminishing immune responses towards the highly variable MSP2(P44) outer membrane protein of Anaplasma phagocytophilum during persistent infection in lambs. Vet Immunol Immunopathol 133, 117–124. 10.1016/j.vetimm.2009.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K. A., Sechman E. V., Skaar E. P., Seifert H. S. (2003). Recombination, repair and replication in the pathogenic Neisseriae: the 3 R’s of molecular genetics of two human-specific bacterial pathogens. Mol Microbiol 50, 3–13. 10.1046/j.1365-2958.2003.03679.x [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A. & other authors (2007). clustal w and clustal_x version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lin Q., Rikihisa Y. (2005). Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect Immun 73, 5106–5114. 10.1128/IAI.73.8.5106-5114.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Zhang C., Rikihisa Y. (2006). Analysis of involvement of the RecF pathway in p44 recombination in Anaplasma phagocytophilum and in Escherichia coli by using a plasmid carrying the p44 expression and p44 donor loci. Infect Immun 74, 2052–2062. 10.1128/IAI.74.4.2052-2062.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsdale J., Tevis L. (1951). The Dusky-Footed Woodrat: a Record of Observations Made on the Hastings Natural History Reservation. Berkeley, CA: University of California Press. [Google Scholar]
- Macleod J., Gordon W. (1933). Studies in tick-borne fever of sheep. I. Transmission by the tick, Ixodes ricinus, with a description of the disease produced. Parasitology 25, 273–285. 10.1017/S0031182000019442 [DOI] [Google Scholar]
- Nei M., Kumar S. (2000). Molecular Evolution and Phylogenetics. Oxford, UK: Oxford University Press. [Google Scholar]
- Nicholson W. L. (1998). Epidemiology of human granulocytic ehrlichiosis, with special reference to the role of wild rodents and Ixodid ticks as natural hosts of Ehrlichia phagocytophila sensu lato. PhD thesis, Johns Hopkins School of Hygiene and Public Health, Baltimore, MD, USA. [Google Scholar]
- Nicholson W. L., Castro M. B., Kramer V. L., Sumner J. W., Childs J. E. (1999). Dusky-footed wood rats (Neotoma fuscipes) as reservoirs of granulocytic Ehrlichiae (Rickettsiales: Ehrlichieae) in northern California. J Clin Microbiol 37, 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Allen K. E., McQuiston J. H., Breitschwerdt E. B., Little S. E. (2010). The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol 26, 205–212. 10.1016/j.pt.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Nieto N. C., Foley J. E. (2008). Evaluation of squirrels (Rodentia: Sciuridae) as ecologically significant hosts for Anaplasma phagocytophilum in California. J Med Entomol 45, 763–769. 10.1603/0022-2585(2008)45[763:EOSRSA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nieto N. C., Foley J. E. (2009). Reservoir competence of the redwood chipmunk (Tamias ochrogenys) for Anaplasma phagocytophilum. Vector Borne Zoonotic Dis 9, 573–577. 10.1089/vbz.2008.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto N. C., Madigan J. E., Foley J. E. (2010). The dusky-footed woodrat (Neotoma fuscipes) is susceptible to infection by Anaplasma phagocytophilum originating from woodrats, horses, and dogs. J Wildl Dis 46, 810–817. [DOI] [PubMed] [Google Scholar]
- Ohashi N., Inayoshi M., Kitamura K., Kawamori F., Kawaguchi D., Nishimura Y., Naitou H., Hiroi M., Masuzawa T. (2005). Anaplasma phagocytophilum-infected ticks, Japan. Emerg Infect Dis 11, 1780–1783. 10.3201/eid1111.050407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmanek D., Bradburd G., Foley J. (2012a). Molecular characterization reveals distinct genospecies of Anaplasma phagocytophilum from diverse North American hosts. J Med Microbiol 61, 204–212. 10.1099/jmm.0.034702-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmanek D., Foley P., Barbet A., Foley J. (2012b). Evolution of antigen variation in the tick-borne pathogen Anaplasma phagocytophilum. Mol Biol Evol 29, 391–400. 10.1093/molbev/msr229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter P. J., Jr, Kimsey R. B., Madigan J. E., Barlough J. E., Dumler J. S., Brooks D. L. (1996). Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae). J Med Entomol 33, 1–5. [DOI] [PubMed] [Google Scholar]
- Scorpio D. G., Leutenegger C., Berger J., Barat N., Madigan J. E., Dumler J. S. (2008). Sequential analysis of Anaplasma phagocytophilum msp2 transcription in murine and equine models of human granulocytic anaplasmosis. Clin Vaccine Immunol 15, 418–424. 10.1128/CVI.00417-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio D. G., Dumler J. S., Barat N. C., Cook J. A., Barat C. E., Stillman B. A., DeBisceglie K. C., Beall M. J., Chandrashekar R. (2011). Comparative strain analysis of Anaplasma phagocytophilum infection and clinical outcomes in a canine model of granulocytic anaplasmosis. Vector Borne Zoonotic Dis 11, 223–229. 10.1089/vbz.2009.0262 [DOI] [PubMed] [Google Scholar]
- Stuen S., Engvall E. O., Artursson K. (1998). Persistence of Ehrlichia phagocytophila infection in lambs in relation to clinical parameters and antibody responses. Vet Rec 143, 553–555. 10.1136/vr.143.20.553 [DOI] [PubMed] [Google Scholar]
- Taylor J. E., Rudenko G. (2006). Switching trypanosome coats: what’s in the wardrobe? Trends Genet 22, 614–620. 10.1016/j.tig.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Telford S. R., III, Dawson J. E., Katavolos P., Warner C. K., Kolbert C. P., Persing D. H. (1996). Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci U S A 93, 6209–6214. 10.1073/pnas.93.12.6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rikihisa Y., Lai T. H., Kumagai Y., Zhi N., Reed S. M. (2004). Rapid sequential changeover of expressed p44 genes during the acute phase of Anaplasma phagocytophilum infection in horses. Infect Immun 72, 6852–6859. 10.1128/IAI.72.12.6852-6859.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. (1990). Genetic Data Analysis. Sunderland, MA: Sinauer. [Google Scholar]
- Zhang J. R., Hardham J. M., Barbour A. G., Norris S. J. (1997). Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89, 275–285. 10.1016/S0092-8674(00)80206-8 [DOI] [PubMed] [Google Scholar]
- Zhi N., Ohashi N., Rikihisa Y. (1999). Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J Biol Chem 274, 17828–17836. 10.1074/jbc.274.25.17828 [DOI] [PubMed] [Google Scholar]


