Abstract
Human milk is a rich source of nutrients and energy, shaped by mammalian evolution to provide all the nutritive requirements of the newborn. In addition, several molecules in breast milk act as bioactive agents, playing an important role in infant protection and guiding a proper development. While major breast milk nutrients such as lactose, lipids and proteins are readily digested and consumed by the infant, other molecules, such as human milk oligosaccharides and glycosylated proteins and lipids, can escape intestinal digestion and transit through the gastrointestinal tract. In this environment, these molecules guide the composition of the developing infant intestinal microbiota by preventing the colonization of enteric pathogens and providing carbon and nitrogen sources for other colonic commensals. Only a few bacteria, in particular Bifidobacterium species, can gain access to the energetic content of milk as it is displayed in the colon, probably contributing to their predominance in the intestinal microbiota in the first year of life. Bifidobacteria deploy exquisite molecular mechanisms to utilize human milk oligosaccharides, and recent evidence indicates that their activities also target other human milk glycoconjugates. Here, we review advances in our understanding of how these microbes have been shaped by breast milk components and the strategies associated with their consumption of milk glycoconjugates.
Introduction
After birth, the profound and intimate connection between a mother and her newborn continues in several ways. Breast milk represents a physical representation of this relationship: an intriguing fluid synthesized at the mother’s expense, shaped throughout evolution to nourish the infant and improve its rate of survival. Human milk is perhaps the most personalized food, where the molecular make-up varies from mother to mother and across lactation, providing the infant all the nutrients needed in a concentrated form (Allen et al., 1991; Mitoulas et al., 2002). Breastfeeding is regarded as the ‘normal way of providing young infants with the nutrients they need for healthy growth and development’ (Fewtrell et al., 2007). Exclusive breastfeeding is recommended for up to 6 months of age (American Academy of Pediatrics Section on Breastfeeding, 2012), and its benefits are multifold and some can last beyond childhood (Hernell, 2011; Le Huërou-Luron et al., 2010).
Human milk is a complex food matrix and its composition reflects all the nutritional and physiological demands of the newborn. Essential nutrients in human milk such as lactose, fatty acids and proteins are absorbed by the small intestine at a rate that is limited by the developing conditions of the gastrointestinal (GI) tract of the infant (Neu, 2007). Other micronutrients such as nucleotides, vitamins and minerals are also highly bioavailable for the infant (Picciano, 2001).
Numerous studies have shown that breastfeeding is associated with a lower risk of infections and diarrhoea. This has been associated with the activity of milk immunoglobulins (Xanthou et al., 1995), antimicrobial agents such as lactoferrin and lysozyme (Håversen et al., 2002; Jollès & Jollès, 1961; Lönnerdal, 2009), and human milk glycoconjugates (Newburg et al., 2005). Several of these molecules are not readily absorbed by the small intestine and transit throughout the GI tract (Dallas et al., 2012), but their impact and biological activities are poorly understood. These bioactive compounds play additional roles in protection and/or stimulate development regardless of their nutritive value (Hamosh, 2001; Lönnerdal, 2010). Bioactives in human milk represent a significant difference between breast milk and bovine milk-based formulas (Hernell, 2011; Le Huërou-Luron et al., 2010).
A common characteristic of these bioactive agents is that they are glycosylated molecules. Glycans in milk can be found as free human milk oligosaccharides (HMO), or conjugated via glycosidic bonds to proteins or lipids. Among other functions, human milk glycans represent the main driving force for bacterial colonization of the distal large intestine of the breast-fed infant (Scholtens et al., 2012). The high concentrations of HMO and conjugated oligosaccharides processed after intestinal digestion are thought to be the main contributors to the predominance of Bifidobacterium species in the infant gut. The genome sequences of bifidobacteria show that these micro-organisms are highly adapted to the intestinal environment (Schell et al., 2002), and the genomes of infant gut-associated bifidobacteria have been shaped by complex carbohydrates (Sela & Mills, 2010). In this review, we examine recent advances in our understanding of how milk oligosaccharides and other glycoconjugates influence the dominance of beneficial micro-organisms in the gut microbiota, especially Bifidobacterium, and of the mechanisms and strategies that these micro-organisms have devised for using milk components as a carbon source.
HMO
Structures of HMO
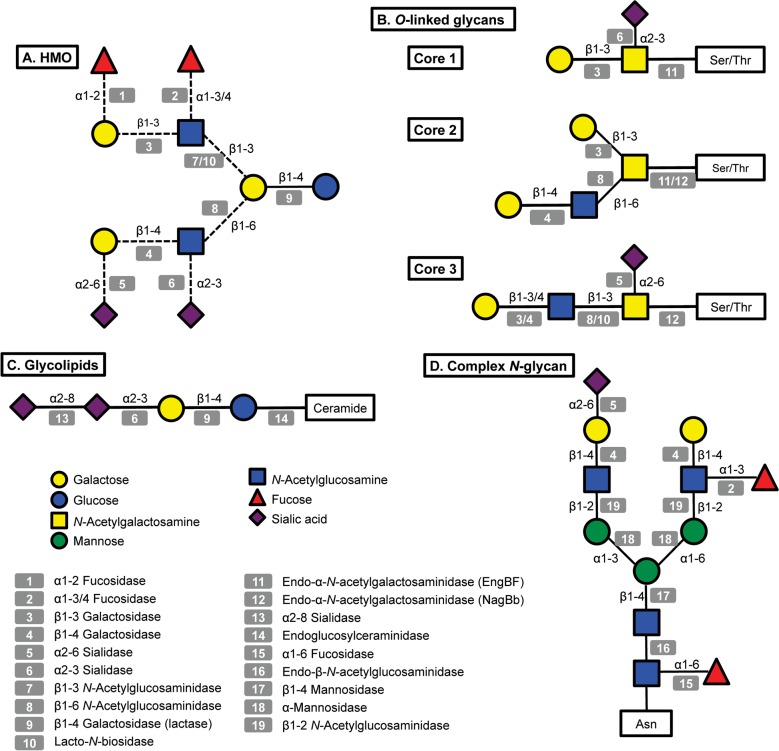
A great amount of the energy invested in human milk production is dedicated to synthesize complex free oligosaccharides. These molecules represent the third most abundant component in breast milk after lactose and fatty acids (Petherick, 2010). HMO consist of a pool of soluble carbohydrates with a degree of polymerization of 3 to 15 and linked through a variety of glycosidic bonds (Kunz et al., 2000; Urashima et al., 2012). HMO are composed of five monosaccharides: glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc) and N-acetylneuraminic acid (NeuAc; sialic acid). Fig. 1 presents a representative HMO structure and the diversity of linkages that can be found. All HMO are characterized by a terminal lactose molecule, modified by fucose or sialic acid in the case of the shorter HMO such as 2′-fucosyllactose (FL), 3FL and sialyllactose (SL), or by repeats of building blocks of lacto-N-biose type 1 (LNB; Galβ1-3GlcNAc) or N-acetyllactosamine (LacNAc; Galβ1-4GlcNAc). These repeats can be further decorated by fucose or sialic acid in α-linkages, adding more complexity and diversity to these molecules (Bode & Jantscher-Krenn, 2012). HMO can be classified as acidic or neutral depending on the presence of the negatively charged sialic acid. Neutral HMO can be further categorized by the presence of fucose on their structures. Over 200 oligosaccharide structures have been identified in human milk (Wu et al., 2010, 2011).
Fig. 1.
Structural diversity of glycans in human milk and corresponding glycosyl hydrolases in infant-gut associated bifidobacteria. Legends at the bottom left indicate monosaccharide composition and the corresponding potential glycolytic enzymes in bifidobacteria acting at specific linkages. A: illustrative structure of HMO; B: three different cores found in human O-linked glycans; C: glycolipids, the structure of ganglioside GD3 is shown; D: a complex N-glycan.
Significant differences exist in HMO abundance and composition among different mothers and across lactation stages (Coppa et al., 1999; De Leoz et al., 2012; Niñonuevo et al., 2008). An important association also exists between HMO and the blood group type of the mother represented by the Lewis system and the secretor genes, which generates four different groups of milks (Totten et al., 2012). Another important characteristic of human milk is the overabundance of type 1 HMO, containing type 1 LNB, over type 2 HMO containing LacNAc (Urashima et al., 2012). Type 1 predominance and large amounts of fucosylated HMO are characteristic of human milk but much less so for other mammals (Tao et al., 2011; Taufik et al., 2012).
Great efforts have been made to elucidate the composition and structures of HMO. As recently reviewed (Kunz, 2012), milk carbohydrates research started in the early 1900s. Despite recent technological advances, structural elucidation of oligosaccharides from breast milk still remains a challenge, mainly due to the variety of possible isomeric forms of any given composition. MS has become a method of choice for oligosaccharide analysis, and current methods allow isomer differentiation with high resolution (Ruhaak & Lebrilla, 2012).
Physiological effects of HMO
GI enzymes in the infant are not capable of breaking down the diversity of HMO linkages synthesized by glycosyltransferases in the mammary gland (Dallas et al., 2012), which emphasizes the role of milk bioactive molecules in functions beyond nutrition. HMO are minimally affected by transit through the stomach and small intestine, reaching a high concentration in infant faeces (Chaturvedi et al., 2001; Engfer et al., 2000; Gnoth et al., 2000). Excreted faecal HMO in breast-fed infants are a reflection of the mother secretor status, and the course of oligosaccharide excretion is apparently individual-specific and intermediate degradation products can be observed (Albrecht et al., 2011). Furthermore, small amounts of certain HMO can be found in urine (Rudloff & Kunz, 2012), suggesting that these molecules can exert physiological effects not only locally in the GI tract but also systemically.
HMO are well known for their ability to prevent adherence and invasion of several pathogens (Imberty & Varrot, 2008; Morrow et al., 2005). This is probably due to the structural similarity between HMO and glycoconjugates present in the intestinal mucosa. Fucose- and sialic acid-containing HMO are particularly important in pathogen deflection as they are found at terminal positions in these molecules. Therefore, the abundance of HMO and other milk glycoconjugates can explain in great part how breast milk helps to prevent infant diarrhoea and GI infections in breast-fed infants (Coppa et al., 2006; Hakkarainen et al., 2005; Hong et al., 2009; Martín-Sosa et al., 2002; Morrow et al., 2004; Newburg et al., 2004; Ruiz-Palacios et al., 2003).
