Abstract
Seeded conversion of tau monomers into fibrils is a central step in the progression of tau pathology in Alzheimer’s disease and other neurodegenerative disorders. Self-assembly is mediated by the microtubule binding repeats in tau of which either three or four are present, depending on the protein isoform. Here we used double electron-electron resonance spectroscopy to investigate the conformational ensemble of four-repeat tau fibrils. We observe that single point mutations at key positions in the protein (ΔK280, P301S, P312I, D314I) markedly change the distribution of fibril conformers after template-assisted growth, whereas other mutations in the protein (I308M, S320F, G323I, G326I, Q336R) do not. These findings provide unprecedented insights into the seed selection of tau disease mutants and establish conformational compatibility as an important driving force in tau fibril propagation.
Keywords: tau protein, fibril, amyloid, seed selection, conformational ensemble, templating, Alzheimer’s disease
Fibrils resulting from the aggregation of microtubule-associated protein tau are the pathological hallmark of numerous neurodegenerative disorders including Alzheimer’s disease and frontotemporal dementia.[1, 2] The propagation of tau fibrils is characterized by template-assisted conversion of monomers[3–5] and cell-to-cell transfer of aggregates[6] leading to prion-like spreading of protein aggregates in the brain.[7] Six different tau isoforms are expressed in the central nervous system, which can be grouped into three-repeat (3R) tau and four-repeat (4R) tau, based on the number of microtubule binding repeats in the amyloidogenic core region.[1] An asymmetric barrier allows 4R tau to grow onto 3R tau seeds, but not vice versa.[8] Conformational differences between tau fibrils may play an important role in determining phenotypic diversity in human tauopathies.[9] In vitro experiments have demonstrated that fibrils of 4R tau are structurally heterogeneous, composed of at least three distinct conformers.[10] Structural heterogeneity is a common feature among amyloid fibrils and a potential basis for conformation-induced strain switching in prion propagation.[11, 12] The investigation of heterogeneous amyloid ensembles, however, is challenging as multiple conformers have to be monitored at the same time.
In order to overcome this problem we have employed double electron-electron resonance (DEER) spectroscopy,[13] a technique that has been used to measure the distances between unpaired electrons of spin labels in proteins[14] and has emerged as a powerful new tool for elucidating fibril structure.[15] Importantly, DEER is capable of describing the relative populations of fibrils in a heterogeneous mixture.[10] Here we report that single point mutations in key positions of 4R tau affect seed selection and thereby alter the populations of fibril conformers. These findings establish conformational compatibility as an important determinant in fibril propagation.
To explore the seeding properties of tau, we used truncated versions of 4R tau (K18) and 3R tau (K19), which contain the repeat region that forms the structural core but not the disordered fuzzy coat.[16] These constructs show properties similar to their full-length counterparts with respect to cross-β structure,[17] strand-registry,[18] seeding,[5] uptake,[19] and transmission,[20] yet have the advantage of greatly accelerated aggregation kinetics.[21] We selected a representative K18 double cysteine mutant, 311/328, with a heterogeneous conformation distribution[10] to examine differences in seed selection initiated by mutated monomers. The following mutations were incorporated into the K18 311/328 core: known disease-related mutants of ΔK280,[22] P301S,[23] S320F,[24] Q336R,[25] and additional mutants of I308M, P312I, D314I, G323I, G326I. The latter mutants are not found in humans, but were chosen based on their potential effects on seed selection because of the position in the protein and the nature of the amino acid change. All mutated monomers grew onto the templates provided by K18 wild-type (WT) seeds. Interactions between spin labels of stacked tau molecules were avoided by diluting samples with a 50-fold molar excess of K18 WT. Resultant DEER data were analyzed analogously for all samples to detect whether the mutants grew preferentially onto specific fibril conformations present in the mixture. Variations in seed selection were determined through comparison to the distance distribution of non-mutated K18 311/328.
