Summary
Recent findings indicate that the role of Th17 cells in the pathogenesis of tissue inflammation and autoimmunity has become rather complicated. While interleukin-17 (IL-17) producing CD4+ T cells are found frequently within the peripheral target tissue during the course of autoimmune disease, these cells may contribute to or protect from inflammation. Accumulating reports have revealed the existence of both pathogenic and non-pathogenic Th17 cells. These Th17 subsets produce the signature cytokines IL-17A and IL-17F yet have distinct and divergent roles in inducing autoimmune tissue inflammation. Comparative genomic sequence analyses between the pathogenic and non-pathogenic Th17 cells have exposed unexpected and extensive population heterogeneity within the Th17 subset. Here we review some of the unexpected factors that may drive pathogenic divergence. Understanding the functional consequences of Th17 cell diversity may allow for the selection of more precise targets for intervention in autoimmune and inflammatory diseases.
Keywords: IL-17, inflammation, autoimmunity, pathogenicity, single cell RNA sequencing, CD4+ T helper cells, Th17 cells, IL-23R
Heterogeneity of Th17 cells
CD4+ T helper cells have evolved to provide host defense against invading pathogens with exquisite specificity (1, 2). Functionally distinct CD4+ T cells can be derived when naïve T cells are activated in the presence of cytokines produced by the innate immune system (3). Initially, all effector CD4+ T cells were thought to be a uniform population that mobilized to elicit an immune response. Then in 1986, Mossman and Coffman published a seminal article that conveyed the heterogeneity of the effector CD4+ T cells and characterized these T cells into distinct subsets, each defined by a unique master transcription factor and signature cytokines (4, 5). The first two subsets were called T helper 1 (Th1) and T helper 2 (Th2). Classically Th1 cells are characterized by the production of the signature cytokine interferon γ(IFNγ) and are thought to regulate immunity to intracellular pathogens while Th2 cells are characterized by the production of interleukin (IL)-4, IL-5, and IL-13 and are thought to regulate immunity to extracellular pathogens like parasites (6, 7). More recently, another T helper subset, called Th17 cells, defined by a distinct transcription factor, RORγt, and by the secretion of IL-17 was discovered (8). While these cells have been shown to be important for host defense against opportunistic pathogenic fungal infections, such as candida albicans, numerous studies have implicated this subset in pathological autoimmune inflammation and tissue destruction (9–12). As such, there has been an intense interest in characterizing the function of Th17 cells to develop potential strategies to inhibit their deleterious effects.
Naive T cells differentiate into Th17 cells when activated in the presence of TGF-β and IL-6 (13). Upon induction of the master transcription factor, RORγt, the cells produce the Th17 signature cytokines IL-17A/F as well as IL-21 (8). The production of IL-21 is significant in that it acts as a feed forward amplification factor to induce expansion of differentiating Th17 cells and it also elicits the expression of IL-23R on the Th17 cells (14, 15). It is upon exposure to IL-23 that Th17 cells have been shown to stabilize their phenotype and also to produce another pro-inflammatory cytokine, IL-22 (16). This unique capacity to express IL-23R by the Th17 cells has been shown to be one of the critical components that promote expansion and pathogenicity of these cells. Multiple studies in mice and humans have revealed a central role for IL-23 in the development of autoimmune disease (17–22). In fact, there is a strong genetic linkage to the IL-23/IL-23R loci in the development of Th17-associated diseases, which include rheumatoid arthritis, psoriasis, insulin dependent diabetes mellitus, inflammatory bowel disease, and multiple sclerosis (MS) (23–26). However, there is growing evidence that not all Th17 cells are pathogenic and that in some autoimmune inflammatory conditions, IL-17 producing Th17 cells may, in fact, be protective (27, 28).
Perhaps the strongest evidence for the protective role of Th17 cells has been in studies in inflammatory bowel diseases (IBD) (27, 29). In mice, T cell mediated colitis is severely exacerbated when CD4+ T cells lack the capacity to produce IL-17 or the mice can not respond to IL-17 due to IL-17R deficiency (28). Additional studies in mice suggest that IL-22, another cytokine produced by Th17 cells, plays a role in maintaining tissue integrity at mucosal sites such as the intestine(30–32). Th17 cells may also play a protective role in humans, as clinical trials of neutralizing antibodies targeting IL-17 in patients with Crohn’s disease resulted in disease exacerbation (33). These data suggest that Th17 cells may protect intestinal sites during inflammatory responses instead of propagating the disease.
