Abstract
Previous studies have documented the roles of transport via the reduced folate carrier, retention via polyglutamylation, and increased levels of the target enzyme, dihydrofolate reductase in sensitivity to methotrexate. Recent studies have shown that the mitochondrial enzymes in the cellular metabolism of serine, folate, and glycine are overexpressed in a subset of human cancers and that their expression is required for tumor maintenance. In this Perspective article, we propose that the expression of mitochondrial enzymes in the metabolism of serine and glycine, in addition to those involved in folate metabolism, are determinants of the response to methotrexate. Furthermore, we show that myc activation in tumors is associated with upregulation of these enzymes. We propose that patients whose tumors show this phenotype will be sensitive to folate antagonists targeting thymidylate or purine biosynthesis.
Introduction
Previous studies have documented the roles of transport via the reduced folate carrier (RFC), retention via polyglutamylation, and increased levels of the target enzyme, dihydrofolate reductase in sensitivity to the folate antagonists methotrexate, pralatrexate, and pemetrexed (1). Although these factors explain in part why tumor cells are sensitive or resistant to antifolates, they do not fully account for selectivity. Two recent publications have now provided insight about why some tumor cells, as compared with normal replicating cells, are sensitive to methotrexate: Cancer cells that undergo metabolic reprogramming and are characterized by rapid proliferation and upregulate glycine consumption and metabolism are potently inhibited by methotrexate, whereas normal cells with similar proliferative rates are not as sensitive.
The contribution by Jain and colleagues (2) showed that metabolic reprogramming that occurs in cancer may lead to higher expression of the mitochondrial glycine biosynthetic pathway and upregulation of mitochondrial folate enzymes that include serine hydroxymethyl transferase (SHMT2), methylenetetrahydrofolate dehydrogenase (MTHFD2), and tetrahydrofolate synthetase (MTHFD1L; Fig. 1). The up-regulation of these mitochondrial enzymes correlated with increased proliferation, whereas the folate cytosolic enzymes were not upregulated. Unlike the trifunctional cytosolic enzyme, MTHFD1, that contains methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase, and formyltetrahydrofolate synthetase activity, the mitochondrial enzyme MTHFD2 is a bifunctional enzyme that contains methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase activity. Formyltetrahydrofolate synthetase activity is encoded by a separate enzyme, MTHFD1L. These enzymes contribute to the synthesis and use of glycine, methylene THF, and N-10 formyl THF for de novo purine and thymidylate biosynthesis (Fig. 1).
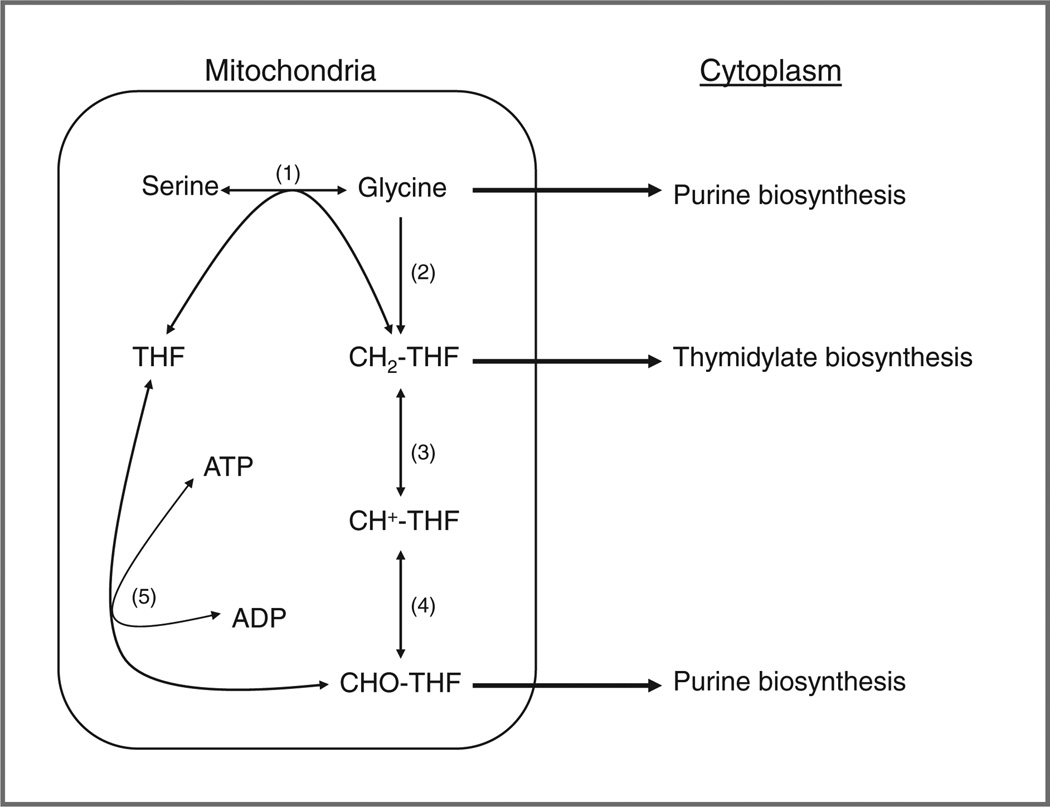
Figure 1.
