Abstract
The disparity in diversity between unencapsulated (non-typable; NT) and encapsulated, serotypable Haemophilus influenzae (Hi) has been recognized for some time. It has previously been suggested that the wider diversity evidenced within NTHi compared with typable lineages may be due to different rates of recombination within the encapsulated and NT populations. To examine whether there is evidence for different levels of recombination within typable and NT lineages of Hi, we performed a statistical genetic analysis of 819 distinct genotypes of Hi to explore the congruence of serotype with population genetic clustering, and to identify patterns of recombination within the Hi population. We find that a significantly larger proportion of NT isolates show evidence of recombination, compared with typable isolates, and also that when admixture is present, the total amount of recombination per strain is greater within NT isolates, compared with the typable population. Furthermore, we demonstrate significant heterogeneity in the number of admixed individuals between NT lineages themselves, while such variation was not observed in typable lineages. This variability suggests that factors other than the presence of capsule are important determinants of recombination rate in the Hi population.
Introduction
Haemophilus influenzae (Hi) is a Gram-negative bacterium commonly isolated from episodes of asymptomatic nasopharyngeal carriage. While the vast majority of Hi exists in carriage rather than disease states, it remains a globally significant pathogen, capable of causing both localized mucosal infections, such as otitis media, and invasive infections, such as meningitis and septicaemia (Turk, 1984). Hi is thought to cause at least three million cases of serious illness every year, mostly in young children (WHO, 2005). To date, six distinct serotypes (designated a–f) of Hi are distinguishable (Pittman, 1930), while a substantial proportion of the population is unencapsulated and so remains non-typable (NT). Of the encapsulated population, serotype b has historically been the most significant pathogen, and is responsible for approximately 95 % of all invasive Hi disease in unvaccinated children (Falla et al., 1993; Peltola, 2000). An effective vaccine against H. influenzae serotype b (Hib) disease has been available since the early 1980s (Anderson, 1983; Chu et al., 1983), and its introduction into childhood immunization programs in the developed world has all but eliminated Hib disease in western countries (for example, see Bisgard et al., 1998; Booy et al., 1997). However, vaccination is rare in many developing countries, where Hib remains a significant cause of mortality, causing an estimated 380 000 deaths each year (WHO, 2005).
While Hib disease is controlled by vaccination in the developed world, Hi possessing the other serotypes, as well as NTHi, are also reported to be capable of causing invasive disease (St Geme, 1993; Tsang et al., 2007; Waggoner-Fountain et al., 1995). NTHi is also a leading cause of economically significant, but rarely fatal, infections such as otitis media (Klein, 1997; Talbird et al., 2010). Phylogenetically, typable Hi have been found to group into lineages generally concordant with their serotype (Meats et al., 2003; Musser et al., 1988a, b, 1990), while NTHi is more phylogenetically diverse (Erwin et al., 2008; Meats et al., 2003; Quentin et al., 1990).
Hi is naturally transformable, taking up DNA from its environment by the recognition of specific Hi uptake sequences in free DNA (Danner et al., 1980) and integrating it into its chromosome (Goodgal & Herriott, 1961; Smith et al., 1981). The precise mechanism of competence in Hi remains incompletely understood, although many proteins important for competence have been identified, including those that make up structures implicated in uptake on the surface of the cells (ComE, PilA), those that are in the periplasm/inner membrane (ComF, ComC, Rec-2) and the cytoplasm (ComA, DprA, ComM) (Gwinn et al., 1998; Karudapuram et al., 1995; Karudapuram & Barcak, 1997; Larson & Goodgal, 1991; McCarthy, 1989; Tomb et al., 1991). Following internalization, the DNA can be inserted into the chromosome by homologous recombination. While Hi isolates are competent and can be readily transformed in the laboratory environment (Alexander & Leidy, 1951), the contribution of homologous recombination to population structure in nature is less clear. One phylogenetic study (Feil et al., 2001) found relatively high congruence between housekeeping loci, suggesting that homologous recombination was not frequent enough to scramble phylogenetic signals. However, later work that specifically compared NT isolates with encapsulated ones using the same approach found a lower degree of congruence between housekeeping loci in NT isolates (Meats et al., 2003). NT strains are more diverse (both genetically and phenotypically) than their encapsulated relatives (Erwin et al., 2008; Musser et al., 1986), and it is not clear whether this reflects differences in the ability or opportunity for NT and typable strains to take up DNA (Meats et al., 2003).