In sharp contrast, in other Th17-associated autoimmune diseases, such as psoriasis, multiple sclerosis, and ankylosing spondylitis, IL-17 neutralization has been shown to ameliorate disease in both mouse models and humans (34–38). This divergence in the therapeutic benefit of IL-17 neutralization is highly suggestive of the duality within the Th17 cells. In fact, any distinguishing factors that exist between the pathogenic and non-pathogenic Th17 cells are difficult to elucidate as both subsets produce the signature cytokines that define Th17 cells, namely IL-17A, IL-17F, and IL-21, and express RORγt, the master Th17 transcription factor (20, 22, 39). This suggests that additional aspects of Th17 biology may be important in determining disease outcome, and that a comparative evaluation of genes expressed by Th17 cells beyond IL-17 may reveal the necessary components that drive pathogenicity.
Uncovering key regulatory genes
While the key cytokines required for Th17 generation in vitro have been uncovered, much remains unknown regarding the extensive circuitry of genes that regulate the differentiation of Th17 cells (40). In our recent study, we combined temporal transcriptional profiling at high density and transcriptional perturbations via a novel nanowire-based short interfering RNA (siRNA) delivery method, and were able to construct a dynamic transcriptional network of Th17 differentiation that included 1291 genes that were differentially, yet specifically, expressed during Th17 differentiation(41). Within the differentially expressed genes, approximately 71 regulatory genes were discovered and validated by “knockdown” with siRNA. These novel regulators reveal that the Th17 differentiation program is a tightly regulated, self-reinforcing module, which is exquisitely balanced to sustain Th17 programing while suppressing other T helper subset lineages (41). In fact, this dynamic process of Th17 differentiation revealed a bifurcation of the network circuit that can be characterized by two antagonistic modules: a module of 22 Th17 positive factors and 5 negative factors. A perturbation in any one of these regulatory modules has the capacity to skew the delicate equilibrium (41).
By clustering the genes based on distinct temporal profiles, we were able to discern that Th17 differentiation undergoes three discrete transcriptional phases, which can be summed up as: induction, amplification, and stabilization by IL-23 signaling (41). However, what was perhaps most interesting from the transcriptional profiling was that as a developing Th17 cells undergoes a transition from the intermediate phase, marked by the induction of RORγt, to the late phase of development, these cells initiate expression of regulatory cytokines from the IL-10 family (such as IL-10 and IL-24) (41). The cells subsequently enter the late phase of development marked by the expression of Il23r, a gene that not only has been shown to stabilize the Th17 phenotype, but also has a strong genetic linkage to inflammation and autoimmune pathologies (24). This may explain the emergence of a natural “switch” of Th17 cells from non-pathogenic to pathogenic Th17 cells as described by our group and others. In fact, it has been shown that two different types of human IL-17 producing cells may have evolved to co-exist. One subtype produce both IL-17/IL-10 and are better at clearing bacterial infections such as Staphylococcus aureus, and another produce IL-17/IFN-γbut not IL-10 and are specific for fungi such as Candida albicans (22, 42). The existence of these subtypes within the Th17 subset may rationalize the functional divergence of IL-17 producing cells in autoimmune diseases. The “non-pathogenic” Th17 population that produces IL-10 and IL-17 may play an important role in places such as the intestine to maintain gut homeostasis, limit invasion of the gut microbiome and promote epithelial barrier function by production of IL-17 and IL-22. On the other hand “pathogenic” Th17 cells may be involved in inducing inflammatory responses to limit fungal infections by the production of IL-17 and IFN-γ, but also have the potential to induce devastating tissue inflammation in multiple Th17-associated autoimmune diseases. This heterogeneity in the Th17 population reveals the need for more precise genomic expression profiling, perhaps at a single cell level to measure the functional diversity within the Th17 lineage (20, 22, 28, 29, 39).
Single cell transcriptomics to uncover bimodality
Pathogenic and non-pathogenic Th17 cells share the expression of many genes at the population level, including expression of signature genes and molecules associated with Th17 cells, such as IL-17A/F, RORγt, as well as IL-23R, although non-pathogenic Th17 cells express IL-23R at a lower level as compared to pathogenic Th17 cells (22). In fact, only 233 genes are differentially expressed between these two populations of Th17 cells when analyzed with whole genome microarray (22). However, this list is comprised only from in vitro differentiated Th17 cells and it is possible that in vivo generated Th17 cells may possess a very distinctive heterogeneous signature based on pathogenicity and tissue environments. A comparison of the gene profiles of in vitro and in vivo Th17 cells could allow generation of a comprehensive transcriptional network to delineate mechanisms that regulate pathogenic and non-pathogenic Th17 responses.