The role of the mitochondria in the generation of purines and thymidylate for DNA synthesis. Reactions 1, 3, 4, and 5 occur in both the mitochondria and the cytoplasm, with reaction 2 limited only to the mitochondria. Reaction 1 and 2 are catalyzed by SHMT2 and the glycine cleavage system, respectively. In the cytoplasm, reactions 3, 4, and 5 are carried out by the trifunctional enzyme (MTHFD1); in the mitochondria, 2 enzymes are required (MTHFD2 catalyzes reactions 3 and 4 and MTHFD1L catalyzes reaction 5). The enzyme 10-formyl tetrahydrofolate dehydrogenase (not shown) has been reported to be absent in most cancer cells (10). Serine hydroxymethytransferase (1); glycine oxidase complex (2); 5–10 methylene tetrahydrofolate dehydrogenase (3); methenyltetrahydrofolate cyclohydrolase (4); and formyltetrahydrofolate synthetase (5).
Supporting the importance of the above pathway is the high expression of mitochondrial 1-carbon pathway components in embryos. A developmental period marked by high cell proliferation rates and embryogenesis requires significant amounts of protein, lipid, and nucleic acid synthesis, of similar nature to neoplastic cells. MTHFD1L was found to be upregulated in mouse embryos, with the mitochondria providing more than 75% of the 1-carbon units present in the cytoplasm used for purine synthesis and other biosynthetic pathways (3). MTHFD1L and MTHFD2 are targets of microRNA miR-9, which may act as a tumor suppressor in regard to these and other genes (4). High expression of miR-9 suppresses levels of MTHFD1L and MTHFD2 and miR-9 has been found to be downregulated in breast cancer cell lines (4). Increasing miR-9 levels or knocking down MTHFD2 both have antiproliferative and proapoptotic effects in tumor cell lines, which suggests that inhibitors of MTHFD2 may show similar effects.
Zhang and colleagues (5) isolated tumor-initiating cells from primary non–small cell lung cancers (NSCLC) and found that upregulation of the enzyme glycine decarboxylase and other glycine–serine enzymes was associated with increased rates of proliferation and poorer outcome in patients with NSCLC. Remarkably, cDNA of the glycine decarboxylase gene transformed 3T3 cells. Large increases in the levels of thymidylate synthase (TS; ref. 6) and dihydrofolate reductase (DHFR; unpublished data) by transfection of immortalized cells can also cause transformation, and these findings are in accord with the idea that upregulation of nucleotide synthesis is associated with metabolic reprogramming and increased tumor cell proliferation. The details of how this increase in enzymes involved in nucleotide biosynthesis is associated with malignant transformation are not clear. In the study by Zhang and colleagues (5), the mitochondrial SHMT2 and MTHFD2 and MTHFD1L enzyme levels were not assessed, and emphasis was focused on the role of glycine decarboxylase to generate methylenetetrahydrofolate, the 1-carbon donor for thymidylate biosynthesis. Because the glycine decarboxylase enzyme complex is also located in mitochondria of mammalian cells (7), this further shows the role of the mitochondria in proliferation by supplying tumor cells with precursors for pyrimidine biosynthesis. In this regard, an earlier report showed that transformation imposes a stress on cancer cells in that the demand for nucleotide formation is difficult for the cell to meet (8). As suggested by both Jain and colleagues and Zhang and colleagues (2, 5), inhibitors of glycine decarboxylase and/or serine hydroxymethyl transferase may be novel targets for tumors in which proliferation is driven by overexpression of these enzymes.