Such differences in recombination rate among different lineages of the same named species have been documented or hypothesized in other bacteria, including Neisseria meningitidis (Bart et al., 2001) and the pneumococcus (Hanage et al., 2009). A history of frequent recombination within a bacterial lineage has been associated with resistance to antibiotics (Hanage et al., 2009) and vaccine escape (Croucher et al., 2011), and a better understanding of the flow of genetic material between bacterial lineages is hence important for public health. We assessed evidence for differences in recombination between serotypable and NTHi, by analysing molecular epidemiological data, and also whether there was evidence of differences in recombination between different NTHi populations.
Methods
Data.
Gene sequences of seven loci from 1624 Hi isolates representing 819 unique genotypes were collected from the Multi Locus Sequence Typing database. Multi-locus sequence typing (MLST) (Maiden et al., 1998) is an unambiguous portable typing methodology that has been widely applied to pathogenic bacterial species. The system works by using a set of predefined housekeeping loci (usually seven) with known start and end points. Each locus is sequenced, and the sequence compared with a central database that stores records of each unique allele identified. Each unique sequence identified at each locus is given an allele number, and every unique combination of allele numbers reported is assigned a sequence type (ST). We used a complete set of the publicly available STs for Hi from www.mlst.net as of March 6 2011. Two STs retrieved (STs 71 and 268) were excluded from the analysis on account of the presence of insertions or deletions within the allelic sequence. This provided a dataset comprising 819 STs, each characterized by seven sequences. An analysis of earlier Hi MLST database contents that were more limited in their extent has been described elsewhere (Meats et al., 2003). The sets of allele sequences corresponding to each ST in the MLST database were concatenated for subsequent analysis.
Analysis.
To examine the population structure and estimate the amounts of admixture between population clusters, we utilized the software package Bayesian Analysis of Population Structure (baps) (Corander & Marttinen, 2006; Corander & Tang, 2007; Corander et al., 2008). baps is freely available online, and implements a number of models to cluster individuals based on shared polymorphisms using a non-reversible stochastic optimization algorithm, and returns as results the number of populations that best fit the model and the genotypes associated with each population. As a result of this, each polymorphic site can be associated with a specific population retrieved as a result of the analysis, and genotypes containing polymorphisms that are characteristic of more than one population can be identified as cases of likely recombination (admixture). baps can then be used to identify cases of individuals containing polymorphisms typical of populations other than the one to which they are assigned, such cases possibly being the result of recombination. To examine differential levels of admixture within the NT and typable populations, we combined serotype information from the MLST database with the baps outputs indicating the cluster assignments for the STs, and the cases with significant patterns of polymorphisms from more than one population. These results enabled us to examine the extent to which the presence of admixture is associated with particular clusters, and the extent to which the presence of admixture is associated with the possession of a serotype. The sequence data were used to construct a phylogeny using RAxML version 7.0.4 for HPC environments, onto which the results of these analyses were superimposed.