Before analysis of mutation effects, it is important to demonstrate that the K18 311/328 system is robust for use as an indicator of conformational variation. The sample was prepared and analyzed in triplicate and the distribution shown to be reproducible (Figure 1a). When prepared with different seed batches, we consistently observe three distances for this system at 3.2, 3.8, and 4.8 nm. The relative populations of fibrils adopting these conformations are also well-conserved (for data analysis, see Experimental Procedures and Figures S1 and S2 in the Supporting Information). We therefore concluded that K18 311/328 is an appropriate starting point to examine variation in conformation selection of mutated monomer grown onto K18 WT seeds.
Figure 1.
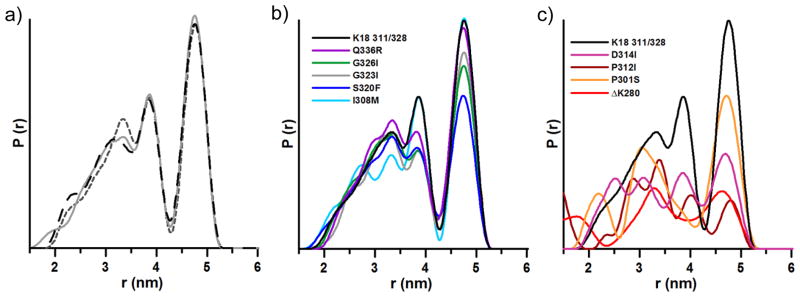
Distance distributions of K18 311/328: a) reproducibility, b) mutants with distance distributions similar to non-mutated K18 311/328, c) mutants with very different distributions compared to non-mutated K18 311/328.
We next examined the distance distributions for the mutant-bearing fibrils, and found that several of the mutations gave distributions similar to that of non-mutated K18 311/328 (Figure 1b). Conversely, some of the mutations at significant positions in the protein (ΔK280, P301S, P312I, and D314I) clearly changed the distribution of fibril conformers following template-assisted growth (Figure 1c). The spin-labeled proteins did not affect overall aggregation kinetics (Figure S3). Neither did the addition of ΔK280 to K18 311/328 alter the underlying distance distribution (Figure S4). This suggests that individual mutants do not confer their growth properties onto K18 WT or K18 311/328, but instead act independently.
It is noteworthy that the labeling efficiencies for all tau mutants were similar (Figure S5) excluding the possibility that incomplete labeling of tau monomers resulted in the different peak patterns observed in Figure 1c. To ensure that the dramatic changes in seed selection were not a result of amorphous monomer aggregation, fibril formation was confirmed by electron microscopy (Figure S6). Furthermore, continuous wave electron paramagnetic resonance measurements verified that the mutant monomers became incorporated into the fibrils (Figure S7). The spectra revealed no major broadening indicating the absence of large subpopulations with spin labels < 2 nm apart. Spin labels separated > 5 nm apart are out of the detectable range for the DEER measurements presented here.[10]
We propose that key mutations play a role in seed selection during fibril growth, whereas mutations with similar distributions have little or no conformational effect. In addition to variation in distance distributions, examination of the DEER data supports these conclusions. When grouped according to degree of similarity to non-mutated K18 311/328, the traces and fit functions for the background-subtracted data align with the grouping of similar and dissimilar distance distributions (Figures S8–S10). The initial drop in signal of the raw data before background subtraction provides a qualitative measure of the interaction between the spin labels; in this area of the data, similar mutants trace together and different mutants trace separately, with the ΔK280 mutant being the furthest outlier (Figure 2).
Figure 2.