Sensitivity to methotrexate in vitro and in vivo
Here, we provide evidence to show that known potent and clinically approved inhibitors of thymidylate and purine biosynthesis, which include DHFR inhibitors methotrexate and pralatrexate and the thymidylate synthase inhibitors 5-fluoro-uracil and pemetrexed, show selectivity to rapidly proliferating tumors in patients with overexpression of genes coding for folate metabolism enzymes. To test this hypothesis, we analyzed in vitro data reported by the Genomics of Drug Sensitivity in Cancer at the Sanger Institute (Hinxton, Cambridge, UK; ref. 9) to determine whether upregulation of the mitochondrial folate enzymes was correlated with sensitivity to methotrexate, a potent inhibitor of dihydrofolate reductase, and when converted intracellularly to polyglutamates (10), an inhibitor of two enzymes of purine synthesis, phosphoribosylglycinamide formyltransferase (GARFT), and phosphoribosylaminoimidazolecarboxamide formyltransferase (AICART). A strong correlation between increased expression of 7 genes and lower IC50 for methotrexate was observed (Fig. 2a), including the genes coding for the folate mitochondrial enzymes SHMT2 (P = 3.8 × 10−4) and GLDC (P = 5.4 × 10−3), as well as SLC19A1 (coding for the RFC, P=4.1×10−3), DHFR (P=3.6× 10−5–5.4 × 10−4), TYMS (TS, P = 6.9 × 10−8), GART (GARFT, P = 3.6 × 10−3−1.7 × 10−2), and PHGDH (P = 1.5 × 10−2). These data support the hypothesis that methotrexate and potentially other inhibitors of thymidylate and purine biosynthesis could be used to target cancers with high rates of proliferation.
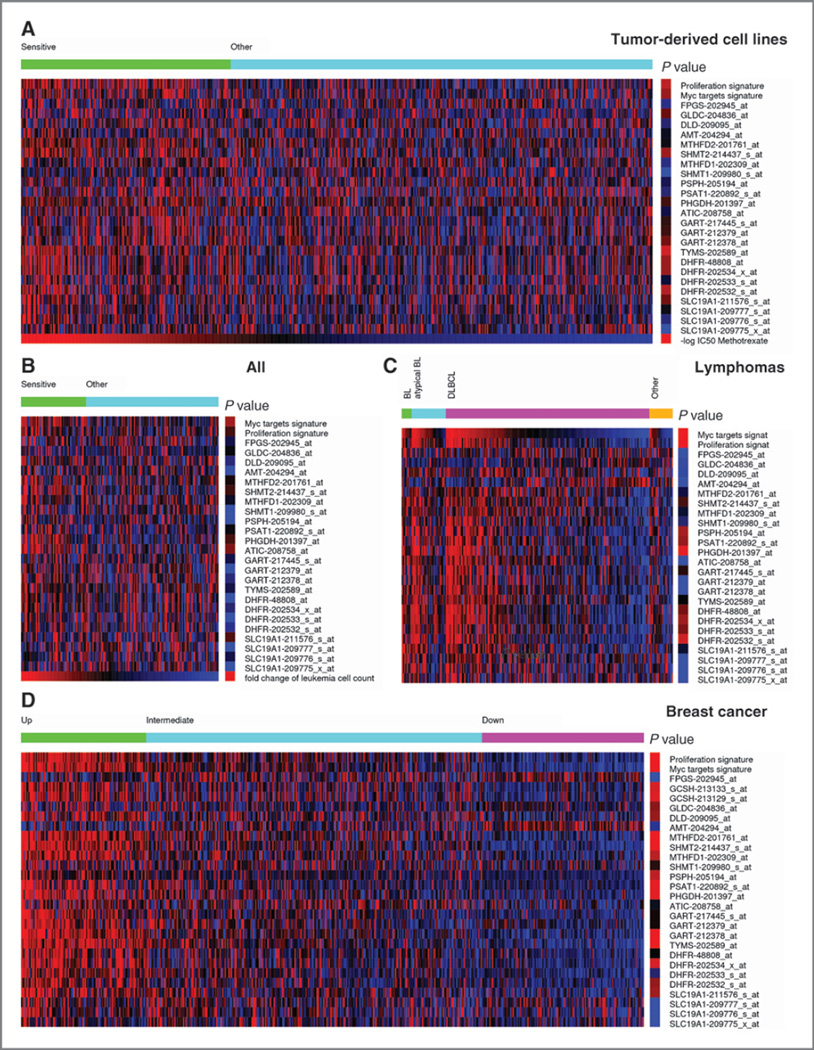
Figure 2.