Results
Concordance of populations with serotypes
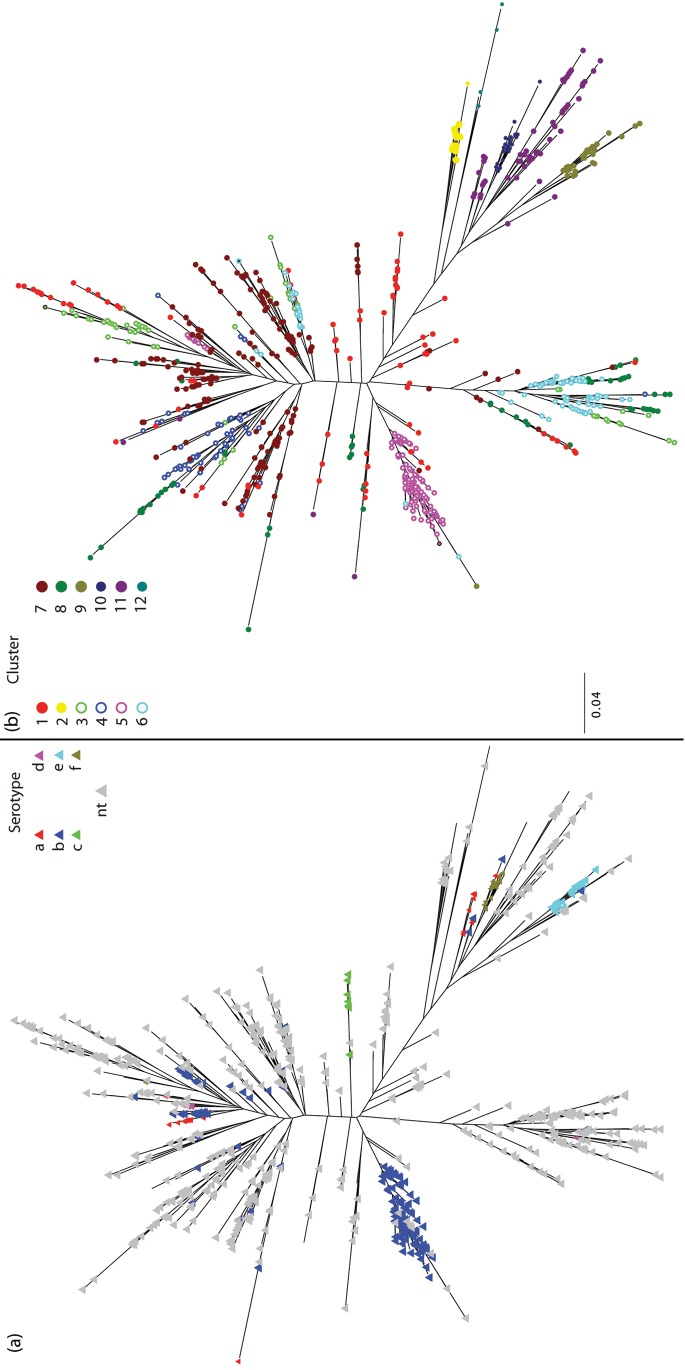
Our analysis subdivided the MLST dataset into 12 clusters. We then matched the serotype reported for each isolate held by the MLST databases to the STs used in the analysis, to identify the distribution of serotypes amongst the clusters (Table 1). In most cases (729 of 738 STs), all of the isolates associated with an ST were reported as having the same serotype (see Table S1, available with the online version of this paper, for exceptions). The clustering results demonstrate broad concordance between cluster and serotype as reported earlier (Erwin et al., 2008; Meats et al., 2003; Musser et al., 1988b, 1990). The greater diversity in the NT population compared with the typable population is evidenced by the wide dispersal of NT STs over the phylogenetic tree (Fig. 1) and the fact that typables form a majority of isolates in only three clusters, 5, 9 and 10, which are associated with serotypes b, e and f, respectively. Other than clusters 7 and 11, where typables make up 27 and 22 % of the clusters, NT STs make up more than 90 % of the individuals in all of the remaining clusters. The baps clusters correlate well with the classical subdivisions of the Hi population (Fig. S1) (Meats et al., 2003; Musser et al., 1986, 1988a, b), despite the addition of considerable diversity in the NT strains. The addition of extra diversity does have an effect on the groupings observed by Erwin et al. (2008): three of the clusters reported in that study fall within the same baps group; however, the remaining clusters are now broken up both in terms of baps clustering and phylogenetic grouping, assessed both by maximum-likelihood methods and by maximum-parsimony methods, such as those used originally by Erwin and co-workers. We suggest that the limited concordance with the Erwin et al. (2008) clusters is due to the effects of recombination on the production of the phylogenies used to determine their clusters. Recombination is known to adversely effect phylogenetic reconstruction (Schierup & Hein, 2000).
Table 1. Distribution of serotypes amongst baps clusters.
Numbers of admixed STs are shown in parentheses.
| baps cluster | Serotype | |||||||
| a | b | c | d | e | f | NT | Unknown | |
| 1 | – | 7 (7) | 1 (1) | – | – | – | 70 (50) | 6 (3) |
| 2 | – | – | – | – | – | – | 10 (0) | 2 (0) |
| 3 | – | – | – | – | – | 1 (0) | 51 (27) | 4 (1) |
| 4 | – | 3 (3) | – | – | – | – | 49 (12) | 7 (3) |
| 5 | – | 130 (32) | – | – | – | – | 6 (1) | 2 (2) |
| 6 | – | 4 (2) | – | 2 (0) | – | – | 69 (9) | 11 (5) |
| 7 | 10 (1) | 29 (7) | 6 (0) | 6 (0) | – | 1 (0) | 120 (36) | 17 (5) |
| 8 | 1 (0) | 2 (1) | 1 (1) | – | – | – | 46 (17) | 9 (6) |
| 9 | – | 1 (0) | – | – | 27 (1) | – | 8 (7) | 1 (0) |
| 10 | 1 (0) | 2 (0) | – | – | – | 15 (2) | – | 6 (0) |
| 11 | 9 (1) | 2 (0) | – | – | – | – | 36 (9) | 4 (1) |
| 12 | – | – | – | – | – | – | 3 (1) | 1 (0) |
| Total | 21 (2) | 180 (52) | 8 (2) | 8 (0) | 27 (1) | 17 (2) | 468 (169) | 70 (26) |
Fig. 1.