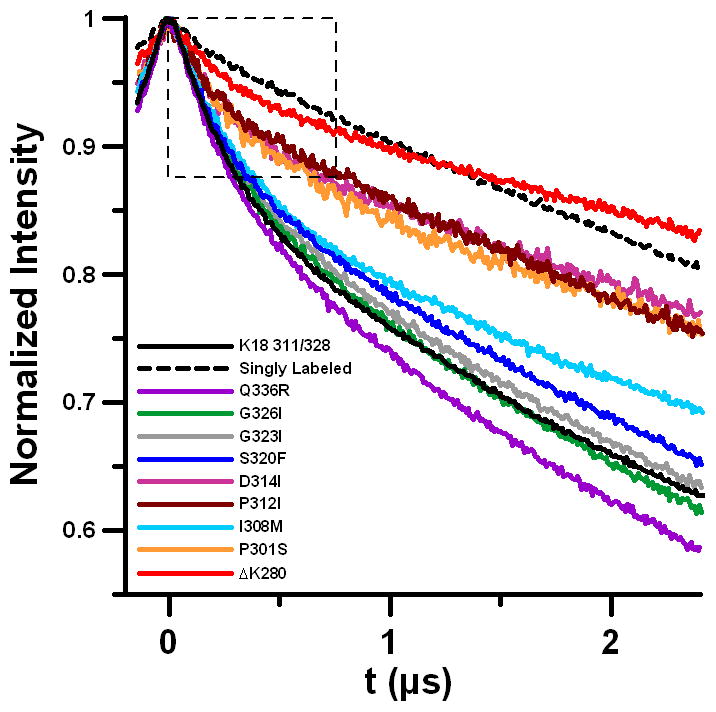
Overlay of raw DEER data for all mutants studied, including the K18 311/328 core and singly labeled monomer (all data are normalized to the maximum signal intensity = 1). The largest difference is apparent from the initial drop region as indicated by the dotted box.
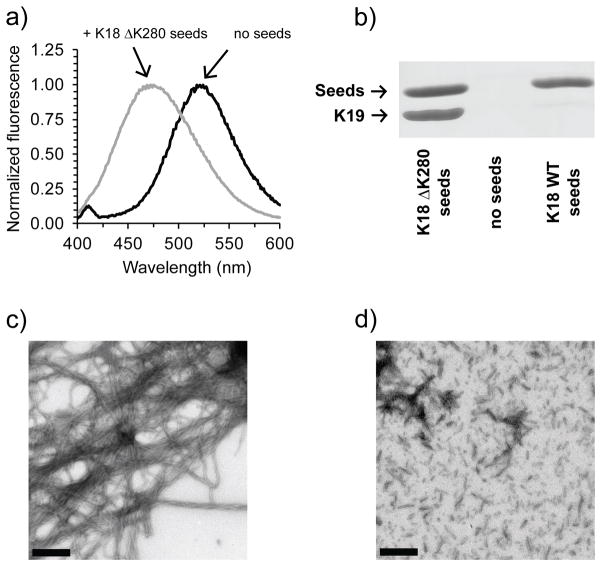
A shallow initial drop in the raw data corresponds to weak spin-spin interactions. This suggests that the mutations with very different population distributions may contain conformers with interspin distances larger than 5 nm. This quality was previously shown for K19 fibrils[10] and now for ΔK280. Note that the ΔK280 mutation resides in the second microtubule binding repeat and hence only occurs in 4R tau. Adoption of extended-monomer conformations may account for the asymmetric seeding barrier demonstrated for tau,[8] for which K18 can grow onto K19 fibrils, but K19 cannot grow onto K18. Since K18 ΔK280 may also adopt a more extended conformation, it was logical to test whether K19 could be grown onto pure K18 ΔK280 seeds. To determine whether K18 ΔK280 seeds can recruit K19 we utilized an intrinsic acrylodan assay[8] to assess K19 aggregation. Acrylodan labeled K19 tau without the introduction of K18 ΔK280 seeds produced a fluorescence spectrum with a λmax of around 522 nm, typical of monomeric tau. When K18 ΔK280 seeds were added to K19 and incubated for 2 hours, the λmax of labeled K19 tau had significantly blue-shifted, indicating the growth of K19 on K18 ΔK280 seeds (Figure 3a). For kinetic traces and comparison with K18- and K19-seeded reactions see Figure S11. In order to independently confirm K19 growth on K18 ΔK280 seeds, we carried out sedimentation experiments of seeded reactions. In contrast with K18 WT seeds, K18 ΔK280 seeds were shown to effectively seed K19 monomer (Figure 3b, see Figure S12 for triplicate quantification). Importantly, the ability of K18 ΔK280 to seed K19 is not due to K18 ΔK280 fibrils aggregating more efficiently, as the sedimented seeds of K18 WT and K18 ΔK280 seeds were found to be of comparable density. Growth of K19 on K18 ΔK280 seeds is furthermore supported by EM images taken after a K18 ΔK280 seeded reaction, revealing an abundance of long fibrils that can be clearly distinguished from the short and fragmented K18 ΔK280 seeds (Figure 3c and d). The new seeding competency of K18 ΔK280 is in line with this protein having a different conformation from K18 WT[3] and emphasizes how a single point mutation can effectively change the functional properties of a fibril.