Single cell RNA sequencing has recently been recognized as a valuable tool that allows for profiling of heterogeneous population of cells. It can be used to perform accurate quantitative and qualitative transcriptomic measurements at the individual cell level and accurately profile interesting subpopulations from a larger heterogeneous population (43–47). By utilizing this high throughput sequencing technology, it may be possible to reveal critical regulatory genes that may be essential in driving the bifurcation of the Th17 subset while providing in depth understanding of the complex regulatory network that drive pathogenicity. Facilitating the use of this single cell RNA sequencing technology is the recent discovery by our group and others that it is possible to generate both pathogenic and non-pathogenic Th17 cells in vitro (20, 22, 39). By comparing in vitro IL-17+ cell single cell sequencing data from in vivo IL-17+ cells sorted at the height of autoimmune inflammation from the target tissue, it may be possible to uncover the key regulatory genes that control pathogenic versus non-pathogenic phenotypes. Utilizing the single cell RNA sequencing will allow us to characterize expression variability on a genomic scale that was could not be distinguished at a population-level expression profiles alone.
Unexpected targets of pathogenicity
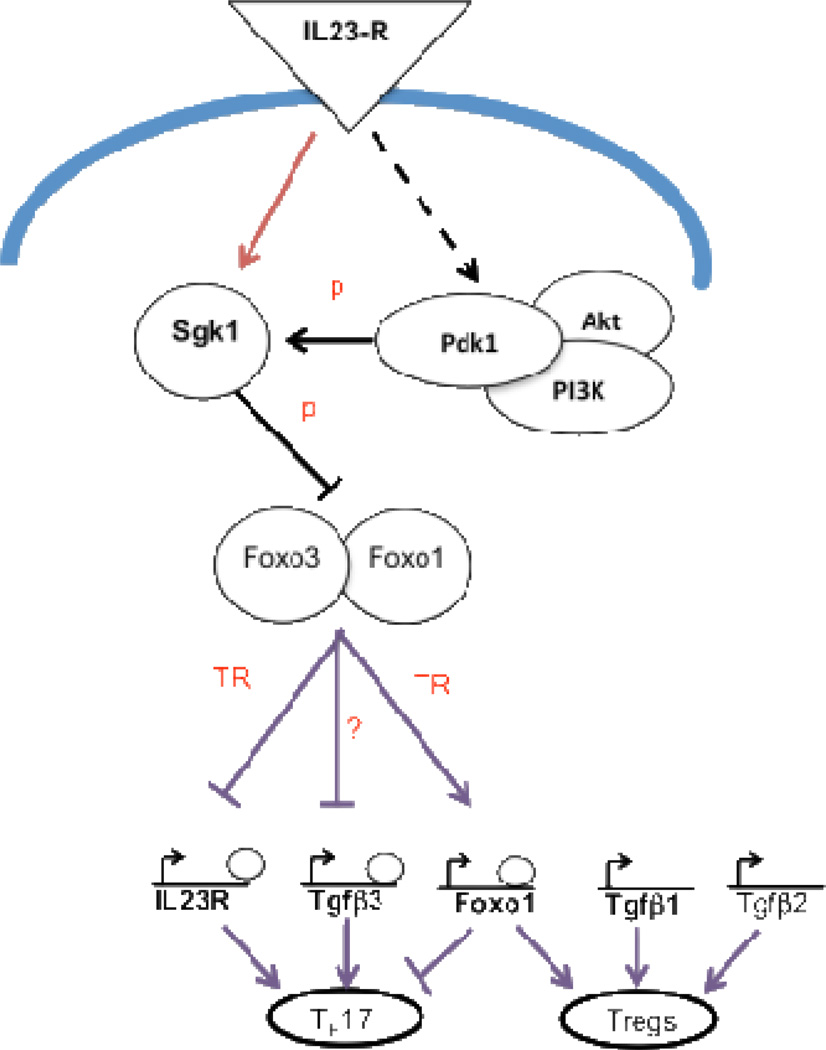
The IL-23R/IL-17 axis is a critical pro-inflammatory pathway that drives the development of pathogenic Th17 cells implicated in autoimmune diseases (18, 19, 21), yet how IL-23 signaling mediates pathogenic in Th17 cells is still poorly understood. From our temporal microarray analysis at 18 different time points during Th17 cell development under the influence of IL-23, our lab developed an extensive transcriptional network and protein-protein network that is downstream of the IL-23R (41). Understanding the genes that are induced downstream of IL-23:IL-23R may be the key in figuring out potential strategies to inhibit progression of destructive inflammation, and to ameliorate autoimmune diseases that have a genetic risk factor associated with the IL-23R-Th17 pathway. Our analysis of IL-23R dependent nodal points revealed two genes of interest: TGF-β3 and serum glucocorticoid kinase 1 (SGK1) (22, 48). Both SGK1 and TGF-β3 are specifically induced in Th17 cells and their expression is dependent on IL-23R signaling (22, 48) (Figure 1). SGK-1 is salt-sensing and regulates absorption of salt by regulating expression of the Epithelial Sodium Channel (EcNaC). Interestingly, our lab has also shown that high concentrations of NaCl induce expression of SGK1 and promote specific differentiation into the Th17 lineage (48). Exposure of human T cells to high salt concentrations also results in skewing of T cell differentiation to the Th17 lineage and increases expression of genes that have been associated with pathogenicity, including SGK1 (49). Supporting studies have shown that the loss of SGK1 in mice prevents the development of experimental autoimmune encephalomyelitis (EAE), a murine model of multiple sclerosis (48). In addition, differentiating Th17 cells endogenously produced TGF-β3 in an IL-23R dependent manner to induce highly pathogenic Th17 cells (22). Uniquely, TGF-β3 is required for the development of pathogenic Th17 cells, but not non-pathogenic Th17 cells, thus