Maximum-likelihood tree produced using RAxML version 7.0.4 from the concatenated sequence for all seven loci (total length of 3057 bp) with a General Time Reversible model and Gamma-distributed rates amongst sites for the 819 genotypes examined. Coloured triangles indicate serotype (a) and coloured circles represent baps cluster assignment (b).
Variance of admixture amounts within the population
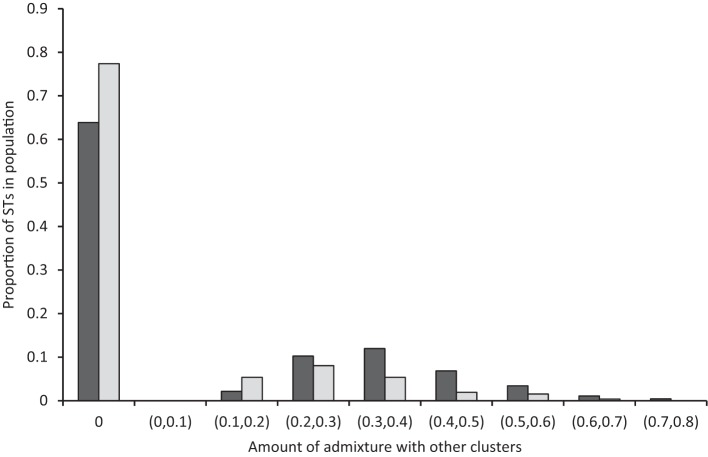
baps was also used to perform an admixture analysis on the dataset, and we asked whether STs associated with NT isolates were significantly more likely to show evidence of admixture. We found a significant difference between the typable and NT populations (χ2 P value 0.00013) in the numbers of STs showing admixture. We also asked whether the amount of admixture, in terms of the numbers of polymorphisms apparently acquired from other populations, was greater in NT isolates. For each ST the baps admixture analysis estimates the amount of sequence that is characteristic of a different cluster (Fig. 2). We found a significant difference in the extent of admixture per ST present in the typable and NT populations when comparing the populations using a Kolmogorov–Smirnov test (Chakravarti et al., 1967), to test the variance in admixture between the populations (P value 8×10−5), and a Mann–Whitney U test (Wilcoxon, 1946), to test the overall difference in the numbers of individuals that show admixture in the populations (P value 9.9×10−6). Therefore, the STs in the NT population appear to be significantly more likely to show evidence of admixture, and that admixture is found to affect a higher proportion of sites than is the case for typable STs. With the exception of cluster 2 (which contains only 10 genotypes), all of the other clusters contain STs exhibiting some polymorphisms inferred to be significantly characteristic of other clusters (Table 1). Clusters that harbour a majority of NT STs show a marked heterogeneity in their degree of admixture; for example, cluster 1 contains a high proportion (>0.7) of STs with evidence of recombination, in contrast to a low proportion (<0.25) in cluster 4. This is reflected by the fact that the number of admixed/unadmixed NT samples varies significantly between NT clusters (χ2 P value 2.9×10−13). In contrasting, the proportion of STs with evidence of a history of admixture among clusters associated with encapsulated strains ranges between 12 and 26 % (see Table S2 for proportions), with no significant differences evident between these clusters (χ2 P value 0.34).
Fig. 2.
Distribution of estimated levels of admixture between NT (dark-grey bars) and typable (light-grey bars) populations.
Discussion
Homologous recombination is capable of confounding phylogenetic reconstruction, blurring the boundaries of a species, and disseminating virulence and resistance determinants into different genomic backgrounds. The amount of homologous recombination has been observed to differ between bacterial species to a marked degree. Some, such as Mycobacterium tuberculosis, are considered to be nearly clonal, whereas others, such as Helicobacter pylori, are almost panmictic. It is beginning to become clear that the amount of recombination that a strain has experienced may not be consistent across a species, with some lineages exhibiting a significantly greater history of admixture than others. In some cases this has been associated with the acquisition of drug resistance. Hence, it is important to identify any lineages in the community that contribute and take up genetic material at a high rate, as they are potential risk populations in the response of pathogens to medical intervention.