Figure 3.
K19 grows on K18 ΔK280 seeds. a) Fibril formation is monitored via acrylodan fluorescence and is accompanied by a blue shift from 522 to 478 nm. b) Sedimentation confirms K19 growth onto K18 ΔK280 but not K18 WT (Quantitation in Figure S4). c) EM images of K19 WT (10 μM) grown onto 10% K18 ΔK280 seeds after 3 hours of incubation at 37 °C, d) and of ΔK280 seeds without K19 WT addition. Bar = 400 nm.
In conclusion, DEER spectroscopy has provided a unique tool to study variations in protein conformation. We utilized this tool to investigate the effects of single point mutations on seed selection. We observe that some mutants of tau, when grown onto WT seeds, retain the composition of the original ensemble, while other mutants significantly alter the populations of conformers (schematically summarized in Figure 4). These effects can be explained by the conformational compatibilities of individual mutants. Mutants that do not interfere with any of the original conformers will be recruited akin to the WT monomer. Mutants that are incompatible with some of the original conformers will change the overall composition of the ensemble. While these selection processes could undoubtedly be modulated by additional factors in the complex cellular environment of the human brain, our findings provide an important general model of how new fibril ensembles might emerge. It is plausible that the conformational composition of tau fibrils could vary for different forms of inherited frontotemporal dementia, where dominant mutations in the tau gene (MAPT) are linked to disease. Also important in the context of the herein presented study, at least in some cases somatic mutations in tau could define the initial ensemble of conformers in sporadic forms of tauopathies and hence influence the selective processes as fibrils spread throughout the brain. Specifically, if different fibril ensembles composed of mutant tau transferred to connected neurons (free of tau mutations) the recruitment of WT tau would vary in a conformation dependent manner. The presented model shows striking similarities to current models of strain mutation and selection in prions.[11] Remarkably, a single amino acid difference at position 226 in the prion proteins of elk and deer, combined with conformational selection of compatible seeds, determines strain mutation in chronic wasting disease.[26] Although transmissibility of tau is confined to neurons within a single human brain, the overall conformational selection processes appear to be governed by the same underlying structural principles. It is likely that other pathological amyloid fibrils share similar selection properties. The recent finding that structural variations in fibrils of the β-amyloid peptide may contribute to variations in Alzheimer’s disease[27] underscores the biological relevance of fibril conformation. Changes in fibril conformation through seed selection could be an important molecular mechanism of diversifying disease phenotype. The existence of different fibril ensembles and their emergence upon spreading must be taken into account when designing new therapeutic strategies for interfering with amyloid diseases.
Figure 4.