Establishment of the infant intestinal microbiota
At birth, the newborn is first exposed to the extrauterine environment, entering a microbial-laden world that results in quick colonization of different body sites, typically in a non-pathogenic fashion (Dominguez-Bello et al., 2010). Of particular interest is how the intestinal microbiota is established, given the potential impact it has on subsequent health and disease (Reinhardt et al., 2009; Scholtens et al., 2012). Patterns of early intestinal colonization can have both short-term and long-term health effects (Bager et al., 2008; Cho et al., 2012; Collado et al., 2012; Kalliomäki et al., 2008; Salvini et al., 2011). Bacterial colonization of the intestine is key in several aspects: bacteria provide essential nutrients for the infant such as vitamins and short-chain fatty acids, they stimulate the development of the immune system, especially adaptive responses, and they provide general protection against pathogen colonization, among several other functions (Hooper et al., 2012; Nicholson et al., 2012). A contribution of the intestinal microbiota has been established in the onset of obesity and type 2 diabetes (Harris et al., 2012; Ley et al., 2005). The establishment of the microbiota in the infant colon has been described as an orchestrated, but chaotic, succession of bacteria (Koenig et al., 2011), wherein the composition depends on a diverse number of factors such as mode of delivery, type of feeding, and genetic, cultural and geographical determinants (Scholtens et al., 2012). The first colonizers of the intestinal tract are facultative anaerobic bacteria, such as Escherichia coli, enterococci and streptococci, which predominate in the first days of life. These bacteria will consume the oxygen in the intestinal lumen, creating an anaerobic environment more favourable for strict anaerobes, such as Bacteroides, Clostridium and Bifidobacterium (Jost et al., 2012).
Mode of delivery is one of the most important factors that dictate the composition of the infant intestinal microbiota in the first months of life. Normal vaginal delivery exposes the infant to the vaginal and faecal microbiota of the mother (Dominguez-Bello et al., 2010; Makino et al., 2011). Human breast milk has been also considered another source of micro-organisms that can potentially contribute to gut colonization (Cabrera-Rubio et al., 2012; Grönlund et al., 2007; Makino et al., 2011); however, this remains controversial, since the microbiota in breast milk can be instead a reflection of the skin or faecal microbiota. In the other hand, the hospital environment (Martirosian et al., 1995) and the skin microbiota (Dominguez-Bello et al., 2010) are considered sources of bacteria for caesarian-born infants. A delay in microbial colonization by prominent members of the intestinal microbiota such as Bifidobacterium, Bacteroides and E. coli has also been observed (Adlerberth et al., 2006; Mitsou et al., 2008), and bifidobacterial counts are also lower in caesarian-born infants (Chen et al., 2007; Penders et al., 2006).
Significant differences are found in the early composition of the infant intestinal microbiota relative to the type of diet. Infant formulas are traditionally based on bovine milk, and great advances have been made to improve their composition by adding supplements such as minerals, vitamins and prebiotics, in order to simulate the essential components in breast milk (Hernell, 2011; Koletzko, 2010). Unfortunately, some of the bioactive molecules in human milk are not found in bovine milk, and therefore replicating their effects is challenging (Dewey et al., 1995; Le Huërou-Luron et al., 2010).
Bifidogenic effect of HMO
Culture-based and current large-scale metagenomic studies show that Bifidobacterium is a dominant genus in the intestinal microbiota of breast-fed infants, in some cases representing approximately 75 % of total bacteria (Harmsen et al., 2000; Roger & McCartney, 2010; Sakata et al., 2005; Yatsunenko et al., 2012). The overrepresentation of bifidobacteria in this environment is less observed in formula-fed infants, who show a more diverse microbiota (Fallani et al., 2010; Penders et al., 2006). Therefore, differences in bacterial colonization between breast-fed and formula-fed infants can be explained in great part by the non-essential components in human milk.
The predominance of bifidobacteria in breast-fed infant faeces was first noticed over 100 years ago (Moro, 1905). Moro and Tissier suggested that breast milk contained certain molecules that stimulated the growth of these bacteria, defined as bifidus factors (Moro, 1905). Gynolactose was later described as a mixture of milk oligosaccharides containing GlcNAc that stimulated the growth of certain Bifidobacterium strains (Polonowski & Lespagnol, 1931). These studies first suggested a prebiotic role for oligosaccharides in breast milk.
The ability of these micro-organisms to metabolize HMO might therefore represent an example of co-evolution with their host, and the physiology and mechanism of this consumption has been addressed. Ward and co-workers first showed that Bifidobacterium longum subsp. infantis (B. infantis) can grow vigorously on HMO in vitro as the sole carbon source (Ward et al., 2006). B. infantis displays a preference for shorter HMO but can use larger oligosaccharides as well (LoCascio et al., 2007). The ability to consume HMO in vitro has been demonstrated for additional strains of B. infantis and also Bifidobacterium bifidum, and to a lesser extent strains of Bifidobacterium breve and Bifidobacterium longum (Asakuma et al., 2011; LoCascio et al., 2009; Turroni et al., 2010). These four species are usually present in breast-fed infant faeces (Turroni et al., 2012; Yatsunenko et al., 2012; Avershina et al., 2013; Boesten et al., 2011; Matsuki et al., 2002).
Hence, the enrichment in bifidobacteria in breast-fed infant faeces can be explained in great part by their ability to consume and metabolize HMO. Moreover, the prebiotic character of HMO seems to be selective for infant bifidobacteria and a few Bacteroides species, and not for adult bifidobacteria or other prominent members of the intestinal microbiota such as Clostridium and enterobacteria (Marcobal et al., 2010). Bottle-fed infants display higher numbers of Firmicutes and Bacteroides and less of Bifidobacterium in their faeces, and the bifidobacteria characteristic of formula-fed infants include additionally Bifidobacterium adolescentis and Bifidobacterium pseudocatenulatum, which are more commonly associated with the adult intestinal microbiota (Haarman & Knol, 2005; Mangin et al., 2006).
Bifidobacterial strategies for HMO consumption
Bifidobacteria possess a fermentative metabolism, and they are almost exclusively associated with the GI tract of animals (Lee et al., 2008; Sela et al., 2010). They are considered to be beneficial for the human host, and several strains of bifidobacteria are commercialized as probiotics. In general they devote a significant portion of their genomes to the consumption of complex oligosaccharides (O’Connell Motherway et al., 2011b; Schell et al., 2002; Sela et al., 2008; Turroni et al., 2010), either of plant origin in the case of adult-associated species or host-derived in the case of species better adapted to the nursing period. Analysis of genome sequences of bifidobacteria isolated from breast-fed infants has enabled predictions regarding the HMO consumption phenotype. In particular, B. infantis ATCC 15697 and B. bifidum PRL2010 are prototypical members of the infant intestinal microbiota that have possibly co-evolved with their host to consume milk or host oligosaccharides (Sela et al., 2008; Turroni et al., 2010).
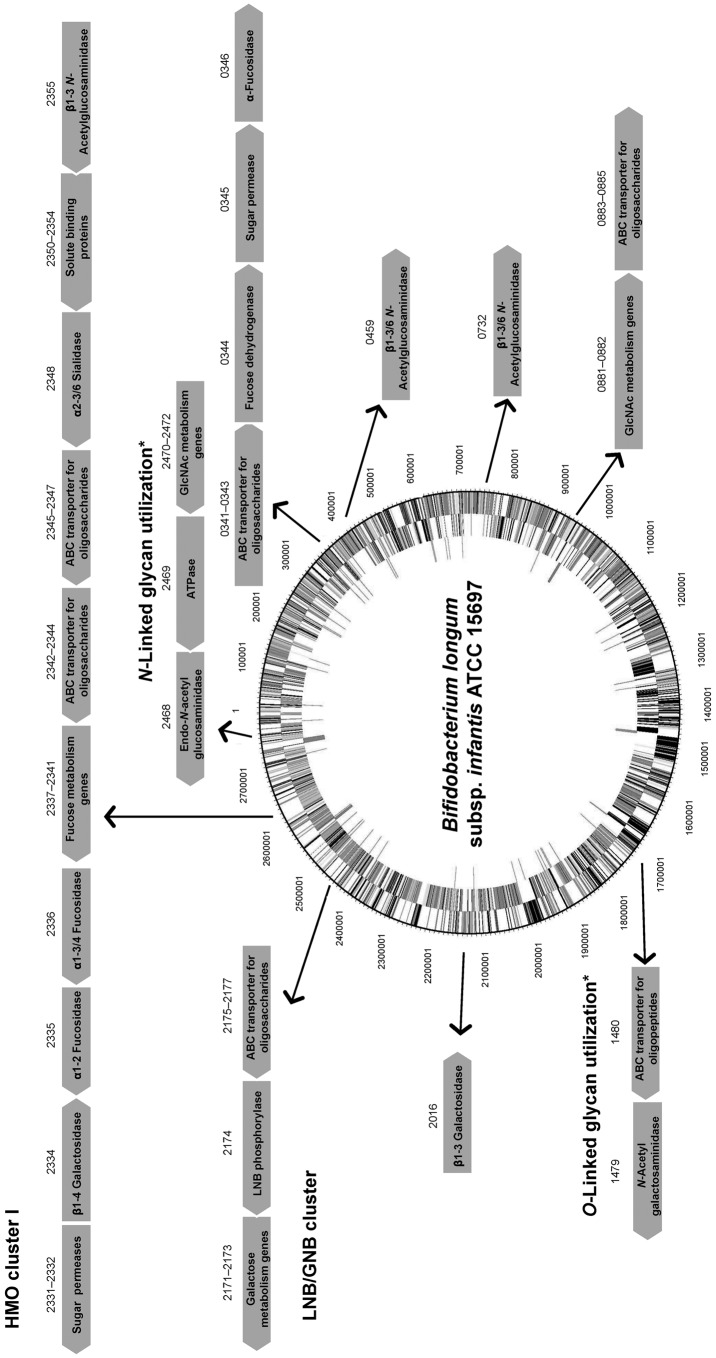
Physiologically, B. infantis can simultaneously consume distinct classes of HMO in vitro with high efficiency, reaching higher cell densities compared to other infant-associated bifidobacteria (Asakuma et al., 2011; Ward et al., 2006). A hallmark of the genome of this species is a conserved cluster of genes, the HMO cluster I (Fig. 2), containing several glycosyl hydrolases and ABC transporters (Sela et al., 2008). Other potentially important clusters for HMO consumption are also conserved among other B. infantis strains but are absent in B. longum strains (LoCascio et al., 2010). The overall overrepresentation of genes encoding family 1 solute binding proteins (SBPs) and also intracellular glycosyl hydrolases (GHs) with putative affinity for or activity on HMO in the genome of B. infantis is suggestive of a consumption strategy based on the import of intact HMO structures and their intracellular enzymic degradation (Zivkovic et al., 2011). Several of these predictions regarding HMO consumption have been addressed and genes encoding functions in HMO import and hydrolysis have been identified (Fig. 2).
Fig. 2.
Clusters of genes in B. infantis ATCC 15697 with assigned or putative functions in the utilization of milk glycans. Numbers above the arrows correspond to the respective locus tags (Blon_xxxx). Genes are not drawn to scale, and the genome circle was adapted from Sela et al. (2008). SBPs from ABC transporters with affinities for HMO and expressed during growth on these substrates were identified by Garrido et al. (2011). An α-sialidase and two α-fucosidases were characterized by Sela et al. (2011, 2012). Two β-galactosidases and three β-hexosaminidases active on different linkages in representative HMO are also included (Garrido et al., 2012c; Yoshida et al., 2012). Finally, potential gene clusters for N-linked and O-linked glycan utilization (*) are depicted (Garrido et al., 2012b; Kiyohara et al., 2012).