While it has long been known that Hi can take up DNA in the laboratory, unlike other naturally competent bacteria such as the pneumococcus or meningococcus this ability had not removed phylogenetic signals, and good concordance of lineages with serogroups was observed. The issue of NTHi has been more confused, with some evidence suggesting a higher rate of recombination in this population (Meats et al., 2003), but based on relatively few (27) isolates. Our results indicate that the population structure of NTHi does indeed exhibit a significantly larger contribution of recombination, but that different NTHi lineages vary significantly in the degree to which recombination has occurred over their history. While the greater genetic diversity of NTHi has been speculated to result from increased recombination (Meats et al., 2003), these results suggest that absence of capsule is not a sufficient cause, and other factors are important in facilitating transformation (Maughan & Redfield, 2009).
In agreement with the previously reported concordance of phylogeny with serogroups, baps clusters appear to provide an effective method for the subdivision of the Hi population into groupings consistent with the classical subdivisions of the species. Typable Hi appear in a relatively small number of clusters, separated into lineages that are grouped together and remain distinct from NT STs, by baps and eburst. However, the notable exception to this is the presence of serogroup b STs in three lineages dominated by NT STs. The globally distributed nature of the clusters (Table S3) suggests that on a population level, genetic interplay between these lineages continues, irrespective of the presence of Hib vaccination in the developed world.
The presence of admixed STs within the population expressing a serotype implies that typable STs have no mechanistic limitation preventing them from taking up DNA; however, in the case of serotypes such as serotype e, where only one ST shows admixture, it is possible that the admixed typable STs are actually NT STs that have recently gained a serotype, rather than recombinogenic STs from a ‘typable’ lineage. This would be an explanation for the serotype b STs found in NT lineages. Another potential explanation is laboratory errors. However, it should be noted that 19 of the 60 serotype b’s outside the main serotype b cluster were serotyped using molecular methods, which should be less open to ambiguity than classical serotyping. Moreover, in six cases the STs in question have been found on multiple occasions (Table S4), which allows us to ask whether they have repeatedly been found with serotype b capsules, which is hard to explain as repeated laboratory errors. In five of the six STs with multiple samples, all isolates were serotype b. In the case of ST12 the records are mixed and on other occasions it has been reported as NTHi. We interpret this as evidence that while serotyping errors can generate this signal, and may well have done so in the case of ST12, typable isolates are indeed found sporadically in NT clusters. The difference in admixture between the two populations may be explained by some unknown difference in the fine-scale niches of the typable and NTHi, such that one of them rarely encounters DNA in the environment or enters a competent state, or it may also be due to the absence of a capsule, which in some way facilitates the uptake of DNA in NTHi. This could affect the operation of the cellular competence machinery itself, or possibly the action of other mechanisms such as phage-mediated exchange of DNA and conjugation. The wide range of admixture levels identified within the NT population also suggests that the reasons for admixture may vary from cluster to cluster. The cause, be it due to mechanistic or genetic factors (or, indeed, a combination of the two), is not a question which it is possible to answer using the MLST data alone, but it is our hope that the causes – and full extent – of the differences in levels of admixture within NTHi and between NTHi and typable Hi will be revealed in future analyses using genomic data.
Acknowledgements
T. R. C. was funded by a BBSRC studentship and is now a postdoctoral fellow at the Wellcome Trust Sanger Institute, funded on grant number 098051, W. P. H. was funded by a Royal Society University Research Fellowship and funding for the MIDAS Center for Communicable Disease Dynamics at the Harvard School of Public Health was from National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM088558-01. The work of J. C. was supported by ERC grant number 239784, Academy of Finland grant number 251170 and a grant from the Sigrid Juselius Foundation.
Abbreviations:
- Hi
Haemophilus influenzae
- Hib
H. influenzae serotype b
- MLST
multi-locus sequence typing
- NT
non-typable
- ST
sequence type
Footnotes
A supplementary figure and five supplementary tables are available with the online version of this paper.