Heatmap showing the expression of genes coding for enzymes in folate metabolism (blue, underexpressed; red, overexpressed). A, a collection of 515 tumor-derived cell lines sorted in decreasing order of their sensitivity for methotrexate (MTX), based on data from ref. (9). The samples were divided into 2 groups: sensitive (1/3 of samples with lowest IC50) and other (remaining samples). The right column shows a color-coded quantification of the P value for enrichment of samples with high expression (2-fold or above) in the sensitive group, whereas red indicates significantly enriched, the brighter the more significant, and blue not significantly enriched. B, a collection of 161 ALL sorted in decreasing order of their sensitivity to MTX, based on data from ref. (11). The samples were divided into 2 groups: sensitive (1/3 of samples with highest fold decrease in the leukemia cell count) and other (remaining samples). The right column shows a color-coded quantification of the P value as in A. C, a collection of 221 lymphomas grouped by subtype and sorted in decreasing order of c-myc gene signature, based on data from ref. (16). The right column shows a color-coded quantification of the P value for enrichment of samples with high expression (2-fold or above) among samples with a significant upregulation of the c-myc signature. D, samples from 508 breast cancers, based on data from ref. (18). The samples were divided in 3 groups with significant upregulation (top), nonsignificant (middle), and significant downregulation (Bottom) of the folate metabolism gene signature. The right column shows a color-coded quantification of the P value for enrichment of samples with high expression in the top group.
To provide further evidence, we analyzed a study reporting the response of patients with acute lymphoblastic leukemia (ALL) to methotrexate treatment (11). This study reports the leukemia cell counts before and after treatment together with the leukemia gene expression profiles before treatment. We used the fold change reduction in leukemia cell count as a quantification of sensitivity to methotrexate treatment. A strong correlation was observed between the sensitivity to methotrexate treatment and increased expression of the genes coding for the folate mitochondrial enzymes SHMT2 (P = 1.6 × 10−2) and MTHFD2 (P = 3.8 × 10−2) as well as the gene SLC19A1 (P = 1.1 × 10−2) coding for the RFC (Fig. 2b). Surprisingly, the expression of DHFR did not correlate with the response to methotrexate, underscoring the need to include the folate mitochondrial enzymes in the investigation of the mechanisms of response to methotrexate.
What malignancies may be expected to be sensitive to methotrexate?