For example, a large array of oligosaccharide-binding SBPs in B. infantis ATCC 15697 is biased towards mammalian glycans, especially HMO (Garrido et al., 2011). Their substrate affinities are diverse and include neutral HMO containing either LNB or LacNAc (type 1 or type 2 HMO), or fucosylated HMO such as 2′FL and Lewis epitopes. Chemical blockage of ABC transporters inhibits the ability of B. infantis to consume lacto-N-tetraose (LNT; type 1 HMO) as the sole carbon source in vitro (Yoshida et al., 2012). In addition, genes encoding SBPs with affinity for HMO are exclusively induced during exponential growth on HMO (Fig. 2). In addition, some of these proteins are able to bind epithelial surfaces in vitro, probably due to the structural similarities between HMO and epithelial glycoconjugates. These results also suggested physiological differences between B. infantis cells growing on either simple lactose or HMO, where epithelium-binding SBPs are induced only during growth on HMO. In agreement with these observations, B. infantis cells growing on HMO but not lactose display increased binding to intestinal epithelial cells, and under these conditions they enhance the production of anti-inflammatory cytokines and tight junction proteins (Chichlowski et al., 2012).
Another relevant aspect of bacterial HMO utilization is the enzymic processing of these molecules by glycosyl hydrolases. Interestingly, the microbiome of breast-fed infants is enriched in fucosidases and sialidases (Yatsunenko et al., 2012). The genome of B. infantis contains an army of glycosyl hydrolases active on these carbohydrates, including two genes encoding α-sialidases, five α-fucosidases, five β-galactosidases and three β-N-acetylglucosaminidases (Garrido et al., 2012a). Recent functional studies on the enzymic properties of these enzymes and their induction by HMO have greatly advanced our understanding of HMO consumption by B. infantis (Fig. 2). Acidic HMO such as 3′SL, 6′SL and sialyl-LNT represent nearly 15 % of total HMO. These are probably imported inside the cells by systems different from ABC transporters, and membrane permeases of the major facilitator family are likely candidates. NanH2 is an α-sialidase in B. infantis (Blon_2348 in HMO cluster I, Fig. 2) that removes sialic acid from α2-3 and α2-6 sialyl linkages found in individual HMO such as mono and disialyl-LNT (Sela et al., 2011). The expression of NanH2, but not a second encoded sialidase NanH1, was significantly increased during bacterial growth on HMO. In addition, two fucosidases in B. infantis have significant activity in fucosylated HMO and blood group oligosaccharides. Blon_2335 and Blon_2336 are located in HMO cluster I and belong to GH families 95 and 29, respectively (Fig. 2). Growth on pooled HMO, LNT or LNnT (a type 2 HMO) upregulates their gene expression (Sela et al., 2012). Blon_2335 is a highly efficient α1-2 fucosidase that has also considerable activity towards α1-3 and α1-4 fucosyl linkages, contrary to the AfcA fucosidase in B. bifidum, which acts exclusively on Fucα1-2 linkages (Ashida et al., 2009). Blon_2335 can release fucose from several HMO such as 2′FL, 3FL and lacto-N-fucopentaoses I and III, and also from fucosylated epitopes found in epithelial glycoconjugates such as Lewisa [Galβ1-3(Fucα1-4)GlcNAc], Lewisx [Galβ1-4(Fucα1-3)GlcNAc] and the H-disaccharide (Fucα1-2Gal). In the other hand Blon_2336 is specific for α1-3/4 linkages, such as those found on 3FL, lacto-N-fucopentaose III and Lewisx (Sela et al., 2012).
Galactose and N-acetylglucosamine constitute the building blocks of HMO, and these monosaccharides are generally fermentable carbon sources for bifidobacteria. Galactose is found in simple carbohydrates such as lactose and complex oligosaccharides of mammalian or plant origin. In B. infantis, two β-galactosidases with glycolytic activity on HMO are constitutively expressed (Yoshida et al., 2012). Bga2A, encoded by Blon_2334 in HMO cluster I (Fig. 2), belongs to GH family 2 and has a preference for lactose, also efficiently removing galactose from type 2 HMO such as LacNAc and LNnT. Complementing this activity is Bga42A (encoded by Blon_2016; Fig. 2), a GH42 β-galactosidase highly specific for LNT, one of the most abundant HMO (Yoshida et al., 2012). Interestingly, this enzyme has considerably less activity on LNB, suggesting that each residue in LNT is crucial for its enzymic activity. Finally, β-N-acetylglucosaminidases participate in this process (Garrido et al., 2012c). Blon_0459, Blon_0732 and also Blon_2355 in the HMO cluster I are expressed mostly during early exponential growth on HMO, and while the three enzymes can cleave the GlcNAcβ1-3Gal linkage found in linear HMO such as LNT or LNnT, only Blon_0459 and Blon_0732 are active on branched HMO, characterized by GlcNAcβ1-6Gal as found in lacto-N-hexaose. These results support the concept of sequential hydrolysis of HMO in B. infantis, releasing large amounts of monosaccharides that can be fermented in central metabolic pathways.
Parallel studies have provided important details on the mechanisms of HMO utilization by B. bifidum, another member of the infant intestinal microbiota. B. bifidum and B. infantis are competitive in HMO consumption but using different strategies (Garrido et al., 2012a). While B. infantis has specialized in the import and intracellular deglycosylation of several HMO, B. bifidum uses an array of membrane-associated glycosyl hydrolases, including extracellular α-sialidases (Kiyohara et al., 2011), α-fucosidases (Ashida et al., 2009), β-galactosidases and β-N-acetylglucosaminidases (Miwa et al., 2010), which efficiently remove monosaccharides from complex HMO. Another important difference between these strategies is the presence of membrane lacto-N-biosidase activity in B. bifidum (Wada et al., 2008). This endoglycosidase generates LNB and lactose from LNT, and some of the mono- and disaccharides released can be internalized and metabolized, especially LNB (Asakuma et al., 2011). The binding protein for this disaccharide is a family 1 SBP, part of a gene cluster found in several bifidobacteria, the LNB/GNB cluster (Kitaoka et al., 2005; Nishimoto & Kitaoka, 2007). Genes encoding ABC permeases, a lacto-N-biose phosphorylase that generates galactose 1-phosphate and glucose from LNB, and two other genes in the Leloir pathway for galactose metabolism (Fig. 2), are adjacent to this SBP. The LNB/GNB cluster is actually conserved across all infant gut-associated bifidobacteria, including B. bifidum, B. infantis, B. longum and B. breve isolates (LoCascio et al., 2010; Xiao et al., 2010).
Relatively little attention has been paid to B. breve and B. longum regarding HMO consumption, even considering that they are normally dominant in infant faeces, and B. breve seems to be found exclusively in this environment (Avershina et al., 2013). B. longum ATCC 15707 and B. breve ATCC 15700 show only modest growth on pooled HMO and apparently can metabolize solely LNT, leaving other HMO unmodified (Asakuma et al., 2011; LoCascio et al., 2007). An association between the LNB/GNB cluster and HMO consumption in B. longum ATCC 15707 was suggested after induction of these genes during growth on human milk (González et al., 2008). Several species of bifidobacteria, including strains of B. breve and B. longum, are able to grow in vitro using LNB as the sole carbon source (Xiao et al., 2010). This consumption can be explained solely by the presence and activity of the LNB/GNB cluster in these strains that can import and metabolize this disaccharide.
Human milk glycoconjugates
The complexity of human milk is far from understood, and one example of this is the multiplicity of functions played by several bioactive agents. While the high concentrations of oligosaccharides in human milk can explain in great part the enrichment in bifidobacteria in breast-fed infant faeces, glycans conjugated to other molecules in milk, such as proteins, peptides or lipids, can also have a prebiotic role. Here we address some of the biological properties of these glycoconjugates and what the mechanisms are that infant bifidobacteria have devised to use these glycoconjugates as a carbon source.
Human milk glycolipids
Lipids make up 3–5 % of the total volume of human milk, of which 98 % are triacylglycerols (Jensen, 1999). A fraction of the remaining fats in human milk consists of glycolipids, mostly associated with the milk fat globule membrane (Newburg & Chaturvedi, 1992). Glycolipids are composed of a lipid chain of ceramide, a fatty acid linked to a sphingoid base, covalently attached to one or more monosaccharides. Milk glycolipids can be classified as neutral, including galactosylceramide (Galβ1-1Cer) and glucosylceramide (Glcβ1-1Cer; Bouhours & Bouhours, 1979), or acidic glycosphingolipids (or gangliosides), containing sialic acid (Laegreid et al., 1986). The most abundant gangliosides in human milk are GD3 (Fig. 1) and GM3 (NeuAcα2-3Galβ1-4Glcβ1-1Cer; Lee et al., 2011).
The glycan portion of milk glycolipids plays an important role in pathogen deflection, similar to other milk glycoconjugates. Binding the epithelium is the first line of entry for pathogens or their toxins, and this process is usually mediated by glycolipids. Therefore, milk-borne glycolipids associated with milk fat globule membranes can prevent bacterial, viral or toxin binding to the intestine (Lindberg et al., 1987; Miller-Podraza et al., 2005; Newburg, 2009; Otnaess et al., 1983). In addition, several intestinal commensals are able to bind glycolipids in vitro (Mukai et al., 2004; Neeser et al., 2000; Strömberg et al., 1988; Yamamoto et al., 1996).
Milk fat globules are degraded by diverse lipases in the GI tract (Lindquist & Hernell, 2010), releasing lipids that are readily absorbed into the small intestine. However, the fate of milk glycolipids is not yet understood, and it is possible that they transit distal portions of the GI tract. Only a few studies have addressed the degradation of milk or epithelial glycolipids by members of the infant intestinal microbiota, and evidence has indicated that they possess enzymes that can hydrolyse in great part these glycoconjugates (Larson et al., 1988). Right after establishment, the intestinal microbiota is responsible for the degradation of glycolipids observed in breast-fed infant faeces (Gustafsson et al., 1986; Midtvedt et al., 1988). The degree of this hydrolytic activity is however lower than that in adults, but higher compared to newborns or germ-free mice (Larson et al., 1987; Larson & Midtvedt, 1989).
It has been observed in vitro that glycosidases from Ruminococcus torque, B. bifidum and B. infantis degrade several glycosides containing certain blood group determinants, including the H disaccharide, Lewisa and Lewisb (Falk et al., 1991; Larson et al., 1988). Lactosylceramide is a common end product of their reactions (Falk et al., 1990). The ability of bifidobacteria to release sialic acid from predominant milk gangliosides such as GD3 and GM3 has been observed (Falk et al., 1990), suggesting that certain bifidobacteria possess α2-8 and α2-3 sialidase activity (Fig. 1).
Human milk glycoproteins
Protein glycosylation is a post-translational modification in which a glycan is covalently linked to predetermined amino acids in the protein structure. There are two major types of oligosaccharides attached to eukaryotic proteins: N-linked and O-linked glycans. These conjugated carbohydrates play several biochemical and physiological roles, for example in protein synthesis, folding, trafficking and secretion, resistance to proteolysis, and prevention of pathogen colonization of epithelial surfaces among several others (Barboza et al., 2012; Gopal & Gill, 2000; Newburg et al., 2005; Peterson et al., 1998; Rudd et al., 1994). In human milk a large number of human milk proteins are glycosylated, including lactoferrin, immunoglobulins and κ-casein among several others (Froehlich et al., 2010).