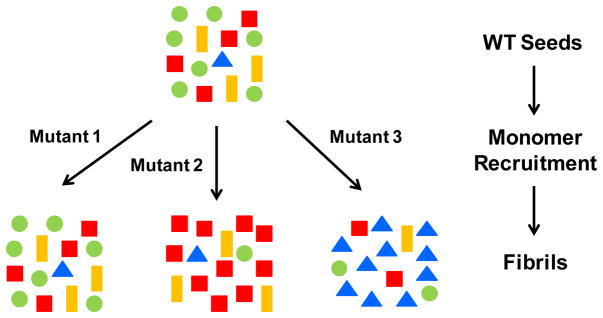
General model for sequence dependent seed selection in tau fibril growth. Tau mutants that are fully compatible with the conformational ensemble of WT tau seeds retain the original composition of conformers (Mutant 1). Mutants that are not compatible cause a switch in the dominant species (Mutant 2) and in some cases may amplify a minor subspecies (Mutant 3). The reverse process in which monomers of WT tau grow onto mutant seeds will follow similar selection rules based on conformational compatibility. Symbols of different color and shape represent different conformers of tau fibrils.
Supplementary Material
Acknowledgments
This project was supported by National Institute of Neurological Disorders and Stroke Grant R01NS076619 (to M. M.). This work was also supported by National Cancer Institute Contract HHSN261200800001E, the intramural research program of the National Cancer Institute Center for Cancer Research, National Science Foundation CAREER Award CBET-0952624 and CBET-1158447 (to J. Z.), National Natural Science Foundation of China Grant 11074047, 11274075, and 91227102 (to G. W.). The authors thank Dr. Eric Hustedt for his assistance using GLADD, and helpful discussions regarding analysis of DEER data.
References
- 1.Spillantini MG, Goedert M. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee VM, Goedert M, Trojanowski JQ. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 3.Frost B, Ollesch J, Wille H, Diamond MI. J Biol Chem. 2009;284:3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nonaka T, Watanabe ST, Iwatsubo T, Hasegawa M. J Biol Chem. 2010;285:34885–34898. doi: 10.1074/jbc.M110.148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo JL, Lee VM. J Biol Chem. 2011;286:15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. J Biol Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Frost B, Jacks RL, Diamond MI. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) de Calignon A, Polydoro M, Suarez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, Spires-Jones TL, Hyman BT. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K. PLoS One. 2012;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Braak H, Del Tredici K. Acta Neuropathol. 2011;121:589–595. doi: 10.1007/s00401-011-0825-z. [DOI] [PubMed] [Google Scholar]; b) Goedert M, Clavaguera F, Tolnay M. Trends Neurosci. 2010;33:317–325. doi: 10.1016/j.tins.2010.04.003. [DOI] [PubMed] [Google Scholar]; c) Aguzzi A, Rajendran L. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]; d) Brundin P, Melki R, Kopito R. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Frost B, Diamond MI. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Jucker M, Walker LC. Nature. 2013;501:45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinkel PD, Siddiqua A, Huynh H, Shah M, Margittai M. Biochemistry. 2011;50:4330–4336. doi: 10.1021/bi2004685. [DOI] [PubMed] [Google Scholar]
- 9.Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, Ghetti B, Goedert M, Tolnay M. Proc Natl Acad Sci USA. 2013;110:9535–9540. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siddiqua A, Luo Y, Meyer V, Swanson MA, Yu X, Wei G, Zheng J, Eaton GR, Ma B, Nussinov R, Eaton SS, Margittai M. J Am Chem Soc. 2012;134:10271–10278. doi: 10.1021/ja303498q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinge J, Clarke AR. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 12.a) Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bateman DA, Wickner RB. PLoS Genet. 2013;9:e1003257. doi: 10.1371/journal.pgen.1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Milov AD, Salikhov KM, Shchirov MD. Sov Phys Solid State. 1981;23:565–569. [Google Scholar]; b) Jeschke G, Pannier M, Spiess HW. Biol Magn Reson. 2000;19:493–512. [Google Scholar]