identifying an important molecule that may allow us to target only pathogenic Th17 cells (22). Interestingly, our network model also predicts that the IL-23R-SGK1 axis specifically controls the transcription of TGF-β3, but not TGF-β1, an essential cytokine for the generation of Foxp3+ T regulatory cells (Figure 1). However, it is unclear how SGK1 and TGF-β3 regulate the generation of pathogenic Th17 cells or whether specifically inhibiting both SGK1 and TGF-β3 can suppress the development of autoimmune diseases. In addition, further study is required to determine whether targeting SGK1 and/or TGF-β3 would have applicability in Th17-associated autoimmune diseases that do not respond to targeting of IL-17. An understanding of how specific nodes in the network of Th17 associated genes link to responses in each autoimmune or inflammatory disease would allow for selection of the right therapeutic strategy for the appropriate stage of disease pathogenesis.
Figure 1. Schematic description of IL-23R-SGK1-Foxo1 axis during Th17 maintenance and its link to Tgfβ3 regulation.
Sgk1 is induced by IL-23R signaling and is a substrate of PDK1 kinase a member of the IL-23R signaling pathway, which also includes Akt a kinase homologous to SGK1. IL-23R signaling transcriptionally induces SGK1 which in turn further induces IL-23R expression by inhibiting Foxo-1. Deactivation by Sgk1 of Foxo and possibly also Foxo3, de-represses IL-23R transcription, and deactivates Foxo1 transcription. Foxo-1 also induces its own transcription and promotes Foxp3-mediated Treg development by acting as a placeholder for Foxp3 for loading onto genomic sites for transcription. Tgfβ3, but not Tgfβ1/2 contains the Foxo1 and Foxo3 binding motifs in its promoter region, suggesting that TGF-β3 is specifically regulated by the Sgk1-Foxo axis. Edges represent interactions between molecules. “p”: phosphorylation. “TR”: transcriptional regulation.
Conclusion
Genome-wide association studies in humans have linked IL-23R and Th17 cells with susceptibility to psoriasis, psoriatic arthritis, rheumatoid arthritis, autoimmune thyroid disease, ankylosing spondylitis, and inflammatory bowel disease (23–26). There is a growing body of empirical evidence in mice that support these genetic linkages of Th17 cells to pathogenic inflammation (16, 17, 24, 50–52). Inhibiting the function and differentiation of Th17 cells may provide the key to restraining autoimmunity. As proof of principle, we and others have shown that by the incorporation of a small molecule inhibitor of RORγt in murine T cells, can specifically disrupt the transcription of Th17 canonical genes while leaving other T cell lineages intact. (53, 54) Furthermore, these RORγt inhibitors can suppress the development of EAE, a murine model of multiple sclerosis (54). However, we now have evidence that Th17 cells are a heterogeneous population and more precise targets within the Th17 paradigm may be required to specifically target pathogenic Th17 cells while leaving the non-pathogenic Th17 cells intact (20, 22, 28, 39). By applying genomic profiling methods to single cells, we may be able to tease apart the key regulatory modules that govern pathogenicity in Th17 cells. The discovery of regulators of potential pathogenic Th17 cells will provide new targets for drug development and for therapeutic targeting of autoimmune diseases.