References
- Alexander H. E., Leidy G. (1951). Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med 93, 345–359. 10.1084/jem.93.4.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. (1983). Antibody responses to Haemophilus influenzae type b and diphtheria toxin induced by conjugates of oligosaccharides of the type b capsule with the nontoxic protein CRM197. Infect Immun 39, 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart A., Barnabé C., Achtman M., Dankert J., van der Ende A., Tibayrenc M. (2001). The population structure of Neisseria meningitidis serogroup A fits the predictions for clonality. Infect Genet Evol 1, 117–122. 10.1016/S1567-1348(01)00011-9 [DOI] [PubMed] [Google Scholar]
- Bisgard K. M., Kao A., Leake J., Strebel P. M., Perkins B. A., Wharton M. (1998). Haemophilus influenzae invasive disease in the United States, 1994–1995: near disappearance of a vaccine-preventable childhood disease. Emerg Infect Dis 4, 229–237. 10.3201/eid0402.980210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy R., Heath P. T., Slack M. P. E., Begg N., Moxon E. R. (1997). Vaccine failures after primary immunisation with Haemophilus influenzae type-b conjugate vaccine without booster. Lancet 349, 1197–1202. 10.1016/S0140-6736(96)06392-1 [DOI] [PubMed] [Google Scholar]
- Chakravarti I. M., Laha R. G., Roy J. (1967). Handbook of Methods of Applied Statistics. New York, London: Wiley. [Google Scholar]
- Chu C., Schneerson R., Robbins J. B., Rastogi S. C. (1983). Further studies on the immunogenicity of Haemophilus influenzae type b and pneumococcal type 6A polysaccharide-protein conjugates. Infect Immun 40, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J., Marttinen P. (2006). Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15, 2833–2843. 10.1111/j.1365-294X.2006.02994.x [DOI] [PubMed] [Google Scholar]
- Corander J., Tang J. (2007). Bayesian analysis of population structure based on linked molecular information. Math Biosci 205, 19–31. 10.1016/j.mbs.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Corander J., Marttinen P., Sirén J., Tang J. (2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9, 539. 10.1186/1471-2105-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J. H. & other authors (2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434. 10.1126/science.1198545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner D. B., Deich R. A., Sisco K. L., Smith H. O. (1980). An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11, 311–318. 10.1016/0378-1119(80)90071-2 [DOI] [PubMed] [Google Scholar]
- Erwin A. L., Sandstedt S. A., Bonthuis P. J., Geelhood J. L., Nelson K. L., Unrath W. C., Diggle M. A., Theodore M. J., Pleatman C. R. & other authors (2008). Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol 190, 1473–1483. 10.1128/JB.01207-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla T. J., Crook D. W. M., Kraak W. A. G., Nichols W. W., Anderson E. C., Jordens J. Z., Slack M. P. E., Dobson S. R. M., Moxon E. R., Mayon-White D. (1993). Population-based study of non-typable Haemophilus influenzae invasive disease in children and neonates. Lancet 341, 851–854. 10.1016/0140-6736(93)93059-A [DOI] [PubMed] [Google Scholar]
- Feil E. J., Holmes E. C., Bessen D. E., Chan M. S., Day N. P., Enright M. C., Goldstein R., Hood D. W., Kalia A. & other authors (2001). Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci U S A 98, 182–187. 10.1073/pnas.98.1.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodgal S. H., Herriott R. M. (1961). Studies on transformations of Hemophilus influenzae. I. Competence. J Gen Physiol 44, 1201–1227. 10.1085/jgp.44.6.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn M. L., Ramanathan R., Smith H. O., Tomb J. F. (1998). A new transformation-deficient mutant of Haemophilus influenzae Rd with normal DNA uptake. J Bacteriol 180, 746–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanage W. P., Fraser C., Tang J., Connor T. R., Corander J. (2009). Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324, 1454–1457. 10.1126/science.1171908 [DOI] [PubMed] [Google Scholar]
- Karudapuram S., Barcak G. J. (1997). The Haemophilus influenzae dprABC genes constitute a competence-inducible operon that requires the product of the tfoX (sxy) gene for transcriptional activation. J Bacteriol 179, 4815–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karudapuram S., Zhao X., Barcak G. J. (1995). DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J Bacteriol 177, 3235–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. O. (1997). Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr Infect Dis J 16 (Suppl. 2), S5–S8. 10.1097/00006454-199702001-00002 [DOI] [PubMed] [Google Scholar]