N-Linked glycans are attached to the protein via specific asparagines in the sequence Asn-xxx-Ser/Thr (Stanley et al., 2009). All N-linked glycans have in common a pentasaccharide with the structure Man3GlcNAc2, where the last GlcNAc is linked to the asparagine via a β-linkage (Fig. 1). This pentasaccharide can sometimes be modified with core α1-6 fucosylation or a bisecting GlcNAc. N-Glycans can be heterogeneous and tissue-specific, but three main classes of N-glycans have been described based on further modifications of the pentasaccharide: (a) high mannose, consisting of branches of α-mannose; (b) complex, characterized by core α1-6 fucosylation of the basal GlcNAc and by two or more antennae (Galβ1-4GlcNAc repeats) that can be additionally decorated by fucose or sialic acid in terminal positions (Fig. 1); and (c) the hybrid type, which consists of a mixture of these two types. The human milk N-glycome has been recently described in detail, and in general human milk N-glycans are largely fucosylated and present in larger concentrations compared to bovine milk (Dallas et al., 2011; Nwosu et al., 2012). In contrast, bovine N-glycans are also more sialylated and characterized by the presence of N-glycolylneuraminic acid (NeuGc) instead of N-acetylneuraminic acid (Nwosu et al., 2012).
O-Linked glycans differ from N-linked glycans by attachment to serine or threonine residues, with no obvious surrounding amino acid consensus sequence. Eight different core structures have been described, each beginning with an α-GalNAc attachment to the amino acid (Brockhausen et al., 2009). Only four of these core structures (cores 1–4) are usually found in humans (Brockhausen et al., 2009). These O-linked structures can be further elongated by N-acetyllactosamine chains and decorated by fucose, sialic acid or GalNAc at terminal positions (Fig. 1).
Bifidobacterial consumption of human milk glycoproteins
In milk, protein glycosylation increases the resistance of proteins to proteolysis (van Berkel et al., 1995), probably contributing to the excretion of considerable amounts of intact or partially degraded milk proteins in breast-fed infant faeces (Davidson & Lönnerdal, 1987; Prentice et al., 1989). Milk proteins vary in their digestibility (Le et al., 2012; Ye et al., 2011), and non-glycosylated proteins such as β-casein and α-lactoglobulin are more digested in comparison to lactoferrin, IgA and milk mucins (Jakobsson et al., 1982; Lindh, 1975; Prentice et al., 1989).
Therefore, breast milk glycoproteins, in conjunction with mucosal secretions and shed epithelial cells, transit the GI tract of the breast-fed infant and can play a role in shaping the developing intestinal microbiota. Evidence indicates that these microbes largely modify host glycoconjugates (Hoskins et al., 1985; Variyam & Hoskins, 1981). Germ-free mice secrete intact mucins in their faeces, while conventionalized animals are able to degrade and metabolize mucins completely (Corfield et al., 1992; Midtvedt et al., 1987). In addition, bacteria extracted en masse from adult and infant stools produce a variety of extracellular glycosidases that degrade the glycans of hog gastric mucin under anaerobic conditions (Midtvedt et al., 1988; Variyam & Hoskins, 1981). In addition, individual members of the infant and adult intestinal microbiota have been well studied for their ability to deglycosylate mucins in order to gain access to the bound oligosaccharides as a carbon source (Derrien et al., 2010; Wright et al., 2000). Several species of Bacteroides deploy exquisite mechanisms for mucin glycan degradation based on membrane-bound glycosyl hydrolases and importers that are crucial for the survival and predominance of these species in the adult microbiota (Bäckhed et al., 2005; Martens et al., 2009). Interestingly, certain Bacteroides species can utilize HMO (Marcobal et al., 2010), and the transcriptional responses elicited during growth in vitro on mucin are highly similar to those witnessed on HMO for Bacteroides thetaiotaomicron, suggesting that these substrates are energetically similar for this species (Marcobal et al., 2011).
Some bifidobacteria are also well-known mucin degraders (Crociani et al., 1994; Hoskins et al., 1985). To date, this phenotype seems to be exclusive to B. bifidum and certain isolates of B. longum (Ruas-Madiedo et al., 2008). Pivotal to the release of O-linked glycans from mucins are endo-α-N-acetylgalactosaminidases (EngBF, glycosyl hydrolase family 101). Functional studies have shown that these extracellular enzymes cleave Core 1 O-linked glycans (Galβ1-3GalNAcα-Ser/Thr) found in mucins (Fig. 1; Ashida et al., 2008; Fujita et al., 2005). This hydrolysis releases galacto-N-biose (Galβ1-3GalNAc; GNB), a disaccharide structurally similar to LNB from HMO that can be directly used as a carbon source by B. longum via an ABC importer and enzymes in the LNB/GNB cluster (Kitaoka et al., 2005; Nishimoto & Kitaoka, 2007). Since EngBFs are highly specific, GNB release and import probably require the previous action of several glycosyl hydrolases on mucin glycans, such as α-fucosidases, α-sialidases and lacto-N-biosidase. These enzymes are also active on HMO, and the genes encoding these activities are highly expressed during growth in vitro on hog gastric mucin as well as on HMO (Turroni et al., 2010).
An alternative mucin utilization pathway has been recently described in bifidobacteria, which might represent a more accurate representation of intestinal mucin degradation by these micro-organisms in vivo (Kiyohara et al., 2012). The glycans on colonic mucins contain mostly Core 3 O-linked glycans based on the structure GlcNAcβ1-3GalNAcα1-Ser/Thr (Fig. 1), which are inaccessible for EngBF. A novel endo-α-N-acetylgalactosaminidase (NagBb, GH129) in B. bifidum is specific for the Tn antigen (GalNAcα-Ser/Thr). This structure is potentially produced after extracellular degradation by glycosyl hydrolases. While this mechanism remains to be validated, this novel enzyme was shown to be present in several genomes of infant gut-associated bifidobacteria including B. infantis (Fig. 2), B. breve and B. longum (Kiyohara et al., 2012), suggesting a common route to degradation of Core 3 O-linked glycans (Fig. 1).
The ability of bifidobacteria to access O-linked glycans in human or bovine milk proteins has been less explored; however, we hypothesize that similar mechanisms to those described above are prevalent for human milk mucins. As mentioned above, milk mucins contain a majority of Core 2 O-glycans (Fig. 1). It is possible that after gastric and intestinal digestive processes a higher concentration of mucin-derived glycopeptides is available for infant-associated bifidobacteria. This is not a new concept, as the bovine κ-casein-derived GMP is a highly sialylated glycopeptide that has been suggested to have prebiotic effects (Gomes et al., 1998; Janer et al., 2004; Petschow & Talbott, 1990).
We recently explored the ability of bifidobacteria to gain access to N-glycans from host glycoproteins using a representative panel of 76 strains of these micro-organisms isolated from infant faeces (Garrido et al., 2012b). Endo-β-N-acetylglucosaminidases (EC 3.2.1.96; endoglycosidases) hydrolyse the N,N′-diacetyl-chitobiose core common to all N-glycans (Fig. 1). Genes encoding these enzymes were found in several isolates of B. longum, B. infantis and B. breve, and their presence correlated with the ability of these micro-organisms to release the N-linked glycan of bovine RNase B. Among these enzymes, those belonging to glycosyl hydrolase family 18 (GH18) were able to remove the N-glycans from bovine and human lactoferrin (Garrido et al., 2012b), containing high mannose and complex N-glycans, respectively (Nwosu et al., 2011; Yu et al., 2011). Further characterization by MALDI-Fourier transform ion cyclotron resonance (FTICR) MS of endoglycosidase EndoBI-1 from B. infantis ATCC 15697 (Blon_2468, Fig. 2) revealed that this enzyme can deglycosylate common host glycoproteins such as IgA and IgG in their native forms in addition to human and bovine lactoferrin. Surprisingly, EndoBI-1 cleavage specificity was wide, releasing N-glycans with a variety of structures including high mannose N-linked glycans, or complex glycans with core α1-6 fucosylation, chain sialylation or fucosylation, and bi- and tri-antennary structures (Fig. 1). Furthermore, incubation of the enzyme with fresh breast milk samples led to a complete removal of milk protein N-glycans.
Replicating the bifidogenic effect of breast milk
For some mothers, breastfeeding is not possible, and therefore there is an increased need for human milk substitutes. Commercial production of synthetic mimics of HMO or other milk glycoconjugates is challenging, given the diversity of complex glycans involved. However, commercial production of more simple HMO species such as LNnT, 2′FL and 6′SL is now possible, as is the ability to test these molecules for bifidogenicity in animal (Marcobal et al., 2011) and human trials. Other prebiotics such as fructooligosaccharides (FOS), galactooligosaccharides (GOS) and inulin (Gibson et al., 2004; Torres et al., 2010) are commonly included in infant formula. GOS are synthetic substrates derived enzymically from the transglycosylation of lactose, with a degree of polymerization of 3–15 (Barboza et al., 2009). It has been suggested that GOS resemble galactan chains found in plant oligosaccharides (O’Connell Motherway et al., 2011a). We recently observed that the consumption of GOS with a large degree of polymerization is strain-dependent in B. infantis isolates (Garrido et al., 2013), and discrete mechanisms for import and intracellular degradation are active in B. infantis strain ATCC 15697. On the other hand, FOS and inulin are naturally found in chicory plants. The wide availability of FOS and GOS has enabled numerous in vitro, animal and human studies of their prebiotic effects, and their bifidogenic effect is currently accepted (Bakker-Zierikzee et al., 2005; Brunser et al., 2006; Davis et al., 2011).
Analysis of the milk of other mammals indicates that they do not possess the high concentration of oligosaccharides combined with a high level of fucosylation witnessed in human milks. For example, mature bovine milk is very low in free oligosaccharides, which are mainly sialylated (Sundekilde et al., 2012; Tao et al., 2008). More efficient analytical tools have recently revealed the presence of low concentrations of several neutral fucosylated oligosaccharides that resemble derivatives of lacto-N-neohexaose present in early milk HMO (Barile et al., 2010; Sundekilde et al., 2012). At present, new approaches are being applied to use dairy streams from cheese production to recover bovine milk oligosaccharides (BMO) in larger quantities (Zivkovic & Barile, 2011).
Conclusion and future directions
The complexity of breast milk is intriguing and still far from being understood. The fundamental role of human milk as a nutrient source for the infant has been the focus of study for decades, with the critical goal of understanding and improving nutritional deficiencies during the neonatal period. However, the influence of breastfeeding beyond nutrition is increasingly being revealed, demonstrating that milk provides much more than protection against pathogens. Breastfeeding has been associated with a variety of long-term health impacts including lowered incidence of obesity (Kalliomäki et al., 2008), diabetes (Mayer et al., 1988; Owen et al., 2006; Pettitt et al., 1997) and allergies (Gdalevich et al., 2001; Snijders et al., 2007). A future challenge will be to identify a mechanistic basis for these benefits.