References
- 1.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy KM, Ouyang W, Farrar JD, Yang J, Ranganath S, Asnagli H, Afkarian M, Murphy TL. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell cloneIDefinition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 5.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez-Santos N, Gaffen SL. Th17 cells in immunity to Candida albicans. Cell Host Microbe. 2012;11:425–435. doi: 10.1016/j.chom.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jager A, Kuchroo VK. Effector and regulatory T-cell subsets in autoimmunity and tissue inflammation. Scand J Immunol. 2010;72:173–184. doi: 10.1111/j.1365-3083.2010.02432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- 12.Gaffen SL, Hernandez-Santos N, Peterson AC. IL-17 signaling in host defense against Candida albicans. Immunol Res. 2011;50:181–187. doi: 10.1007/s12026-011-8226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 17.McGeachy MJ, Cua DJ. The link between IL-23 and Th17 cell-mediated immune pathologies. Semin Immunol. 2007;19:372–376. doi: 10.1016/j.smim.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 18.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O'Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Croxford AL, Mair F, Becher B. IL-23: one cytokine in control of autoimmunity. Eur J Immunol. 2012;42:2263–2273. doi: 10.1002/eji.201242598. [DOI] [PubMed] [Google Scholar]
- 25.Duvallet E, Semerano L, Assier E, Falgarone G, Boissier MC. Interleukin-23: a key cytokine in inflammatory diseases. Ann Med. 2011;43:503–511. doi: 10.3109/07853890.2011.577093. [DOI] [PubMed] [Google Scholar]
- 26.Hazlett J, Stamp LK, Merriman T, Highton J, Hessian PA. IL-23R rs11209026 polymorphism modulates IL-17A expression in patients with rheumatoid arthritis. Genes Immun. 2012;13:282–287. doi: 10.1038/gene.2011.80. [DOI] [PubMed] [Google Scholar]
- 27.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O'Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor W, Jr, Zenewicz LA, Flavell RA. The dual nature of T(H)17 cells: shifting the focus to function. Nat Immunol. 2010;11:471–476. doi: 10.1038/ni.1882. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor W, Jr, Kamanaka M, Booth CJ, Town T, Nakae S, Iwakura Y, Kolls JK, Flavell RA. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diepolder H, Marquardt A, Jagla W, Popp A, et al. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, Wehkamp J, Feagan BG, Yao MD, Karczewski M, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar S, Fox DA. Targeting IL-17 and Th17 cells in rheumatoid arthritis. Rheum Dis Clin North Am. 2010;36:345–366. doi: 10.1016/j.rdc.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Hueber W, Patel DD, Dryja T, Wright AM, Koroleva I, Bruin G, Antoni C, Draelos Z, Gold MH, Psoriasis Study G, et al. Effects of AIN457, a fully human antibody to interleukin-17A, on psoriasis, rheumatoid arthritis, and uveitis. Sci Transl Med. 2010;2:52ra72. doi: 10.1126/scitranslmed.3001107. [DOI] [PubMed] [Google Scholar]
- 36.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 37.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, Edson-Heredia E, Braun D, Banerjee S. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–1199. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 38.Genovese MC, Van den Bosch F, Roberson SA, Bojin S, Biagini IM, Ryan P, Sloan- Lancaster J. LY2439821, a humanized anti-interleukin-17 monoclonal antibody, in the treatment of patients with rheumatoid arthritis: A phase I randomized, double-blind, placebocontrolled, proof-of-concept study. Arthritis Rheum. 2010;62:929–939. doi: 10.1002/art.27334. [DOI] [PubMed] [Google Scholar]
- 39.McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 40.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zielinski CE, Zuberbier T, Maurer M. Immunoregulation in cutaneous allergy: prevention and control. Curr Opin Allergy Clin Immunol. 2012;12:498–503. doi: 10.1097/ACI.0b013e3283574ccb. [DOI] [PubMed] [Google Scholar]
- 43.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eberwine J, Sul JY, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2014;11:25–27. doi: 10.1038/nmeth.2769. [DOI] [PubMed] [Google Scholar]
- 46.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 47.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–517. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–522. doi: 10.1038/nature11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Langrish CL, McKenzie B, Joyce-Shaikh B, Stumhofer JS, McClanahan T, Blumenschein W, Churakovsa T, Low J, Presta L, et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest. 2006;116:1317–1326. doi: 10.1172/JCI25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wakashin H, Hirose K, Iwamoto I, Nakajima H. Role of IL-23-Th17 cell axis in allergic airway inflammation. Int Arch Allergy Immunol. 2009;149 Suppl 1:108–112. doi: 10.1159/000211382. [DOI] [PubMed] [Google Scholar]
- 53.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Baloglu E, Schmidt D, Ramesh R, Lobera M, et al. Chemical Inhibition of the RORγt-dependent Transcriptional Network in Th17 cells. Immunity. 2014 (In Press). [Google Scholar]