- 14.Jeschke G. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 15.a) Karyagina I, Becker S, Giller K, Riedel D, Jovin TM, Griesinger C, Bennati M. Biophys J. 2011;101:L1-3. doi: 10.1016/j.bpj.2011.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bedrood S, Li Y, Isas JM, Hegde BG, Baxa U, Haworth IS, Langen R. J Biol Chem. 2012;287:5235–5241. doi: 10.1074/jbc.M111.327817. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pornsuwan S, Giller K, Riedel D, Becker S, Griesinger C, Bennati M. Angew Chem. 2013;125:10480–10484. doi: 10.1002/anie.201304747. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2013;52:10290–10294. doi: 10.1002/anie.201304747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Wischik CM, Novak M, Thogersen HC, Edwards PC, Runswick MJ, Jakes R, Walker JE, Milstein C, Roth M, Klug A. Proc Natl Acad Sci USA. 1988;85:4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wegmann S, Medalsy ID, Mandelkow E, Muller DJ. Proc Natl Acad Sci USA. 2013;110:E313–321. doi: 10.1073/pnas.1212100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) von Bergen M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow EM, Mandelkow E. J Biol Chem. 2001;276:48165–48174. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]; b) Berriman J, Serpell LC, Oberg KA, Fink AL, Goedert M, Crowther RA. Proc Natl Acad Sci USA. 2003;100:9034–9038. doi: 10.1073/pnas.1530287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Margittai M, Langen R. Proc Natl Acad Sci USA. 2004;101:10278–10283. doi: 10.1073/pnas.0401911101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Siddiqua A, Margittai M. J Biol Chem. 2010;285:37920–37926. doi: 10.1074/jbc.M110.185728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes BB, Devos SL, Kfoury N, Li M, Jacks R, Yanamandra K, Ouidja MO, Brodsky FM, Marasa J, Bagchi DP, Kotzbauer PT, Miller TM, Papy-Garcia D, Diamond MI. Proc Natl Acad Sci USA. 2013;110:E3138–3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iba M, Guo JL, McBride JD, Zhang B, Trojanowski JQ, Lee VM. J Neurosci. 2013;33:1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barghorn S, Mandelkow E. Biochemistry. 2002;41:14885–14896. doi: 10.1021/bi026469j. [DOI] [PubMed] [Google Scholar]
- 22.Rizzu P, Van Swieten JC, Joosse M, Hasegawa M, Stevens M, Tibben A, Niermeijer MF, Hillebrand M, Ravid R, Oostra BA, Goedert M, van Duijn CM, Heutink P. Am J Hum Genet. 1999;64:414–421. doi: 10.1086/302256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugiani O, Murrell JR, Giaccone G, Hasegawa M, Ghigo G, Tabaton M, Morbin M, Primavera A, Carella F, Solaro C, Grisoli M, Savoiardo M, Spillantini MG, Tagliavini F, Goedert M, Ghetti B. J Neuropathol Exp Neurol. 1999;58:667–677. doi: 10.1097/00005072-199906000-00011. [DOI] [PubMed] [Google Scholar]
- 24.Rosso SM, van Herpen E, Deelen W, Kamphorst W, Severijnen LA, Willemsen R, Ravid R, Niermeijer MF, Dooijes D, Smith MJ, Goedert M, Heutink P, van Swieten JC. Ann Neurol. 2002;51:373–376. doi: 10.1002/ana.10140. [DOI] [PubMed] [Google Scholar]
- 25.Pickering-Brown SM, Baker M, Nonaka T, Ikeda K, Sharma S, Mackenzie J, Simpson SA, Moore JW, Snowden JS, de Silva R, Revesz T, Hasegawa M, Hutton M, Mann DM. Brain. 2004;127:1415–1426. doi: 10.1093/brain/awh147. [DOI] [PubMed] [Google Scholar]
- 26.Angers RC, Kang HE, Napier D, Browning S, Seward T, Mathiason C, Balachandran A, McKenzie D, Castilla J, Soto C, Jewell J, Graham C, Hoover EA, Telling GC. Science. 2010;328:1154–1158. doi: 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.