An infant gut microbiota dominated by bifidobacteria has long been associated with health; however, our understanding of this process is still unclear. Recently the protective role of production of short-chain fatty acids by certain species of bifidobacteria against pathogenic E. coli challenge (Fukuda et al., 2011) has been demonstrated. This work highlighted the importance of in situ metabolism of complex carbohydrates by bifidobacteria in host–microbe interactions. However, the amounts of protective acetate and lactate produced by bifidobacteria can be different depending on the growth substrate, which might have direct consequences for the host (Garrido et al., 2013). Consumption of certain HMO can also be a selective colonization factor; for example, B. infantis grows vigorously on lacto-N-neotetraose, and this ability enables the bacterium to outcompete Bacteroides thetaiotamicron in a mouse model, emphasizing the selectivity and bifidogenic activities of these unique glycans (Marcobal et al., 2011).
A clear benefit of mechanistic research is the rapid nature by which this information can be translated to address a range of intestinal maladies (Gordon et al., 2012). The ability to purify, or synthesize, HMO-like oligosaccharides and/or glycoconjugates at commercial scales is increasingly becoming a reality. This ability, combined with an expanding number of well-characterized bifidobacterial strains that grow on these complex milk glycans, will help to design tailored synbiotic formulations to target specific at-risk populations such as premature and malnourished infants. Given that milk is the product of millions of years of mammalian evolution, it is not surprising that it displays a constellation of health benefits for the infant. With the advances in nanotechnology and systems biology perhaps this ‘constellation’ will become more comprehensible and inspire new opportunities for protective modulation of the human GI tract.
Acknowledgements
We acknowledge all the researchers in the UC Davis Foods for Health Institute and the Milk Bioactives Program for their enthusiasm, imagination and collective contribution to this subject matter. Work by the Milk Bioactives Program has been supported by University of California Discovery Grant Program, the California Dairy Research Foundation, the Bill and Melinda Gates Foundation and National Institutes of Health awards R01HD059127, R01HD065122, R01HD061923, R21AT006180, R01AT007079. D. G. has been supported in part by a Fulbright-Conicyt Chile scholarship and a National Milk Producers Federation scholarship. D. A. M. acknowledges support as the Peter J. Shields Endowed Chair in Dairy Food Science.
References
- Adlerberth I., Lindberg E., Aberg N., Hesselmar B., Saalman R., Strannegård I. L., Wold A. E. (2006). Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res 59, 96–101. 10.1203/01.pdr.0000191137.12774.b2 [DOI] [PubMed] [Google Scholar]
- Albrecht S., Schols H. A., van den Heuvel E. G., Voragen A. G., Gruppen H. (2011). Occurrence of oligosaccharides in feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carbohydr Res 346, 2540–2550. 10.1016/j.carres.2011.08.009 [DOI] [PubMed] [Google Scholar]
- Allen J. C., Keller R. P., Archer P., Neville M. C. (1991). Studies in human lactation: milk composition and daily secretion rates of macronutrients in the first year of lactation. Am J Clin Nutr 54, 69–80. [DOI] [PubMed] [Google Scholar]
- American Academy of Pediatrics Section on Breastfeeding (2012). Breastfeeding and the use of human milk. Pediatrics 129, e827–e841. 10.1542/peds.2011-3552 [DOI] [PubMed] [Google Scholar]
- Asakuma S., Hatakeyama E., Urashima T., Yoshida E., Katayama T., Yamamoto K., Kumagai H., Ashida H., Hirose J., Kitaoka M. (2011). Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286, 34583–34592. 10.1074/jbc.M111.248138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H., Maki R., Ozawa H., Tani Y., Kiyohara M., Fujita M., Imamura A., Ishida H., Kiso M., Yamamoto K. (2008). Characterization of two different endo-α-N-acetylgalactosaminidases from probiotic and pathogenic enterobacteria, Bifidobacterium longum and Clostridium perfringens. Glycobiology 18, 727–734. 10.1093/glycob/cwn053 [DOI] [PubMed] [Google Scholar]
- Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., Yamamoto K. (2009). Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19, 1010–1017. 10.1093/glycob/cwp082 [DOI] [PubMed] [Google Scholar]
- Avershina E., Storrø O., Oien T., Johnsen R., Wilson R., Egeland T., Rudi K. (2013). Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl Environ Microbiol 79, 497–507. 10.1128/AEM.02359-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A., Gordon J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- Bager P., Wohlfahrt J., Westergaard T. (2008). Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 38, 634–642. 10.1111/j.1365-2222.2008.02939.x [DOI] [PubMed] [Google Scholar]
- Bakker-Zierikzee A. M., Alles M. S., Knol J., Kok F. J., Tolboom J. J., Bindels J. G. (2005). Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br J Nutr 94, 783–790. 10.1079/BJN20051451 [DOI] [PubMed] [Google Scholar]
- Barboza M., Sela D. A., Pirim C., Locascio R. G., Freeman S. L., German J. B., Mills D. A., Lebrilla C. B. (2009). Glycoprofiling bifidobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specific, preferential consumption of glycans. Appl Environ Microbiol 75, 7319–7325. 10.1128/AEM.00842-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza M., Pinzon J., Wickramasinghe S., Froehlich J. W., Moeller I., Smilowitz J. T., Ruhaak L. R., Huang J., Lönnerdal B. & other authors (2012). Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol Cell Proteomics 11, 015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile D., Marotta M., Chu C., Mehra R., Grimm R., Lebrilla C. B., German J. B. (2010). Neutral and acidic oligosaccharides in Holstein-Friesian colostrum during the first 3 days of lactation measured by high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. J Dairy Sci 93, 3940–3949. 10.3168/jds.2010-3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L., Jantscher-Krenn E. (2012). Structure-function relationships of human milk oligosaccharides. Adv Nutr 3, 383S–391S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten R., Schuren F., Ben Amor K., Haarman M., Knol J., de Vos W. M. (2011). Bifidobacterium population analysis in the infant gut by direct mapping of genomic hybridization patterns: potential for monitoring temporal development and effects of dietary regimens. Microb Biotechnol 4, 417–427. 10.1111/j.1751-7915.2010.00216.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhours J. F., Bouhours D. (1979). Galactosylceramide is the major cerebroside of human milk fat globule membrane. Biochem Biophys Res Commun 88, 1217–1222. 10.1016/0006-291X(79)91109-4 [DOI] [PubMed] [Google Scholar]
- Brockhausen I., Schachter H., Stanley P. (2009). O-GalNAc glycans. In Essentials of Glycobiology, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [PubMed] [Google Scholar]
- Brunser O., Gotteland M., Cruchet S., Figueroa G., Garrido D., Steenhout P. (2006). Effect of a milk formula with prebiotics on the intestinal microbiota of infants after an antibiotic treatment. Pediatr Res 59, 451–456. 10.1203/01.pdr.0000198773.40937.61 [DOI] [PubMed] [Google Scholar]
- Cabrera-Rubio R., Collado M. C., Laitinen K., Salminen S., Isolauri E., Mira A. (2012). The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr 96, 544–551. 10.3945/ajcn.112.037382 [DOI] [PubMed] [Google Scholar]
- Chaturvedi P., Warren C. D., Buescher C. R., Pickering L. K., Newburg D. S. (2001). Survival of human milk oligosaccharides in the intestine of infants. Adv Exp Med Biol 501, 315–323. 10.1007/978-1-4615-1371-1_39 [DOI] [PubMed] [Google Scholar]
- Chen J., Cai W., Feng Y. (2007). Development of intestinal bifidobacteria and lactobacilli in breast-fed neonates. Clin Nutr 26, 559–566. 10.1016/j.clnu.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Chichlowski M., De Lartigue G., German J. B., Raybould H. E., Mills D. A. (2012). Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr 55, 321–327. 10.1097/MPG.0b013e31824fb899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., Yamanishi S., Cox L., Methé B. A., Zavadil J., Li K., Gao Z., Mahana D., Raju K. & other authors (2012). Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626. 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M. C., Laitinen K., Salminen S., Isolauri E. (2012). Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res 72, 77–85. 10.1038/pr.2012.42 [DOI] [PubMed] [Google Scholar]
- Coppa G. V., Pierani P., Zampini L., Carloni I., Carlucci A., Gabrielli O. (1999). Oligosaccharides in human milk during different phases of lactation. Acta Paediatr Suppl 88, 89–94. 10.1111/j.1651-2227.1999.tb01307.x [DOI] [PubMed] [Google Scholar]
- Coppa G. V., Zampini L., Galeazzi T., Facinelli B., Ferrante L., Capretti R., Orazio G. (2006). Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res 59, 377–382. 10.1203/01.pdr.0000200805.45593.17 [DOI] [PubMed] [Google Scholar]
- Corfield A. P., Wagner S. A., Clamp J. R., Kriaris M. S., Hoskins L. C. (1992). Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun 60, 3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crociani F., Alessandrini A., Mucci M. M., Biavati B. (1994). Degradation of complex carbohydrates by Bifidobacterium spp. Int J Food Microbiol 24, 199–210. 10.1016/0168-1605(94)90119-8 [DOI] [PubMed] [Google Scholar]
- Dallas D. C., Martin W. F., Strum J. S., Zivkovic A. M., Smilowitz J. T., Underwood M. A., Affolter M., Lebrilla C. B., German J. B. (2011). N-Linked glycan profiling of mature human milk by high-performance microfluidic chip liquid chromatography time-of-flight tandem mass spectrometry. J Agric Food Chem 59, 4255–4263. 10.1021/jf104681p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas D. C., Sela D., Underwood M. A., German J. B., Lebrilla C. B. (2012). Protein-linked glycan degradation in infants fed human milk. J Glycomics Lipidomics S1, 002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L. A., Lönnerdal B. (1987). Persistence of human milk proteins in the breast-fed infant. Acta Paediatr Scand 76, 733–740. 10.1111/j.1651-2227.1987.tb10557.x [DOI] [PubMed] [Google Scholar]
- Davis L. M., Martínez I., Walter J., Goin C., Hutkins R. W. (2011). Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS ONE 6, e25200. 10.1371/journal.pone.0025200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz M. L., Gaerlan S. C., Strum J. S., Dimapasoc L. M., Mirmiran M., Tancredi D. J., Smilowitz J. T., Kalanetra K. M., Mills D. A. & other authors (2012). Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res 11, 4662–4672. 10.1021/pr3004979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., van Passel M. W., van de Bovenkamp J. H., Schipper R. G., de Vos W. M., Dekker J. (2010). Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1, 254–268. 10.4161/gmic.1.4.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey K. G., Heinig M. J., Nommsen-Rivers L. A. (1995). Differences in morbidity between breast-fed and formula-fed infants. J Pediatr 126, 696–702. 10.1016/S0022-3476(95)70395-0 [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello M. G., Costello E. K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107, 11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engfer M. B., Stahl B., Finke B., Sawatzki G., Daniel H. (2000). Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr 71, 1589–1596. [DOI] [PubMed] [Google Scholar]