Another key aspect is to have an estimate of the patient population size that could be targeted using this approach. To this end, we used, as a proxy, a set of genes coding for folate/serine/glycine metabolism enzymes (folate metabolism: SLC19A1, DHFR, TYMS, GART, ATIC, FPGS; serine synthesis: PHGDH, PSAT1, PSPH; cytosolic serine/glycine/folate metabolism: SHMT1, MTHFD1; and mitochondrial serine/glycine/folate metabolism: SHMT2, MTHFD2, MTHFD2L, AMT, DLD, GLDC). Wehypothesize that cancers overexpressing a subset of these genes are more dependent on folate metabolism and, therefore, could benefit from treatment with methotrexate. However, we note that this signature is not a predictor of sensitivity to methotrexate, but rather a rough quantification of the potential benefit from stratified therapy with methotrexate. For example, Fig. 2D shows the expression of the folate metabolism genes in a cohort of patients with breast cancer. The patients with breast cancer can be divided into 3 groups. Fig. 2D shows the subset of cancers with a significant upregulation (top, left) of the folate metabolism gene signature, characterized by the concomitant expression of most genes in the gene signature. The color-coded P value on the right emphasizes in red those genes that tend to be coexpressed. This group of patients is likely to benefit from methotrexate therapy because their cancers seem to be dependent on folate metabolism. In contrast, the subset of cancers on the right manifest a significant downregulation of the folate metabolism gene signature (Fig. 2D, bottom) and the corresponding patients are not expected to benefit from methotrexate therapy. Finally, an intermediate group of patients has a mixed phenotype (Fig. 2D, middle), and the response of these patients cannot be anticipated unless we know which specific enzyme mediates the response to methotrexate in this specific cancer subtype.
Extending this type of analysis to other cancer types, we observe that about 25% of cancers manifest a significant upregulation of the folate metabolism gene signature and that patients with this feature are likely to benefit from treatment with methotrexate (Table 1). Some variations exist depending on the cancer subtype, with colorectal cancers and lymphomas reaching slightly more than 30% to the lowest value of about 10% in prostate cancers.
Table 1.
Prevalence of samples with concomitant expression of folate metabolism genes across different cancer types
| Type | Sample size |
Sensitivity (%) |
PMID | GEO |
|---|---|---|---|---|
| Brain | 180 | 39 | 16616334 | GSE4290 |
| 100 | 22 | 16530701 | GSE4271 | |
| Breast | 508 | 21 | 21558518 | GSE25066 |
| 266 | 27 | 22110708 | GSE21653 | |
| 251 | 22 | 16141321 | GSE3494 | |
| Colorectal | 290 | 32 | 19996206 | GSE14333 |
| 177 | 34 | 19914252 | GSE17536 | |
| 145 | 36 | 20957034 | GSE20916 | |
| Lung | 246 | 27 | 22080568 | GSE31210 |
| 163 | 22 | 16549822 | GSE11969 | |
| 156 | 34 | 20421987 | GSE19188 | |
| Lymphoma | 420 | 30 | 19038878 | GSE10846 |
| 221 | 30 | 16760442 | GSE4475 | |
| Ovarian | 295 | 30 | 18698038 | GSE9899 |
| 103 | 24 | 17418409 | GSE6008 | |
| Pancreatic | 132 | 20 | 20644708 | GSE21501 |
| Prostate | 281 | 12 | 20233430 | GSE16560 |
NOTE: The PubMed reference (PMID) and Gene Expression Omnibus (GEO) of the gene expression profile sources are shown in their respective columns.
The concomitant expression of genes encoding for enzymes in folate metabolism indicates a common regulatory mechanism. It is known that several serine, folate, and glycine metabolism genes are targets of c-myc (PHGDH, PSPH, SLC19A1, DHFR, TYMS, GART, SHMT1, MTHFD1, MTHFD2, FGPS and GCSH; refs. 12, 13), and that their expression is required for cell proliferation. Once again, we used gene signatures as a proxy to quantify the activation of c-myc targets (12) and cell proliferation (14). Burkitt lymphoma (BL) is a typical example of a myc-driven cancer (15). Indeed, from our analysis of a published dataset (16), 88% and 68% of lymphoma classified as BL and atypical BL, respectively, manifest a significant upregulation of the c-myc gene signature (Fig. 2C). Furthermore, as previously noticed (16), there is a group of diffuse large B-cell lymphomas (DLBCL) that is also Myc driven. Indeed, 44% of the DLBCLs manifest a significant upregulation of the c-myc signature. Focusing on the folate metabolism genes, the P column in Fig. 2C highlights in red those genes that are frequently overexpressed (2-fold or higher) in the group of lymphomas with a significant Myc signature upregulation. In addition to DHFR (P = 1.5 × 10−7–2.4 × 10−3 depending on the microarray probe), we note the serine synthesis genes PHGDH (P = 9.7 × 10−13), PSAT1 (P = 6.1 × 10−4), PSPH (P = 1.5 × 10−4), and the mitochondrial gene SHMT2 (P = 1.4 × 10−2). The correlation between the folate/serine/glycine metabolism, c-Myc, and proliferation gene signatures is actually a universal feature of human cancers (17). We also note a significant correlation between the upregulation of the c-myc and proliferation signatures and the response to methotrexate in both the in vitro and in vivo studies (Fig. 2A and B, respectively). More precisely, in the sensitive group, a significant number of samples manifests an upregulation of the c-myc (in vitro, P = 5.1 × 10−4; in vivo, P = 2.6 × 10−4) and proliferation (in vitro, P = 6.4 × 10−6; in vivo, P = 1.5 × 10−2) signature.