- Larson T. G., Goodgal S. H. (1991). Sequence and transcriptional regulation of com101A, a locus required for genetic transformation in Haemophilus influenzae. J Bacteriol 173, 4683–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M. C., Bygraves J. A., Feil E., Morelli G., Russell J. E., Urwin R., Zhang Q., Zhou J., Zurth K. & other authors (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95, 3140–3145. 10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan H., Redfield R. J. (2009). Extensive variation in natural competence in Haemophilus influenzae. Evolution 63, 1852–1866. 10.1111/j.1558-5646.2009.00658.x [DOI] [PubMed] [Google Scholar]
- McCarthy D. (1989). Cloning of the rec-2 locus of Haemophilus influenzae. Gene 75, 135–143. 10.1016/0378-1119(89)90390-9 [DOI] [PubMed] [Google Scholar]
- Meats E., Feil E. J., Stringer S., Cody A. J., Goldstein R., Kroll J. S., Popovic T., Spratt B. G. (2003). Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41, 1623–1636. 10.1128/JCM.41.4.1623-1636.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Barenkamp S. J., Granoff D. M., Selander R. K. (1986). Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun 52, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kroll J. S., Moxon E. R., Selander R. K. (1988a). Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci U S A 85, 7758–7762. 10.1073/pnas.85.20.7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kroll J. S., Moxon E. R., Selander R. K. (1988b). Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun 56, 1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kroll J. S., Granoff D. M., Moxon E. R., Brodeur B. R., Campos J., Dabernat H., Frederiksen W., Hamel J. & other authors (1990). Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis 12, 75–111. 10.1093/clinids/12.1.75 [DOI] [PubMed] [Google Scholar]
- Peltola H. (2000). Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev 13, 302–317. 10.1128/CMR.13.2.302-317.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M. (1930). The S and R Forms of Haemophilus influenzae. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine (New York) 27, 299–301. [Google Scholar]
- Quentin R., Goudeau A., Wallace R. J., Jr, Smith A. L., Selander R. K., Musser J. M. (1990). Urogenital, maternal and neonatal isolates of Haemophilus influenzae: identification of unusually virulent serologically non-typable clone families and evidence for a new Haemophilus species. J Gen Microbiol 136, 1203–1209. 10.1099/00221287-136-7-1203 [DOI] [PubMed] [Google Scholar]
- Schierup M. H., Hein J. (2000). Consequences of recombination on traditional phylogenetic analysis. Genetics 156, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. (1981). Genetic transformation. Annu Rev Biochem 50, 41–68. 10.1146/annurev.bi.50.070181.000353 [DOI] [PubMed] [Google Scholar]
- St Geme J. W., III (1993). Nontypeable Haemophilus influenzae disease: epidemiology, pathogenesis, and prospects for prevention. Infect Agents Dis 2, 1–16. [PubMed] [Google Scholar]
- Talbird S. E., Taylor T. N., Caporale J., Ismaila A. S., Gomez J. (2010). Residual economic burden of Streptococcus pneumoniae- and nontypeable Haemophilus influenzae-associated disease following vaccination with PCV-7: a multicountry analysis. Vaccine 28 (Suppl. 6), G14–G22. 10.1016/j.vaccine.2010.06.080 [DOI] [PubMed] [Google Scholar]
- Tomb J. F., el-Hajj H., Smith H. O. (1991). Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene 104, 1–10. 10.1016/0378-1119(91)90457-M [DOI] [PubMed] [Google Scholar]
- Tsang R. S., Sill M. L., Skinner S. J., Law D. K., Zhou J., Wylie J. (2007). Characterization of invasive Haemophilus influenzae disease in Manitoba, Canada, 2000–2006: invasive disease due to non-type b strains. Clin Infect Dis 44, 1611–1614. 10.1086/518283 [DOI] [PubMed] [Google Scholar]
- Turk D. C. (1984). The pathogenicity of Haemophilus influenzae. J Med Microbiol 18, 1–16. 10.1099/00222615-18-1-1 [DOI] [PubMed] [Google Scholar]
- Waggoner-Fountain L. A., Hendley J. O., Cody E. J., Perriello V. A., Donowitz L. G. (1995). The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin Infect Dis 21, 1322–1324. 10.1093/clinids/21.5.1322 [DOI] [PubMed] [Google Scholar]
- WHO (2005). Haemophilus influenzae type B (HiB). WHO Factsheet Number 294.
- Wilcoxon F. (1946). Individual comparisons of grouped data by ranking methods. J Econ Entomol 39, 269. [DOI] [PubMed] [Google Scholar]