- Falk P., Hoskins L. C., Larson G. (1990). Bacteria of the human intestinal microbiota produce glycosidases specific for lacto-series glycosphingolipids. J Biochem 108, 466–474. [DOI] [PubMed] [Google Scholar]
- Falk P., Hoskins L. C., Larson G. (1991). Enhancing effects of bile salts on the degradation of glycosphingolipids by glycosidases from bacteria of the human fecal flora. Biochim Biophys Acta 1084, 139–148. 10.1016/0005-2760(91)90212-Z [DOI] [PubMed] [Google Scholar]
- Fallani M., Young D., Scott J., Norin E., Amarri S., Adam R., Aguilera M., Khanna S., Gil A. & other authors (2010). Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 51, 77–84. 10.1097/MPG.0b013e3181d1b11e [DOI] [PubMed] [Google Scholar]
- Fewtrell M. S., Morgan J. B., Duggan C., Gunnlaugsson G., Hibberd P. L., Lucas A., Kleinman R. E. (2007). Optimal duration of exclusive breastfeeding: what is the evidence to support current recommendations? Am J Clin Nutr 85, 635S–638S. [DOI] [PubMed] [Google Scholar]
- Froehlich J. W., Dodds E. D., Barboza M., McJimpsey E. L., Seipert R. R., Francis J., An H. J., Freeman S., German J. B., Lebrilla C. B. (2010). Glycoprotein expression in human milk during lactation. J Agric Food Chem 58, 6440–6448. 10.1021/jf100112x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Oura F., Nagamine N., Katayama T., Hiratake J., Sakata K., Kumagai H., Yamamoto K. (2005). Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem 280, 37415–37422. 10.1074/jbc.M506874200 [DOI] [PubMed] [Google Scholar]
- Fukuda S., Toh H., Hase K., Oshima K., Nakanishi Y., Yoshimura K., Tobe T., Clarke J. M., Topping D. L. & other authors (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. 10.1038/nature09646 [DOI] [PubMed] [Google Scholar]
- Garrido D., Kim J. H., German J. B., Raybould H. E., Mills D. A. (2011). Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE 6, e17315. 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Barile D., Mills D. A. (2012a). A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr 3, 415S–421S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Nwosu C., Ruiz-Moyano S., Aldredge D., German J. B., Lebrilla C. B., Mills D. A. (2012b). Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics 11, 775–785. 10.1074/mcp.M112.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Ruiz-Moyano S., Mills D. A. (2012c). Release and utilization of N-acetyl-d-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe 18, 430–435. 10.1016/j.anaerobe.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Ruiz-Moyano S., Jimenez-Espinoza R., Eom H. J., Block D. E., Mills D. A. (2013). Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol 33, 262–270. 10.1016/j.fm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdalevich M., Mimouni D., David M., Mimouni M. (2001). Breast-feeding and the onset of atopic dermatitis in childhood: a systematic review and meta-analysis of prospective studies. J Am Acad Dermatol 45, 520–527. 10.1067/mjd.2001.114741 [DOI] [PubMed] [Google Scholar]
- Gibson G. R., Probert H. M., Loo J. V., Rastall R. A., Roberfroid M. B. (2004). Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17, 259–275. 10.1079/NRR200479 [DOI] [PubMed] [Google Scholar]
- Gnoth M. J., Kunz C., Kinne-Saffran E., Rudloff S. (2000). Human milk oligosaccharides are minimally digested in vitro. J Nutr 130, 3014–3020. [DOI] [PubMed] [Google Scholar]
- Gomes A. M., Malcata F. X., Klaver F. A. (1998). Growth enhancement of Bifidobacterium lactis Bo and Lactobacillus acidophilus Ki by milk hydrolyzates. J Dairy Sci 81, 2817–2825. 10.3168/jds.S0022-0302(98)75840-0 [DOI] [PubMed] [Google Scholar]
- González R., Klaassens E. S., Malinen E., de Vos W. M., Vaughan E. E. (2008). Differential transcriptional response of Bifidobacterium longum to human milk, formula milk, and galactooligosaccharide. Appl Environ Microbiol 74, 4686–4694. 10.1128/AEM.00122-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal P. K., Gill H. S. (2000). Oligosaccharides and glycoconjugates in bovine milk and colostrum. Br J Nutr 84 (Suppl. 1), S69–S74. 10.1017/S0007114500002270 [DOI] [PubMed] [Google Scholar]
- Gordon J. I., Dewey K. G., Mills D. A., Medzhitov R. M. (2012). The human gut microbiota and undernutrition. Sci Transl Med 4, 37ps12. [DOI] [PubMed] [Google Scholar]
- Grönlund M. M., Gueimonde M., Laitinen K., Kociubinski G., Grönroos T., Salminen S., Isolauri E. (2007). Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy 37, 1764–1772. 10.1111/j.1365-2222.2007.02849.x [DOI] [PubMed] [Google Scholar]
- Gustafsson B. E., Karlsson K. A., Larson G., Midtvedt T., Strömberg N., Teneberg S., Thurin J. (1986). Glycosphingolipid patterns of the gastrointestinal tract and feces of germ-free and conventional rats. J Biol Chem 261, 15294–15300. [PubMed] [Google Scholar]
- Haarman M., Knol J. (2005). Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Environ Microbiol 71, 2318–2324. 10.1128/AEM.71.5.2318-2324.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkarainen J., Toivanen M., Leinonen A., Frängsmyr L., Strömberg N., Lapinjoki S., Nassif X., Tikkanen-Kaukanen C. (2005). Human and bovine milk oligosaccharides inhibit Neisseria meningitidis pili attachment in vitro. J Nutr 135, 2445–2448. [DOI] [PubMed] [Google Scholar]
- Hamosh M. (2001). Bioactive factors in human milk. Pediatr Clin North Am 48, 69–86. 10.1016/S0031-3955(05)70286-8 [DOI] [PubMed] [Google Scholar]
- Harmsen H. J., Wildeboer-Veloo A. C., Raangs G. C., Wagendorp A. A., Klijn N., Bindels J. G., Welling G. W. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 30, 61–67. 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- Harris K., Kassis A., Major G., Chou C. J. (2012). Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J Obes 2012, 879151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håversen L., Ohlsson B. G., Hahn-Zoric M., Hanson L. A., Mattsby-Baltzer I. (2002). Lactoferrin down-regulates the LPS-induced cytokine production in monocytic cells via NF-κB. Cell Immunol 220, 83–95. 10.1016/S0008-8749(03)00006-6 [DOI] [PubMed] [Google Scholar]
- Hernell O. (2011). Human milk vs. cow’s milk and the evolution of infant formulas. Nestle Nutr Workshop Ser Pediatr Program 67, 17–28. 10.1159/000325572 [DOI] [PubMed] [Google Scholar]
- Hong P., Ninonuevo M. R., Lee B., Lebrilla C., Bode L. (2009). Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br J Nutr 101, 482–486. 10.1017/S0007114508025804 [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Littman D. R., Macpherson A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins L. C., Agustines M., McKee W. B., Boulding E. T., Kriaris M., Niedermeyer G. (1985). Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75, 944–953. 10.1172/JCI111795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A., Varrot A. (2008). Microbial recognition of human cell surface glycoconjugates. Curr Opin Struct Biol 18, 567–576. 10.1016/j.sbi.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Jakobsson I., Lindberg T., Benediktsson B. (1982). In vitro digestion of cow’s milk proteins by duodenal juice from infants with various gastrointestinal disorders. J Pediatr Gastroenterol Nutr 1, 183–192. 10.1097/00005176-198201020-00006 [DOI] [PubMed] [Google Scholar]
- Janer C., Pelaez C., Requena T. (2004). Caseinomacropeptide and whey protein concentrate enhance Bifidobacterium lactis growth in milk. Food Chem 86, 263–267. 10.1016/j.foodchem.2003.09.034 [DOI] [Google Scholar]
- Jensen R. G. (1999). Lipids in human milk. Lipids 34, 1243–1271. 10.1007/s11745-999-0477-2 [DOI] [PubMed] [Google Scholar]
- Jollès P., Jollès J. (1961). Lysozyme from human milk. Nature 192, 1187–1188. 10.1038/1921187b013892505 [DOI] [Google Scholar]
- Jost T., Lacroix C., Braegger C. P., Chassard C. (2012). New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 7, e44595. 10.1371/journal.pone.0044595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomäki M., Collado M. C., Salminen S., Isolauri E. (2008). Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87, 534–538. [DOI] [PubMed] [Google Scholar]
- Kitaoka M., Tian J., Nishimoto M. (2005). Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol 71, 3158–3162. 10.1128/AEM.71.6.3158-3162.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., Yamamoto K. (2011). An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology 21, 437–447. 10.1093/glycob/cwq175 [DOI] [PubMed] [Google Scholar]
- Kiyohara M., Nakatomi T., Kurihara S., Fushinobu S., Suzuki H., Tanaka T., Shoda S., Kitaoka M., Katayama T. & other authors (2012). α-N-Acetylgalactosaminidase from infant-associated bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem 287, 693–700. 10.1074/jbc.M111.277384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J. E., Spor A., Scalfone N., Fricker A. D., Stombaugh J., Knight R., Angenent L. T., Ley R. E. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 (Suppl. 1), 4578–4585. 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koletzko B. (2010). Innovations in infant milk feeding: from the past to the future. Nestle Nutr Workshop Ser Pediatr Program 66, 1–17. 10.1159/000318944 [DOI] [PubMed] [Google Scholar]
- Kunz C. (2012). Historical aspects of human milk oligosaccharides. Adv Nutr 3, 430S–439S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C., Rudloff S., Baier W., Klein N., Strobel S. (2000). Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20, 699–722. 10.1146/annurev.nutr.20.1.699 [DOI] [PubMed] [Google Scholar]
- Laegreid A., Kolstø Otnaess A. B., Bryn K. (1986). Purification of human milk gangliosides by silica gel chromatography and analysis of trifluoroacetate derivatives by gas chromatography. J Chromatogr A 377, 59–67. [PubMed] [Google Scholar]
- Larson G., Midtvedt T. (1989). Glycosphingolipids in feces of germ-free rats as a source for studies of developmental changes of intestinal epithelial cell surface carbohydrates. Glycoconj J 6, 285–292. 10.1007/BF01047848 [DOI] [PubMed] [Google Scholar]
- Larson G., Watsfeldt P., Falk P., Leffler H., Koprowski H. (1987). Fecal excretion of intestinal glycosphingolipids by newborns and young children. FEBS Lett 214, 41–44. 10.1016/0014-5793(87)80009-1 [DOI] [PubMed] [Google Scholar]
- Larson G., Falk P., Hoskins L. C. (1988). Degradation of human intestinal glycosphingolipids by extracellular glycosidases from mucin-degrading bacteria of the human fecal flora. J Biol Chem 263, 10790–10798. [PubMed] [Google Scholar]
- Le T. T., Van de Wiele T., Do T. N., Debyser G., Struijs K., Devreese B., Dewettinck K., Van Camp J. (2012). Stability of milk fat globule membrane proteins toward human enzymatic gastrointestinal digestion. J Dairy Sci 95, 2307–2318. 10.3168/jds.2011-4947 [DOI] [PubMed] [Google Scholar]