Conclusions
It will be important to learn whether known potent and clinically approved inhibitors of thymidylate and purine biosynthesis, which include DHFR inhibitors methotrexate and pralatrexate and the thymidylate synthase inhibitors 5-fluorouracil and pemetrexed, show selectivity to rapidly proliferating tumors in patients with overexpression of genes coding for mitochondrial folate metabolism enzymes. Based on the concomitant overexpression of folate metabolism genes, we estimate that around 25% of patients with cancer manifest this phenotype, and determination of this phenotype will allow patients to be selected for treatment with inhibitors of thymidylate and purine biosynthesis.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: J.R. Bertino, A. Vazquez
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J.R. Bertino, A. Vazquez, P.M. Tedeschi
Writing, review, and/or revision of the manuscript: J.R. Bertino, A. Vazquez, P.M. Tedeschi
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): P.M. Tedeschi Study supervision: J.R. Bertino
References
- 1.Cole PD, Bertino JR, Kamen BA. Folate antagonists. In: Kufe DW, Bast RC Jr, Hait WN, Hong WK, Pollock RE, Weichselbaum, et al., editors. Cancer medicine. Vol. 7. Hamilton, Ontario, Canada: Decker; 2008. pp. 648–660. [Google Scholar]
- 2.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, et al. MicroRNA-9 Inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 6.Rahman L, Voeller D, Rahman M, Lipkowitz S, Allegra C, Barrett JC, et al. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer Cell. 2004;5:341–351. doi: 10.1016/s1535-6108(04)00080-7. [DOI] [PubMed] [Google Scholar]
- 7.Motokawa Y, Kikuchi G. Glycine metabolism in rat liver mitochondria. V. Intramitochondrial localization of the reversible glycine cleavage system and serine hydroxymethyltransferase. Arch Biochem Biophys. 1971;146:461–464. doi: 10.1016/0003-9861(71)90149-4. [DOI] [PubMed] [Google Scholar]
- 8.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chabner BA, Allegra CJ, Curt GA, Clendeninn NJ, Baram J, Koizumi S, et al. Polyglutamation of methotrexate. Is methotrexate a prodrug? J Clin Invest. 1985;76:907–912. doi: 10.1172/JCI112088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorich MJ, Pottier N, Pei D, Yang W, Kager L, Stocco G, et al. In vivo response to methotrexate forecasts outcome of acute lymphoblastic leukemia and has a distinct gene expression profile. PLoS Med. 2008;5:e83. doi: 10.1371/journal.pmed.0050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vazquez A, Markert EK, Oltvai ZN. Serine biosynthesis with one carbon catabolism and the glycine cleavage system represents a novel pathway for ATP generation. PLoS ONE. 2011;6:e25881. doi: 10.1371/journal.pone.0025881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yustein JT, Dang CV. Biology and treatment of Burkitt's lymphoma. Curr Opin Hematol. 2007;14:375–381. doi: 10.1097/MOH.0b013e3281bccdee. [DOI] [PubMed] [Google Scholar]
- 16.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 17.Markert EK, Levine AJ, Vazquez A. Proliferation and tissue remodeling in cancer: the hallmarks revisited. Cell Death Dis. 2012;3:e397. doi: 10.1038/cddis.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]