- Le Huërou-Luron I., Blat S., Boudry G. (2010). Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 23, 23–36. 10.1017/S0954422410000065 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Karamychev V. N., Kozyavkin S. A., Mills D., Pavlov A. R., Pavlova N. V., Polouchine N. N., Richardson P. M., Shakhova V. V. & other authors (2008). Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9, 247. 10.1186/1471-2164-9-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., An H. J., Lerno L. A., Jr, German J. B., Lebrilla C. B. (2011). Rapid profiling of bovine and human milk gangliosides by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance mass spectrometry. Int J Mass Spectrom 305, 138–150. 10.1016/j.ijms.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102, 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A., Brown J. E., Strömberg N., Westling-Ryd M., Schultz J. E., Karlsson K. A. (1987). Identification of the carbohydrate receptor for Shiga toxin produced by Shigella dysenteriae type 1. J Biol Chem 262, 1779–1785. [PubMed] [Google Scholar]
- Lindh E. (1975). Increased resistance of immunoglobulin A dimers to proteolytic degradation after binding of secretory component. J Immunol 114, 284–286. [PubMed] [Google Scholar]
- Lindquist S., Hernell O. (2010). Lipid digestion and absorption in early life: an update. Curr Opin Clin Nutr Metab Care 13, 314–320. 10.1097/MCO.0b013e328337bbf0 [DOI] [PubMed] [Google Scholar]
- LoCascio R. G., Ninonuevo M. R., Freeman S. L., Sela D. A., Grimm R., Lebrilla C. B., Mills D. A., German J. B. (2007). Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55, 8914–8919. 10.1021/jf0710480 [DOI] [PubMed] [Google Scholar]
- LoCascio R. G., Niñonuevo M. R., Kronewitter S. R., Freeman S. L., German J. B., Lebrilla C. B., Mills D. A. (2009). A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol 2, 333–342. 10.1111/j.1751-7915.2008.00072.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoCascio R. G., Desai P., Sela D. A., Weimer B., Mills D. A. (2010). Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol 76, 7373–7381. 10.1128/AEM.00675-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönnerdal B. (2009). Nutritional roles of lactoferrin. Curr Opin Clin Nutr Metab Care 12, 293–297. 10.1097/MCO.0b013e328328d13e [DOI] [PubMed] [Google Scholar]
- Lönnerdal B. (2010). Bioactive proteins in human milk: mechanisms of action. J Pediatr 156 (Suppl.), S26–S30. 10.1016/j.jpeds.2009.11.017 [DOI] [PubMed] [Google Scholar]
- Makino H., Kushiro A., Ishikawa E., Muylaert D., Kubota H., Sakai T., Oishi K., Martin R., Ben Amor K. & other authors (2011). Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol 77, 6788–6793. 10.1128/AEM.05346-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin I., Suau A., Magne F., Garrido D., Gotteland M., Neut C., Pochart P. (2006). Characterization of human intestinal bifidobacteria using competitive PCR and PCR-TTGE. FEMS Microbiol Ecol 55, 28–37. 10.1111/j.1574-6941.2005.00005.x [DOI] [PubMed] [Google Scholar]
- Marcobal A., Barboza M., Froehlich J. W., Block D. E., German J. B., Lebrilla C. B., Mills D. A. (2010). Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem 58, 5334–5340. 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A., Barboza M., Sonnenburg E. D., Pudlo N., Martens E. C., Desai P., Lebrilla C. B., Weimer B. C., Mills D. A. & other authors (2011). Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10, 507–514. 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens E. C., Roth R., Heuser J. E., Gordon J. I. (2009). Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem 284, 18445–18457. 10.1074/jbc.M109.008094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Sosa S., Martín M. J., Hueso P. (2002). The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J Nutr 132, 3067–3072. [DOI] [PubMed] [Google Scholar]
- Martirosian G., Kuipers S., Verbrugh H., van Belkum A., Meisel-Mikolajczyk F. (1995). PCR ribotyping and arbitrarily primed PCR for typing strains of Clostridium difficile from a Polish maternity hospital. J Clin Microbiol 33, 2016–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T., Watanabe K., Fujimoto J., Miyamoto Y., Takada T., Matsumoto K., Oyaizu H., Tanaka R. (2002). Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 68, 5445–5451. 10.1128/AEM.68.11.5445-5451.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer E. J., Hamman R. F., Gay E. C., Lezotte D. C., Savitz D. A., Klingensmith G. J. (1988). Reduced risk of IDDM among breast-fed children. The Colorado IDDM Registry. Diabetes 37, 1625–1632. 10.2337/diabetes.37.12.1625 [DOI] [PubMed] [Google Scholar]
- Midtvedt T., Carlstedt-Duke B., Höverstad T., Midtvedt A. C., Norin K. E., Saxerholt H. (1987). Establishment of a biochemically active intestinal ecosystem in ex-germfree rats. Appl Environ Microbiol 53, 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtvedt A. C., Carlstedt-Duke B., Norin K. E., Saxerholt H., Midtvedt T. (1988). Development of five metabolic activities associated with the intestinal microflora of healthy infants. J Pediatr Gastroenterol Nutr 7, 559–567. 10.1097/00005176-198807000-00014 [DOI] [PubMed] [Google Scholar]
- Miller-Podraza H., Lanne B., Angström J., Teneberg S., Milh M. A., Jovall P. A., Karlsson H., Karlsson K. A. (2005). Novel binding epitope for Helicobacter pylori found in neolacto carbohydrate chains: structure and cross-binding properties. J Biol Chem 280, 19695–19703. 10.1074/jbc.M412688200 [DOI] [PubMed] [Google Scholar]
- Mitoulas L. R., Kent J. C., Cox D. B., Owens R. A., Sherriff J. L., Hartmann P. E. (2002). Variation in fat, lactose and protein in human milk over 24 h and throughout the first year of lactation. Br J Nutr 88, 29–37. 10.1079/BJN2002579 [DOI] [PubMed] [Google Scholar]
- Mitsou E. K., Kirtzalidou E., Oikonomou I., Liosis G., Kyriacou A. (2008). Fecal microflora of Greek healthy neonates. Anaerobe 14, 94–101. 10.1016/j.anaerobe.2007.11.002 [DOI] [PubMed] [Google Scholar]
- Miwa M., Horimoto T., Kiyohara M., Katayama T., Kitaoka M., Ashida H., Yamamoto K. (2010). Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20, 1402–1409. 10.1093/glycob/cwq101 [DOI] [PubMed] [Google Scholar]
- Moro E. (1905). Morphologische und biologische Untersuchung über die Darmbakterien des Säuglings. Jahrb f Kinderh 61, 687–734. [Google Scholar]
- Morrow A. L., Ruiz-Palacios G. M., Altaye M., Jiang X., Guerrero M. L., Meinzen-Derr J. K., Farkas T., Chaturvedi P., Pickering L. K., Newburg D. S. (2004). Human milk oligosaccharide blood group epitopes and innate immune protection against Campylobacter and calicivirus diarrhea in breastfed infants. Adv Exp Med Biol 554, 443–446. [DOI] [PubMed] [Google Scholar]
- Morrow A. L., Ruiz-Palacios G. M., Jiang X., Newburg D. S. (2005). Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr 135, 1304–1307. [DOI] [PubMed] [Google Scholar]
- Mukai T., Kaneko S., Matsumoto M., Ohori H. (2004). Binding of Bifidobacterium bifidum and Lactobacillus reuteri to the carbohydrate moieties of intestinal glycolipids recognized by peanut agglutinin. Int J Food Microbiol 90, 357–362. 10.1016/S0168-1605(03)00317-9 [DOI] [PubMed] [Google Scholar]
- Neeser J. R., Granato D., Rouvet M., Servin A., Teneberg S., Karlsson K. A. (2000). Lactobacillus johnsonii La1 shares carbohydrate-binding specificities with several enteropathogenic bacteria. Glycobiology 10, 1193–1199. 10.1093/glycob/10.11.1193 [DOI] [PubMed] [Google Scholar]
- Neu J. (2007). Gastrointestinal development and meeting the nutritional needs of premature infants. Am J Clin Nutr 85, 629S–634S. [DOI] [PubMed] [Google Scholar]
- Newburg D. S. (2009). Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J Anim Sci 87 (Suppl.), 26–34. 10.2527/jas.2008-1347 [DOI] [PubMed] [Google Scholar]
- Newburg D. S., Chaturvedi P. (1992). Neutral glycolipids of human and bovine milk. Lipids 27, 923–927. 10.1007/BF02535874 [DOI] [PubMed] [Google Scholar]
- Newburg D. S., Ruiz-Palacios G. M., Altaye M., Chaturvedi P., Meinzen-Derr J., Guerrero M. L., Morrow A. L. (2004). Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology 14, 253–263. 10.1093/glycob/cwh020 [DOI] [PubMed] [Google Scholar]
- Newburg D. S., Ruiz-Palacios G. M., Morrow A. L. (2005). Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr 25, 37–58. 10.1146/annurev.nutr.25.050304.092553 [DOI] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- Niñonuevo M. R., Perkins P. D., Francis J., Lamotte L. M., LoCascio R. G., Freeman S. L., Mills D. A., German J. B., Grimm R., Lebrilla C. B. (2008). Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agric Food Chem 56, 618–626. 10.1021/jf071972u [DOI] [PubMed] [Google Scholar]
- Nishimoto M., Kitaoka M. (2007). Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol 73, 6444–6449. 10.1128/AEM.01425-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu C. C., Seipert R. R., Strum J. S., Hua S. S., An H. J., Zivkovic A. M., German B. J., Lebrilla C. B. (2011). Simultaneous and extensive site-specific N- and O-glycosylation analysis in protein mixtures. J Proteome Res 10, 2612–2624. 10.1021/pr2001429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu C. C., Aldredge D. L., Lee H., Lerno L. A., Zivkovic A. M., German J. B., Lebrilla C. B. (2012). Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res 11, 2912–2924. 10.1021/pr300008u [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell Motherway M., Fitzgerald G. F., van Sinderen D. (2011a). Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol 4, 403–416. 10.1111/j.1751-7915.2010.00218.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell Motherway M., Zomer A., Leahy S. C., Reunanen J., Bottacini F., Claesson M. J., O’Brien F., Flynn K., Casey P. G. & other authors (2011b). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108, 11217–11222. 10.1073/pnas.1105380108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otnaess A. B., Laegreid A., Ertresvåg K. (1983). Inhibition of enterotoxin from Escherichia coli and Vibrio cholerae by gangliosides from human milk. Infect Immun 40, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C. G., Martin R. M., Whincup P. H., Smith G. D., Cook D. G. (2006). Does breastfeeding influence risk of type 2 diabetes in later life? A quantitative analysis of published evidence. Am J Clin Nutr 84, 1043–1054. [DOI] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I., van den Brandt P. A., Stobberingh E. E. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521. 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- Peterson J. A., Patton S., Hamosh M. (1998). Glycoproteins of the human milk fat globule in the protection of the breast-fed infant against infections. Biol Neonate 74, 143–162. 10.1159/000014020 [DOI] [PubMed] [Google Scholar]
- Petherick A. (2010). Development: mother’s milk: a rich opportunity. Nature 468, S5–S7. 10.1038/468S5a [DOI] [PubMed] [Google Scholar]
- Petschow B. W., Talbott R. D. (1990). Growth promotion of Bifidobacterium species by whey and casein fractions from human and bovine milk. J Clin Microbiol 28, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt D. J., Forman M. R., Hanson R. L., Knowler W. C., Bennett P. H. (1997). Breastfeeding and incidence of non-insulin-dependent diabetes mellitus in Pima Indians. Lancet 350, 166–168. 10.1016/S0140-6736(96)12103-6 [DOI] [PubMed] [Google Scholar]
- Picciano M. F. (2001). Nutrient composition of human milk. Pediatr Clin North Am 48, 53–67. 10.1016/S0031-3955(05)70285-6 [DOI] [PubMed] [Google Scholar]
- Polonowski M., Lespagnol A. (1931). Sur deux nouveaux sucres du lait de femme, le gynolactose et l’allolactose. C R Acad Sci 192, 1319. [Google Scholar]
- Prentice A., MacCarthy A., Stirling D. M., Vasquez-Velasquez L., Ceesay S. M. (1989). Breast-milk IgA and lactoferrin survival in the gastrointestinal tract – a study in rural Gambian children. Acta Paediatr Scand 78, 505–512. 10.1111/j.1651-2227.1989.tb17928.x [DOI] [PubMed] [Google Scholar]
- Reinhardt C., Reigstad C. S., Bäckhed F. (2009). Intestinal microbiota during infancy and its implications for obesity. J Pediatr Gastroenterol Nutr 48, 249–256. 10.1097/MPG.0b013e318183187c [DOI] [PubMed] [Google Scholar]
- Roger L. C., McCartney A. L. (2010). Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiology 156, 3317–3328. 10.1099/mic.0.041913-0 [DOI] [PubMed] [Google Scholar]
- Ruas-Madiedo P., Gueimonde M., Fernández-García M., de los Reyes-Gavilán C. G., Margolles A. (2008). Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol 74, 1936–1940. 10.1128/AEM.02509-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P. M., Joao H. C., Coghill E., Fiten P., Saunders M. R., Opdenakker G., Dwek R. A. (1994). Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry 33, 17–22. 10.1021/bi00167a003 [DOI] [PubMed] [Google Scholar]
- Rudloff S., Kunz C. (2012). Milk oligosaccharides and metabolism in infants. Adv Nutr 3, 398S–405S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak L. R., Lebrilla C. B. (2012). Advances in analysis of human milk oligosaccharides. Adv Nutr 3, 406S–414S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M., Cervantes L. E., Ramos P., Chavez-Munguia B., Newburg D. S. (2003). Campylobacter jejuni binds intestinal H(O) antigen (Fucα1, 2Galβ1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 278, 14112–14120. 10.1074/jbc.M207744200 [DOI] [PubMed] [Google Scholar]
- Sakata S., Tonooka T., Ishizeki S., Takada M., Sakamoto M., Fukuyama M., Benno Y. (2005). Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol Lett 243, 417–423. 10.1016/j.femsle.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Salvini F., Riva E., Salvatici E., Boehm G., Jelinek J., Banderali G., Giovannini M. (2011). A specific prebiotic mixture added to starting infant formula has long-lasting bifidogenic effects. J Nutr 141, 1335–1339. 10.3945/jn.110.136747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G., Zwahlen M. C., Desiere F., Bork P. & other authors (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99, 14422–14427. 10.1073/pnas.212527599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens P. A., Oozeer R., Martin R., Amor K. B., Knol J. (2012). The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol 3, 425–447. 10.1146/annurev-food-022811-101120 [DOI] [PubMed] [Google Scholar]
- Sela D. A., Mills D. A. (2010). Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol 18, 298–307. 10.1016/j.tim.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R., Lapidus A., Rokhsar D. S., Lebrilla C. B. & other authors (2008). The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105, 18964–18969. 10.1073/pnas.0809584105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A., Price N. P., Mills D. A. (2010). Metabolism of bifidobacteria. In Bifidobacteria: Genomics and Molecular Aspects, pp. 45–70. Edited by Mayo B., van Sinderen D. Norwich, UK: Caister Academic Press. [Google Scholar]
- Sela D. A., Li Y., Lerno L., Wu S., Marcobal A. M., German J. B., Chen X., Lebrilla C. B., Mills D. A. (2011). An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 286, 11909–11918. 10.1074/jbc.M110.193359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela D. A., Garrido D., Lerno L., Wu S., Tan K., Eom H. J., Joachimiak A., Lebrilla C. B., Mills D. A. (2012). Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78, 795–803. 10.1128/AEM.06762-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders B. E., Thijs C., Dagnelie P. C., Stelma F. F., Mommers M., Kummeling I., Penders J., van Ree R., van den Brandt P. A. (2007). Breast-feeding duration and infant atopic manifestations, by maternal allergic status, in the first 2 years of life (KOALA study). J Pediatr 151, 347–351, e1–e2. 10.1016/j.jpeds.2007.03.022 [DOI] [PubMed] [Google Scholar]
- Stanley P., Schachter H., Taniguchi N. (2009). N-Glycans. In Essentials of Glycobiology, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [PubMed] [Google Scholar]
- Strömberg N., Ryd M., Lindberg A. A., Karlsson K.-A. (1988). Studies on the binding of bacteria to glycolipids. Two species of Propionibacterium apparently recognize separate epitopes on lactose of lactosylceramide. FEBS Lett 232, 193–198. 10.1016/0014-5793(88)80415-0 [DOI] [PubMed] [Google Scholar]
- Sundekilde U. K., Barile D., Meyrand M., Poulsen N. A., Larsen L. B., Lebrilla C. B., German J. B., Bertram H. C. (2012). Natural variability in bovine milk oligosaccharides from Danish Jersey and Holstein-Friesian breeds. J Agric Food Chem 60, 6188–6196. 10.1021/jf300015j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao N., DePeters E. J., Freeman S., German J. B., Grimm R., Lebrilla C. B. (2008). Bovine milk glycome. J Dairy Sci 91, 3768–3778. 10.3168/jds.2008-1305 [DOI] [PubMed] [Google Scholar]
- Tao N., Wu S., Kim J., An H. J., Hinde K., Power M. L., Gagneux P., German J. B., Lebrilla C. B. (2011). Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res 10, 1548–1557. 10.1021/pr1009367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taufik E., Fukuda K., Senda A., Saito T., Williams C., Tilden C., Eisert R., Oftedal O., Urashima T. (2012). Structural characterization of neutral and acidic oligosaccharides in the milks of strepsirrhine primates: greater galago, aye-aye, Coquerel’s sifaka and mongoose lemur. Glycoconj J 29, 119–134. 10.1007/s10719-012-9370-9 [DOI] [PubMed] [Google Scholar]
- Torres D. P. M., Gonçalves M. P. F., Teixeira J. A., Rodrigues L. R. (2010). Galacto-oligosaccharides: production, properties, applications, and significance as prebiotics. Comprehensive Reviews in Food Science and Food Safety 9, 438–454. 10.1111/j.1541-4337.2010.00119.x [DOI] [PubMed] [Google Scholar]
- Totten S. M., Zivkovic A. M., Wu S., Ngyuen U., Freeman S. L., Ruhaak L. R., Darboe M. K., German J. B., Prentice A. M., Lebrilla C. B. (2012). Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res 11, 6124–6133. [DOI] [PubMed] [Google Scholar]
- Turroni F., Bottacini F., Foroni E., Mulder I., Kim J. H., Zomer A., Sánchez B., Bidossi A., Ferrarini A. & other authors (2010). Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107, 19514–19519. 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Peano C., Pass D. A., Foroni E., Severgnini M., Claesson M. J., Kerr C., Hourihane J., Murray D. & other authors (2012). Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7, e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., Oftedal O. T. (2012). The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv Nutr 3, 473S–482S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel P. H., Geerts M. E., van Veen H. A., Kooiman P. M., Pieper F. R., de Boer H. A., Nuijens J. H. (1995). Glycosylated and unglycosylated human lactoferrins both bind iron and show identical affinities towards human lysozyme and bacterial lipopolysaccharide, but differ in their susceptibilities towards tryptic proteolysis. Biochem J 312, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Variyam E. P., Hoskins L. C. (1981). Mucin degradation in human colon ecosystems. Degradation of hog gastric mucin by fecal extracts and fecal cultures. Gastroenterology 81, 751–758. [PubMed] [Google Scholar]
- Wada J., Ando T., Kiyohara M., Ashida H., Kitaoka M., Yamaguchi M., Kumagai H., Katayama T., Yamamoto K. (2008). Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol 74, 3996–4004. 10.1128/AEM.00149-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R. E., Niñonuevo M., Mills D. A., Lebrilla C. B., German J. B. (2006). In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol 72, 4497–4499. 10.1128/AEM.02515-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. P., Rosendale D. I., Robertson A. M. (2000). Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett 190, 73–79. 10.1111/j.1574-6968.2000.tb09265.x [DOI] [PubMed] [Google Scholar]
- Wu S., Tao N., German J. B., Grimm R., Lebrilla C. B. (2010). Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res 9, 4138–4151. 10.1021/pr100362f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Grimm R., German J. B., Lebrilla C. B. (2011). Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res 10, 856–868. 10.1021/pr101006u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthou M., Bines J., Walker W. A. (1995). Human milk and intestinal host defense in newborns: an update. Adv Pediatr 42, 171–208. [PubMed] [Google Scholar]
- Xiao J. Z., Takahashi S., Nishimoto M., Odamaki T., Yaeshima T., Iwatsuki K., Kitaoka M. (2010). Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol 76, 54–59. 10.1128/AEM.01683-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Miwa T., Taniguchi H., Nagano T., Shimamura K., Tanaka T., Kumagai H. (1996). Binding specificity of Lactobacillus to glycolipids. Biochem Biophys Res Commun 228, 148–152. 10.1006/bbrc.1996.1630 [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N. & other authors (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye A., Cui J., Singh H. (2011). Proteolysis of milk fat globule membrane proteins during in vitro gastric digestion of milk. J Dairy Sci 94, 2762–2770. 10.3168/jds.2010-4099 [DOI] [PubMed] [Google Scholar]
- Yoshida E., Sakurama H., Kiyohara M., Nakajima M., Kitaoka M., Ashida H., Hirose J., Katayama T., Yamamoto K., Kumagai H. (2012). Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22, 361–368. 10.1093/glycob/cwr116 [DOI] [PubMed] [Google Scholar]
- Yu T., Guo C., Wang J., Hao P., Sui S., Chen X., Zhang R., Wang P., Yu G. & other authors (2011). Comprehensive characterization of the site-specific N-glycosylation of wild-type and recombinant human lactoferrin expressed in the milk of transgenic cloned cattle. Glycobiology 21, 206–224. 10.1093/glycob/cwq151 [DOI] [PubMed] [Google Scholar]
- Zivkovic A. M., Barile D. (2011). Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr 2, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic A. M., German J. B., Lebrilla C. B., Mills D. A. (2011). Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A 108 (Suppl. 1), 4653–4658. 10.1073/pnas.1000083107 [DOI] [PMC free article] [PubMed] [Google Scholar]